Although phospholipid:diacylglycerol acyltransferase (PDAT) activity is presumed to exist in prokaryotic oleaginous bacteria, the corresponding gene has not been identified yet. In this article, we have demonstrated that an acyl-CoA-independent pathway can divert phospholipid flux into TAG formation in Escherichia coli mediated by exogenous CrPDAT from Chlamydomonas reinhardtii without interfering with membrane functions. In addition, the acyl-CoA-independent pathway and the acyl-CoA-dependent pathway had the synergistic effect on TAG accumulation. Overexpression of CrPDAT led to synchronous TAG accumulation during cell growth. In particular, CrPDAT possessed multiple catalytic activities, and the rational mutation of CrPDAT led to the decrease of TAG lipase activity without impairing acyltransferase activity. The present findings suggested that applying PDAT in E. coli or other prokaryotic microbes may be a promising strategy for accumulation of TAGs and their derivatives.
KEYWORDS: phospholipid:diacylglycerol acyltransferase, protein engineering, triacylglycerol, wax ester synthase/acyl-CoA:diacylglycerol acyltransferase
ABSTRACT
Researchers have long endeavored to accumulate triacylglycerols (TAGs) or their derivatives in easily managed microbes. The attempted production of TAGs in Escherichia coli has revealed barriers to the broad applications of this technology, including low TAG productivity and slow cell growth. We have demonstrated that an acyl-CoA-independent pathway can divert phospholipid flux into TAG formation in E. coli mediated by Chlamydomonas reinhardtii phospholipid:diacylglycerol acyltransferase (CrPDAT) without interfering with membrane functions. We then showed the synergistic effect on TAG accumulation via the acyl-CoA-independent pathway mediated by PDAT and the acyl-CoA-dependent pathway mediated by wax ester synthase/acyl-CoA:diacylglycerol acyltransferase (WS/DGAT). Furthermore, CrPDAT led to synchronous TAG accumulation during cell growth, and this could be enhanced by supplementation of arbutin. We also showed that rationally mutated CrPDAT was capable of decreasing TAG lipase activity without impairing PDAT activity. Finally, ScPDAT from Saccharomyces cerevisiae exhibited similar activities as CrPDAT in E. coli. Our results suggest that the improvement in accumulation of TAGs and their derivatives can be achieved by fine-tuning of phospholipid metabolism in E. coli. Understanding the roles of PDAT in the conversion of phospholipids into TAGs during the logarithmic growth phase may enable a novel strategy for the production of microbial oils.
IMPORTANCE Although phospholipid:diacylglycerol acyltransferase (PDAT) activity is presumed to exist in prokaryotic oleaginous bacteria, the corresponding gene has not been identified yet. In this article, we have demonstrated that an acyl-CoA-independent pathway can divert phospholipid flux into TAG formation in Escherichia coli mediated by exogenous CrPDAT from Chlamydomonas reinhardtii without interfering with membrane functions. In addition, the acyl-CoA-independent pathway and the acyl-CoA-dependent pathway had the synergistic effect on TAG accumulation. Overexpression of CrPDAT led to synchronous TAG accumulation during cell growth. In particular, CrPDAT possessed multiple catalytic activities, and the rational mutation of CrPDAT led to the decrease of TAG lipase activity without impairing acyltransferase activity. The present findings suggested that applying PDAT in E. coli or other prokaryotic microbes may be a promising strategy for accumulation of TAGs and their derivatives.
INTRODUCTION
The global demand for oils and oil-based products has been increasing constantly over recent decades. Renewable microbial oils, such as microbial triacylglycerol (TAG), are expected to play a crucial role in providing a sustainable source for oils and their derivatives. TAGs are the important energy storage lipids with highly reduced carbon molecules found in eukaryotes and a few oleaginous bacteria. TAGs can be synthesized via multiple pathways (1–4). TAG synthesis via the acyl-CoA-dependent Kennedy pathway is sequentially catalyzed by glycerol-3-phosphate acyltransferase (GPAT, EC 2.3.1.15), lysophosphatidic acid acyltransferase (LPAAT, EC 2.3.1.51), phosphatidic acid phosphatase (PAP, EC 3.1.3.4), and diacylglycerol acyltransferase (DGAT, EC 3.12.1.20) (5). Nearly 20 years ago, the acyl-CoA-independent pathway mediated by the enzyme phospholipid:diacylglycerol acyltransferase (PDAT, EC 2.3.1.158) was characterized in yeast (Saccharomyces cerevisiae) and plants (6). PDAT catalyzes the formation of TAG by transferring an acyl group from the sn-2 position of phospholipid (PL) to the sn-3 position of diacylglycerol (DAG). Evolutionary analysis reveals that PDATs are present in all examined green plants, including algae, mosses, and lycophytes, as well as fungi (7). Although PDAT activity is presumed to exist in prokaryotic oleaginous bacteria, the corresponding gene has not been identified yet (8, 9).
PDATs participate in regulation of membrane lipid turnover, degradation, and TAG synthesis (10–12). Previous investigations indicate that DGAT and PDAT play overlapping roles in TAG synthesis. In yeast and the unicellular green microalga Chlamydomonas reinhardtii, PDATs contribute to TAG production under favorable growth conditions or during the log phase, while DGATs seem to be more essential for TAG synthesis during the stationary phase (10, 13). AtPDAT1 from Arabidopsis thaliana has a much higher impact than AtDGAT1 on TAG synthesis in leaves, whereas AtDGAT1 is a major contributor to TAG synthesis in developing seeds (14). In addition, neither AtPDAT1 overexpression nor the knockout change lipid content or fatty acid composition of seed oils, suggesting that AtDGAT1 compensates for the function of AtPDAT1 (15).
Despite the fact that the physiological functions of PDAT seem conserved among yeasts, algae, and plants, its biochemical characteristics vary in some aspects. Of all the discovered PDATs, the biochemical characteristics of CrPDAT from C. reinhardtii have been investigated in the most detail (10). CrPDAT exhibits less activity with phosphatidylethanolamine (PE) compared to phosphatidylcholine (PC). Specifically, CrPDAT has a preference for anionic PL, including phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidylglycerol (PG), over the cationic PC and PE (10). In contrast, AtPDAT and ScPDAT from Saccharomyces cerevisiae show preference for PE over PC as the acyl donor in vitro (13, 15). In addition to typical PDAT activity, CrPDAT also possesses lipase activity toward both TAGs and various PLs, whereas ScPDAT shows phospholipase activity (10, 13). However, no lipase activity has been described for AtPDAT (15). Although the motif (G/A/S-X-S-X-G) conserved among the α/β hydrolase family is also essential for the lipase activity (15), the catalytic site for lipase activity has not been confirmed for any PDAT (16). Precise information about the lipase active or binding sites is limited mainly due to the lack of structure information of any membrane-bound PDATs.
Escherichia coli is an ideal bacterial cell factory because it is a highly studied model system with well-developed genetic systems that is also easy to culture in the laboratory. E. coli has been employed to produce high levels of fatty acids and their derivatives (17). E. coli does not produce any TAGs naturally (18), mainly due to the lack of wax ester synthase/acyl-CoA:diacylglycerol acyltransferase (WS/DGAT), which catalyzes the last and rate-limiting step of the acyl-CoA-dependent TAG synthesis pathway (19). The WS/DGAT-mediated acyl-CoA-dependent pathway has been successfully introduced into E. coli for the production of regular TAGs or TAGs rich in medium-chain fatty acids (20, 21). Early strategies attempting the accumulation of TAGs were focused on the enhancement of fatty acid and/or acyl-CoA synthesis or on blocking the degradation of fatty acid and/or TAGs (20, 22–24).
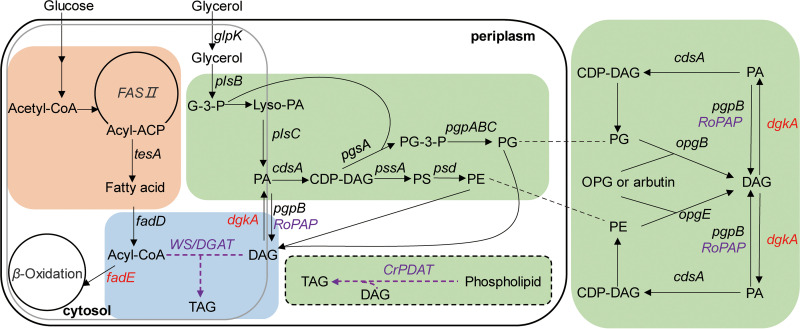
Membrane PLs and storage TAGs compete for acyl flux; thus, we hypothesized that TAG accumulation might be achieved by diverting fatty acids from PLs to TAGs in E. coli. E. coli cells have the ability to accumulate abundant PEs (about 75% of total PLs) and PGs (20% of total PLs) (25), which are available substrates for TAG formation catalyzed by CrPDAT. In addition, DAGs, which are by-products generated by the osmo-regulated periplasmic glucan (OPG) biosynthesis pathway, are strictly regulated (26), but they can be greatly enhanced by overexpression of the phosphatidylglycerol phosphatase gene pgpB or deletion of the diacylglycerol kinase gene dgkA (18, 27). Alternatively, DAG availability may be enhanced by supplementing with arbutin via tuning of the membrane-derived oligosaccharide (MDO) biosynthesis pathway (18). Arbutin is a hydroquinone glucoside compound existing mainly in plants, including bearberry, wheat, and pear (28). In E. coli, arbutin can be used as an artificial β-glucoside acceptor for glycerolphosphate, which is transferred from PG and mediated by OpgB (29), or from PE mediated by OpgE (26) (Fig. 1). Previous results revealed that dgkA-knockout E. coli cells were unable to grow on medium containing arbutin due to the lethal accumulation of DAGs (30), suggesting that DAG levels are probably stimulated by the supplementation of arbutin.
FIG 1.
Engineering of E. coli cells for TAG accumulation. Heterologous genes are highlighted in purple. Deleted genes are shown in red. G-3-P, glycerol-3-phosphate; Lyso-PA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; CDP-DAG, cytidine-diphosphate-diacylglycerol; PG-3-P, phosphatidylglycerol-3-phosphate; PG, phosphatidylglycerol; PS, phosphatidylserine; PE, phosphatidylethanolamine; WS/DGAT, wax ester synthase/acyl-CoA:diacylglycerol acyltransferase gene; RoPAP, phosphatidic acid phosphatase gene from R. opacus PD630; CrPDAT, phospholipid:diacylglycerol acyltransferase gene from C. reinhardtii; tesA, acyl-CoA thioesterase I gene; fadD, fatty acyl-CoA synthetase gene; fadE, acyl-CoA dehydrogenase gene; glpK, glycerol kinase gene; plsB, glycerol-3-phosphate acyltransferase gene; plsC, 1-acylglycerol-3-phosphate O-acyltransferase gene; dgkA, diacylglycerol kinase gene; pgpB, phosphatidylglycerophosphatase B gene; cdsA, CTP:2,3,4-saturated l-phosphatidate cytidylyltransferase gene; pgsA, phosphatidylglycerophosphate synthase gene; pgpABC, phosphatidylglycerophosphatase ABC gene; pssA, phosphatidylserine synthase gene; psd, phosphatidylserine decarboxylase gene; opgE, phosphoethanolamine transferase gene; opgB, phosphoglycerol transferase I gene; OPG, osmoregulated periplasmic glucans.
In this report, we first reconstructed the CrPDAT-mediated acyl-CoA-independent TAG synthesis pathway in E. coli, even though such a pathway has not been identified in any prokaryotic microbes (Fig. 1). We then demonstrated the synergistic effects of CrPDAT and the WS/DGAT of Acinetobacter baylyi ADP1 (AtfA) on TAG accumulation. Furthermore, CrPDAT led to synchronous TAG accumulation during cell growth with enhancement following the supplementation of arbutin. Finally, we showed that rationally mutated CrPDAT possessed the ability to decrease TAG lipase activity without impairing PDAT activity.
RESULTS AND DISCUSSION
Overexpression of a solo CrPDAT in an E. coli double mutant led to TAG accumulation.
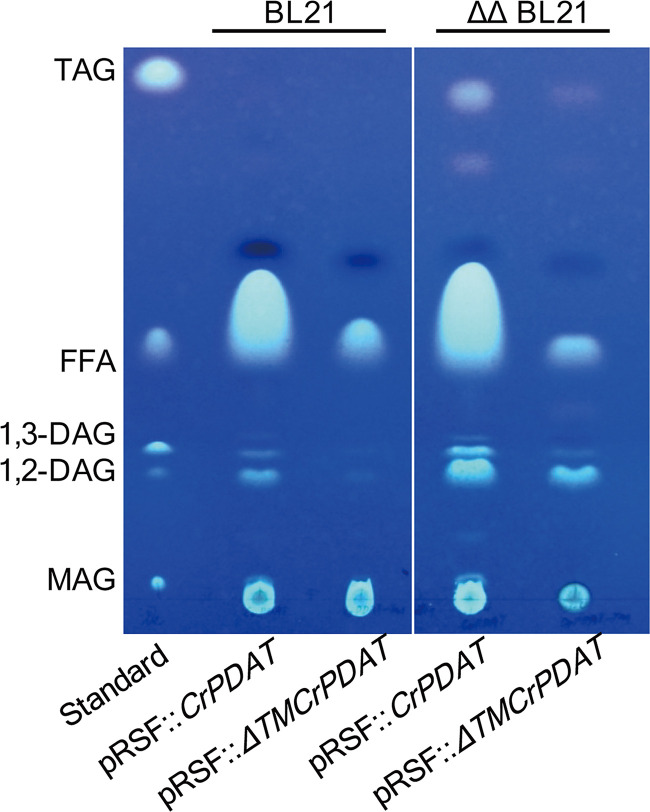
To the best of our knowledge, no PDAT-encoding gene has been identified from any prokaryotic oleaginous microorganism to date. Thus, we were interested in testing whether heterologous PDAT could convert PLs into TAGs in E. coli. An earlier report showed that the truncated ΔTMCrPDAT (CrPDAT without a transmembrane domain) had higher PDAT activity than a full-length CrPDAT in vitro enzyme test (10). Unfortunately, overexpression of a solo CrPDAT or ΔTMCrPDAT did not result in any detectable TAG accumulation in E. coli BL21(DE3) (Fig. 2). We next sought to delete the acyl-CoA dehydrogenase encoded by the gene fadE and the diacylglycerol kinase encoded by the gene dgkA to generate a double mutant strain designated ΔΔBL21 to elevate the DAG level and prevent the degradation of acyl-CoA (see Fig. S1 in the supplemental material). The results of thin-layer chromatography (TLC) showed that overexpressing CrPDAT in ΔΔBL21 led to obvious TAG accumulation (Fig. 2). Compared to wild-type E. coli, the DAG levels in ΔΔBL21 cells were greatly enhanced, especially 1,2-DAG. This suggested that the double deletion was beneficial for circumventing the DAG bottleneck and thus promoting TAG synthesis. More importantly, we demonstrated that TAGs could be produced from PL pools via the acyl-CoA-independent pathway by CrPDAT in this E. coli double mutant. Transmission electron microscopy observation showed no obvious difference in cell shape between the control and the engineered cells harboring CrPDAT during their growth, suggesting that overexpressing CrPDAT did not interfere with membrane functions (Fig. S2). In addition, no abnormal phenotype was observed during the growth (data not shown).
FIG 2.
Overexpression of a solo CrPDAT or ΔTMCrPDAT in E. coli BL21 and ΔΔBL21. Cells were cultured in auto-induction medium supplemented with 3% glycerol with shaking at 200 rpm at 37°C for 48 h. TLC was carried out on lipids extracted from 20 mg of dried recombinant cells. ΔΔBL21, knockout of dgkA and fadE in E. coli BL21(DE3); CrPDAT, PDAT from C. reinhardtii; ΔTMCrPDAT, CrPDAT without a transmembrane domain.
Only full-length CrPDAT resulted in obvious TAG synthesis in E. coli (Fig. 2). In contrast, previous in vitro results revealed that the truncated ΔTMCrPDAT exhibited much higher activities than full-length CrPDAT (10). In addition, an N-terminal deletion version of ScPDAT was highly active in vitro, suggesting that the membrane-spanning region was not essential for ScPDAT activity as well (13). The results presented here could be explained by the differences between in vivo and in vitro conditions. We predicted that the truncated CrPDAT might result in inappropriate folding or instability in E. coli. Western blot analysis with anti-His-tag antibody showed that ΔTMCrPDAT exhibited a lower steady-state level of protein than that of full-length CrPDAT (Fig. S3), suggesting the potential instability of the truncated form.
Synergistic effect on TAG production by coexpressing WS/DGAT and CrPDAT.
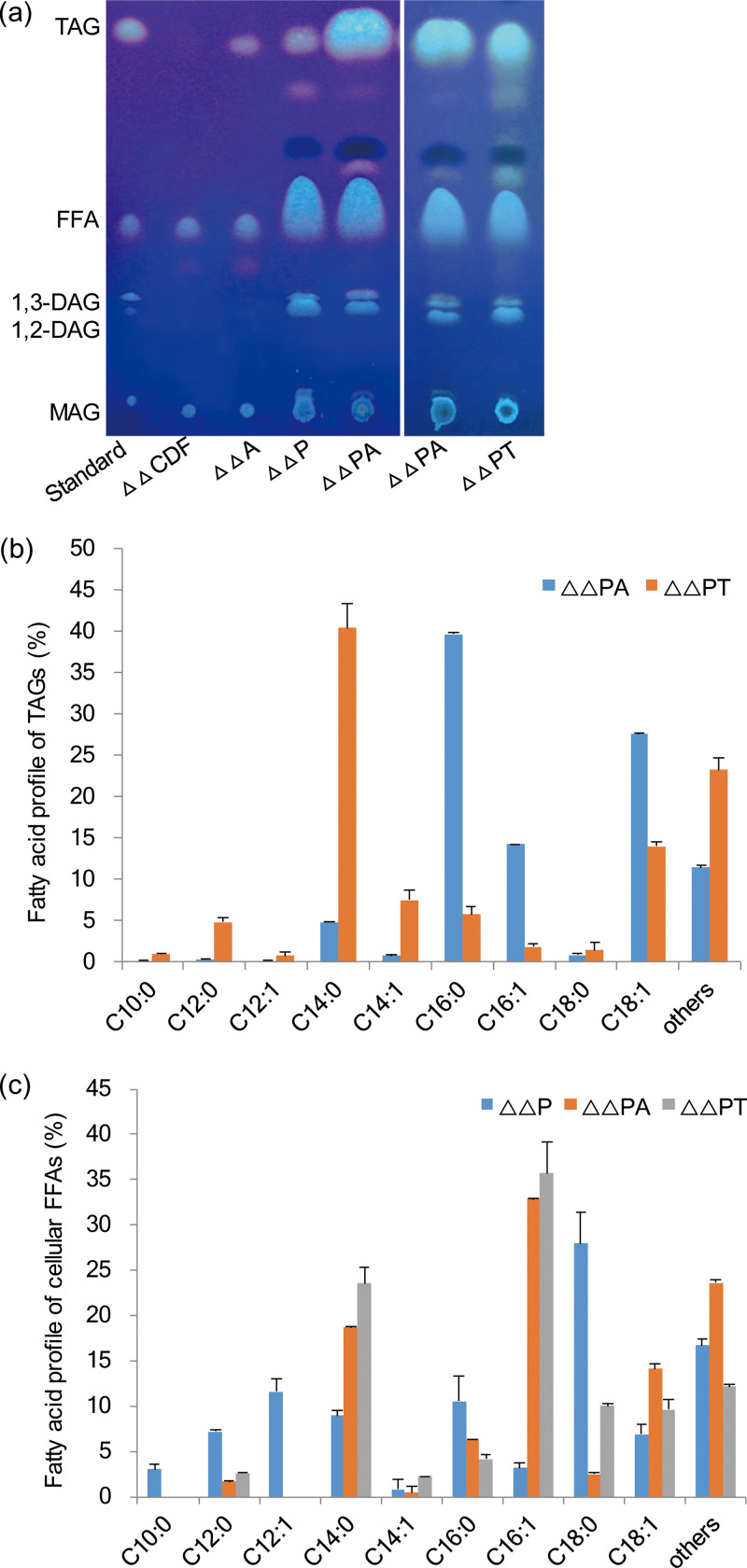
PDAT and DGAT play overlapping roles in TAG synthesis in algae and plants (10, 14). In addition, we previously constructed TAG-producing E. coli by overexpressing the bacterial WS/DGAT- and PAP-encoding genes atfA (from Acinetobacter baylyi) and RoPAP (from Rhodococcus opacus) (21). Therefore, we investigated if expressing CrPDAT would further increase TAG levels. The results indicated that coexpressing atfA and RoPAP along with CrPDAT in ΔΔBL21 indeed enhanced the cellular TAG levels further, from 0.1% TAG (wt/wt cell dry weight [CDW]) in the cells without CrPDAT to 2.3% TAG (corresponding to 64.4 mg/liter) (Fig. 3a and 4a). Of interest, a strong synergistic effect was observed in the strain ΔΔPA (ΔΔBL21 harboring atfA, RoPAP, and CrPDAT), as only less than 0.1% TAG was produced by expressing either a solo CrPDAT or the combination of atfA and RoPAP in ΔΔBL21 (Fig. 3a).
FIG 3.
Synergistic effects of CrPDAT and WS/DGAT on TAG content and fatty acid profile. (a) Thin-layer chromatography (TLC) result of neutral lipid profile. (b) Fatty acid profiles of cellular TAGs extracted from ΔΔPA and ΔΔPT. (c) Fatty acid profiles of cellular FFAs extracted from ΔΔP, ΔΔPA, and ΔΔPT. Cells were cultured in auto-induction medium with 3% glycerol at 37°C with shaking at 200 rpm for 48 h. Lipids were extracted from 20 mg of dried recombinant cells. ΔΔCDF, ΔΔBL21 harboring pCDFDuet-1; ΔΔA, ΔΔBL21 harboring pCDFDuet::atfA::RoPAP; ΔΔP, ΔΔBL21 harboring CrPDAT; ΔΔPA, ΔΔBL21 harboring CrPDAT, atfA, and RoPAP; ΔΔPT, ΔΔBL21 harboring CrPDAT, tDGAT, and RoPAP. All data are the mean ± standard deviation (shown as error bar) from triplicate samples.
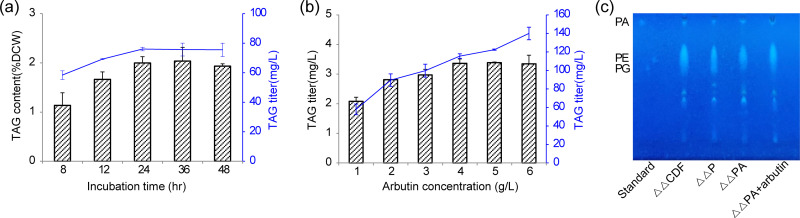
FIG 4.
Effects of incubation time (a) and arbutin concentration (b) on TAG content and TAG titer of the ΔΔPA strain and the TLC showing the phospholipid profile compared to its parental strains (c). Cells were cultured in auto-induction medium with 3% glycerol at 37°C with shaking at 200 rpm. All data are the mean ± standard deviation (shown as error bar) from triplicate samples.
Moreover, overexpression of a solo CrPDAT in ΔΔBL21 led to a 1.8-fold increase in biomass (6.8 g/liter) compared to that in ΔΔBL21 harboring empty vector (3.7 g/liter). In contrast, individual overexpression of atfA or RoPAP did not obviously affect the biomass (3.8 g/liter). A similar observation was obtained in Arabidopsis, but the mechanism was not characterized (31). We also noticed that the cellular total fatty acid levels were greatly upregulated in ΔΔBL21 harboring CrPDAT, leading to a 2.3-fold increase compared to the control strain (208.9 versus 92.8 mg/liter). In addition, CrPDAT did not alter the fatty acid profile (data not shown). Accumulation of large amounts of TAGs with a concomitant increase in total fatty acids (TFAs) suggested an increase in the rate of fatty acid synthesis. This result was in accordance with the phenomenon of AtPDAT1 overexpression (14). Compared to the combination of atfA and RoPAP, CrPDAT alone also caused drastic increases in free fatty acids (FFAs) and DAGs (Fig. 3a). Such increases were probably caused by the TAG lipase activity of CrPDAT (10).
Our previous study and reports by other labs demonstrated that a thermophilic tDGAT (DGAT from Thermomonospora curvata) was more efficient than AtfA for TAG synthesis in the engineered E. coli (21, 32). However, the combined expression of tDGAT and RoPAP along with CrPDAT (designated strain ΔΔPT) yielded 16.3 mg/liter of TAGs, which was only one-third of the levels in ΔΔPA (Fig. 3a). We also observed that ΔΔPT led to the TAGs being dominated by C10-C14 fatty acids (47.2% of total), whereas the proportion of long-chain fatty acids, including C16:0, C16:1, and C18:1, had significantly dropped (Fig. 3b). The fatty acid composition in cellular FFA was changed as well, with increases in the proportions of both C14:0 and the monounsaturated fatty acids C16:1 and C18:1 (Fig. 3c). We previously observed that tDGAT also had the ability to facilitate incorporation of large amounts of C14 fatty acids into glycerol backbones (21). Therefore, tDGAT might exhibit a preference for tailored acyl-CoAs in E. coli. The reduced proportion of TAG in the strain ΔΔPT might be caused by differential substrate selectivity of tDGAT.
To alleviate the metabolic burden, all three candidate genes, CrPDAT, atfA, and RoPAP, were then rearranged into pRSFDuet-1 or pCDFDuet-1 (Table 1) to generate the strains ΔΔRSF-PA or ΔΔCDF-PA. However, assembly of the three genes into one vector (pRSFDuet-1 or pCDFDuet-1) did not lead to higher TAG production than the strain ΔΔPA harboring two separate vectors (Fig. S4A). Although the biomass of ΔΔRSF-PA was higher than that of the other strains, its corresponding TAGs dropped substantially (Fig. S4B). Therefore, the ΔΔPA strain was used in the following tests.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pCDFDuet-1 | Sper ori CDF lacl T7 | Novagen |
| pRSFDuet-1 | Kanr ori RSF lacl T7 | Novagen |
| pRSF::CrPDAT | pRSFDuet-1 containing CrPDAT | This study |
| pRSF::RcPDAT | pRSFDuet-1 containing RcPDAT | |
| pRSF::ScPDAT | pRSFDuet-1 containing ScPDAT | |
| pRSF::ΔTMCrPDAT | pCDFDuet-1 containing CrPDAT without a transmembrane domain | This study |
| pCas9 | repA101(Ts) kan pCas-CAS9 ParaB-Red lacIq Ptrc-sgRNA-pMB | 37 |
| pTargetF | pMB1::aadA::sgRNA | 37 |
| pTargetF::fadE-sgRNA | pMB1::aadA::fadE-sgRNA | This study |
| pTargetF::dgkA-sgRNA | pMB1::aadA::dgkA-sgRNA | This study |
| pTargetT::fadE | pMB1::aadA::fadE-sgRNA containing deletion cassette of fadE | This study |
| pTargetT::dgkA | pMB1::aadA::dgkA-sgRNA containing deletion cassette of dgkA | This study |
| pCDF::atfA::RoPAP | pCDFDuet-1 containing atfA and RoPAP | 21 |
| pRSF::CrPDAT::atfA-RoPAP | pRSFDuet-1 containing CrPDAT and atfA-RBS-RoPAP | This study |
| pCDF::atfA-RoPAP::CrPDAT | pCDFDuet-1 containing atfA-RBS-RoPAP and CrPDAT | This study |
| pRSF::CrPDAT-Phe-262-Gly | pRSFDuet-1 containing CrPDAT-Phe-262-Gly | This study |
| pRSF::CrPDAT-Phe-262-Ala | pRSFDuet-1 containing CrPDAT-Phe-262-Ala | This study |
| pRSF::CrPDAT-Ser-401-Asn | pRSFDuet-1 containing CrPDAT-Ser-401-Asn | This study |
| pRSF::CrPDAT-Lys-461-Arg | pRSFDuet-1 containing CrPDAT-Lys-461-Arg | This study |
| pRSF::CrPDAT-Lys-461-Thr | pRSFDuet-1 containing CrPDAT-Lys-461-Thr | This study |
| pRSF::CrPDAT-Gly-941-Ile | pRSFDuet-1 containing CrPDAT-Gly-941-Ile | This study |
| pRSF::CrPDAT-Thr-942-Lys | pRSFDuet-1 containing CrPDAT-Thr-942-Lys | This study |
| pRSF::CrPDAT-His-994-Asp | pRSFDuet-1 containing CrPDAT-His-994-Asp | This study |
| E. coli strains | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) phoA supE44 λ thi-1 gyrA96 relA | TaKaRa Bio |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ (DE3) | TaKaRa Bio |
| BL21ΔfadE | Knockout of fadE in E. coli BL21(DE3) | This study |
| BL21/pRSF::CrPDAT | E. coli BL21(DE3) harboring pRSF::CrPDAT | This study |
| BL21/pRSF::ΔTMCrPDAT | E. coli BL21(DE3) harboring pRSF::ΔTMCrPDAT | This study |
| ΔΔBL21 | Knockout of dgkA and fadE in E. coli BL21(DE3) | This study |
| ΔΔRSF | ΔΔBL21 harboring pRSFDuet-1 | This study |
| ΔΔCDF | ΔΔBL21 harboring pCDFDuet-1 | This study |
| ΔΔP | ΔΔBL21 harboring pRSF::CrPDAT | This study |
| ΔΔBL21/pRSF::ΔTMCrPDAT | ΔΔBL21 harboring pRSF::ΔTMCrPDAT | This study |
| ΔΔA | ΔΔBL21 harboring pCDFDuet::atfA::RoPAP | This study |
| ΔΔPA | ΔΔBL21 harboring pRSF::CrPDAT and pCDFDuet::atfA::RoPAP | This study |
| ΔΔPT | ΔΔBL21 harboring pRSF::CrPDAT and pCDFDuet::tDGAT::RoPAP | This study |
| ΔΔRSF-PA | ΔΔBL21 harboring pRSF::CrPDAT::atfA-RoPAP | This study |
| ΔΔCDF-PA | ΔΔBL21 harboring pCDF::atfA-RoPAP::CrPDAT | This study |
Shortening the incubation time and supplementation of the medium with arbutin-enhanced TAG synthesis in the engineered E. coli.
Considering that the contribution of CrPDAT to TAG synthesis is mainly during the logarithmic growth phase in C. reinhardtii, and that CrPDAT also plays a role in TAG degradation, as it possesses TAG lipase activity (10), the time course for TAG production in the strain ΔΔPA was then investigated. The results showed that the TAG yield from the strain ΔΔPA reached 66.6 mg/liter by 24 h, and extension of the incubation time to 36 or 48 h did not further increase the TAG yield (Fig. 4a). This observation demonstrated that the combinational expression of CrPDAT and AtfA led to higher TAG productivity due to the synchronous TAG accumulation with cell growth.
Arbutin as an artificial β-glucoside acceptor that participates in turnover of PG and PE, and supplementation of arbutin into the medium is reported to redirect PG and PE into DAG (26). Thus, we speculated that TAG synthesis might be enhanced by supplementation of arbutin. The effects of arbutin at different concentrations on TAG accumulation in the ΔΔPA strain were investigated. Results showed that the supplementation of medium with 4.6 g/liter arbutin led to a 61.7% increase in TAG yield, reaching up to 108.3 mg/liter of TAGs after the cells were incubated for 5 h (Fig. 4b). Although the supplementation of a higher concentration of arbutin (9.3 g/liter) resulted in the highest percentage of TAG, the cell biomass was drastically decreased. Indeed, the TAG yield was not further enhanced with increased arbutin concentrations (Fig. 4b). Meanwhile, the PL content and profiles from the ΔΔPA strain did not change when incubated with 4.6 g/liter of arbutin (Fig. 4c). In addition, the fatty acid profile of the TAG fraction was not obviously affected by the supplementation of arbutin. These results suggested that in addition to pushing PAs toward DAG synthesis and downregulation of DAG turnover, redirection of osmo-regulated periplasmic glucan (OPG) synthesis into DAGs was an alternative way for enhancement of TAG accumulation. However, supplementation of arbutin into the medium to enhance the TAG production is not economically feasible. Recently, the arbutin biosynthetic pathway has been reconstructed in E. coli (28). Additional expression of MNX1 and AS, which are two essential genes involved in arbutin biosynthesis might enhance TAG production by the de novo-produced arbutin.
Rational mutagenesis of CrPDAT for decreasing TAG lipase activity without impairing acyl transferase activity.
We noticed that expression of CrPDAT alone or a combined expression of CrPDAT and atfA in ΔΔBL21 greatly elevated the cellular FFA levels. In contrast to CrPDAT, individual expression of atfA did not lead to an obvious increase in FFA (Fig. 2). CrPDAT has been demonstrated to be a multifunctional enzyme possessing TAG lipase activity, which probably causes the TAG degradation, in turn, and an increase in FFAs, especially when CrPDAT has been overexpressed (10). Therefore, we carried out the rational mutagenesis of CrPDAT to specifically inhibit the TAG lipase activity.
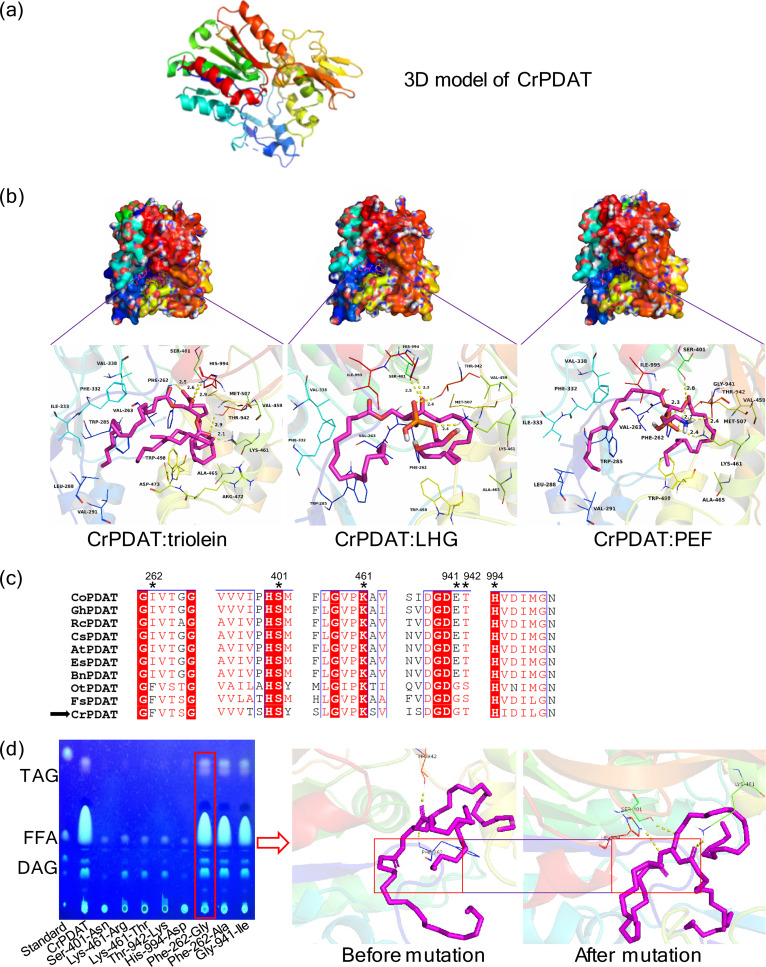
A three-dimensional (3D) model of CrPDAT (Fig. 5a) was generated using Phyre2 according to the crystal structure of lysosomal phospholipase A2 in complex with isopropyl dodec-11-enylfluorophosphonate (IDFP; PDB: 4X91) as the template. The triolein compound was docked into the binding site of CrPDAT, and the theoretical binding mode of the triolein in the CrPDAT binding site is illustrated in Fig. 5b. Triolein adopted a compact conformation to bind inside the CrPDAT pocket (Fig. 5b). Two of the aliphatic chains of triolein were positioned at the hydrophobic pocket, one surrounded by the residues Val-263, Trp-285, Leu-288, Val-291, Phe-332, Ile-333, and Val-338 and the other surrounded by the residues Phe-262, Val-263, Ala-465, Trp-498, and Met-507, forming a strong hydrophobic binding domain (Fig. 5b). The 3D docking model further suggested that three hydrogen bonds could be formed between the triolein and the residues Phe-262, Ser-401, and His-994, with bond lengths of 2.5, 2.9, and 2.6 Å, respectively. Importantly, the carbonyl “O” of the triolein formed two hydrogen bonds with the residues Lys-461 and Thr-942, with bond lengths of 2.1 and 2.9 Å, respectively. All these interactions probably help triolein to anchor in CrPDAT.
FIG 5.
Schematic of the rational mutagenesis system workflow. The aim of this system is to decrease TAG lipase activity without impairing acyltransferase activity of CrPDAT (a). Three docking models of triolein (left), LHG (middle), and PEF (right) to CrPDAT were simulated (b). The essential amino acids involved in hydrogen bond formation were predicted. Sequence alignment of 10 selected PDATs was carried out to identify the distribution of those predicted essential amino acids (c). Identical residues are shown in white on a red background, while similar residues are shown in red. The predicted binding sites are indicated by black asterisks. The ability of the rational designed mutants to accumulate TAG was determined by TLC (d). The binding model of the positive mutant (Phe-262-Gly) was reconstructed by molecular docking to confirm the loss of the hydrogen bond between CrPDAT and triolein (d).
The compounds di-palmitoyl-3-sn-phosphatidylethanolamine (PEF) and 1,2-dipalmitoyl-phosphatidyl-glycerole (LHG) were then docked into CrPDAT in the 3D model (Fig. 5b). One of the aliphatic chains of PEF was positioned at hydrophobic pocket 1, surrounded by the residues Val-263, Trp-285, Leu-288, Val-291, Phe-332, Ile-333, Val-338, and Ile-995, while the other PEF aliphatic chain was located at hydrophobic pocket 2, surrounded by the residues Phe-262, Val-459, Ala-465, Trp-498, and Met-507, forming a strong hydrophobic binding domain (Fig. 5b). In the case of LHG, the predicted hydrophobic pocket 1 was surrounded by Val-263, Trp-285, Phe-332, Val-338, and Ile-995, while hydrophobic pocket 2 had the same amino acids as in the CrPDAT:PEF model.
In addition, the residues Lys-461 and Thr-942 might be crucial for formation of the three hydrogen bonds between the phosphate groups of all three candidates (PEF/LHG/triolein). Furthermore, one of the “O” atoms of the PEF formed a hydrogen bond with the residue Ser-401, and the polar hydrogen of the NH2 group of PEF formed a hydrogen bond with the residue Gly-941 (Fig. 5b). In contrast, Ser-401 and His-994, with bond lengths of 2.8 and 2.4 Å, respectively, participated in hydrogen bond formation with LHG according to the three-dimensional model.
The above-described molecular simulations in the 3D model gave us a rational explanation of the interactions between CrPDAT and PEF/LHG/triolein. Although five putative amino acid residues, Ser-401, Lys-461, Thr-942, His-994, and Phe-262, were predicted to play key roles in the CrPDAT:triolein model, Phe-262 was the only differential amino acid among three models. Phe-262 was not essential for hydrogen bond formation in either the CrPDAT:PEF or CrPDAT:LHG model. Therefore, mutation of Phe-262 might alter the hydrogen bond between CrPDAT and triolein. We also noticed that Phe-262 was involved in hydrophobic pocket formation in both the CrPDAT:PEF and CrPDAT:LHG models (Fig. 5b); thus, mutation of Phe-262 might also affect acyl transferase activity.
Moreover, sequence alignment of selected PDAT proteins was carried out. Among the six predicted amino acid residues, four of them, Ser-401, Lys-461, Thr-942, and His-994, were conserved among PDATs, while Phe-262 and Gly-941 were less conservative (Fig. 5c). Notably, Ser-401 and His-994 were also part of the catalytic triad (Ser-Asp-His) (7).
Based on the above-described prediction, we constructed a series of mutants (Table 1). We first investigated the neutral lipid profile of these mutants by using TLC. Five mutants, Ser-401-Asn, Lys-461-Arg, Lys-461-Thr, Thr-942-Lys, and His-994-Asp, were unable to synthesize any detectable TAGs (Fig. 5d), suggesting that these amino acid residues were essential to PDAT activity. Then, we further analyzed the other three mutants which could produce TAGs. Only mutant Phe-262-Gly led to a 17.5% decrease in FFA levels without affecting TAG synthesis (Table 2 and Fig. 5d). An in vitro enzyme test also demonstrated that the TAG lipase activity of mutant Phe-262-Gly decreased, leading to more stable TAG and less produced FFA compared with that of CrPDAT (Fig. S5). In contrast, although the FFA levels in the Phe-262-Ala and Gly-941-Ile mutants decreased by 39.7% and 59.5%, TAGs from these two mutants were also reduced by 32.7% and 27.5%, respectively (Table 2 and Fig. 5d). There was also a side effect of the reduced biomass in the mutants Phe-262-Ala and Gly-941-Ile, so these were not the preferable choices for enhancing TAG production. In addition, we found that the Gly-941-Ile mutant exhibited a much lower growth rate than the wild-type E. coli and other mutants during the exponential growth phase (0 to 8 h), although the difference became insignificant after reaching the stationary growth phase. Nevertheless, mutant Phe-262-Gly was a beneficial candidate for TAG accumulation. Mutation of Phe-262 to other residues might lead to further reduced lipase activity.
TABLE 2.
| Strain | Biomass (g/liter) | TAG (μg/mg CDW) | TAG (mg/liter) | FFA (mg/liter) | TFA (mg/liter) |
|---|---|---|---|---|---|
| ΔΔP | 6.8 ± 0.2 | 0.9 ± 0.2 | 6.1 ± 0.8 | 155.9 ± 15.2 | 208.9 ± 9.6 |
| Mutant phe-262-Gly | 6.3 ± 0.3 | 1.0 ± 0.1 | 6.2 ± 0.5 | 128.6 ± 25.5c | 241.4 ± 27.3 |
| Mutant Phe-262-Ala | 5.1 ± 0.8 | 0.8 ± 0.1 | 4.1 ± 0.1 | 93.9 ± 30.6c | 131.4 ± 68.1 |
| Mutant Gly-941-Ile | 4.9 ± 0.4 | 0.9 ± 0.1 | 4.4 ± 0.3 | 63.1 ± 17.2c | 158.9 ± 11.4 |
ΔΔP, ΔΔBL21 harboring pRSF::CrPDAT; TAG, triacylglycerol; FFA, free fatty acid; TFA, total fatty acid; CDW, cell dry weight.
Cells were cultivated in ZYP-5052 auto-induction medium with 3% glycerol at 37°C and shaking at 200 rpm for 48 h. All data are the means ± standard deviations from triplicates.
The significance between ΔΔP and each mutant (Student’s t test; P < 0.05).
CrPDAT orthologues resulted in TAG accumulation in E. coli.
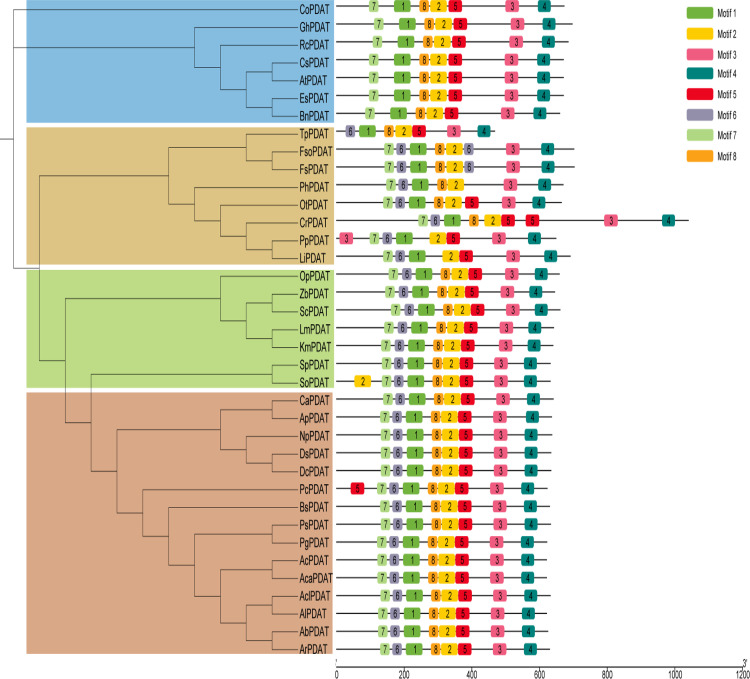
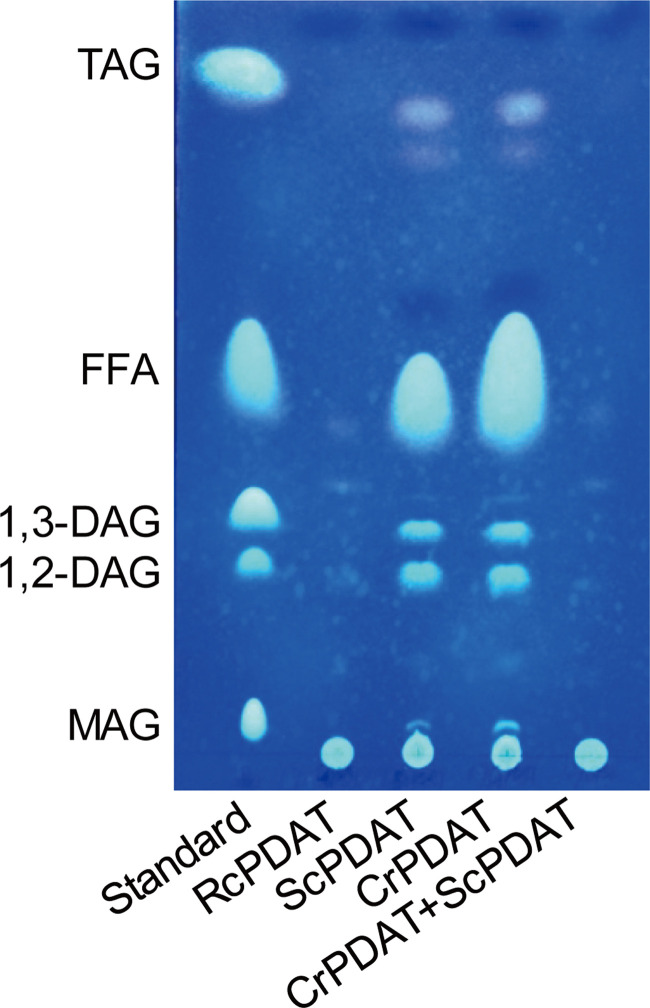
We next investigated if other PDATs could play similar roles in diverting fatty acids from PLs into TAGs in strain ΔΔBL21. According to the motif pattern of the PDAT proteins from plants, fungi, and microalgae, the major difference among CrPDAT and other PDATs is the existence of three long linkers (Fig. 6). Cladogram analysis indicated that plant source RcPDAT (PDAT from Ricinus communis) and yeast ScPDAT reside in the different groups from CrPDAT (Fig. 6). TLC results indicated that overexpression of ScPDAT in ΔΔBL21 led to obvious DAG and TAG accumulation, whereas RcPDAT did not (Fig. 7). In addition, either RcPDAT or ScPDAT overexpression caused an increase in biomass, to the same level of 5.4 g/liter, relative to the 3.7 g/liter of biomass from the ΔΔBL21 strain harboring empty vector (Table 3). This was similar to our above-described observation that biomass was increased to 6.8 g/liter by CrPDAT overexpression and again confirmed our previous hypothesis that overexpression of PDAT in the engineered E. coli could lead to synchronous TAG production with cell growth.
FIG 6.
Cladogram (left) and motif patterns (right) of PDAT proteins from representative plants, fungi, and algae. The detailed information on each PDAT protein is listed in Table S2.
FIG 7.
Evaluation of the ability of RcPDAT and ScPDAT for the production of TAG in ΔΔBL21 by using thin-layer chromatography (TLC). RcPDAT, PDAT from R. communis; ScPDAT, PDAT from S. cerevisiae; CrPDAT, PDAT from C. reinhardtii. Cells were cultured in auto-induction medium with 3% glycerol at 37°C with shaking at 200 rpm for 48 h. Lipids were extracted from 20 mg of dried recombinant cells. All data are the mean ± standard deviation from triplicate samples.
TABLE 3.
| Strain/ΔΔBL21 harboring: | Biomass (g/liter) | TAG (μg/mg CDW) | TAG (mg/liter) |
|---|---|---|---|
| pRSF::CrPDAT | 6.8 ± 0.1 | 0.9 ± 0.1 | 6.1 ± 0.1 |
| pRSF::RcPDAT | 5.4 ± 0.7 | 0.1 ± 0.0 | 0.5 ± 0.0 |
| pRSF::ScPDAT | 5.4 ± 0.1 | 1.0 ± 0.1 | 5.4 ± 0.1 |
CrPDAT, PDAT from C. reinhardtii; RcPDAT, PDAT from R. communis; ScPDAT, PDAT from S. cerevisiae; TAG, triacylglycerol; CDW, cell dry weight.
Cells were cultivated in ZYP-5052 auto-induction medium with 3% glycerol at 37°C and shaking at 200 rpm for 48 h. All data are the means ± standard deviations from triplicates.
Considering that ScPDAT shows a preference for PE over PC, while CrPDAT prefers anionic PLs (10, 13), we speculated that the combinational expression of ScPDAT and CrPDAT might further increase TAG levels via full utilization of PLs. However, coexpressing CrPDAT and ScPDAT drastically decreased TAG levels to less than 0.01% (Fig. 7), suggesting that the two enzymes did not work coordinately or that PLs were strictly regulated in E. coli. In the next step, further downregulation of PL synthesis may be achieved by blocking the expression of the phosphatidylserine decarboxylase gene, psd, and/or the phosphatidylserine synthase gene, pssA, based on CRISPR interference (CRISPRi) (33) (Fig. 1).
PDATs from castor bean, Crepis palaestina, and flax are involved in accumulation of unusual fatty acids, such as ricinoleic acid, vernolic acid, and α-linolenic acid (6, 34, 35). Notably, AtPDAT and ScPDAT also show strong specificity for acyl groups with multiple double bonds or a functional group (13, 15). These findings indicate the that PDATs probably play a vital role in removal of unusual fatty acids from PLs to TAGs. We speculate that these PDATs may be employed to produce TAGs rich in unusual fatty acids via metabolic engineering of microbes.
Conclusions. We demonstrated that a CrPDAT-mediated acyl-CoA-independent pathway could divert fatty acid flux from PLs into TAGs in E. coli without impairing membrane functions. Of interest, CrPDAT and AtfA had a synergistic effect on TAG production. Combined expression of CrPDAT and tDGAT altered TAG composition with increased C10 to C14 fatty acids. Overexpression of CrPDAT alone or the CrPDAT and atfA combination led to synchronous TAG production with cell growth, which resulted in a high level of TAG productivity in 24 h. Supplementation of arbutin into the medium further increased TAG production by altering MDO metabolism. In addition, our experiments with rationally designed CrPDAT mutants revealed that many essential amino acid residues played overlapping roles in both acyltransferase and TAG lipase activity, while only the Phe-262-Gly mutant showed decreased TAG lipase activity without interfering acyltransferase activity. Finally, ScPDAT exhibited functions in the engineered E. coli similar to CrPDAT. The investigation of the roles of CrPDAT in E. coli opens the possibility of utilizing this enzyme for the synthesis of TAGs and other functional lipids in prokaryotic microbes.
MATERIALS AND METHODS
Materials.
Chemicals were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China) or Sigma-Aldrich Co. (St. Louis, MO, USA). Arbutin was purchased from Adamas Reagent, Ltd. (Shanghai, China). Hexane, chloroform, and methanol were purchased from Merck KGaA (Darmstadt, Germany). T4 DNA ligase and restriction enzymes were purchased from Thermo Fisher Scientific (Beverly, USA). The plasmid miniprep kit, total DNA extraction kit, PCR purification kit, SoSoo cloning kit, and gel extraction kit were purchased from Beijing TsingKe Biotech Co., Ltd. (Beijing, China). E. coli DH5α and E. coli BL21(DE3) chemically competent cells were purchased from TransGen Biotech (Beijing, China). Standards, including FFA, DAG, monoacylglycerol (MAG), TAG, PA, PE, PG, and GLC-411 were purchased from Nu-Chek (Elysian, MN, USA) or Larodan (Stockholm, Sweden).
Strains, medium, and growth conditions.
E. coli DH5α was employed for plasmid construction and propagation, while E. coli BL21(DE3) was used as the expression strain for TAG accumulation. E. coli DH5α cells were cultivated in lysogeny broth (LB; 1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 1% [wt/vol] NaCl) at 37°C with shaking at 200 rpm. E. coli BL21(DE3) cells were cultivated in ZYP-5052 auto-induction medium (36) with 3% glycerol at 37°C and shaking at 200 rpm. Appropriate antibiotics were added at the following concentrations: 50 mg/liter kanamycin or 50 mg/liter spectinomycin.
Plasmid construction.
All plasmids used in this study are listed in Table 1. The CrPDAT gene from C. reinhardtii (GenBank accession number XM_001699696), ScPDAT (accession number NP_014405) from S. cerevisiae, and RcPDAT (accession number GU989637) from Ricinus communis were codon-optimized and chemically synthesized (Table S1). Full-length CrPDAT, ScPDAT, RcPDAT, and CrPDAT without the transmembrane domain (ΔTMCrPDAT) were inserted separately into pRSFDuet-1 via the EcoRI/HindIII restriction sites. The plasmid pCDF::atfA::RoPAP was constructed previously (21). Both the A. baylyi WS/DGAT gene (atfA) and the Rhodococcus opacus PD630 PAP gene (RoPAP) were inserted into pRSF::CrPDAT via KpnI/XhoI to generate pRSF::CrPDAT::atfA-RoPAP. Then, the CrPDAT gene was inserted into pCDFDuet::atfA-RoPAP via KpnI/XhoI to generate pCDF::atfA-RoPAP::CrPDAT. According to the instructions of the SoSoo cloning kit, site-directed mutagenesis of DNA fragments of CrPDAT and linearized pRSFDuet-1 were fused by the mixed recombination enzyme in 2× SoSoo mix. The reaction system was incubated for 15 min at 50°C, and then the mixture was transformed into E. coli DH5α. Positive mutants were confirmed by sequencing. Primers are listed in Table 4.
TABLE 4.
Primers used in this study
| Primer | Nucleotide sequence (5′–3′)a |
|---|---|
| CrPDAT-F | CACAGCCAGGATCCGAATTCATGACCACACCGACC |
| CrPDAT-R | AGCATTATGCGGCCGCAAGCTTTTAGGCTGCCAGCGC |
| CrPDAT-TMD-F | CACAGCCAGGATCCGAATTCATGGTTGAAGAAGGTCCG |
| fadE-IF | CCGATTGCCATCACCGTT |
| fadE-IR | GCGAACTTTGTTGCTACCG |
| dgkA-IF | CGGTACACTGATATTGACGCTC |
| dgkA-IR | GCGTCGGCGGCATACCTGT |
| dgkA-OF | GGGAAATTCTGTGGTATCCGCTC |
| dgkA-OR | CGGCGGCATACCTGTCTGG |
| CrPDAT-Phe-262-Gly-LR | TCCAGACCGCTGGTAACACCACCCGGAACAATAACAA |
| CrPDAT-Phe-262-Gly-RF | TTGTTATTGTTCCGGGTGGTGTTACCAGCGGTCTGGA |
| CrPDAT-Phe-262-Ala-LR | TCCAGACCGCTGGTAACAGCACCCGGAACAATAACAA |
| CrPDAT-Phe-262-Ala-RF | TTGTTATTGTTCCGGGTGCTGTTACCAGCGGTCTGGA |
| CrPDAT-Ser-401-Asn-LR | AAAACATTTTCACCATAATTATGGCTGGTCACAACAA |
| CrPDAT-Ser-401-Asn-RF | TTGTTGTGACCAGCCATAATTATGGTGAAAATGTTTT |
| CrPDAT-Lys-461-Arg-LR | AGCAGTGCGCTAACACTTCTCGGAACACCCAGGCTGG |
| CrPDAT-Lys-461-Arg-RF | CCAGCCTGGGTGTTCCGAGAAGTGTTAGCGCACTGCT |
| CrPDAT-Lys-461-Thr-LR | AGCAGTGCGCTAACACTTGTCGGAACACCCAGGCTGG |
| CrPDAT-Lys-461-Thr-RF | CCAGCCTGGGTGTTCCGACAAGTGTTAGCGCACTGCT |
| CrPDAT-Gly-941-Ila-LR | ACTCAGCAGAGGAACCGTAATATCACCGTCGCTAATAT |
| CrPDAT-Gly-941-Ila-RF | ATATTAGCGACGGTGATATTACGGTTCCTCTGCTGAG |
| CrPDAT-Thr-942-Lys-LR | AGACTCAGCAGAGGAACCTTACCATCACCGTCGCTAA |
| CrPDAT-Thr-942-Lys-RF | TTAGCGACGGTGATGGTAAGGTTCCTCTGCTGAGTCT |
| CrPDAT-His-994-Asp-LR | ATTACCCAGAATATCAATATCTGCGGCTGCTGCTGGA |
| CrPDAT-His-994-Asp-RF | TCCAGCAGCAGCCGCAGATATTGATATTCTGGGTAAT |
Underlining indicates restriction enzyme sites.
Gene deletion with the CRISPR/Cas9 system.
Deletion of both the fadE gene encoding acyl-CoA dehydrogenase that catalyzes the first step of β-oxidation and the diacylglycerol kinase gene dgkA in E. coli BL21(DE3) was conducted according to a modified CRISPR/Cas9 system (37). The two-plasmid system comprising pCas and pTarget was applied to delete fadE or dgkA from the genome of E. coli BL21(DE3). A protospacer-adjacent motif (PAM) was chosen by using online software (https://www.atum.bio/eCommerce/cas9/input). The chosen single guide RNA (sgRNA) was then further analyzed with Cas-OFFinder (http://www.rgenome.net/cas-offinder/) to avoid any potential off-target effects. The second structure of the designed sequence was checked by using mfold (http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form). The N20 sequences of sgRNA-fadE and sgRNA-dgkA were designed as follows: 5′-GATGTCTCGATGGCTGTGCT-3′ and 5′-TAAAGATATGGGATCCGCCG-3′.
For the knockout of fadE and dgkA, the deletion cassettes were fused with 200-bp up- and downstream flanking regions of the target genes. The sgRNA fragments of dgkA and fadE were chemically synthesized and subcloned into SpeI/XhoI sites of pTargetF to generate pTargetF::dgkA-sgRNA and pTargetF::fadE-sgRNA. pTargetT::dgkA and pTargetT::fadE were constructed by inserting the corresponding deletion cassette into the HindIII/XhoI-digested pTargetF::dgkA-sgRNA and pTargetF::fadE-sgRNA, respectively.
BL21(DE3) electro-competent cells harboring pCas were prepared as described previously (37, 38). Arabinose (10 mM) was added to the medium for λ-Red induction. Then, 200 ng of pTargetT::fadE was mixed with 100 μl of electro-competent induced cells. The mixture of plasmid and electro-competent cells was chilled on ice for 10 min. After electroporation, the mixture was transferred to 1.5-ml sterile tubes with 1 ml of LB medium and incubated at 30°C for 1 h. Positive mutants were identified by colony PCR and sequencing. pTargetT::fadE was cured by incubation with spectinomycin (50 mg/liter) at 30°C.
In the first round of deletion, BL21ΔfadE was obtained. Then, BL21ΔfadE harboring pCas was used for the deletion of dgkA. After a second round of electroporation, pTargetT::dgkA was cured by incubation with spectinomycin (50 mg/liter) at 30°C. Then, pCas9 was cured by incubating it overnight at 37°C. The obtained double mutant BL21ΔfadE ΔdgkA, designated ΔΔBL21, was confirmed by sequencing.
Bioinformatic analysis.
TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used for transmembrane prediction. Protein sequence alignment was conducted with MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/) (39). Phylogenetic analysis of PDATs was conducted by using MEGA 6.06 with the neighbor-joining method (N-J) (40). Online iTOL (http://itol.embl.de) software was used to visualize the phylogenetic tree (41). Motif occurrences were predicted using the MEME program (http://meme-suite.org/tools/meme) and visualized with TBtools (42).
The three-dimensional model of the CrPDAT was built with Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) (43). AutoDock Vina 1.1.2 was used to search for potential binding sites of LHG, PEF, and triolein within CrPDAT (44). The 2D structures of LHG, PEF, and triolein were drawn with ChemBioDraw Ultra 14.0 and converted to a 3D structure with the ChemBio3D Ultra 14.0 package. The AutoDockTools 1.5.6 package was employed to generate the docking input files (45). The best-scoring pose judged by the Vina docking score was chosen and visualized with PyMOL 1.7.6 software (http://www.pymol.org/).
Lipid analysis.
Cells were collected by centrifugation and vacuum-dried overnight. Total lipids were extracted from freeze-dried cells as previously described (21). The extracted lipids were separated by TLC using a solvent system (hexane:diethyl ester:acetic acid = 70:30:1, vol/vol/vol) and visualized by spraying with primuline. For separation of PLs, a solvent system consisting of chloroform/methanol/acetic acid/water (90:15:10:3, vol/vol/vol/vol) was used. The bands corresponding to TAG or FFA were scraped from TLC plates into vials and methylated to generate fatty acid methyl esters (FAMEs) (46). FAMEs were then analyzed by using a 7890A gas chromatograph (GC; Agilent Technologies, USA) equipped with a flame ionization detector (FID) and an HP-FFAP column (30 m by 250 μm inside diameter [i.d.], 0.25 μm thickness). For quantification, either 5 μg of C15:0-TAG and 5 μg of C15:0-FFA (for low levels of TAG or FFA) or 20 μg of C15:0-TAG and 10 μg of C15:0-FFA were added to the samples as internal standards before lipid extraction. To quantify cellular TFAs, lipids from 2 mg of cell powder were directly methylated to produce corresponding FAMEs and were analyzed with GC as described above.
Microscopy analysis.
For detection of the membrane integrity of engineered cells, cells were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) under vacuum for at least 24 h. Then, the samples were treated with 50, 70, 80, and 95% ethanol sequentially and processed with a mixed solution of acetone and epoxy resin (1:1 for 1 h, then 1:3 for 3 h). After infiltration with epoxy resin and ultrathin-section treatment, the samples were stained with uranyl acetate and lead citrate. The samples were observed with a HT7700 transmission electron microscope (Hitachi High-Tech).
Data availability.
All data generated or analyzed during this study are included in this article and its supplemental files.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Harley Edwards for providing help with English.
We declare that we have no competing interests.
This work was financially supported by the Chinese Academy of Agricultural Sciences (grant numbers Y2020XK25 and CAAS-ASTIP-2016-OCRI), the Ministry of Science and Technology of the People’s Republic of China (grant number 2016YFD0501209), and the “3551” Innovative Talent Project of Optics Valley of China (K159).
L.W., S.J., W.-C.C., and X.-R.Z. conceived the project. L.W., S.J., and T.-X.H. carried out experimental work. L.W., W.-C.C., X.-R.Z., and X.W. analyzed and interpreted the data. X.-R.Z., F.-H.H., and X.W. wrote the paper, with all authors assisting in the process.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Waltermann M, Luftmann H, Baumeister D, Kalscheuer R, Steinbuchel A. 2000. Rhodococcus opacus strain PD630 as a new source of high-value single-cell oil? Isolation and characterization of triacylglycerols and other storage lipids. Microbiology 146:1143–1149. doi: 10.1099/00221287-146-5-1143. [DOI] [PubMed] [Google Scholar]
- 2.Olukoshi ER, Packter NM. 1994. Importance of stored triacylglycerols in Streptomyces: possible carbon source for antibiotics. Microbiology 140:931–943. doi: 10.1099/00221287-140-4-931. [DOI] [PubMed] [Google Scholar]
- 3.Theodoulou FL, Eastmond PJ. 2012. Seed storage oil catabolism: a story of give and take. Curr Opin Plant Biol 15:322–328. doi: 10.1016/j.pbi.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez HM, Steinbuchel A. 2002. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol 60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy EP. 1961. Biosynthesis of complex lipids. Fed Proc 20:934–940. [PubMed] [Google Scholar]
- 6.Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. 2000. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A 97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan X, Peng FY, Weselake RJ. 2015. Genome-wide analysis of phospholipid:diacylglycerol acyltransferase (PDAT) genes in plants reveals the eudicot-wide PDAT gene expansion and altered selective pressures acting on the core eudicot PDAT paralogs. Plant Physiol 167:887–904. doi: 10.1104/pp.114.253658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rottig A, Strittmatter CS, Schauer J, Hiessl S, Poehlein A, Daniel R, Steinbuchel A. 2016. Role of wax ester synthase/acyl coenzyme A:diacylglycerol acyltransferase in oleaginous Streptomyces sp. strain G25. Appl Environ Microbiol 82:5969–5981. doi: 10.1128/AEM.01719-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabolaza A, Rodriguez E, Altabe S, Alvarez H, Gramajo H. 2008. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl Environ Microbiol 74:2573–2582. doi: 10.1128/AEM.02638-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon K, Han D, Li Y, Sommerfeld M, Hu Q. 2012. Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24:3708–3724. doi: 10.1105/tpc.112.100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J, Yan C, Roston R, Shanklin J, Xu C. 2014. Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26:4119–4134. doi: 10.1105/tpc.114.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu XY, Ouyang LL, Zhou ZG. 2016. Phospholipid: diacylglycerol acyltransferase contributes to the conversion of membrane lipids into triacylglycerol in Myrmecia incisa during the nitrogen starvation stress. Sci Rep 6:26610. doi: 10.1038/srep26610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosal A, Banas A, Stahl U, Dahlqvist A, Lindqvist Y, Stymne S. 2007. Saccharomyces cerevisiae phospholipid:diacylglycerol acyl transferase (PDAT) devoid of its membrane anchor region is a soluble and active enzyme retaining its substrate specificities. Biochim Biophys Acta 1771:1457–1463. doi: 10.1016/j.bbalip.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Fan J, Yan C, Zhang X, Xu C. 2013. Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25:3506–3518. doi: 10.1105/tpc.113.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stahl U, Carlsson AS, Lenman M, Dahlqvist A, Huang B, Banas W, Banas A, Stymne S. 2004. Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol 135:1324–1335. doi: 10.1104/pp.104.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrag JD, Cygler M. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol 284:85–107. doi: 10.1016/s0076-6879(97)84006-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Pfleger BF, Kim SW. 2017. Reassessing Escherichia coli as a cell factory for biofuel production. Curr Opin Biotechnol 45:92–103. doi: 10.1016/j.copbio.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Janßen HJ, Steinbüchel A. 2014. Production of triacylglycerols in Escherichia coli by deletion of the diacylglycerol kinase gene and heterologous overexpression of atfA from Acinetobacter baylyi ADP1. Appl Microbiol Biotechnol 98:1913–1924. doi: 10.1007/s00253-013-5460-2. [DOI] [PubMed] [Google Scholar]
- 19.Uthoff S, Stoveken T, Weber N, Vosmann K, Klein E, Kalscheuer R, Steinbuchel A. 2005. Thio wax ester biosynthesis utilizing the unspecific bifunctional wax ester synthase/acyl coenzyme A:diacylglycerol acyltransferase of Acinetobacter sp. strain ADP1. Appl Environ Microbiol 71:790–796. doi: 10.1128/AEM.71.2.790-796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rottig A, Zurek PJ, Steinbuchel A. 2015. Assessment of bacterial acyltransferases for an efficient lipid production in metabolically engineered strains of E. coli. Metab Eng 32:195–206. doi: 10.1016/j.ymben.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, Wang L, Zhou XR, Chen WC, Singh S, Hu Z, Huang FH, Wan X. 2018. Stepwise metabolic engineering of Escherichia coli to produce triacylglycerol rich in medium-chain fatty acids. Biotechnol Biofuels 11:177. doi: 10.1186/s13068-018-1177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang MH, Jiang JG. 2013. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog Lipid Res 52:395–408. doi: 10.1016/j.plipres.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Marella ER, Holkenbrink C, Siewers V, Borodina I. 2018. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr Opin Biotechnol 50:39–46. doi: 10.1016/j.copbio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM. 2015. Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol 99:2191–2207. doi: 10.1007/s00253-014-6002-2. [DOI] [PubMed] [Google Scholar]
- 25.Rowlett VW, Mallampalli V, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H. 2017. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol 199. doi: 10.1128/JB.00849-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bontemps-Gallo S, Cogez V, Robbe-Masselot C, Quintard K, Dondeyne J, Madec E, Lacroix JM. 2013. Biosynthesis of osmoregulated periplasmic glucans in Escherichia coli: the phosphoethanolamine transferase is encoded by opgE. Biomed Res Int 2013:371429. doi: 10.1155/2013/371429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin F, Chen Y, Levine R, Lee K, Yuan Y, Lin XN. 2013. Improving fatty acid availability for bio-hydrocarbon production in Escherichia coli by metabolic engineering. PLoS One 8:e78595. doi: 10.1371/journal.pone.0078595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X, Wang J, Wang J, Chen Z, Yuan Q, Yan Y. 2017. High-level de novo biosynthesis of arbutin in engineered Escherichia coli. Metab Eng 42:52–58. doi: 10.1016/j.ymben.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Jackson BJ, Bohin JP, Kennedy EP. 1984. Biosynthesis of membrane-derived oligosaccharides: characterization of mdoB mutants defective in phosphoglycerol transferase I activity. J Bacteriol 160:976–981. doi: 10.1128/JB.160.3.976-981.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedler W, Rotering H. 1985. Characterization of an Escherichia coli mdoB mutant strain unable to transfer sn-1-phosphoglycerol to membrane-derived oligosaccharides. J Biol Chem 260:4799–4806. [PubMed] [Google Scholar]
- 31.Banas W, Carlsson AS, Banas A. 2014. Effect of overexpression of PDAT gene on Arabidopsis growth rate and seed oil content. J Agric Sci 6. doi: 10.5539/jas.v6n5p65. [DOI] [Google Scholar]
- 32.Lázaro B, Villa JA, Santín O, Cabezas M, Milagre CDF, de la Cruz F, Moncalián G. 2017. Heterologous expression of a thermophilic diacylglycerol acyltransferase triggers triglyceride accumulation in Escherichia coli. PLoS One 12:e0176520. doi: 10.1371/journal.pone.0176520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Du G, Chen J, Zhou J. 2015. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci Rep 5:13477. doi: 10.1038/srep13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marmon S, Sturtevant D, Herrfurth C, Chapman K, Stymne S, Feussner I. 2017. Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol 173:2081–2095. doi: 10.1104/pp.16.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banaś W, Sanchez Garcia A, Banaś A, Stymne S. 2013. Activities of acyl-CoA:diacylglycerol acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds. Planta 237:1627–1636. doi: 10.1007/s00425-013-1870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siedler S, Schendzielorz G, Binder S, Eggeling L, Bringer S, Bott M. 2014. SoxR as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli. ACS Synth Biol 3:41–47. doi: 10.1021/sb400110j. [DOI] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Xia R, Chen H, He Y. 2018. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv doi: 10.1101/289660. [DOI]
- 43.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao K, Yue XH, Chen WC, Zhou XR, Wang L, Xu L, Huang FH, Wan X. 2018. Metabolic engineering for enhanced medium chain omega hydroxy fatty acid production in Escherichia coli. Front Microbiol 9:139. doi: 10.3389/fmicb.2018.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplemental files.