REPLY
We appreciate the additional data analysis and comments on our paper (1), confirming our finding that the GC bias in whole-genome sequence data will impact Salmonella in silico serotyping prediction with antigen-mapping-based tools. A host of studies has shown that the use of the Nextera XT (XT) library preparation kit (Illumina) causes a GC bias in the resulting sequence data (2–7). This leads to a decreased coverage depth of the characteristically low-GC O antigen sequences, which has the potential to disrupt serovar prediction by mapping-based tools (8–10). In addition to finding that the TruSeq library preparation kit (Illumina) is superior to the Nextera XT library preparation kit, Li and colleagues quantified GC-associated coverage bias in different data sets. Specifically, they compared XT sequence data from our previously submitted data set (PRJEB31846) used in an independent in silico serotyping tool evaluation study (11) with TruSeq sequence data used in the SeqSero2 validation study (National Antimicrobial Resistance Monitoring System for Enteric Bacteria [NARMS] subset of PRJNA230403) (12). They found the extent of the GC bias associated with our data to be greater than in comparable XT sequence data sets, concluding that SeqSero2 tool performance was overly challenged in our study (the overall evaluation data set in our study was comprised of 59% XT data and 41% Nextera Flex data). Assessing and interpreting sequence quality are essential for all research purposes, and we were interested in analyzing additional data to explore the prevalence and extent of GC biases in different data sets.
For this purpose, we randomly sampled short-read Illumina sequencing data from 13 publicly available BioProjects, taking care to only include sequence data prepared with the XT library preparation kit (with the exception of PRJEB31846). An overview of the selected BioProjects is given in Table S1 in the supplemental material (NCBI Sequence Read Archive [SRA] accession numbers are listed in Table S3). We then calculated the GC bias according to the method of Benjamini and Speed (13) with an in-house Python script (https://gitlab.com/bfr_bioinformatics/calculate_gc_bias, v0.9) with parameters –windowsize 200 –upperbound 35 and –lowerbound 25. For read mapping, we obtained complete genome sequences from the NCBI reference database for all respective serovars, manually excluding plasmid sequences (NCBI accession numbers are provided in Table S2).
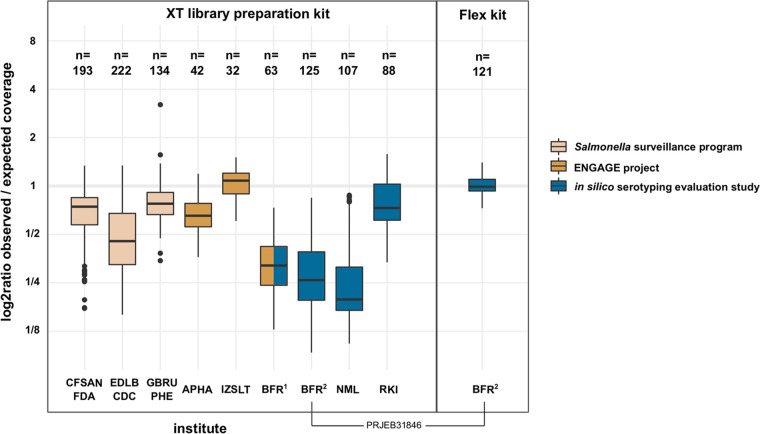
Our results, visualized in Fig. 1 (numerical data in Table S3), confirm that most XT sequencing data are associated with a GC bias but that the different data sets had considerably different extents of GC bias. Besides statistical effects (number of analyzed isolates and serovar constitution), this may reflect other influential parameters, such as the chosen library normalization method. From our combined results, we conclude that the extent of the GC bias associated with different data sets should be taken into consideration when evaluating antigen detection-based in silico serotyping tools. Equally, users of serotyping tools should be conscious of GC bias in their data and if in doubt choose a sequence type- or cluster-based tool for a more robust analysis of GC-biased sequencing data. Positively, library preparation kits and sequencing procedures are constantly updated and developed, as exemplified by the improved Nextera Flex kit (Illumina), which allows the generation of largely GC bias-free sequence data.
FIG 1.
Comparison of GC-associated coverage bias between sequence data from different institutes. Sequence data of major Salmonella serovars (Agona, Bareilly, Berta, Braenderup, Brandenburg, Cerro, Choleraesuis, Corvallis, Derby, Dublin, Enteritidis, Gallinarum, Give, Goldcoast, Hadar, Heidelberg, Indiana, Infantis, Java, Javiana, Johannesburg, Kentucky, Manhattan, Mbandaka, Mikawasima, Montevideo, Muenchen, Muenster, Newport, Ohio, Oranienburg, Panama, Pullorum, Rissen, Saintpaul, Schwarzengrund, Senftenberg, Stanleyville, Tennessee, Thompson, Typhimurium, Virchow, Waycross, Worthington) were randomly sampled from 13 BioProjects: Center for Food Safety and Applied Nutrition, Food and Drug Administration (United States) (CFSAN-FDA), PRJNA186035; Enteric Diseases Laboratory Branch, Centers for Disease Control and Prevention (United States) (EDLB-CDC), PRJNA230403; Gastrointestinal Bacteria Reference Unit, Public Health England (United Kingdom) (GBRU-PHE), PRJNA248792; Animal and Plant Health Agency (United Kingdom) (APHA), PRJEB24097, PRJEB24103, PRJEB24107, and PRJEB24311; Istituto Zooprofilattico Sperimentale del Lazio e la Toscana (Italy) (IZSLT), PRJEB23728 and PRJEB23778; German Federal Institute for Risk Assessment, PRJEB23094 (BFR1) and PRJEB31846 (BFR2); National Microbiology Laboratory (Canada) (NML), PRJNA353625; and Robert Koch-Institut (Germany) (RKI), PRJEB30317. The total number of isolates sampled per data set is displayed above each box plot. The type of Nextera sequencing library preparation kit is indicated (left, XT; right, Flex). Fill colors indicate the type of sequencing project.
Lastly, as there was some lack of clarity about which version of the SeqSero2 program was used in our previous study, we want to point out that the information about all programs and versions can be found in Table S1 in the accompanying supplemental material (https://aem.asm.org/content/aem/suppl/2020/02/06/AEM.02265-19.DCSupplemental/AEM.02265-19-s0001.pdf). As stated, all analyses were performed with SeqSero2 version v.1.0.0, as version v.1.0.2 was not released before 30 September 2019 and the manuscript was submitted to Applied and Environmental Microbiology (AEM) on 2 October 2019.
Li et al. further criticized that the k-mer-based and microassembly workflows of SeqSero2 were addressed to as “k-mer mode” and “allele-mode” in our publication. We want to emphasize that we chose to refer to the different workflows implemented in SeqSero2 as k-mer and allele-mode because these are the terms used in the official documentation of SeqSero2 on GitHub (latest version of the read me, current commit: 70dc513, 29 April 2020).
Supplementary Material
Footnotes
Supplemental material is available online only.
This is a response to a letter by Li et al. (https://doi.org/10.1128/AEM.00614-20).
REFERENCES
- 1.Li S, Zhang S, Deng X. 2020. GC content-associated sequencing bias caused by library preparation method may infrequently affect Salmonella serotype prediction using SeqSero2. Appl Environ Microbiol 86:e00614-20. doi: 10.1128/AEM.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan JH, Yin Y, Reed EF, Moua K, Thomas K, Zhang Q. 2015. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum Immunol 76:166–175. doi: 10.1016/j.humimm.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyler AD, Christianson S, Knox NC, Mabon P, Wolfe J, Van Domselaar G, Graham M, Sharma M. 2016. Comparison of sample preparation methods used for the next-generation sequencing of Mycobacterium tuberculosis. PLoS One 11:e0148676. doi: 10.1371/journal.pone.0148676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grützke J, Malorny B, Hammerl JA, Busch A, Tausch SH, Tomaso H, Deneke C. 2019. Fishing in the soup—pathogen detection in food safety using metabarcoding and metagenomic sequencing. Front Microbiol 10:1805. doi: 10.3389/fmicb.2019.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato MP, Ogura Y, Nakamura K, Nishida R, Gotoh Y, Hayashi M, Hisatsune J, Sugai M, Takehiko I, Hayashi T. 2019. Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes. DNA Res 26:391–398. doi: 10.1093/dnares/dsz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne PD, Nielsen TK, Kot W, Aggerholm A, Gilbert MTP, Puetz L, Rasmussen M, Zervas A, Hansen LH. 2020. GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms. Gigascience 9:giaa008. doi: 10.1093/gigascience/giaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uelze L, Borowiak M, Brinks E, Deneke C, Stingl K, Kleta S, Tausch SH, Szabo K, Wöhlke A, Malorny B. 2020. German-wide interlaboratory study compares consistency, accuracy and reproducibility of whole-genome short read sequencing. bioRxiv doi: 10.1101/2020.04.22.054759. [DOI] [PMC free article] [PubMed]
- 8.Zhang S, Yin Y, Jones MB, Zhang Z, Kaiser BLD, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yachison CA, Yoshida C, Robertson J, Nash JHE, Kruczkiewicz P, Taboada EN, Walker M, Reimer A, Christianson S, Nichani A, PulseNet Canada Steering Committee, Nadon C. 2017. The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a National Salmonella Reference Laboratory. Front Microbiol 8:1044. doi: 10.3389/fmicb.2017.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerji S, Simon S, Tille A, Fruth A, Flieger A. 2020. Genome-based Salmonella serotyping as the new gold standard. Sci Rep 10:4333. doi: 10.1038/s41598-020-61254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uelze L, Borowiak M, Deneke C, Szabó I, Fischer J, Tausch SH, Malorny B. 2020. Performance and accuracy of four open-source tools for in silico serotyping of Salmonella spp. based on whole-genome short-read sequencing data. Appl Environ Microbiol 86:e02265-19. doi: 10.1128/AEM.02265-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, Lauer AC, Fields PI, Deng X. 2019. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol 85:e01746-19. doi: 10.1128/AEM.01746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Speed TP. 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.