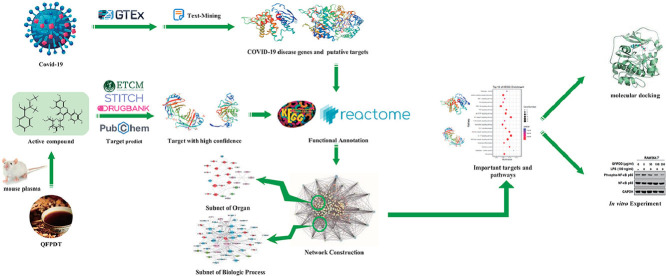
Abstract
Background
The traditional Chinese medicine (TCM) formula Qing-Fei-Pai-Du decoction (QFPDD) was the most widely used prescription in China's campaign to contain COVID-19, which has exhibited positive effects. However, the underlying mode of action is largely unknown.
Purpose
A systems pharmacology strategy was proposed to investigate the mechanisms of QFPDD against COVID-19 from molecule, pathway and network levels.
Study design and methods
The systems pharmacological approach consisted of text mining, target prediction, data integration, network study, bioinformatics analysis, molecular docking, and pharmacological validation. Especially, we proposed a scoring method to measure the confidence of targets identified by prediction and text mining, while a novel scheme was used to identify important targets from 4 aspects.
Results
623 high-confidence targets of QFPDD's 12 active compounds were identified, 88 of which were overlapped with genes affected by SARS-CoV-2 infection. These targets were found to be involved in biological processes related with the development of COVID-19, such as pattern recognition receptor signaling, interleukin signaling, cell growth and death, hemostasis, and injuries of the nervous, sensory, circulatory, and digestive systems. Comprehensive network and pathway analysis were used to identify 55 important targets, which regulated 5 functional modules corresponding to QFPDD's effects in immune regulation, anti-infection, anti-inflammation, and multi-organ protection, respectively. Four compounds (baicalin, glycyrrhizic acid, hesperidin, and hyperoside) and 7 targets (AKT1, TNF-α, IL6, PTGS2, HMOX1, IL10, and TP53) were key molecules related to QFPDD's effects. Molecular docking verified that QFPDD's compounds may bind to 6 host proteins that interact with SARS-CoV-2 proteins, further supported the anti-virus effect of QFPDD. At last, in intro experiments validated QFPDD's important effects, including the inhibition of IL6, CCL2, TNF-α, NF-κB, PTGS1/2, CYP1A1, CYP3A4 activity, the up-regulation of IL10 expression, and repressing platelet aggregation.
Conclusion
This work illustrated that QFPDD could exhibit immune regulation, anti-infection, anti-inflammation, and multi-organ protection. It may strengthen the understanding of QFPDD and facilitate more application of this formula in the campaign to SARS-CoV-2.
Keywords: Network pharmacology, Traditional Chinese medicine, COVID-19, SARS-CoV-2, Drug target, Pathway
Abbreviations: ACE2, Angiotensin-converting enzyme 2; AP-MS, affinity purification mass spectrometry; COVID-19, Coronavirus disease 2019; ELISA, enzyme linked immunosorbent assay; ETCM, the Encyclopedia of Traditional Chinese medicine; GTEx, Genotype-Tissue Expression Portal; HLMs, human liver microsomes; LPS, lipopolysaccharide; QFPDD, Qing-Fei-Pai-Du decoction; NEN, N-ethyl-1,8-naphthalimide; PGE2, prostaglanding E2; PPP, platelet poor plasma; PRP, rich platelet plasma; PRR, pattern recognition receptor; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; STITCH, Search Tool for Interactions of Chemicals; STRING, the Search Tool for the Retrieval of Interacting Genes; SymMap, Symptom Mapping; TCM, traditional Chinese medicine; TPM, transcripts per million
Graphical abstract
Introduction
The newly emerged coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2) has spread worldwide since the end of 2019. Its most common symptoms are fever, cough, and shortness of breath. Other symptoms include nasal obstruction, headache, myalgia, fatigue, diarrhea, loss of appetite, and sputum production (Huang et al., 2020). In addition, coagulation dysfunction was observed in COVID-19 patients (Campbell and Kahwash, 2020), while olfactory and gustatory dysfunctions were also reported as a clinical presentation of mild-to-moderate forms of the disease (Lechien et al., 2020). Inflammatory cytokine storm was very common in patients with severe COVID-19 (Zhang et al., 2020). The virus infection not only led to pneumonia and lung injury, but also attacked many tissues and organs in the body, including the heart, the liver, the brain, the kidneys, the intestines, eyes, and blood vessels (Wadman et al., 2020). Therefore, COVID-19 is not just a respiratory disease, but a systemic disease which damages multiple body systems and organs of human beings. However, currently no specific drug has been discovered for this pandemic and the vaccines are still under development.
In this hard campaign to SARS-CoV-2, traditional Chinese medicine (TCM) has been playing an active role for China to contain the spread of the virus in two months (Ren et al., 2020). In the trial 6th edition of the "Guideline on Diagnosis and Treatment of COVID-19" issued by the National Health Commission of China on Feb 18, 2020, Qing-Fei-Pai-Du decoction (QFPDD) was recommended as a general TCM formula for the treatment of mild, moderate and severe cases of COVID-19 (National Health Commission & State Administration of Traditional Chinese Medicine, 2020). Clinical observations suggested that QFPDD could significantly improve the abnormal laboratory indexes and clinical symptoms of patients, reduce the adverse reactions, and improve the therapeutic effect (Kuang-yu et al., 2020; Wang et al., 2020b). Since then, QFPDD has been the most widely used TCM prescription in China. Therefore, understanding the mode of action for QFPDD's treatment to COVID-19 could be very helpful for the campaign against SARS-CoV-2.
At the molecular level, TCM formulas regulate the body system through multi-component and multi-target in a holistic way. Network pharmacology is an effective method to study TCM's mode of action (Li et al., 2014; Mou et al., 2020; Zhao et al., 2019). Earlier network pharmacological studies on QFPDD have preliminarily illustrated its mechanism against COVID-19 (Wu et al., 2020a; Zhao et al., 2020). It could inhibit the replication of virus by acting on a group of host ribosomal proteins (Zhao et al., 2020). By regulating immune associated pathways and cytokine signaling pathways, QFPDD may suppress over-active immune response and diminish inflammation (Wu et al., 2020a; Zhao et al., 2020). However, these earlier works included all known ingredients of the herbs in the analysis, while all targets were from predictions and lacked experimental validations, thus reducing the confidence of the results. In fact, since the TCM formula is taken orally, only the compounds that eventually appear in blood may have the chance of exerting their effects. In addition, in the early stage of COVID-19’s outbreak, human's knowledge about this new disease was extremely limited, which hindered the research for therapeutic mechanism of drugs to the disease. Thanks to the tireless efforts of scientific community, more knowledge and data about the disease and the TCM formula accumulated in these months. For example, 332 human proteins that physically interact with SARS-CoV-2 proteins were identified by affinity purification mass spectrometry (AP-MS), which could be putative targets for drug discovery of COVID-19 (Gordon et al., 2020). Recently, the material basis of QFPDD was revealed by UHPLC-Q-Orbitrap HRMS experiments, which identified 30 compounds included in QFPDD and 12 compounds appeared in the mouse plasma following QFPDD administration (Liu et al., 2020). These results afford new research basis for our better understanding of the mechanism of QFPDD's treatment on COVID-19.
This work aims to systematically investigate the mechanism of action for QFPDD to treat COVID-19. Following extensive data mining, we collected a list of COVID-19 related disease genes and targets, QFPDD's compounds that appear in blood after treatment, and their corresponding targets. We conducted network and pathway analysis to explore biological processes and organ systems influenced by COVID-19 and QFPDD. After identifying important targets and pathways regulated by QFPDD, we constructed a compound-target network and investigated its functional modules. The molecule docking verified some important compound-target interactions. At last, a series of in vitro experiments were conducted to validate some important targets.
Materials and methods
Plasma absorbed QFPDD compounds and their putative targets
The 12 compounds identified by Liu et al in the mouse plasma following QFPDD administration were considered as potential active ingredients of QFPDD (Liu et al., 2020). These compounds i.e., amygdalin, baicalin, ephedrine, hesperidin, hyperoside, irisflorentin, isochlorogenic acid A, liquiritin, prunasin, pseudoephedrine, alisol B 23-acetate, and glycyrrhizic acid, were identified by UHPLC-Q-Orbitrap HRMS experiments. For each compound, putative targets were acquired from four databases: DrugBank (Wishart et al., 2018), PubChem (Kim et al., 2016), STITCH (Search Tool for Interactions of Chemicals) (Kuhn et al., 2008), and ETCM (Xu et al., 2018). ETCM predicts targets by MedChem Studio and only outputs targets with confidence score at least 0.8. We also set the threshold of confidence score as 0.8 to filter results predicted by STITCH. We collected compound-protein interactions from 4 sections of PubChem database: Chemical-Gene Co-Occurrences in Literature, Protein Bound 3-D Structures, Drug-Gene Interactions, and BioAssay Results. In the BioAssay Results section of the PubChem database, we only accepted interactions between compound and human proteins with activity value at most 100 μmol. Finally, we obtained 1289 compound-protein interactions between the 12 compounds and 623 distinct targets (Supplementary Table S1).
To measure the confidence of the interactions, we first defined a source score for each data collected from the sources. If a compound-target interaction was predicted by ETCM or STITCH, the source score was set as 0.8. Considering that targets from literatures generally have higher reliability, to distinguish these compound-target interactions from predicted ones, we gave them higher source scores. Specifically, each publication about the compound-target interaction in the section of Chemical-Gene Co-Occurrences in Literature of PubChem contributed 1 to the source score (Supplementary Table S2), and data from the other 3 sections of PubChem and from DrugBank got source score 2. Then, for each compound-target pair, we defined a confidence score which summed up all of its source scores. In this way, 712 of the collected 1289 compound-protein pairs have confidence scores at least 2, taking 55% of all the data.
Disease genes and putative targets of COVID-19
Considering that ACE2 (Angiotensin-converting enzyme 2) is the host receptor for the virus SARS-CoV-2, we collected genes co-expressed with ACE2 from two sources. First, we downloaded the supplementary material of the reference (Wang et al., 2020a), which included 5556 genes co-expressed with ACE2 in the colonic epithelial cells. Then, the expression data of 288 normal lung tissue samples were downloaded from the Genotype-Tissue Expression Portal (GTEx) database (Consortium, 2015). The co-expression Pearson correlation analysis was conducted between ACE2 and all protein-coding genes. Setting the threshold as , we obtained 5685 genes co-expressed with ACE2 in the lung cells. In the end, the intersection of the two sets resulted in 1158 common genes. Next, we collected genes reported to be involved in the disease process of COVID-2019 from literatures (Huang et al., 2020; Li et al., 2020), and then combined them with the 1158 ACE2 co-expressed genes. Finally, we obtained 1202 genes associated with COVID-19 (Supplementary Table S3).
Besides, host proteins that directly interact with virus proteins were considered as potential targets for antiviral drugs. Combining literature reported human proteins which interact with SARS-CoV-2 and coronavirus proteins (Gordon et al., 2020; Zhou et al., 2020), we obtained 445 distinct proteins, which were set as putative COVID-19 targets in our study (Supplementary Table S4).
Network construction and analysis
Protein-protein interaction networks were constructed by STRING (The Search Tool for the Retrieval of Interacting Genes) platform (von Mering et al., 2005). Interaction confidence score was set as 0.7 or 0.9. For networks constructed under the confidence score of 0.7 and 0.9, the importance of nodes was measured by strength and degree, respectively. Node strength is the sum of weights for links of the node, and degree is the number of links of this node.
It is noted that two thresholds of confidence score were helpful for accurately identifying hub nodes. The STRING platform sets protein-protein interactions with confidence score greater than 0.7 as high confidence and those greater than 0.9 as the highest confidence. Therefore, a network constructed with confidence score greater than 0.7 is actually a highly reliable network. In this work, some target subsets contain proteins that are highly functionally associated so that they form a very densely connected sub-network with confidence threshold 0.7. In such cases, to identify hub nodes which play key roles in such sub-network, we increased the threshold of confidence score to 0.9 to construct network with the highest confidence.
Network visualization and analysis were performed by software Cytoscape 3.6.0 (Shannon et al., 2003). The algorithm of Community Cluster (Glay) in the Clustermaker2 app of Cytoscape was applied to decompose network into modules.
Functional enrichment analysis for proteins
Functional enrichment analysis was conducted by STRING. The threshold for statistical significance was set as FDR < 10-4. The results for KEGG pathway (Kanehisa and Goto, 2000) and Reactome pathway (Fabregat et al., 2018) enrichment were used to annotate the functions of proteins.
Molecular docking
The crystal structures of protein receptors were downloaded from RCSB Protein Data Bank (Rose et al., 2015). In each crystal structure, the ligand and protein were remained and other molecules like waters or metals were removed by PyMol (Version 2.1.1). Every protein receptor was prepared by “Protein Preparation Wizard” module in Maestro (Version 10.2.010). With the prepared ligand-receptor complex in the workspace, the “Receptor Grid Generation Panel” was used to generate the receptor grid files. And the binding site of the original ligand in its crystal structure was specified as the docking site of each protein. The chemical structures of compounds were extracted from PubChem, and then optimized using “LigPrep” module. Finally, each compound was docked into the protein receptors using “Glide Docking” module in Maestro.
Reagents and antibodies
Baicalin, hyperoside and alisol B 23-acetate were purchased from Shanghai Standard Technology Co., Ltd. (Shanghai, China). QFPDD dry powder was obtained from Shanghai University of Traditional Chinese Medicine (Shanghai, China). Arachidonic acid (AA) was obtained from Cayman Chemicals (Ann Arbor, MI, USA). Pam3CSK4 was purchased from InvivoGen (San Diego, CA, USA). The lipopolysaccharide (LPS), NS-398, aspirin, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St Louis, MO, USA).
Baicalin, hyperoside, alisol B 23-acetate, and QFPDD were dissolved in DMSO, and then stored frozen as stock solutions and diluted to the indicated concentrations before use.
The probe substrates for cytochrome P450 enzymes, including 7-ethoxyresorufin (for CYP1A1) and N-ethyl-1,8-naphthalimide (NEN, for CYP3A4) were synthesized by the authors according to previously reported literatures (Ning et al., 2019; Yamaori et al., 2010). D-glucose-6-phosphate, glucose-6-phosphate dehydrogenase, NADP+, resorufin were obtained from meilunbio (Dalian, China). MgCl2 was from Sinopharm Chemical Reagent (Shanghai, China). Recombinant human CYP1A1 and the pooled human liver microsomes (HLMs, from 50 donors, lot no. X008067) were obtained from Bioreclamation IVT (Baltimore, MD, USA). LC grade acetonitrile was ordered from Tedia Company (USA). Ultrapure water purified by Milli-Q® Integral Water Purification System (Millipore, USA) was used throughout.
Cell culture
RAW 264.7 murine macrophages were obtained from the Shanghai Institute of Cell Biology and Biochemistry (Shanghai, China) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 Units/ml), and streptomycin (100 µg/ml) in a humidified atmosphere containing 5% CO2 at 37 °C.
Animals
Male New Zealand rabbits (2–2.5 kg) were purchased from Shanghai Shengwang Experimental Animal Breeding Co, Ltd. Animals were housed in standard pathogen-free cages in a climate-controlled environment with a 12-h light/dark cycle. Sterile food and water were naturally provided. All procedures were conducted in accordance with the guidelines for care and use of laboratory animals of the university and the government.
RNA quantification of cytokines and chemokine
To quantify RNA expression of cytokines and chemokine in the cells stimulated with Pam3CSK4, RAW264.7 (2 × 105/well) were seeded into 24-well cell culture plates one day before. Cells were treated with different concentration of compound and stimulated with Pam3CSK4 (100 ng/ml) for 3 h or 6 h. The expression of IL-6, CCL2 and TNF-α mRNA were examined by qRT-PCR. The primers were described as following. Data were normalized by the expression of β-Actin mRNA in each sample.
IL-6: forward 5’-TAGTCCTTCCTACCCCAATTTCC-3’
reverse 5’-TTGGTCCTTAGCCACTCCTTC-3’
TNF-α: forward 5’-AAGCCTGTAGCCCACGTCGTA-3’
reverse 5’-GGCACCACTAGTTGGTTGTCTTTG-3’
CCL2: forward 5’-TTTTTGTCACCAAGCTCAAGAG-3’
reverse 5’-TTCTGATCTCATTTGGTTCCGA-3’
β-Actin: forward 5’-AGTGTGACGTTGACATCCGT-3’
reverse 5’-GCAGCTCAGTAACAGTCCGC-3’
In the other set of experiments, to quantify RNA expression of cytokines in cells stimulated with LPS, RAW264.7 cells were pre-incubated with indicated concentrations of QFPDD for 30 min, and then co-treated with 100 ng/ml of LPS for 5 hours. Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from 0.5 μg RNA by reverse transcriptase (Takara, Dalian, China). The levels of IL-6 and IL-10 in the cells were assessed by qRT-PCR as previously described (Li et al., 2020b). Data were normalized by the expression of GAPDH mRNA in each sample. The primers used were as following:
IL-6: forward 5’- TCGTGGAAATGAGAAAAGAGTTG-3’
reverse 5’-AGTGCATCATCGTTGTTCATACA-3’
IL-10: forward 5’- CCAAGCCTTATCGGAAATGA-3’
reverse 5’- TTTTCACAGGGGAGAAATCG-3’
GAPDH: forward 5’- AGGTCGGTGTGAACGGATTTG-3’
reverse 5’-TCCACCACCCTGTTGCTGTA-3’.
Immunoblotting
RAW264.7 cells were pre-incubated with indicated concentrations of QFPDD for 30 min, and then co-treated with 100 ng/ml LPS for 4 h. The cell lysates were prepared as described previously (Xu et al., 2019). After separated by SDS-PAGE and transferred onto nitrocellulose membrane, the immunoblottings were carried out using primary antibodies against phospho-NF-κB p65, NF-κB p65, and GAPDH (Cell Signaling Technology, Danvers, MA), and horseradish peroxidase conjugated anti–mouse IgG and anti–rabbit IgG antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). GAPDH was used as a loading control.
Cytochrome P450 enzyme inhibition assay
For CYP1A1 inhibition(He et al., 2015), the reaction mixture (200 µl) consisted of recombinant human CYP1A1, the substrate 7-ethoxyresorufin (0.2 µM), a NADPH-generating solution (containing 1 mM NADP+, 10 mM glucose-6-phosohate, 1 unit/ml glucose-6-phosohate dehydrogenase, and 4 mM MgCl2), 100 mM potassium phosphate buffer and hyperoside at various concentrations. Meanwhile, for the negative incubations without hyperoside, 1 µl of LC grade acetonitrile was added as negative controls. The percentage of organic solvent in all incubation system was 1% (v/v). After 3 min pre-incubation at 37 ºC, the reaction was initiated by adding NADPH-generating system, the mixture of the reaction was taken into multi-Mode microplate reader for continuous analysis for 10 min. After that, the fluorescence signals of resorufin were analyzed by a microplate reader using excitation and emission wavelengths of 550 and 590 nm, respectively (Gain = 500). The residual activities of CYP1A1 were expressed as percent decrease in fluorescence intensities.
For CYP3A4 inhibition, the incubation system (200 μl) included potassium phosphate buffer (pH 7.4, 100 mM), HLM (0.05 mg/ml), NADPH-generating system, the probe substrate (NEN, 10 μM) and alisol B 23-acetate (as inhibitor). The percentage of organic solvent in the reaction system was 1% (v/v). After 3 min pre-incubation at 37 ºC, the reaction was initiated by adding NADPH-generating system, the mixture of the reaction was taken into multi-Mode microplate reader for continuous analysis for 30 min. The excitation wavelength of NEHN (N-ethyl-4-hydroxyl-1, 8-naphthalimide, the metabolite of NEN in CYP3A4 or HLM) was set at 450 nm, and the emission wavelength was 558 nm. The residual activities of CYP3A4 were expressed as percent decrease in fluorescence intensities.
The effect on PTGS1/2 enzymatic activity
Ovine PTGS1 and human recombinant PTGS2 were obtained from Cayman Chemicals (Ann Arbor, MI, USA) and used to assay the inhibitory effect on enzymatic activity. According to the protocol of the manufacture, the inhibitors were incubated with assay buffer containing pure enzymes for 5 min at 25 ℃, then, Absorbance at 590 nm representing the enzymatic activity were detected precisely 2 min after initiation of the reaction by the colorimetric substrate and AA.
The effect on prostaglandin E2 derived from PTGS2
The cell-based assay system to observe the effect on prostaglanding E2 (PGE2) derived from PTGS2 was established according to previous reports (Li et al., 2012). RAW 264.7 macrophages were plated in 96-well plates (1 × 104 cells per well) and pre-treated with aspirin (200 mM) for 2 h to deactivate PTGS1. The cells were incubated with the compounds for 24 h, and stimulated simultaneously by LPS 2 μg/ml to induce PTGS2 expression. The prostaglandin E2 content in supernatants was assayed with enzyme linked immunosorbent assay (ELISA) according to the protocol of manufacture (Cayman Chemicals, Ann Arbor, MI, USA).
The effect on platelet aggregation induced by AA
The Microtiter Plate Method was used to observe platelet aggregation induced by AA via PTGS1 (Wu et al., 2020b). Briefly, Blood was collected from the ear artery of rabbits by syringe and anticoagulated with 3.8% sodium citrate solution (1:10, vol/vol). Rich platelet plasma (PRP) and platelet poor plasma (PPP) were prepared by successive centrifugation of rabbit blood for 10 min at 100 and 2300 g, respectively. The compounds or vehicle solution DMSO was added into the 96-well microplate containing PRP (200 mL per well and DMSO, 0.5% vol/vol), and the microplate was read at 570 nm to obtain the absorbance before AA stimulation. PRP was incubated at 37 ℃ for 5 min and vibrated at 1000 rpm with stirrer, then AA was added to induce platelet aggregation by shaking for 5 min. The plate was read at 570 nm to obtain absorbance after AA stimulation. The absorbance of PPP was set as background value, and the absorbance before and after AA stimulation was used to calculate the platelet aggregation rate.
Statistical analysis
In Cytochrome P450 enzyme inhibition assays, the dose-inhibition curves were fitted according to nonlinear regression using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, USA) and IC50 values were calculated. All data are expressed as the mean±SD and analyzed by one-way analysis of variance (ANOVA) followed by a Dunnett's test for multiple comparisons. The results were statistically significant at p < 0.05.
Results and discussions
QFPDD's targets acting on genes affected by SARS-CoV-2 infection
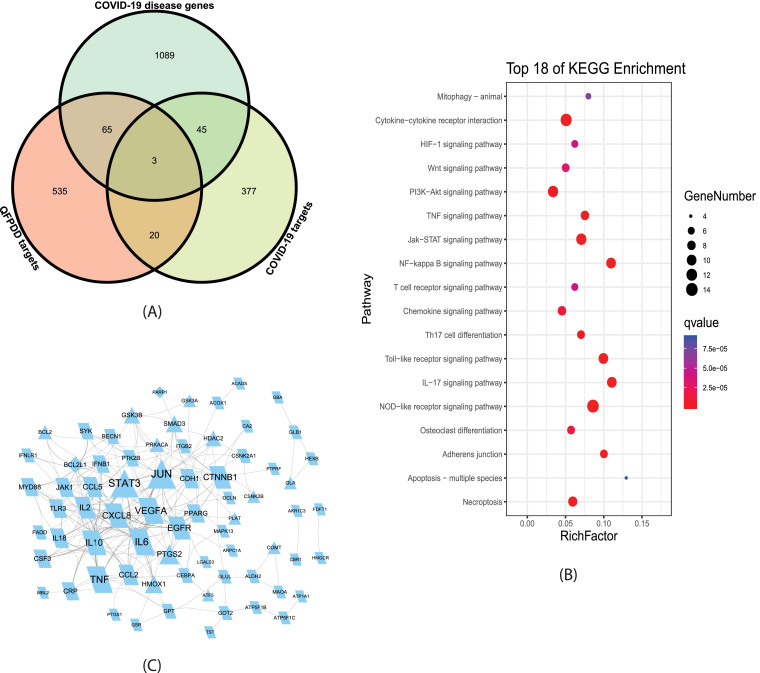
We identified 623 putative targets for 12 compounds appeared in mouse blood after the administration of QFPDD. The overlaps of QFPDD target genes with COVID-19 associated diseases genes and potential target genes were shown in Fig 1 A. Among QFPDD's target genes, 68 were COVID-19 disease genes, 23 were potential COVID-19 target genes. Taking COVID-19 associated diseases genes and potential target genes as targets affected by SARS-CoV-2 infection, the 88 distinct overlap genes were QFPDD's targets that regulate the targets affected by SARS-CoV-2 infection. These 88 targets may play an important role in the treatment of COVID-19. Fig 1B shows the top KEGG pathways significantly enriched with the 88 genes (FDR < 10-4). These pathways indicated that the major functions of QFPDD included the regulation of innate immune (NOD-like and Toll-like receptor signaling), cytokine activities (Cytokine-cytokine receptor interaction, Chemokine signaling, Th17 cell differentiation, IL-17 signaling pathway, NF-kappa B signaling pathway, TNF signaling pathway), cell growth and death (Necroptosis, Apoptosis, HIF-1 signaling pathway), and the degradation of damaged cells (Mitophagy). Fig 1C shows that 71 of the 88 gene-coded proteins interact with each other to form a protein-protein interaction network, which includes a large number of cytokines, such as TNF, VEGFA, IL6, IL10, CXCL8, CCL5, IFNLR1 and so on.
Fig. 1.
Analysis of QFPDD's targets acting on genes affected by SARS-CoV-2 infection (A) Intersection analysis between COVID-19 diseases genes, potential target genes, and putative QFPDD target genes identified 88 QFPDD's targets acting on genes affected by SARS-CoV-2 infection. (B) Top KEGG pathways significantly enriched with the 88 genes. (C) Protein-protein interaction between the 88 gene-coded proteins. Triangles and parallelograms represent COVID-19 targets and disease genes, respectively. Diamonds represent overlap of COVID-19 targets and disease genes. The size of node is proportional to its strength.
Interferons, a family of cytokines with antiviral properties, have been suggested as a potential treatment for COVID-19 because of their in vitro and in vivo antiviral properties. According to our data, some compounds in QFPDD, including prunasin, hesperidin, glycyrrhizic acid, isochlorogenic acid A and baicalin, could target interferon IFNB1 and interferon receptor IFNLR1 (Supplementary Table S1), both of which were involved in SARS-CoV-2 infection (Fig 1C). In fact, earlier studies have picked up low levels of IFNB1 in human small intestinal organoids infected by SARS-CoV-2 (Lamers et al., 2020), while increased expression level of IFNB1 were likely responsible for the distinct host immune response patterns of SARS-CoV-2 infection (Sun et al., 2020; Wyler et al., 2020). Baicalin was found to alleviate lung injury induced by H1N1 influenza virus A in mice by positively regulating the expression of IFNB1 (Li and Wang, 2019). On the other hand, IFNLR1 (interferon lambda receptor 1) is a component of the IFNLR1/IL10RB dimer, which mediates antiviral host defense of IFN-λ (interferon lambda). Recent study on SARS-CoV-2 shows that SARS-CoV-2 is sensitive to pre-treatment with IFN-λ (Park and Iwasaki, 2020). Thus the expression of IFNLR1 may facilitate the antiviral activity of IFN-λ (Prokunina-Olsson et al., 2020). These results indicate that the therapeutic effects of QFPDD may be partly achieved by regulating interferon and their receptors.
In addition, we explored the effects of the QFPDD from perspective of TCM symptoms. According to the theory of Traditional Chinese Medicine, COVID-19 belongs to wind cold dampness plague, with clinical manifestations of cold and hot symptoms at different courses of the disease (Yan et al., 2020). We extracted hub genes from the cold syndrome network and hot syndrome network constructed by Li et al, respectively (Li et al., 2007). It was found that two targets of QFPDD (IL6 and TNF) belonged to the hot syndrome network, while another two targets (POMC and AVP) belonged to the cold syndrome network, indicating that QFPDD could treat COVID-19 at different courses of the disease.
Biological processes and organ systems regulated by QFPDD's putative targets
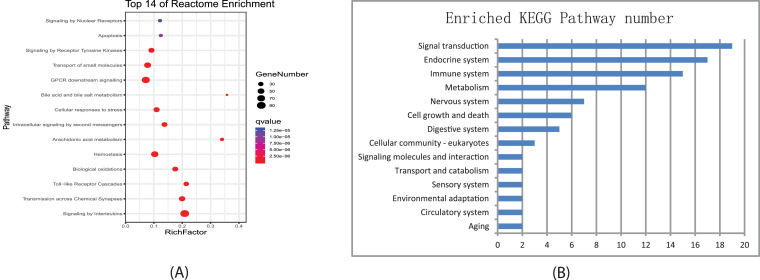
We conducted functional enrichment analysis for QFPDD's 623 putative targets based on Reactome and KEGG pathways. Fig 2 A showed the top Reactome pathways or superpathways significantly enriched with QFPDD's targets, and Fig 2B listed the number of KEGG pathways significantly enriched with QFPDD's targets that were classified in different biological systems (Supplementary Table S5, S6). Then we integrated information of the enriched pathways in the two databases and constructed sub-networks for QFPDD's targets in different biological processes (Fig. 3 ) and different organ systems (Fig. 4 ).
Fig. 2.
Pathways enriched with putative targets of QFPDD. (A) Top Reactome pathways/ superpathways significantly enriched with QFPDD's targets. (B) Number of KEGG pathways significantly enriched with QFPDD's targets which belong to different biological systems.
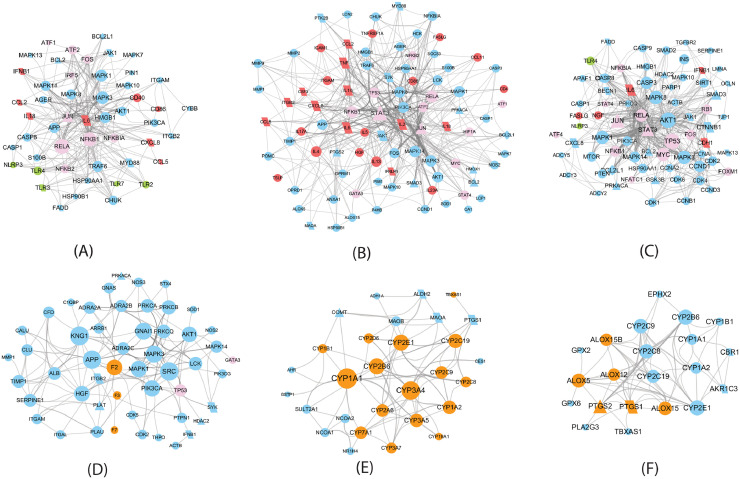
Fig. 3.
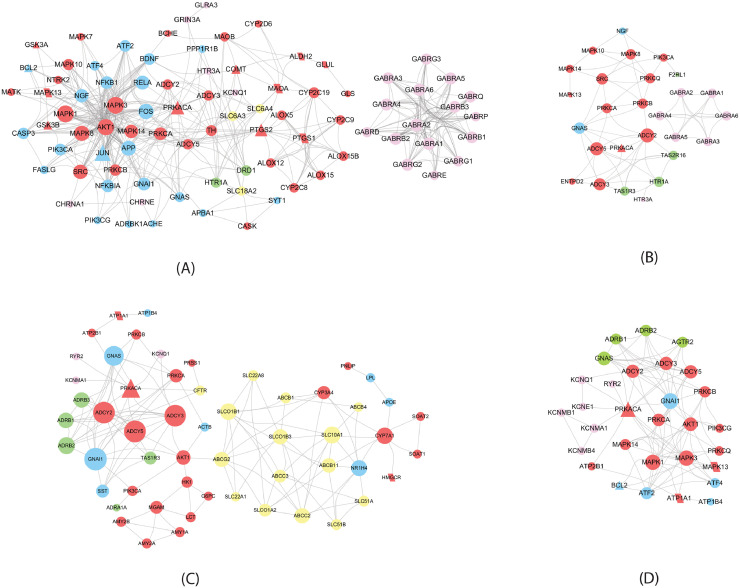
QFPDD's target sub-networks in different biological processes. Triangles and parallelograms represent COVID-19 targets and disease genes, respectively. Diamonds represent overlap of COVID-19 targets and disease genes. The size of node is proportional to its degree. Red nodes represent cytokines, cytokine receptors, or CD molecules; pink nodes represent transcription factors; green nodes are PRRs; Brown nodes in (D), (E) and (F) represent coagulation factors, cytochrome P450, and dioxygenases, respectively. (A) Pattern recognition receptor signaling; (B) Signaling by interleukins (C) Cell growth and death (D) Hemostasis (E) Biological oxidations (F) Arachidonic acid metabolism.
Fig. 4.
QFPDD's target sub-networks in different organ systems. Triangles and parallelograms represent COVID-19 targets and disease genes, respectively. The size of node is proportional to its strength. Red, green, pink, and yellow nodes represent enzymes, G protein-coupled receptors, ion channels, and transporters, respectively. (A) Nervous system; (B) Sensory system; (C) Digestive system; (D) Circulatory system.
Fig. 3 shows that QFPDD acts on some targets specific for the corresponding biological process, such as toll-like receptors (TLR2, TLR3, TLR7) and NOD-like receptor (NLRP3) in the pattern recognition receptor (PRR) signaling, cytokines (IL6, IL10, TNF) in the interleukin signaling, coagulation factors (F2, F3) in hemostasis, cytochrome P450 enzymes in the biological oxidations, and dioxygenases in arachidonic acid metabolism, so as to regulate these biological processes. All the first 3 sub-networks include transcript factors and cytokines, while the first 4 sub-network owns same protein kinases (MAPK1, MAPK3). The last two sub-networks share some cytochrome P450 enzymes. These features suggest the interplays between these biological processes.
In the subnetwork of PRR signaling (Fig. 3A), QFPDD acts on 4 PRRs (TLR2, TLR3, TLR7 and NLRP3). PRRs are proteins expressed by cells of the innate immune system to identify pathogen-associated molecular patterns, which are associated with microbial pathogens or cellular stress. This subnetwork suggests QFPDD's effects in anti-infection.
The target sub-network of interleukin signaling (Fig. 3B) includes 20 cytokines or cytokine receptors, such as IL6, IL10, and TNF-α, implying QFPDD's effects in anti-inflammation, preventing or relieving cytokine storm and organ protection. Cytokine storm refers to the phenomenon that a variety of cytokines are produced rapidly and in large quantities in body fluids because of the overacting of immune system to the virus. It is an important cause of acute respiratory distress syndrome (ARDS) and multiple organ failure in the progress of COVID-19.
During viral infections, apoptosis is induced as one of the host antiviral responses to limit virus replication and production. Many viruses have evolved distinct strategies to subvert host cell apoptosis and autophagy so as to prevent premature death of infected cells (Lim et al., 2016). Bcl2-like proteins are anti-apoptotic factors that inhibit apoptosis process. The expressions of BCL2 and BCL2L1 in different types of cells were regulated by 10 and 2 compounds of QFPDD, respectively. Thus, the target sub-network of cell growth and death (Fig. 3C) suggests that QFPDD may inhibit the replication of virus by triggering the apoptosis and autophagy of host infected cells.
Coagulopathy was reported as a complication of severe COVID-19 (Tang et al., 2020), and the application of anticoagulants for patients with severe COVID-19 has been recommended by the guide for the prevention and treatment of COVID-19 in China (Chinese Thoracic Society, 2020). The hemostasis sub-network (Fig. 3D) shows that QFPDD targets on coagulation factors F2, F3 and F7, indicating QFPDD's effect in regulating coagulation.
The target sub-network of biological oxidations (Fig. 3E) suggests QFPDD's antioxidative properties. This network includes a group of cytochrome P450 enzymes involved in the metabolism of various endogenous substrates, which are targeted by 11 compounds in QFPDD.
The sub-network of arachidonic acid metabolism (Fig. 3F) implies QFPDD's anti-inflammation, antioxidative and anti-platelet aggregation function. QFPDD's targets in this network include a group of dioxygenases such as PTGS1/2, ALOX5 and cytochrome P450 enzymes.
The oxidation stress and cytokine storm caused by SARS-CoV-2 infection could result in injuries of multiple organs. Fig. 4 shows that QFPDD acts on various enzymes, G protein-coupled receptors, ion channels, and transporters specific for nervous, sensory, circulatory, and digestive systems, so as to regulate these organ systems and relief symptoms caused by SARS-CoV-2 infection. Especially, enzyme PRKACA and its interactions with ADCY2, ADCY3, and ADCY5 are involved in the regulation of QFPDD to all the 4 body systems. The formula also targets on G protein-coupled receptors in all the 4 systems, in which the circulatory and digestive systems are targeted on beta-adrenergic receptors such as ADRB1 and ADRB3, and the nervous and sensory system are targeted on serotonin receptor HTR1A. In the nervous and sensory systems, QFPDD acts on a group of chloride ion channels for GABA (pink nodes in Fig. 4A and B), the major inhibitory neurotransmitter in the brain. The formula targets on a group of transporters in the digestive system, while the nervous system also has several transporter targets.
QFPDD's important targets and pathways
To investigate which targets were important for QFPDD's treatment to COVID-19, we selected important targets from 4 aspects as follows:
-
(1)
Overlap targets: targets that are both COVID-19 disease genes and putative targets (Fig. 1A). Three targets, BCL2L1, PARP1, and F2RL1, belong to this category.
-
(2)
Global hubs: hub genes in the protein-protein interaction network between QFPDD's targets acting on genes affected by SARS-CoV-2 infection (Fig. 1C). We chose 15 genes that have the highest strength, coreness and betweenness in this network, taking about 20% nodes of the network. These targets are JUN, STAT3, TNF, VEGFA, EGFR, IL6, CRP, PTGS2, HMOX1, IL10, CXCL8, IL2, TLR3, MYD88, IFNB1.
-
(3)
System hubs: hub genes in the sub-networks of QFPDD's targets in different biological processes and organ systems (Figs. 3 and 4).
-
(4)
System specific targets: QFPDD's targets that play specific roles in different biological processes and organ systems (Figs. 3 and 4) as illustrate in the last section.
For each sub-network in Figs. 3 and 4, we limited that the total number of selected targets in the 3rd and 4th classes did not exceed 10% of the total nodes. In this way, we identified 55 important targets for QFPDD's treatment to COVID-19.
Based on the results in the last section, we integrated the identified enrichment pathways/superpathways in the Reactome and KEGG databases and listed the 24 important pathways/superpathways that related to the treatment of COVID-19 in Table 1 .
Table 1.
Important pathways regulated by QFPDD's putative targets.
| Database | Pathway Class | Pathway/superpathway | Targets | Total genes | FDR |
|---|---|---|---|---|---|
| Reactome | Cell growth and death | Apoptosis | 20 | 162 | 9.02E-06 |
| KEGG | Cell growth and death | Necroptosis | 20 | 155 | 3.49E-07 |
| KEGG | Cell growth and death | Autophagy - animal | 21 | 125 | 3.87E-09 |
| Reactome | Immune system | Signaling by Interleukins | 90 | 439 | 3.79E-39 |
| Reactome | Immune system | Toll-like Receptor Cascades | 32 | 151 | 6.07E-14 |
| KEGG | Immune system | NOD-like receptor signaling pathway | 27 | 166 | 4.18E-11 |
| KEGG | Immune system | RIG-I-like receptor signaling pathway | 13 | 70 | 1.41E-06 |
| Reactome | Hemostasis | 60 | 601 | 1.38E-12 | |
| Reactome | Metabolism | Biological oxidations | 37 | 214 | 9.23E-14 |
| Reactome | Metabolism | Arachidonic acid metabolism | 20 | 59 | 7.34E-12 |
| KEGG | Metabolism | Steroid hormone biosynthesis | 15 | 58 | 5.97E-09 |
| KEGG | Signal transduction | cAMP signaling pathway | 36 | 195 | 1.23E-15 |
| KEGG | Signal transduction | TNF signaling pathway | 27 | 108 | 1.28E-14 |
| KEGG | Signal transduction | HIF-1 signaling pathway | 24 | 98 | 5.38E-13 |
| KEGG | Signal transduction | NF-kappa B signaling pathway | 20 | 93 | 2.62E-10 |
| KEGG | Signal transduction | Calcium signaling pathway | 20 | 179 | 2.46E-06 |
| KEGG | Environmental adaptation | Thermogenesis | 21 | 228 | 1.94E-05 |
| KEGG | Digestive system | Bile secretion | 29 | 71 | 6.28E-20 |
| KEGG | Digestive system | Salivary secretion | 17 | 86 | 1.54E-08 |
| KEGG | Digestive system | Gastric acid secretion | 15 | 72 | 6.11E-08 |
| Reactome | Nervous system | Transmission across Chemical Synapses | 43 | 218 | 8.86E-18 |
| KEGG | Sensory system | Inflammatory mediator regulation of TRP channels | 17 | 92 | 3.61E-08 |
| KEGG | Sensory system | Taste transduction | 13 | 81 | 5.40E-06 |
| KEGG | Circulatory system | Adrenergic signaling in cardiomyocytes | 26 | 139 | 7.74E-12 |
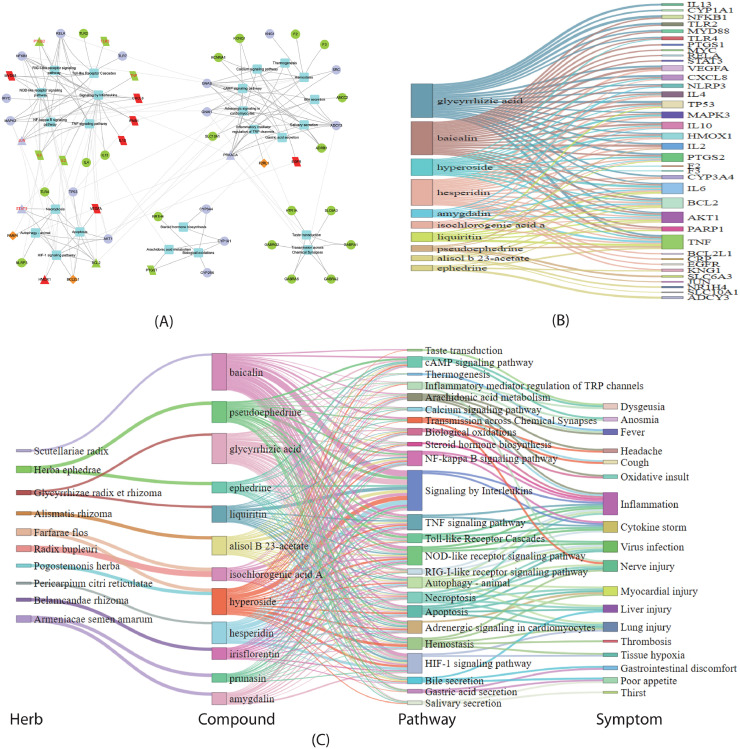
We mapped the 55 important targets to the 24 pathways and constructed the target-pathway network. Applying the algorithm of community cluster (Glay) in the clustermaker2 app of Cytoscape software, we decomposed the network into 5 densely linked topological modules as shown in Fig. 5 A. The 1st module consists of pathways in immune system and related signaling pathways. The 2nd module includes pathways in 3 organ systems (digestive, circulatory and sensory systems) and related pathways. Due to their common targets of chloride ion channels for GABA, the nervous system and one pathway of sensory system were clustered in the 5th module, which only linked to the 2nd module. The 3rd module is related to cell death and degradation, while the 4th module includes metabolism pathways related to oxidation stress and inflammation. According to the theory of network biology that bridges the topology of the biological network to its function (Barabasi and Oltvai, 2004), these modules reflected the effects of QFPDD in immune regulation, organ protection, damaged cell clearance, and anti-inflammation, respectively.
Fig. 5.
QFPDD's important targets and pathways. (A) The target-pathway network of QFPDD's important targets and pathways. Box nodes are pathways. Triangles and parallelograms represent COVID-19 targets and disease genes, respectively. Diamonds represent overlap of COVID-19 targets and disease genes. Brown, red, purple, green nodes corresponds the 1st, 2nd, 3rd and 4th class of important targets, respectively, and nodes with red label are also 2nd class of important targets. (B) Compound-target network of the important targets with confidence score at least 3. The width of the line is proportional to the confidence score. (C) Herb-compound-pathway-symptom relationships for QFPDD. The width of the line for herb-compound links is proportional to the dose of the herb in the formula, and that for compound-pathway links is proportional to the number of targets for the compound that map to the pathway.
To verify the confidence of the 55 important targets, we checked the confidence scores of their corresponding compound-target interactions. The 55 targets were involved in 172 interactions with the 12 compounds, in which 123 and 103 interactions had confidence scores at least 2 and 3, respectively, taking 71% and 59% of all the interactions, respectively. The result suggests the high confidence of the important targets. Fig. 7B shows the compound-target interactions with scores at least 3 between 10 compounds and 37 important targets. Baicalin, glycyrrhizic acid, hesperidin, and hyperoside target on most targets, while targets that are targeted by most compounds are AKT1, TNF-α, IL6, PTGS2, HMOX1, IL10, and TP53. This suggests key roles of these molecules for QFPDD's treatment on COVID-19.
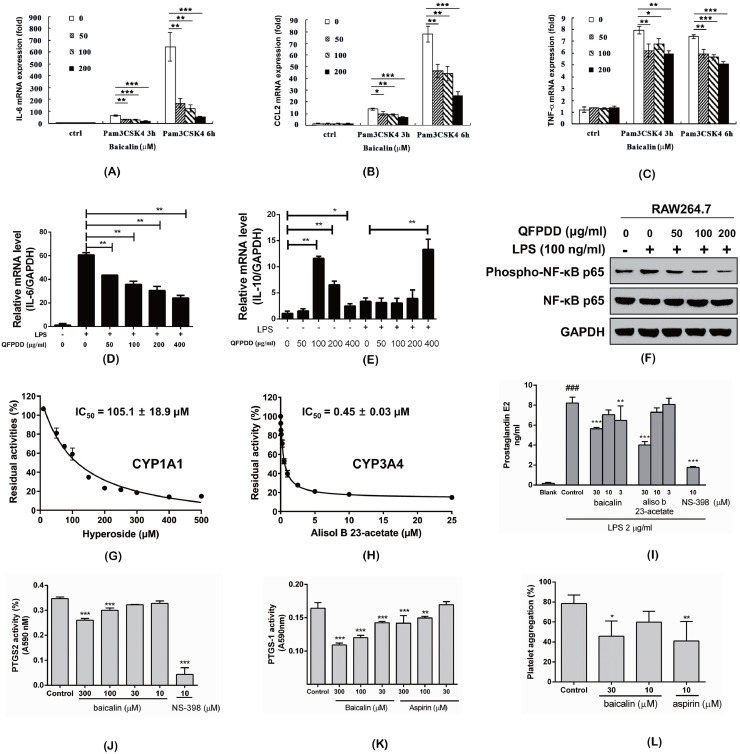
Fig. 7.
Experimental validation of important targets and effects. (A~C) Baicalin suppressed mRNA expression of IL-6 (A), CCL2 (B), and TNF-α (C) with the stimulation of Pam3CSK4 in RAW264.7.cells. (D~F) QFPDD down-regulated IL-6 (D), upregulated IL-10 (E) mRNA expression, and inhibited NF-κB activity (F) in RAW264.7 cells co-stimulated with LPS. (G) Inhibition of CYP1A1 by hyperoside. (H) Inhibition of CYP3A4 by alisol B 23-acetate in HLM. (I) Effects of baicalin and aliso b 23-acetate on PGE2 generation in RAW264.7 cells stimulated with LPS. (J~K) Effects of baicalin on recombinant human PTGS2 and ovine PTGS1 activities. (L) Effects of baicalin on AA induced platelet aggregation. All values are means of more than 3 replicate determinantion (n ≥ 3), and expression as mean ± SD. Significant differences between groups indicated or from the control groups were designated as * p < 0.05, ** p < 0.01, *** p < 0.001. Sinigicant difference between blank group and control (I) was designated as ### p < 0.001.
At last, we drew the herb-compound-pathway-symptom relationship diagram to globally depict the mechanism of QFPDD's treatment to COVID-19 (Fig. 5C), where the pathway-symptom relationships were from literatures. Fig. 5C shows that QFPDD's 12 compounds included in 10 herbs regulate 24 pathways to treat 18 symptoms related to COVID-19, such as virus infection, inflammation, organism injury and cytokine storm.
Validation of QFPDD's important targets by molecular docking
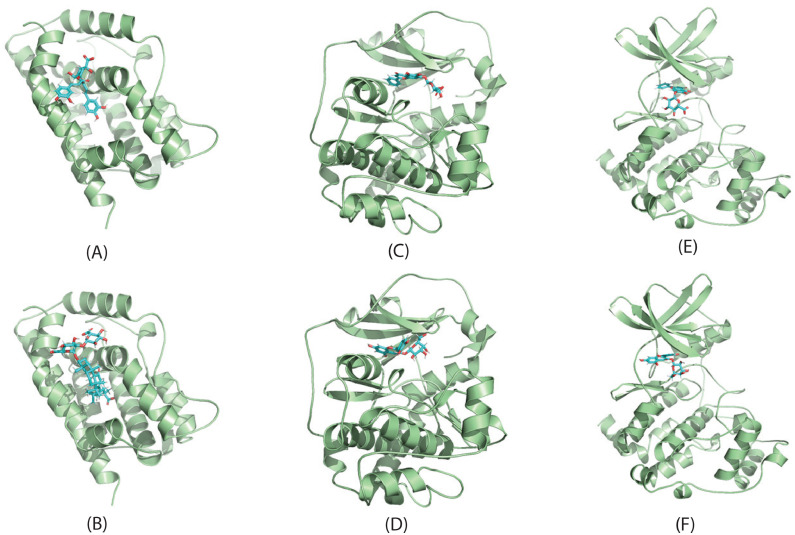
Among the 623 putative targets of QFPDD's compounds, 8 have interactions with different proteins of SARS-CoV-2. We conducted molecular docking to validate if QFPDD's compounds could bind to these targets. The crystal structures of the 8 targets, i.e., HMOX1 (PDB ID: 1N45), F2RL1 (PDB ID: 5NDZ), HDAC2 (PDB ID: 3MAX), PRKACA (PDB ID: 2GU8), GLA (PDB ID: 3GXP), COMT (PDB ID: 3A7E), CSNK2B (PDB ID: 1DS5), and PLAT (PDB ID: 1A5H), were downloaded from RCSB Protein Data Bank. The chemical structures of the compounds were extracted from PubChem. The binding mode with the best docking score was selected to analyze the interaction between the protein receptor and its predicted active compounds. The docking score represents the affinity between the protein receptor and the docking ligand. The lower docking score reflects the better affinity. As shown in Table 2 , the docking scores between 14 pairs of compound-target are lower than -5 kcal/mol (bold numbers), suggesting better binding affinity between the 7 compounds and 6 targets. Hence these compounds may bind to the 6 targets so as to inhibit the host-virus protein interactions they involved in. In this way, the activities of the virus could be repressed. In Fig. 6 we show binding poses of 6 pairs of compound-target interactions.
Table 2.
The docking scores between QFPDD's targets and compounds (kcal/mol).
| Target Compound | HMOX1 | F2RL1 | HDAC2 | PRKACA | GLA | COMT | CSNK2B | PLAT |
|---|---|---|---|---|---|---|---|---|
| baicalin | -5.614 | / | / | -7.02 | / | -5.395 | -8.557 | / |
| ephedrine | / | / | / | / | / | -7.04 | / | -5.731 |
| hesperidin | -6.892 | / | / | / | / | / | / | / |
| hyperoside | -5.201 | / | / | -7.361 | -4.536 | -5.336 | -9.343 | / |
| isochlorogenic acid A | -9.492 | -4.829 | / | / | / | / | / | / |
| pseudoephedrine | / | / | / | / | -6.426 | / | / | / |
| alisol b 23-acetate | / | / | -2.34 | / | / | / | / | / |
| glycyrrhizic acid | -6.923 | / | / | / | / | / | / | / |
Fig. 6.
Molecular docking of QPFPDD's three targets with corresponding compounds. The binding poses of HMOX1 complexed with isochlorogenic acid A (A) and glycyrrhizic acid (B); The binding poses of PRKACA complexed with baicailin (C) and hyperoside (D); The binding poses of CSNK2B complexed with baicailin (E) and hyperoside (F).
In vitro validation of QFPDD's important targets and associated effects
Baicalin is the most important bioactive flavonoid isolated from the roots of traditional Chinese medicine Scutellaria baicalensis. It is the most abundant component in the serum of QFPDD-gavaged mice. Macrophages are one of the main contributors of cytokine secretion in multiple infective diseases. Thus, we checked whether baicalin could influence the secretion of inflammatory mediators. As we know, Pam3CSK4 is a synthetic tripalmitoylated lipopeptide which mimics the acetylated amino terminus of bacterial lipoproteins and a well-known ligand of Toll-like receptor 1 (TLR1)/TLR2. The ligation of Pam3CSK4 leads to the production of pro-inflammatory cytokines (IL-6, TNF-α, etc.) and chemokines (e.g. CCL2). Mouse macrophage-like cell line RAW264.7 were plated in 24-well plate and stimulated with Pam3CSK4 and different concentrations of baicalin. As predicted, baicalin down-regulated the expression of the pro-inflammatory cytokines (IL-6, TNF-α) and chemokine (CCL2) in Pam3CSK4-induced RAW264.7 in a dose-dependent manner (Fig. 7 A~C).
Then we examined whether QFPDD could influence the secretion of inflammatory mediators. As we know, LPS is the main component of the cell wall of Gram-negative bacteria, which can cause the immune response of mammalian cells and lead to the production of pro-inflammatory cytokines (e.g. IL-6), and anti-inflammatory cytokine (e.g. IL-10). Mouse macrophage-like cell line RAW264.7 were plated in 24-well plate and stimulated with different concentrations of QFPDD and LPS. It was found that QFPDD down-regulated the expression of the pro-inflammatory cytokine IL-6 induced by LPS (Fig. 7D), but up-regulated the expression of anti-inflammatory IL-10 in LPS-induced RAW264.7 cells (Fig. 7E).
In recent years, a large number of evidences show that LPS/NF-κB signaling pathway is closely related to biological inflammation. Thus, we checked whether QFPDD could influence the activation of NF-κB signaling. As shown in Fig. 7F, QFPDD inhibited LPS-induced NF-κB activation in RAW264.7 cells. We concluded that QFPDD may display its inhibitory effects by suppressing NF-κB signaling in RAW264.7 cells.
Next, we verified the inhibition effects of QFPDD's compounds on cytochrome P450 enzymes. As shown in Fig. 7G, hyperoside could inhibit CYP1A1 mediated 7-ethoxyresorufin O-deethylation in a dose-dependent manner, with the IC50 value of 105.1 ± 18.9 μM. Fig. 7H illustrated that alisol B 23-acetate could potently inhibit CYP3A4 mediated NEN-hydroxylation in HLM via a dose-dependent manner, with the IC50 value of 0.45 ± 0.03 μM. This result suggested that alisol B 23-acetate might block CYP3A4-catalyzed biotransformation in the human body via inhibiting CYP3A4.
Subsequently, to evaluate PTGS2 activities, we measured the PGE2 levels in RAW264.7 cells. After the irreversible inhibition of aspirin (200 μM) on COX, only low amounts of PGE2 was detected in RAW264.7. Markedly increased PGE2 level was stimulated by LPS (2 μg/ml) and mostly inhibited by selective PTGS2 inhibitor NS-398 at 10 μM, which suggest the increase of PGE2 was due to newly induced PTGS2. Baicalin and alisol b 23-acetate were evaluated in the cell-based assay, and both compounds significantly inhibited PGE2 production at 30 μM (Fig. 7I). Additional experiments on pure human recombinant PTGS2 confirmed that baicalin exhibited a significant direct inhibition on PTGS2 enzymatic activity at 100 μM (Fig. 7J). The result partly explained the inhibition of baicalin on PGE2 formation in cell-based assay. However, alisol B 23-acetate itself had absorbance at 590 nm and interfered pure enzyme-based assay.
At last, to observe the effects of baicalin on PTGS1 activity, several other experiments were performed on pure ovine PTGS1 and platelets which contained PTGS1. Baicalin inhibited pure PTGS1 and AA induced platelet aggregation at the concentration as shown (Fig. 7K, L). Aspirin, as positive drug, inhibited pure PTGS1 and AA induced platelet aggregation at concentrations similar to those of baicalin.
It is noted that the expression of cytokines is usually regulated at transcriptional level and translational level. Although the secretion of some cytokines (such as IL-1 and IL-18) might be quite different from their transcriptional mRNA levels because they are also controlled at the post-transcriptional level by proteasome, for the majority of cytokines studied here (e.g., IL-6, TNF-α, CCL2 and IL-10), these two levels of expression are usually proportional. Thus the mRNA expression of cytokines was examined in our current study.
Conclusions
Herein, a systems pharmacology strategy was proposed to investigate the mechanism of QFPDD for the treatment of COVID-19 from molecule, pathway and network level. From the molecule perspective, 55 targets of QFPDD's 12 plasma compounds were found to play import roles in QFPDD's treatment to COVID-19, in which four compounds (baicalin, glycyrrhizic acid, hesperidin, and hyperoside) and 7 targets (AKT1, TNF-α, IL6, PTGS2, HMOX1, IL10, and TP53) were key molecules related to QFPDD's effects. From pathway level, 24 important pathways interacted with the 55 important targets to form 5 functional modules, corresponding to QFPDD's effects in immune regulation, organ protection, damaged cell clearance, and anti-inflammation. From network point of view, By regulating 10 target subnetworks corresponding to 6 biological processes (pattern recognition receptor signaling, signaling by interleukins, cell growth and death, hemostasis, biological oxidations and arachidonic acid metabolism) and 4 organ systems (nervous, sensory, circulatory, and digestive system), QFPDD could exhibit the effects of immune regulation, anti-infection, anti-inflammation and multi-organ protection. The deductions drawn from the systems study were in part validated by our subsequent molecule docking and in vitro experiments. One limitation of this research is that we could not construct target network for the lung or respiratory system, although the lung is the first organ that is attacked by SARS-CoV-2 infection. This is because our analysis was based on available data, in which the functional annotation was according to KEGG and Reactome pathway database. Currently these two databases have not included pathway category of respiratory system. We believe that further integration of some new data in the future may facilitate better understanding the mechanism of TCM formulas.
Funding
The work was supported by the National Key Research and Development Program of China (2020YFC0845400, 2017YFC1700200), Professor of Chang Jiang Scholars Program, National Natural Science Foundation of China (81520108030, 81572378, 81973706), Shanghai Engineering Research Center for the Preparation of Bioactive Natural Products (16DZ2280200), the Scientific Foundation of Shanghai China (13401900103, 13401900101), Shanghai Scientific Research Program on Prevention and Treatment of COVID-19 in Traditional Chinese Medicine (2020XGKY10), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Education, Natural Science Foundation of Jiangsu Province (BK20171231), CAMS Initiative for Innovative Medicine (CAMS-I2M, 2016-I2M-1-005), the Special Research Fund for Central Universities, Peking Union Medical College (2016ZX310194), the Natural Science Foundation of Chongqing (cstc2018jcyjAX0090).
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available in the supplementary material.
Authors’ contributions
WDZ and JZ designed the study. JZ, SST and JY collected the data. JZ and SST analyzed the data. DL conducted molecular docking. HWZ, FZ, DZT, GBG, YJZ, TS, XX, SZ, YLY performed in vitro experiments. JZ, SST, HWZ, GBG, YJZ, YLY wrote the manuscript. WDZ and JZ revised the manuscript. WDZ provided technical support and advices toward study.
Ethics approval and consent to participate
All animal studies were conducted in accordance with the guidelines for care and use of laboratory animals of the Second Military Medical University.
Declaration of Competing Interest
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phymed.2020.153315.
Appendix. Supplementary materials
References
- Barabasi A.L., Oltvai Z.N. Network biology: Understanding the cells's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Campbell C.M., Kahwash R. Will complement inhibition be the new target in treating COVID-19 related systemic thrombosis? Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.047419. [DOI] [PubMed] [Google Scholar]
- Chinese Thoracic Society, C.A.o.C.P. Guide for the prevention and treatment of coronavirus disease 2019. Chin. J. Tuberc. Respir. Dis. 2020;43 doi: 10.3760/cma.j.cn112147-112147-20200321-00392. [DOI] [PubMed] [Google Scholar]
- Consortium G. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C.D., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Viteri G., Weiser J., Wu G., Stein L., Hermjakob H., D'Eustachio P. The reactome pathway knowledgebase. Nucl. Acids Res. 2018;46:D649–d655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O'Meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Naing Z.Z.C., Zhou Y., Peng S., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Shen W., Shi Y., Zhang Z., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Ramachandran R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Wankowicz S.A., Bohn M., Sharp P.P., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Roesch F., Vallet T., Meyer B., White K.M., Miorin L., Rosenberg O.S., Verba K.A., Agard D., Ott M., Emerman M., Ruggero D., García-Sastre A., Jura N., von Zastrow M., Taunton J., Ashworth A., Schwartz O., Vignuzzi M., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S., Fraser J.S., Gross J., Sali A., Kortemme T., Beltrao P., Shokat K., Shoichet B.K., Krogan N.J. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Wu J.-J., Ning J., Hou J., Xin H., He Y.-Q., Ge G.-B., Xu W. Inhibition of human cytochrome P450 enzymes by licochalcone A, a naturally occurring constituent of licorice. Toxicol. In Vitro. 2015;29:1569–1576. doi: 10.1016/j.tiv.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., Wang J., Yu B., Zhang J., Bryant S.H. Pub chem substance and compound databases. Nucl. Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang-yu L., Wei A., Fei X., Min C., Ping Y., Ya-ling L., Xin X., Qin Z., Sha-sha F., Ming-wei Z. Observation on clinical effect of modified Qingfeipaidu decoction in treatment of COVID-19. Chin. Tradit. Herb. Drugs. 2020:1–4. [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucl. Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L., Chekkoury-Idrissi Y., Hans S., Delgado I.L., Calvo-Henriquez C., Lavigne P., Falanga C., Barillari M.R., Cammaroto G., Khalife M., Leich P., Souchay C., Rossi C., Journe F., Hsieh J., Edjlali M., Carlier R., Ris L., Lovato A., De Filippis C., Coppee F., Fakhry N., Ayad T., Saussez S. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020:1–11. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zeng H., Shan L., Yuan X., Li Y., Liu R., Zhang W. The different inhibitory effects of Huang-Lian-Jie-Du-Tang on cyclooxygenase 2 and 5-lipoxygenase. J. Ethnopharmacol. 2012;143:732–739. doi: 10.1016/j.jep.2012.07.037. [DOI] [PubMed] [Google Scholar]
- Li R., Wang L. Baicalin inhibits influenza virus A replication via activation of type I IFN signaling by reducing miR-146a. Mol. Med. Rep. 2019;20:5041–5049. doi: 10.3892/mmr.2019.10743. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li S., Fan T.-P., Jia W., Lu A., Zhang W. Network pharmacology in traditional Chinese medicine. Evid. Based Complement. Alternat. Med. 2014;2014 doi: 10.1155/2014/138460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Z.Q., Wu L.J., Zhang X.G., Li Y.D., Wang Y.Y. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol. 2007;1:51–60. doi: 10.1049/iet-syb:20060032. [DOI] [PubMed] [Google Scholar]
- Li, T., Qian, Y., Miao, Z., Zheng, P., Shi, T., Jiang, X., Pan, L., Qian, F., Yang, G., An, H., Zheng, Y., Xuebijing injection alleviates Pam3CSK4-induced inflammatory response and protects mice from sepsis caused by methicillin-resistant staphylococcus aureus. Front Pharmacol.11. [DOI] [PMC free article] [PubMed]
- Li X., Yu J., Zhang Z., Ren J., Peluffo A., Zhang W., Zhao Y., Yan K., Cohen D., Wang W. Network bioinformatics analysis provides insight into drug repurposing for COVID-2019. Prepr. - Am. Chem. Soc., Div. Pet. Chem. 2020 doi: 10.1016/j.medidd.2021.100090. 2020030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4 doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Ge G., Wang Y., Huang K., Chen J., Wang C., Liu P. Chemical composition identification and tissue distribution in mice of Qingfei Paidu decoction based on UHPLC-Q-Orbitrap HRMS. Chin. Tradit. Herb. Drugs. 2020;51:2035–2045. [Google Scholar]
- Mou X., Zhou D.Y., Zhou D., Liu K., Chen L.J., Liu W.H. A bioinformatics and network pharmacology approach to the mechanisms of action of Shenxiao decoction for the treatment of diabetic nephropathy. Phytomed. : Int. J. Phytother. Phytopharmacol. 2020;69 doi: 10.1016/j.phymed.2020.153192. [DOI] [PubMed] [Google Scholar]
- Ning J., Wang W., Ge G., Chu P., Long F., Yang Y., Peng Y., Feng L., Ma X., James T.D. Targeted enzyme activated two-photon fluorescent probes: a case study of CYP3A4 using a two-dimensional design strategy. Angew. Chem. Int. Ed. Engl. 2019;58:9959–9963. doi: 10.1002/anie.201903683. [DOI] [PubMed] [Google Scholar]
- National Health Commission & State Administration of Traditional Chinese Medicine. Guideline on diagnosis and treatment of COVID-19 (Trial 6th edition), Feb 19th, 2020.
- Park, A., Iwasaki, A.J.C.H., 2020. Microbe. Type I and Type III interferons–induction, signaling, evasion, and application to combat COVID-19. [DOI] [PMC free article] [PubMed]
- Prokunina-Olsson, L., Alphonse, N., Dickenson, R.E., Durbin, J.E., Glenn, J.S., Hartmann, R., Kotenko, S.V., Lazear, H.M., O'Brien, T.R., Odendall, C.J.J.o.E.M., 2020. COVID-19 and emerging viral infections: the case for interferon lambda. 217. [DOI] [PMC free article] [PubMed]
- Ren J.L., Zhang A.H., Wang X.J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020;155 doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., Young J., Zardecki C., Berman H.M., Bourne P.E., Burley S.K. The RCSB protein data bank: views of structural biology for basic and applied research and education. Nucl. Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genom. Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucl. Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M., Couzin-Frankel J., Kaiser J., Matacic C. How does coronavirus kill? Clinicians trace a ferocious rampage through the body, from brain to toes. Science. 2020 doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- Wang, J., Zhao, S., Liu, M., Zhao, Z., Xu, Y., Wang, P., Lin, M., Xu, Y., Huang, B., Zuo, X., Chen, Z., Bai, F., Cui, J., Lew, A.M., Zhao, J., Zhang, Y., Luo, H., Zhang, Y., 2020a. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv, 2020.2002.2005.20020545.
- Wang R., Yang S., Xie C., Shen Q., Li M., Lei X., Li J., Huang M. Clinical observation of qingfeipaidu decoction in the treatment of COVID-19. Pharmacol. Clin. Chin. Mater. Med. 2020 doi: 10.13412/j.cnki.zyyl.20200303.20200002. [DOI] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. Drug Bank 5.0: a major update to the Drug Bank database for 2018. Nucl. Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Wang J., Yang Y., Li T., Cao Y., Qu Y., Jin Y., Zhang C., Sun Y. Preliminary exploration of the mechanism of Qingfei Paidu decoction against novel coronavirus pneumonia based on network pharmacology and molecular docking technology. Acta Pharm. Sinica. 2020;55:374–383. [Google Scholar]
- Wu Q.L., Dong J., Zeng H.W., Lv C., Liu A.J., Zhang W.D. Monitoring antiplatelet aggregation in vivo and in vitro by microtiter plate method. J. Cardiovasc. Pharmacol. 2020;75:314–320. doi: 10.1097/FJC.0000000000000801. [DOI] [PubMed] [Google Scholar]
- Xu H.-Y., Zhang Y.-Q., Liu Z.-M., Chen T., Lv C.-Y., Tang S.-H., Zhang X.-B., Zhang W., Li Z.-Y., Zhou R.-R., Yang H.-J., Wang X.-J., Huang L.-Q. ETCM: an encyclopaedia of traditional Chinese medicine. Nucl. Acids Res. 2018;47:D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Huang A., Cui X., Han K., Hou X., Wang Q., Cui L., Yang Y. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208–4220. doi: 10.7150/thno.33803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaori S., Kushihara M., Yamamoto I., Watanabe K. Characterization of major phytocannabinoids, cannabidiol and cannabinol, as isoform-selective and potent inhibitors of human CYP1 enzymes. Biochem. Pharmacol. 2010;79:1691–1698. doi: 10.1016/j.bcp.2010.01.028. [DOI] [PubMed] [Google Scholar]
- Yan S., He G., Liang Z., Changchuan B. Preliminary discussion for COVID-19 and wind cold dampness plague. Chin. Arch. Tradit. Chin. Med. 2020;3 [Google Scholar]
- Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., Wang J., Qin Y., Zhang X., Yan X., Zeng X., Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020;214 doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Lv C., Wu Q., Zeng H., Guo X., Yang J., Tian S., Zhang W. Computational systems pharmacology reveals an antiplatelet and neuroprotective mechanism of Deng-Zhan-Xi-Xin injection in the treatment of ischemic stroke. Pharmacol. Res. 2019;147 doi: 10.1016/j.phrs.2019.104365. [DOI] [PubMed] [Google Scholar]
- Zhao J., Tian S., Yang J., Liu J., Zhang W. Investigating mechanism of Qing-Fei-Pai-Du-Tang for treatment of COVID-19 by network pharmacology. Chin. Tradit. Herb. Drugs. 2020;51:829–835. [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the supplementary material.