Abstract
Background:
Plasma cfDNA evaluation at acquired resistance to targeted therapies in lung cancer is routine, however, reports of extended clinical application and pitfalls in laboratory practice are still limited. In this study we describe our experience with cfDNA testing using EGFR T790M as a prototype.
Methods:
Patients with metastatic EGFR-mutant NSCLC patients who underwent plasma EGFR T790M testing at acquired resistance to EGFR tyrosine kinase inhibitors (EGFR-TKI) from January 2016 through August 2017 were identified. Molecular laboratory records were reviewed to assess performance of testing by digital PCR, concordance between plasma and tissue testing, turnaround time (TAT), plasma T790M variant allele frequency (VAF), and its correlations with metastatic sites and clinical outcomes.
Results:
177 patients underwent T790M cfDNA testing during this period. Plasma T790M was positive in 32% of patients. The median TAT was shorter for plasma T790M compared to tissue PCR (9 vs. 15 days, P<.0001), and led to osimertinib use in 84% of positive patients. In 52 patients with plasma and tissue T790M evaluation, the concordance was 77%. Plasma T790M VAF did not correlate with time to osimertinib discontinuation (P=.4). Plasma T790M status correlated with a higher number of metastatic sites (4 vs. 3, P<.001) and bone metastases (P=.0002).
Conclusion:
Plasma EGFR T790M testing had shorter TAT compared to tissue testing, however, it was longer than anticipated. Test sensitivity is higher in patients with osseous metastases and with higher metastatic burden suggesting a more limited role for early detection. T790M VAF was not associated with clinical outcomes.
Keywords: Lung cancer, Liquid Biopsy, EGFR, Erlotinib, Osimertinib
Introduction
Non-small cell lung cancer (NSCLC) represents a burgeoning field for the development of molecularly targeted therapies [1]. With this aim, oncologists have relied on tumor biopsies for genomic profiling, however, biopsies of the primary tumor or metastatic sites are subject to sampling bias due to tumoral heterogeneity [2]. In recent years, technological advances have enabled the identification of tumor derived somatic alterations in plasma due to the analysis of cell-free DNA (cfDNA), RNA (cfRNA) and exosomes [3]. cfDNA is primarily a derivative of ruptured leucocytes, with a very small fraction derived from tumor cells called circulating-tumor DNA (ctDNA). The concentration of ctDNA varies based on the type, location, and staging of the tumor. In most advanced cancers, including NSCLC, ctDNA is detectable in >60% of patients [4,5]. Among the multiple potential clinical applications of this technology are: (1) primary tumor genotyping if tumor biopsies are not available, (2) early detection of relapse, (3) monitoring of patient’s response to systemic therapy and (4) detection of resistance mutations in patients treated with targeted therapies [6]. The latter application has been developed successfully in EGFR-mutant NSCLC during the recent years. The most frequent on-target resistance mechanism to first/second generation EGFR tyrosine-kinase inhibitors (TKIs; erlotinib, afatinib, gefitinib) is the EGFR T790M mutation, accounting for approximately 60% of cases [7]. Several PCR based techniques can be used for the detection of EGFR mutations in cfDNA including Cobas, BEAMing (beads, emulsion, amplification, and magnetics), digital polymerase chain reaction (dPCR) and next generation sequencing (NGS), with variable sensitivities, quantification capabilities and reported concordance with concurrent tissue biopsy results [8–10]. Among many of the platforms, digital PCR techniques have been the most extensively used showing high specificity and higher sensitivity than other platforms with a detection limit of 0.02% [10–14]. Compared to NGS, dPCR only provides targeted assessment of a single or a few known alterations but turnaround times are significantly shorter facilitating more rapid treatment decisions. Quantification and dynamic monitoring of EGFR T790M plays an important role in determining the treatment strategy. The third generation EGFR-TKI osimertinib has demonstrated potent and irreversible efficacy to acquired EGFR T790M mutations after treatment with first/second generation EGFR-TKI with a response rate above 60% [15,16]. The detection of T790M in cfDNA can guide the indication of osimertinib leading to similar outcomes to those of patients positive by a tissue-based assay [17]. Subsequently, the US Food and Drug Administration (FDA) approved a blood-based companion diagnostic for osimertinib to detect EGFR T790M in plasma [18,19]. However, biologic factors influencing ctDNA detection are poorly understood. Furthermore, the clinical performance of this strategy has not been adequately characterized in the real-world clinical setting, particularly the relevance of the variant allele frequency (VAF) for T790M.
In this study, we aim to assess the clinical performance of cfDNA samples using the EGFR T790M mutation as a prototype. We evaluate the clinical utility of testing with a dPCR platform at the time of clinical progression to first/second-generation EGFR-TKI and explore its correlation with clinical outcomes in EGFR-mutant NSCLC patients treated with the third-generation EGFR-TKI osimertinib. We also outline some of the laboratory aspects that are important in the overall incorporation of a cfDNA assay in clinical practice.
Material and Methods
Patients
This is a single center retrospective study of patients treated at Memorial Sloan Kettering Cancer Center (MSK). We included all patients with EGFR-mutant lung cancers who were treated with EGFR-TKIs between January 2016 - August 2017 and underwent ctDNA T790M testing at the time of acquired resistance. This study was approved by the MSK Institutional Review Board and conducted in accordance with the Declaration of Helsinki. Biospecimens in this study were obtained under the informed consent of a banking protocol.
Data collection
Clinical data was retrieved from the electronic medical records. Extracted data included gender, age at diagnosis, stage, metastatic sites, smoking status, treatment history, and clinical outcomes. Staging was based on the seventh edition of American Joint Commission on Cancer staging criteria.
ctDNA T790M evaluation
For all patients, peripheral blood was collected in 2 Streck DNA BCT tubes (Streck, La Vista, NE) at the time of clinical resistance, which was defined by the treating oncologist based on the radiological assessment of progression of disease. The tubes were centrifuged to isolate plasma and cfDNA was extracted using a chaotropic silica-bead-based chemistry, MagMax Cell-Free DNA isolation kit, (Thermo Fisher Scientific, Waltham, MA USA) according to the manufacturer’s instructions. cfDNA concentration was determined by 2200 TapeStation (Agilent, Santa Clara, CA, USA). EGFR T790M testing was performed using picodroplet dPCR as previously described [9]. In brief, this technique was performed to amplify part of EGFR exon 20 in the presence of fluorescent probes specific to the wild-type and mutant alleles. Fluorescent droplet counts were analyzed on a RainDance Sense instrument (Bio-Rad, Hercules, CA, USA); results were compared to control counts. VAF was calculated as: mutant / (wild-type + mutant) allele. Concordance was calculated to tissue in patients who underwent tissue analysis with targeted next generation sequencing (NGS) MSK-IMPACT™ (MSK-Integrated Mutation Profiling of Actionable Cancer Targets) [20,21] and/or dPCR within 90 days of plasma blood draw. Turnaround time (TAT) for ctDNA T790M testing was measured from the time of blood draw to the time of receipt of report and measured in calendar days. TAT of tissue T790M evaluation was measured from the time of the biopsy to the time of report.
Statistical Analysis
Fisher exact test was used to identify significant associations between categorical variables. Time to treatment discontinuation (TTD) was used as a surrogate for progression free survival (PFS) [22,23]. TTD was defined as the time from start of EGFR-TKI to last dose administered. TTD and overall survival (OS) were estimated from treatment start date using the Kaplan-Meier method. Log-rank test was used to evaluate for associations between categorical variables and survival. Patients without complete survival data were censored at date of last follow-up. For patients with both plasma ctDNA testing and tissue testing for EGFR T790M, concordance along with an exact 95% confidence interval was evaluated. All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant. The Bonferroni method was used to adjust for multiple comparisons. Statistical software (SAS version 9.3) was used for statistical analyses.
Results
Patients and clinical outcomes
During the study period, 177 applicable EGFR-mutant NSCLC patients were identified of whom 56 (32%) had the T790M mutation detected by plasma dPCR. T790M status was detected in tissue biopsy in 12 (21%) of ctDNA T790M(+) patients and in 40 (33%) of ctDNA T790M(−) patients (Figure 1). In this cohort, median age of patients was 66 years (range, 38 to 91) and most patients were non-smokers (53%). The most commonly used initial EGFR-TKI was erlotinib (88%). There was no significant difference in baseline characteristics between patients in whom T790M was detected and not detected by ctDNA evaluation (Table 1). The average total cfDNA yield in the 177 analyzed samples was 46 ng (range, 3.94 to 660 ng) (Supplementary Figure 1). Using the total cfDNA recovered to the maximum of 60 ng input, the occupied droplet count per sample averaged 9275 droplets (range, 1268 to 53272 droplets). The failure rate was 1.1% (2/177 samples) with those samples generating less than 500 droplets. In 31% of samples (55/177), the number of droplets generated was <5,000.
Figure 1.

Flow of patients.
Abbreviation: NSCLC, non-small cell lung cancer; ctDNA, circulating-tumor DNA; PCR, polymerase chain reaction; NGS, next generation sequencing.
Table 1:
Baseline characteristics of patients.
| All patients (n=177) | ctDNA T790M detected (n=56) | ctDNA T790M not detected (n=121) | |
|---|---|---|---|
| Age (y, range) | 66 (38 – 91) | 66 (38 – 91) | 66 (42 – 90) |
| Sex (%) | |||
| Female | 115 (65) | 35 (62) | 80 (66) |
| Male | 62 (35) | 21 (38) | 41 (34) |
| Smoking status (%) | |||
| Never | 93 (53) | 31 (55) | 62 (51) |
| Ever | 84 (47) | 25 (45) | 59 (49) |
| EGFR-mutant allele (%) | |||
| exon 21 | 59 (33) | 17 (30) | 42 (35) |
| exon 19 | 107 (60) | 37 (66) | 70 (58) |
| exon 18 | 11 (6) | 2 (4) | 9 (7) |
| Initial EGFR-TKI (%) | |||
| Erlotinib | 155 (88) | 53 (95) | 102 (84) |
| Afatinib | 13 (7) | 2 (4) | 11 (9) |
| Gefitinib | 3 (2) | 1 (1) | 2 (2) |
| Third-generationa | 6 (3) | 0 | 6 (5) |
| Median time EGFR-TKI to T790M ctDNA testing (y) | 1.4 | 1.4 | 1.4 |
Abbreviation: y, years; ctDNA, circulating-tumor DNA; TKI, tyrosine-kinase inhibitor.
Osimertinib (n=4), Rociletinib (n=1), Nazartinib (n=1)
The TTD of first-line EGFR-TKI in ctDNA T790M(+) and (−) patients was 15.7 months and 16.9 months, respectively (HR 1.32, 95% CI 0.93 – 1.88, P=.09) (Figure 2). There was no difference in OS between these two groups (median OS 4.6 vs. 7.6 years, HR 1.39, 95% CI 0.77 – 2.48, P=.24). Median plasma EGFR T790M VAF was 0.98% (range, 0.1 to 49.5%), lower than tissue T790M VAF (12.8%, range, 2.6 to 27.8%, P<.0001). In 47 patients with ctDNA T790M(+) treated with osimertinib, patients with lower and higher than median VAF had a similar TTD (HR 1.38, 95% CI 0.64 – 3, P=.4) and OS (HR 1.17, 95% CI 0.42 – 3.2, P=.76) (Figure 3).
Figure 2.
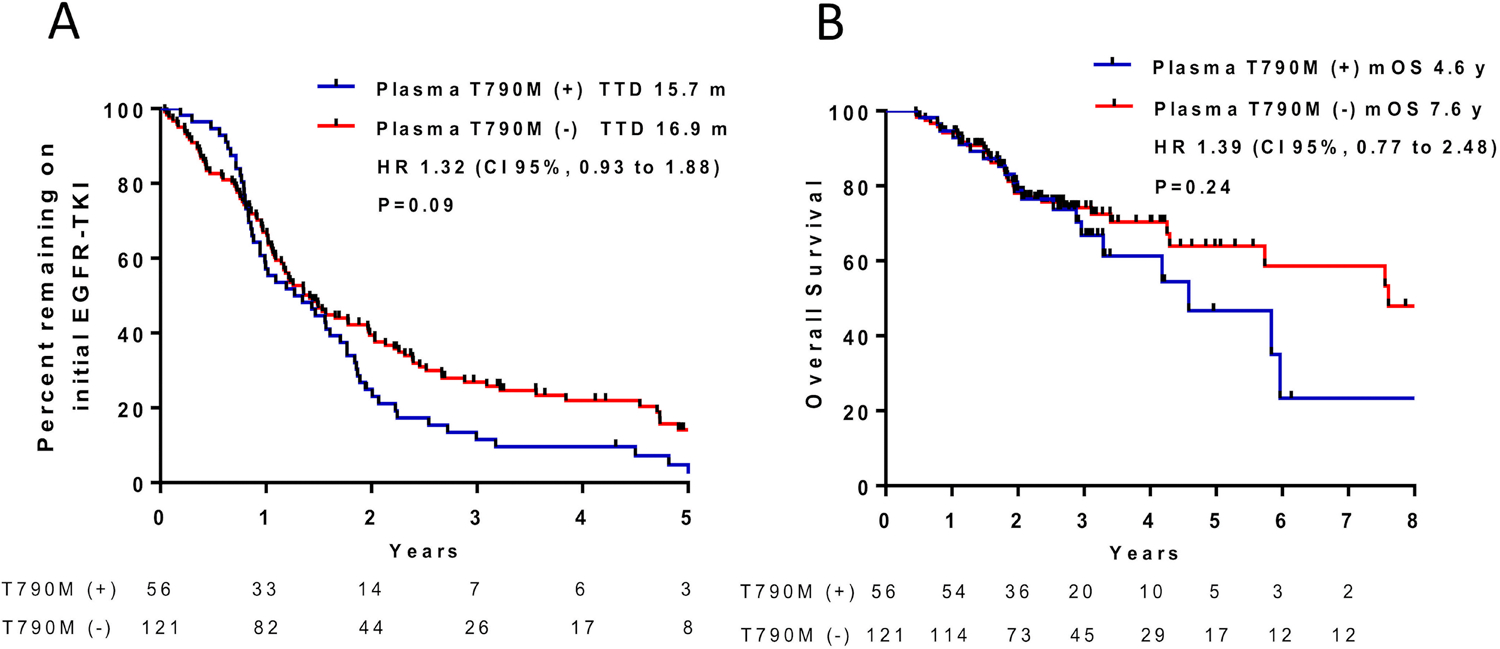
Clinical outcomes in ctDNA T790M(+) and (−) patients A) Time to initial EGFR-TKI discontinuation B) Overall survival.
Abbreviation: TKI, tyrosine kinase inhibitor; TTD, time to treatment discontinuation; OS, overall survival; m, months; y, years; HR, hazard ratio.
Figure 3.
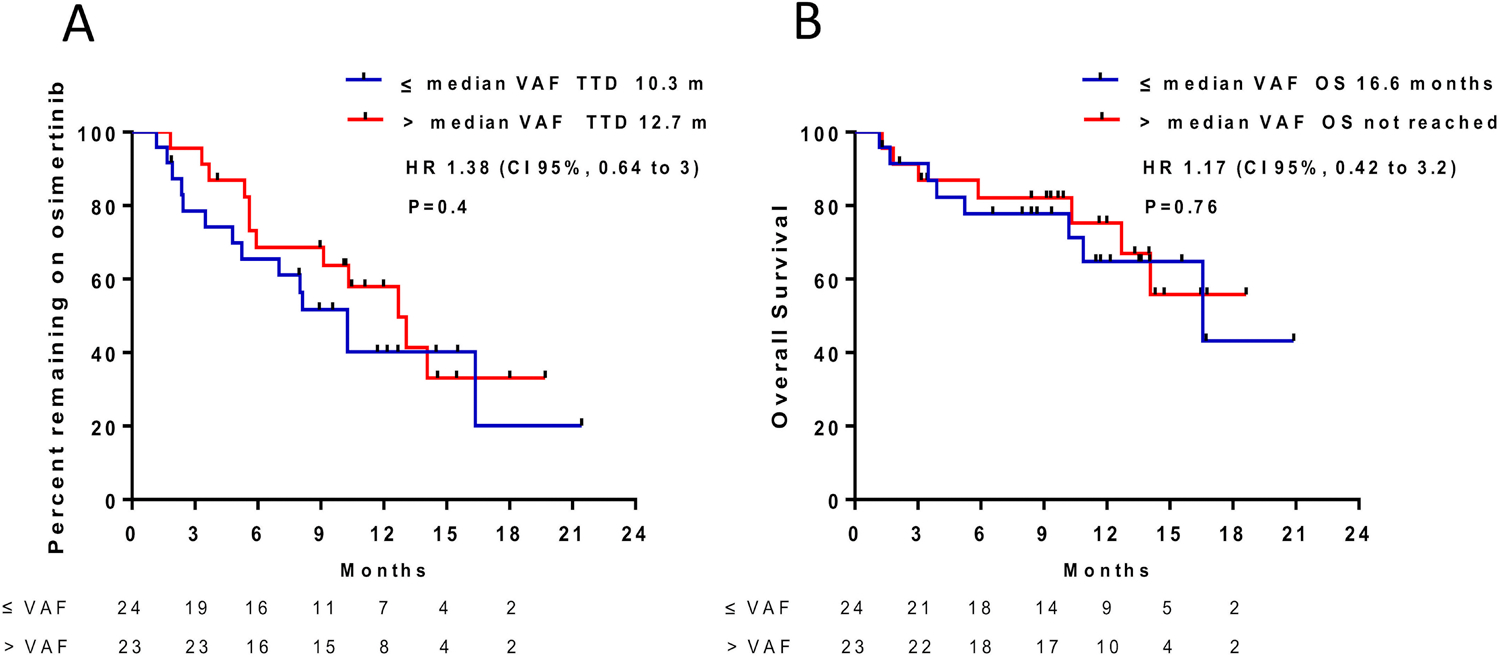
Clinical outcomes by plasma T790M VAF in patients treated with osimertinib A) Time to osimertinib discontinuation B) Overall survival.
Abbreviation: TTD, time to treatment discontinuation; OS, overall survival; VAF, variant allele frequency; m, months; HR, hazard ratio.
Concordance ctDNA and tissue evaluation
Fifty-two patients had evaluation of T790M both in plasma and tissue with an overall concordance of 77% (40/52, 95% CI 63% – 87%). Among patients who tested positive for plasma T790M, 83% (10/12, 95% CI 52% – 98%) were concordant on tissue evaluation. Among patients who tested tissue positive, 50% (10/20, 95% CI 27% – 73%) were concordant on plasma evaluation (Supplementary Table 1). The median TAT was shorter for plasma T790M compared to tissue PCR (9 vs. 15 days, P<.0001), and led to osimertinib use in 84% (47/56) of positive patients. Eighteen percent of the patients (32/177) had more than 1 cfDNA assessment (range, 2 to 4). All these patients had tested negative or equivocal for T790M in the initial assessment; 4 patients who were previously ctDNA negative and 2 who were equivocal were ctDNA T790M(+) in a subsequent evaluation.
Metastatic sites and correlation with ctDNA
Patients with ctDNA T790M(+) had a higher median number of metastatic sites compared to those who were ctDNA T790M(−) (4 vs. 3, P<.001) (Supplementary Figure 2). There was a higher percentage of ctDNA T790M(+) in patients with bone metastases compared to those without osseous lesions (P=.0002). No association was found with other metastatic sites (Table 2).
Table 2.
Metastatic site correlation with positive ctDNA.
| Metastatic site | ctDNA T790M positive | Unadjusted P-value |
|---|---|---|
| Pleura | 37% (36/98) with metastases vs. | 0.14 |
| 25% (20/79) without | ||
| Lymph node | 38% (35/92) with metastases vs. | 0.07 |
| 25% (21/85) without | ||
| Bone | 45% (41/92) with metastases vs. | 0.0002 |
| 17% (15/85) without | ||
| Liver | 38% (19/50) with metastases vs. | 0.28 |
| 29% (37/127) without | ||
| Adrenal | 39% (7/18) with metastases vs. | 0.60 |
| 31% (49/159) without | ||
| Brain | 42% (22/53) with metastases vs. | 0.08 |
| 27% (34/124) without | ||
Abbreviation: ctDNA, circulating-tumor DNA.
Note: The Bonferroni-adjusted significance threshold was set at P=0.008.
Discussion
In this study we have shown that in EGFR-mutant NSCLC patients, ctDNA T790M testing at the time of clinical resistance to first/second generation EGFR-TKIs had high overall concordance (77%) with corresponding tumor tissue-based testing. This is in keeping with prior cross-platform studies using methods of similar sensitivity [10]. Importantly, however, when patients are stratified into subsets, marked differences could be seen. For patients with positive cfDNA results, concordance with tissue biopsy status was 83% while for those with positive tumor tissue results, concordance with cfDNA dropped to 50%. These findings are in line with the recommendation to retest using tumor tissue in patients with negative ctDNA testing [2]. Our study also supports the notion that shedding of ctDNA is not equal for all tumors and mutation detection can be affected by numerous factors such as mitotic rate, necrosis, degree of vascularization and renal clearance, among many others. For metastatic cancers, overall disease burden, bone or liver involvement are significant factors that may favor ctDNA detection [24]. In our study, we confirm that ctDNA T790M was more likely to be positive in patients with more metastatic sites, particularly in patients with osseous metastases, however, no significant associations were identified with liver involvement or metastasis to other sites.
In a small proportion of patients, EGFR T790M status was positive in cfDNA but negative by tissue biopsy. This highlights the unique capabilities of cfDNA in addressing the sampling of tumors that may be highly heterogeneous. This is particularly relevant in the setting of acquired resistance where only some metastatic lesions may harbor the mutation.
At a technical level, our study highlights important challenges and limitations one should consider when implementing cfDNA testing in routine clinical practice. Although platforms like dPCR and BEAMing have been well established among the existing technologies providing the highest sensitivity levels with limits of detection as low as 0.1 to 0.01% (1 in 1000 to 1 in 10000), the ultimate sensitivity that can be attained by any assay is dependent on the quantity of cfDNA recovered from the sample. For NGS assays interrogating multiple genomic targets, the sensitivity can also be highly variable from target to target, often not reaching beyond 1%. In our clinical cohort, the total cfDNA recovered varied widely from patient to patient and 31% samples had total yields that limited the sensitivity of detection in the range of 1 to 0.1%. While this can be circumvented by cfDNA extraction from multiple tubes, there are also technical limitations in the handling of samples with very low cfDNA content. In these cases, additional steps for sample concentration are required, adding to the total turnaround time and often affecting the performance of the assays downstream. Importantly, in the setting of resistance, while the detection of the sensitizing EGFR mutation is often facilitated by the common co-occurrence of EGFR amplification, the detection of the resistance mutation can be compromised by their subclonal nature.
In our cohort, while the median TAT of nine days was shorter for plasma T790M compared to tissue PCR, this was longer than expected based on the hands-on time for the performance of the assay (3 days). The ultimate TAT for any assay in clinical practice is dependent on many factors including volume, on the need for batching, the failure rate and the need for further manipulation of DNA to match the requirements of the assay. These variables explain why often in real pathology practice, TAT described in prospective trials cannot be replicated. Commercial laboratories that only concentrate in one type of cfDNA assay, have overcome some of these challenges reporting impressive TAT even for NGS testing [25]. Continued optimization of internal NGS and targeted sequencing platforms, workflows and reporting is of paramount importance as our understanding of acquired resistance mechanisms, and their actionability, continue to expand.
Recently, the introduction of osimertinib as a first-line option in EGFR-mutant NSCLC has challenged the validity of the treatment algorithm used in the present study. The FLAURA trial compared first-line osimertinib with the standard choice of erlotinib or gefitinib in patients with NSCLC harboring EGFR exon 19 deletions or L858R point mutation [26]. This trial reached its primary outcome showing a PFS of 18.9 months vs. 10.2 months (HR 0.46, 95% CI 0.37 – 0.57, P<.001). There was also a trend towards improved survival in the osimertinib arm (HR 0.63, 95% CI, 0.45 to 0.88, P=.007 [nonsignificant in the interim analysis]). However, among the 129 patients who received treatment after disease progression in the control arm, only 48 patients (37%) crossed over to receive osimertinib [27]. These aforementioned findings led to the FDA approval of the treatment regimen in the United States in 2018. Importantly, there are some caveats for the worldwide adoption of this therapy as a first-line standard, particularly in the absence of definitive benefit in overall survival compared to sequential treatment. The cost-effectiveness of osimertinib has been suggested specifically in the second-line setting in patients with advanced EGFR T790M-positive NSCLC [28], whereas in first-line has not been found cost-effective by World Health Organization criteria [29].
The present study has some limitations. Firstly, it was designed retrospectively, thus we could not assess PFS and we used TTD as real-world surrogate. However, emerging evidence has shown good correlation between TTD and PFS, particularly in NSCLC patients receiving TKIs [22]. Secondly, given our limited sample size the association of positive ctDNA evaluation with metastatic sites was not adjusted by other covariates such as performance status or number of metastatic sites. Thirdly, our dPCR platform only evaluated T790M and not the original known EGFR mutation. These data could have been used as an internal control to better understand false negative cases.
Conclusion
The analysis of ctDNA offers unique analytical opportunities for the assessment of resistance mechanisms in EGFR-mutant NSCLC, particularly those patients with high burden of disease and bone metastases. Plasma EGFR T790M testing had shorter TAT compared to tissue testing, however, it was longer than previous prospective studies likely owing to real world clinical variables not fully accounted for in prospective clinical trial designs. The median VAF for T790M in ctDNA does not appear to correlate with time to treatment discontinuation in patients treated with osimertinib after progression on first/second generation EGFR-TKIs. Liquid biopsy for plasma cfDNA by in-house platform faces relevant challenges in a real world pathology practice, which need to be taken into account while searching for the most pragmatic approach using this technology.
Supplementary Material
Acknowledgements:
This research was supported in part by the National Cancer Institute of the National Institutes of Health (T32 CA009207, P30 CA008748).
Funding statement
This work was supported by the National Cancer Institute of the National Institutes of Health (T32 CA009207, P30 CA008748).
Footnotes
Disclosure of interest
MO received consulting fees from PharmaMar. GJR received research funding from Novartis, Roche, Genentech, Millenium, GlaxoSmithKline, Pfizer, Infinity Pharmaceuticals and ARIAD. He has received travel expenses from Merck Sharp & Dohme. He is listed as inventor on a patent application submitted for pulsatile use of erlotinib to treat or prevent brain metastases. CR is a consultant for Abbvie, Amgen, Ascentage, AstraZeneca, Bicycle, Celgene, Chugai, Daiichi Sankyo, Genentech/Roche, GI Therapeutics, Loxo, Novartis, Pharmamar, and Seattle Genetics, and serves on the Scientific Advisory Boards of Elucida and Harpoon. ML is a consultant for NCCN/Boehringer-Ingelheim Afatinib Targeted Therapy, is on the advisory committee for Foundation Medicine, and has research grant support from LOXO Pharmaceuticals. HY is a consultant for AstraZeneca and has received travel support from Lilly. Her institution, Memorial Sloan Kettering has received research funding from Astellas Pharma, AstraZeneca, Daiichi, Lilly, Novartis and Pfizer for clinical trials she is involved in. BTL is consultant for Genentech, Thermo Fisher Scientific, Guardant Health. MEA received speaker’s fees from Raindance Technologies, Invivoscribe, Archer Dx and Biocartis. All other authors have no relevant disclosures to report.
Data sharing statement
S.M. and M.E.A. had full access to the data in the study and take responsibility for the integrity of the data analysis. Data requests may be submitted to M.E.A., which will be submitted for ethical approval (arcilam@mskcc.org).
References
- 1.Buettner R, Wolf J, Thomas RK. Lessons learned from lung cancer genomics: the emerging concept of individualized diagnostics and treatment. J Clin Oncol 2013. May 20;31(15):1858–65. [DOI] [PubMed] [Google Scholar]
- 2.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017. September;14(9):531–548. [DOI] [PubMed] [Google Scholar]
- 3.Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med 2012;63:199–215. [DOI] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014. February 19;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li BT, Drilon A, Johnson ML, et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol 2016. January;27(1):154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz LA, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012. June;486(7404):537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013. April 15;19(8):2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011. December 15;17(24):7808–15. [DOI] [PubMed] [Google Scholar]
- 9.Borsu L, Intrieri J, Thampi L, et al. Clinical Application of Picodroplet Digital PCR Technology for Rapid Detection of EGFR T790M in Next-Generation Sequencing Libraries and DNA from Limited Tumor Samples. J Mol Diagn 2016. November;18(6):903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015. December;90(3):509–15. [DOI] [PubMed] [Google Scholar]
- 11.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009. March 15;15(6):2076–84. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One. 2014;9(11):e110780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer. 2007. September 17;97(6):778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu C, Liu X, Chen Y, et al. Direct serum and tissue assay for EGFR mutation in non-small cell lung cancer by high-resolution melting analysis. Oncol Rep 2012. November;28(5):1815–21. [DOI] [PubMed] [Google Scholar]
- 15.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015. April 30;372(18):1689–99. [DOI] [PubMed] [Google Scholar]
- 16.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017. February 16;376(7):629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016. October 1;34(28):3375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016. December;17(12):1643–1652. [DOI] [PubMed] [Google Scholar]
- 19.cobas EGFR Mutation Test v2. 2016. https://www.accessdata.fda.gov/cdrh_docs/pdf12/P120019S007c.pdf.
- 20.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015. May;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017. June;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenthal GM, Gong Y, Kehl K, et al. Analysis of Time to Treatment Discontinuation of Targeted Therapy, Immunotherapy, and Chemotherapy in clinical trials of patients with non-small cell lung cancer. Ann Oncol 2019. February 22. [DOI] [PubMed] [Google Scholar]
- 23.Offin M, Rizvi H, Tenet M, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2018. July 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008. September;14(9):985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabari JK, Offin M, Stephens D, et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J Natl Cancer Inst 2018. November 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018. January 11;378(2):113–125. [DOI] [PubMed] [Google Scholar]
- 27.Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018. November;15(11):694–708. [DOI] [PubMed] [Google Scholar]
- 28.Bertranou E, Bodnar C, Dansk V, et al. Cost-effectiveness of osimertinib in the UK for advanced EGFR-T790M non-small cell lung cancer. J Med Econ 2018. February;21(2):113–121. [DOI] [PubMed] [Google Scholar]
- 29.Aguiar PN Jr., Haaland B, Park W, et al. Cost-effectiveness of Osimertinib in the First-Line Treatment of Patients With EGFR-Mutated Advanced Non-Small Cell Lung Cancer. JAMA Oncol 2018. August 1;4(8):1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


