Abstract
Objective
Development of a risk-stratification model to predict severe Covid-19 related illness, using only presenting symptoms, comorbidities and demographic data.
Materials and methods
We performed a case-control study with cases being those with severe disease, defined as ICU admission, mechanical ventilation, death or discharge to hospice, and controls being those with non-severe disease. Predictor variables included patient demographics, symptoms and past medical history. Participants were 556 patients with laboratory confirmed Covid-19 and were included consecutively after presenting to the emergency department at a tertiary care center from March 1, 2020 to April 21, 2020
Results
Most common symptoms included cough (82%), dyspnea (75%), and fever/chills (77%), with 96% reporting at least one of these. Multivariable logistic regression analysis found that increasing age (adjusted odds ratio [OR], 1.05; 95% confidence interval [CI], 1.03–1.06), dyspnea (OR, 2.56; 95% CI: 1.51–4.33), male sex (OR, 1.70; 95% CI: 1.10–2.64), immunocompromised status (OR, 2.22; 95% CI: 1.17–4.16) and CKD (OR, 1.76; 95% CI: 1.01–3.06) were significant predictors of severe Covid-19 infection. Hyperlipidemia was found to be negatively associated with severe disease (OR, 0.54; 95% CI: 0.33–0.90). A predictive equation based on these variables demonstrated fair ability to discriminate severe vs non-severe outcomes using only this historical information (AUC: 0.76).
Conclusions
Severe Covid-19 illness can be predicted using data that could be obtained from a remote screening. With validation, this model could possibly be used for remote triage to prioritize evaluation based on susceptibility to severe disease while avoiding unnecessary waiting room exposure.
Keywords: Covid-19, Severe, Remote triage, Symptoms
1. Introduction
1.1. Background
Since early reports of an unexplained pneumonia in Wuhan, China in December 2019, SARS-CoV-2 has spread to more than 200 countries and has become a global pandemic. There have been more than 24 million confirmed cases of Covid-19 as of August 28th, 2020 with over 800,000 case fatalities in 188 nations [1]. Currently, there are more than fifty thousand new cases of Covid-19 reported daily during July 2020 in the United States [2] and with states lifting strict lockdown measures, there is significant concern that there will be a resurgence of cases [3,4]. Early in the pandemic, the number of cases rose rapidly and emergency departments were inundated with patients in geographic “hot spots” like New York, New York and Detroit, Michigan [5], and new hotspots such as Florida, Arizona and Texas have been emerging [6]. Principles likely underlying this rapid progression of disease include the frequency of encounters between susceptible and infected individuals and the fidelity of transmission with each interaction [7].
1.2. Importance
Given that most patients presenting with symptoms concerning for Covid-19 infection have ultimately been negative [8], there is a concern that a large number of uninfected individuals may come into contact with infected patients at testing centers or emergency departments and thus propagate the infection due to the contagious nature of the virus [9,10]. There is also potential for transmission without direct contact. Viral particles have been shown to persist on surfaces for several days and new evidence suggests that aerosolized particles can be detected up to 29 ft from the infected individual and linger in the environment and may have contributed to superspreading events [[11], [12], [13]]. Crowded hospitals and clinics may therefore present ideal conditions for viral transmission. As such, minimizing crowding in hospital waiting areas and clinics is critical to mitigate nosocomial spread.
Remote triage tools have the potential to facilitate this by evaluating patients without physical contact. Existing web-based triage tools employ branched logic questions based on early recommendations from the CDC [14] and preliminary data from China [15,16] to advise patients, but lack statistical validation [[17], [18], [19]]. More recently, statistical models have been developed to predict potential Covid-19 infections, which represents an innovative step forward. However, these models either necessitate in-person collection of vital signs, imaging, and/or laboratory studies in order to risk stratify patients [[20], [21], [22], [23]] or do not predict disease severity and therefore do not determine which patients should be evaluated most urgently [24]. A validated model for predicting the risk of severe Covid-19 illness using only predictors that do not require in-person evaluation could be utilized as part of a remote triage strategy to prioritize the in-person ED evaluations of those at risk for severe disease while lower risk individuals could await their evaluation at home.
1.3. Goals of this investigation
Our proof-of-concept study aims to demonstrate the value of information that can be collected remotely, prior to presentation at a healthcare facility, such as symptoms and comorbidities, in predicting a patient's risk of developing severe disease from Covid-19 infection. Remote risk-stratification has the potential to facilitate clinical decision making without necessitating in-person evaluation, preventing unnecessary nosocomial spread.
2. Methods
2.1. Study design and setting
We performed a retrospective case-control study using the electronic medical records of an Emergency Department at single large tertiary academic medical center. Included patients presented from March 1, 2020 to April 21, 2020. Outcomes of admission were followed through May 16, 2020. The study was approved by the institutional review board.
2.2. Participants
We obtained data from 556 patients who tested positive for Covid-19 and presented to the Adult Emergency Department (ED), either directly or via ED-to-ED transfer, between March 01, 2020 and April 21, 2020. Testing was performed on patients based on their presentation according institutional guidelines at the time or if they had known exposure. Testing was done on nasopharyngeal swab samples via reverse transcriptase polymerase chain reaction (RT-PCR) assay. Patients were considered Covid-19 positive if they had a positive test anytime 21 days prior to presentation to the ED or had a positive test up to 14 days after the ED presentation. This timeframe was determined by institutional guideline at the time of testing. Patients were excluded if they tested negative for Covid-19 or if test results were inconclusive. During the study period, our institution primarily utilized the Simplexa SARS-CoV-2 EUA Assay (Diasorin Molecular, Cypress, CA) to determine Covid-19 status. Accuracy in comparison to reference laboratory samples was determined to be 100% by our clinical microbiology laboratory and in 400 patients unrelated to this study, repeat testing was performed within 72 h and raw concordance between serial tests was determined to be 95% by our clinical microbiology lab.
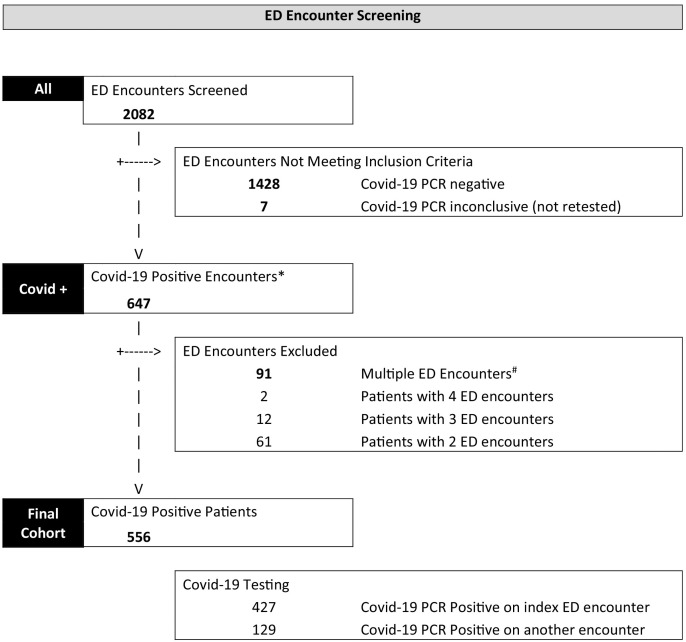
For patients with multiple presentations, the encounter concluding with the highest level of care (ICU admission/mechanical ventilation) was selected for determination of the primary outcome (Fig. 1 ). Cases were those with severe illness, defined as admission to the intensive care unit (ICU), requirement for mechanical ventilation, in-hospital death, or discharge to hospice at any point during hospital admission. Controls were those with non-severe outcomes, defined as either admission to the hospital without the aforementioned outcomes or discharge from the ED without hospital admission.
Fig. 1.
Schematic representation of the patient encounter inclusion process.
*Covid-19 positive encounter window was defined as:
-Positive PCR up to 3 weeks BEFORE index ED encounter
-Positive PCR up to 2 weeks AFTER index ED encounter
-Outside health system positive PCR up to 3 weeks before or 2 weeks after the index ED encounter
#For patients with multiple ED encounters during the Covid-19 positive window, the encounter resulting in the highest level of care or most recent encounter (if all same level of care) during this window was used. Level of care was defined as ICU > Hospital Admission > ED encounter only.
2.3. Variables
Definitions for all collected predictor variables may be found in Supplemental Table 1. Primary endpoints of severe and non-severe disease are defined above.
2.4. Data sources and measurement
Patient data was obtained using the Electronic Medical Record Search Engine (EMERSE) [25] data retrieval tool from the electronic medical record (EMR; Epic Systems, Madison, WI). Symptoms documented by a clinician in the ED H&P, admission H&P, progress/transfer notes, or triage documentation were included if the documentation was describing symptoms at the time of initial presentation. Absence of documentation of and specific denial of symptoms (negative symptoms) were considered equal. Comorbidities, medications, and smoking status for each patient were gathered from the patient's history. If these data were not documented by clinicians during the encounter related to Covid-19, thorough inspection of the remainder of the patient's chart and external records was conducted to obtain said information. Ten patients were found to have no smoking status listed in their EMR and were treated as non-smokers in our analysis due to relatively low prevalence of smoking. Primary endpoints, assorting patients into cases and controls based on presence or absence of severe disease, were collected on May 16, 2020. Data collection was completed with consensus between two senior medical students at time of entry into a spreadsheet instrument. Variables were defined prior to data acquisition (Supplemental Table 1).
2.5. Bias
We attempted to eliminate bias by utilizing consecutive sampling by including all Covid-19 positive patients presenting to the ED to avoid gathering a skewed sample relative to our hospital's population while increasing statistical power.
2.6. Study size
Our study population of 556 patients was determined by including all Covid-19 positive patients presenting through the ED at our institution within the study date range. During the study period, a total of 7238 adult emergency patient encounters occurred and of these, 2082 encounters were screened for inclusion (Fig. 1) because they had received Covid-19 laboratory testing.
2.7. Statistical analysis
We used descriptive statistics to characterize our full patient population as well as severe and non-severe cohorts. Univariate analysis and chi-squared tests were performed on all collected variables, and p-values represent the result of Chi-square analysis with continuity correction. We performed continuity correction to prevent Type-1 error caused by assuming a chi-square distribution on data with small event occurrence.
We fitted a multivariable logistic regression model with severe outcomes as dependent variables and patient characteristics as independent variables. To determine variables for inclusion in logistic regression analyses we first selected 25 variables, each with at least 40 event occurrences, that were the most clinically relevant for predicting severe disease based on previous reports [15,26]. These variables included age (in years), sex, dyspnea, cough, sputum production, fevers/chills, vomiting, diarrhea, anorexia, dizziness/syncope, immunocompromised status, obesity, obstructive sleep apnea (OSA), asthma, chronic obstructive pulmonary disease (COPD), congestive heart failure (CHF)/cardiomyopathy, coronary artery disease (CAD)/myocardial infarction (MI), diabetes mellitus (DM), chronic kidney disease (CDK), hypertension (HTN), hyperlipidemia (HLD), smoking history and home medications of ACE-inhibitors, angiotensin receptor blockers (ARBs) or blood thinners. These 25 variables were then subjected to categorical regression with LASSO elimination, and the top 15 contributing variables were subsequently run in a binary logistic regression (Table 2). The resulting odds ratios and p-values from logistic regression analysis were used to form the final predictive equation. Variables with p < 0.05 were included in the final predictive equation. We applied our predictive equation to our cohort to generate probabilities of severe disease for each patient and generated a receiver operating characteristic (ROC) curve to assess the discriminative ability of this model (Fig. 2 ). All analyses were conducted using SPSS v26 (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp). This study was approved by the Institutional Review Board.
Table 2.
Adjusted Predictors of a Composite of ICU Admission, Mechanical Ventilation, and Death from Multivariable Logistic Regression Analysis in Patients Testing Positive for Covid-19 (n = 556).
| Risk Factor | Adjusted Odds Ratio | 95% Confidence Interval | p-value |
|---|---|---|---|
| Age | 1.05 | 1.03–1.06 | <0.0001 |
| Immunocompromised status | 2.21 | 1.17–4.16 | 0.01 |
| Dyspnea | 2.56 | 1.51–4.33 | 0.004 |
| Vomiting | 1.49 | 0.73–3.02 | 0.27 |
| Chronic kidney disease | 1.76 | 1.01–3.06 | 0.05 |
| COPD | 1.47 | 0.68–3.18 | 0.33 |
| Diabetes Mellitus | 1.32 | 0.83–2.09 | 0.25 |
| ACE inhibitor | 1.40 | 0.81–2.44 | 0.23 |
| Male sex | 1.70 | 1.10–2.64 | 0.02 |
| Obesity | 1.37 | 0.86–2.18 | 0.19 |
| Current or former smoker | 1.32 | 0.83–2.08 | 0.24 |
| Obstructive sleep apnea | 1.54 | 0.89–2.67 | 0.12 |
| Cardiovascular diseasea | 1.41 | 0.77–2.58 | 0.27 |
| Hypertension | 1.15 | 0.69–1.92 | 0.59 |
| Hyperlipidemia | 0.54 | 0.33–0.90 | 0.02 |
Abbreviations: ICU, intensive care unit; COPD, chronic obstructive pulmonary disease; ACE, angiotensin converting enzyme.
Bolded p-values indicate significance at a p < 0.05.
Includes patients with a history of coronary artery disease or a history of myocardial infarction.
Fig. 2.
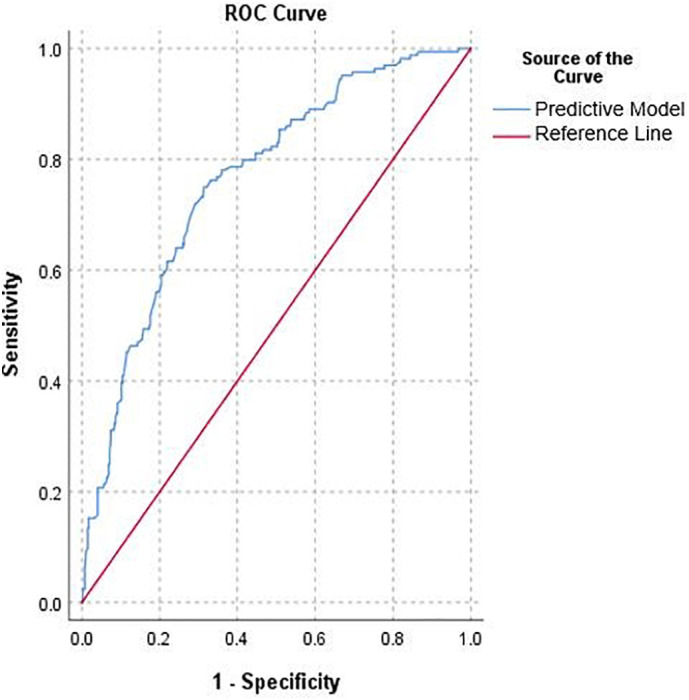
Receiver operator characteristic (ROC) curve demonstrating the discriminative ability of our model applied to our total cohort of patients.
3. Results
3.1. Participants
All 556 patients presenting during the study date range meeting our criteria for Covid-19 positive status were included throughout all aspects of our study. Upon conclusion of data acquisition on 05/16/2020, there were 17 patients who were still admitted to the hospital. However, 16/17 of these patients had already met our criteria for the primary endpoint. 113/556 (20.3%) patients tested positive for Covid-19 on a health system encounter other than the index ED encounter that was studied.
3.2. Descriptive data
Of the 556 patients included in the study, 371 (66.7%) were admitted from their ED encounter and 164 (29.4%) experienced the study outcome of severe illness. A total of 146 (26.2%) patients were transferred to the ICU during their hospital stay. Eighty-three (15.2%) patients in total required mechanical ventilation. Forty-six (8.4%) died during the hospital stay or were discharged to hospice.
The mean age of the study population was 57 ± 17 years (range, 21–95). A total of 296 (53%) were male. Our cohort consisted of 225 (40%) self-identifying as White and 245 (44%) as Black (Table 1 ). Symptoms most commonly reported by Covid-19-positive patients were cough (82%), dyspnea (75%), and fever or chills (77%). In total, 536 (96%) experienced one or more of these symptoms.
Table 1.
Demographic characteristics, Comorbidities, Medications, and associated Symptoms of Covid-19 Positive Patients.
| Variablea | Total (n = 556) | Non-Severe (n = 392) | Severeb (n = 164) | p-value |
|---|---|---|---|---|
| Age, mean (SD) | 57 (17) | 53 (17) | 66 (14) | <0.0001 |
| Male Sex, n (% total) | 296 (53) | 193 (49) | 103 (63) | 0.005 |
| Race | ||||
| White | 225 (40) | 146 (37) | 79 (48) | 0.02 |
| Black | 245 (44) | 176 (45) | 69 (42) | 0.60 |
| Hispanic | 5 (0.9) | 4 (1.0) | 1 (0.6) | 1.0 |
| Asian | 30 (5.4) | 24 (6.1) | 6 (3.7) | 0.33 |
| Other or Unknownc | 56 (10) | 46 (12) | 10 (6.1) | 0.06 |
| Smoking Status | ||||
| Current or Former | 182 (33) | 108 (28) | 74 (45) | <0.0001 |
| Comorbidities | ||||
| Asthma | 99 (18) | 71 (18) | 28 (17) | 0.87 |
| COPD | 40 (7.2) | 16 (4.1) | 24 (15) | <0.0001 |
| Interstitial Lung Disease | 11 (2.0) | 4 (1.0) | 7 (4.3) | 0.03 |
| Obesity | 274 (49) | 195 (50) | 79 (48) | 0.81 |
| Obstructive Sleep Apnea | 93 (17) | 54 (14) | 39 (24) | 0.006 |
| Neuromuscular Disease | 8 (1.4) | 6 (1.5) | 2 (1.2) | 1.0 |
| Heart Failure or Cardiomyopathy | 56 (10) | 31 (7.9) | 25 (15) | 0.01 |
| Cardiovascular Disease | 71 (13) | 33 (8.4) | 38 (23) | <0.0001 |
| Cerebrovascular Disease | 39 (7.0) | 21 (5.4) | 18 (11) | 0.03 |
| Hypertension | 290 (52) | 178 (45) | 112 (68) | <0.0001 |
| Hyperlipidemia | 155 (28) | 104 (27) | 51 (31) | 0.32 |
| Diabetes Mellitus | 172 (31) | 100 (26) | 72 (44) | <0.0001 |
| Hypothyroidism | 36 (6.5) | 21 (5.4) | 15 (9.1) | 0.14 |
| HIV Infection | 6 (1.1) | 5 (1.3) | 1 (0.6) | 0.81 |
| Immunocompromised Status | 57 (10) | 28 (7.1) | 29 (18) | <0.0001 |
| Active Malignancy | 21 (3.8) | 10 (2.6) | 11 (6.7) | 0.04 |
| Active Pregnancy | 9 (1.6) | 7 (1.8) | 2 (1.2) | 0.91 |
| Chronic Kidney Disease | 91 (16) | 42 (11) | 49 (30) | <0.0001 |
| Seizure Disorder | 12 (2.2) | 4 (1.0) | 8 (4.9) | 0.01 |
| Liver Disease | 16 (2.9) | 15 (3.8) | 1 (0.6) | 0.07 |
| Dementia | 27 (4.9) | 11 (2.8) | 16 (9.8) | 0.001 |
| Pulmonary Hypertension | 7 (1.3) | 3 (0.8) | 4 (2.4) | 0.23 |
| Previous Pulmonary Embolism | 14 (2.5) | 9 (2.3) | 5 (3.0) | 0.83 |
| Medication | ||||
| ACE inhibitor | 95 (17) | 56 (14) | 39 (24) | 0.01 |
| ARB | 75 (13) | 49 (13) | 26 (16) | 0.36 |
| Anti-coagulation | 42 (7.6) | 26 (6.6) | 16 (9.8) | 0.27 |
| Symptoms | ||||
| Dyspnea | 419 (75) | 281 (72) | 138 (84) | 0.003 |
| Wheezing | 13 (2.3) | 10 (2.6) | 3 (1.8) | 0.84 |
| Cough | 457 (82) | 325 (83) | 132 (80) | 0.58 |
| Sputum production | 64 (12) | 46 (12) | 18 (11) | 0.91 |
| Blood in sputum | 10 (1.8) | 6 (1.5) | 4 (2.4) | 0.70 |
| Sore throat | 57 (10) | 42 (11) | 15 (9.1) | 0.69 |
| Fever or chills | 429 (77) | 303 (77) | 126 (77) | 0.99 |
| Rhinorrhea or congestion | 107 (19) | 85 (22) | 22 (13) | 0.03 |
| Myalgia or arthralgia | 202 (36) | 160 (41) | 42 (26) | 0.001 |
| Fatigue or malaise | 211 (38) | 148 (38) | 63 (38) | 0.96 |
| Headache | 94 (17) | 80 (20) | 14 (8.5) | 0.001 |
| Loss of appetite | 127 (23) | 87 (22) | 40 (24) | 0.65 |
| Diarrhea | 183 (33) | 131 (33) | 52 (32) | 0.77 |
| Nausea | 115 (21) | 84 (21) | 31 (19) | 0.58 |
| Vomiting | 51 (9.2) | 33 (8.4) | 18 (11) | 0.43 |
| Abdominal pain | 42 (7.6) | 32 (8.2) | 10 (6.1) | 0.51 |
| Chest pain or tightness | 123 (22) | 104 (27) | 19 (12) | <0.0001 |
| Loss of smell or taste | 42 (7.2) | 39 (10) | 3 (1.8) | 0.002 |
| Altered mental status | 37 (6.7) | 15 (3.8) | 22 (13) | <0.0001 |
| Weakness | 71 (13) | 45 (11) | 26 (16) | 0.20 |
| Lightheadedness or syncope | 47 (8.5) | 33 (8.4) | 14 (8.5) | 1.0 |
Data presented as mean (standard deviation) for continuous variables and number (percent) for categorical variables. Univariate comparisons between the severe and non-severe groups were performed using a Student's t-test for continuous data and chi-square tests for categorical data.
Abbreviations: ICU, intensive care unit; ACE, angiotensin converting enzyme; ARB, angiotensin-receptor blocker.
Bolded p-values indicate significance at a p < 0.05.
Variables are defined in Supplemental Table 1.
Includes ICU admission, ventilator status, death during hospitalization, or discharge to hospice due to Covid-19.
Includes patients who did not belong to in any of the above categories or patients with an unknown race.
3.3. Outcome data
Patients with severe outcomes were found to have a higher mean age than those with non-severe outcomes (66 vs 53 years) and were more likely to be male (63% vs 49%). Those with severe outcomes were additionally more likely to have comorbid illnesses. This was especially true with regard to chronic obstructive pulmonary disease (COPD), interstitial lung disease (ILD), obstructive sleep apnea (OSA), heart failure/cardiomyopathy, cardiovascular disease, cerebrovascular disease, history of seizure, hypertension, diabetes mellitus (DM), immunocompromised status, active malignancy, chronic kidney disease (CKD), and dementia. Patients with severe outcomes were also more likely to have ACE inhibitors (ACE-Is) on their home medication list (Table 1). Current or former smokers were also at increased risk. There were also prominent differences in the symptoms reported by patients with severe versus non-severe outcomes, with dyspnea being most significantly associated with severe outcomes. (Table 1).
3.4. Main results
Significant predictors associated with severe outcomes included increasing age (adjusted odds ratio [OR], 1.05; 95% confidence interval [CI], 1.03–1.06), dyspnea (OR, 2.56; 95% CI: 1.51–4.33), male sex (OR, 1.70; 95% CI: 1.10–2.64), immunocompromised status (OR, 2.22; 95% CI: 1.17–4.16) and CKD (OR, 1.76; 95% CI: 1.01–3.06) (Table 2 ). Hyperlipidemia was found to be negatively associated with severe disease (OR, 0.54; 95% CI: 0.33–0.90). These variables were used to generate an equation to predict the probability of severe disease related to Covid-19 infection, where Xvariable is entered as 1 or 0 depending on the presence or absence of that variable (Eq. 1). The model appeared well calibrated (Hosmer-Lemeshow p = 0.60). The area under the ROC curve of the model (Fig. 2) showed fair ability to discriminate between patients with severe and non-severe outcomes (AUC: 0.76). Treatment of the 10 patients with missing smoking status as current or former smokers (rather than non-smokers) did not alter the results of univariate or multivariable analyses.
Equation 1: Predictive model for patients with cough
4. Discussion
Age, male sex, dyspnea, immunocompromised status, and chronic kidney disease were found to be the strongest predictors of disease severity, and hyperlipidemia was found to be a protective factor. Consistent with previous reports, advanced age was found to be the most significant predictor of severe outcomes [27]. Male sex has also been previously reported as a risk factor for poor outcomes in Covid-19 patients [28]. Immunocompromised status has had inconsistent associations with severe illness in the literature, depending on the underlying cause of immune suppression. A review of recent studies indicates that immunosuppression secondary to treatment of solid tumors is associated with severe disease while use of biologic agents for autoimmune diseases may not be [29]. Our results suggest that patients with CKD may be at high risk of severe outcomes, consistent with other studies [27,30]. The protective effect of hyperlipidemia in our analysis has not been previously reported and may be due to the indirect effect of statins, which have been implicated in the modulation of virus replication and degradation, contributing to control of infection [31,32]. Statin use was not a component of our primary data collection. Univariate analysis in our study suggests a higher incidence of severe disease from Covid-19 infection among whites which is discordant with other reports [33]. This is likely explained by the significant differences in mean age between white (mean 60, SD 18) and non-white (mean 55, SD 17) patients in our cohort (p = 0.006).
4.1. Interpretation
Here, we demonstrate a novel approach to predict a patient's risk of severe disease from Covid-19 infection based exclusively on information that can be collected remotely. We focused on predicting which Covid-19 positive patients would have poor clinical outcomes, rather than trying to determine their risk for infection. Not all individuals infected with Covid-19 need to be seen, and unnecessarily bringing those with the virus into public spaces for evaluation will lead to preventable transmission. Our fear in simply having those infected with Covid-19 self-quarantine is, of course, that this disease can be life-threatening for some. Thus, we propose that the most useful aspect of Covid-19 risk-stratification is not the probability of infection, but rather the probability of an individual's susceptibility to that infection. We recommend that all individuals with fever, cough, or other Covid-19 related symptoms quarantine, however only those likely to experience poor outcomes need to present for in-person evaluation. This eliminates the need for a purely diagnostic model such as the one proposed by Menni et al.24. By facilitating clinical prognostication without unnecessary exposure to patients and providers in screening facilities and clinics, our model has the capability of minimizing nosocomial spread while ensuring that the most at risk patients receive appropriate care.
Our predictive model was also designed to minimize overfitting by carefully selecting candidate variables of high clinical relevance and sufficient event occurrence as recommended by Babyak et al. [34] Use of automatic selection methods with a large number of clinically insignificant variables with low event frequency, as described in previous predictive models, results in high risk of overfitting results and may lead to misleading prognostications [20]. In addition, other models excluded patients who failed to experience the primary outcome by the end of the study period, potentially introducing systematic bias [35].
As countries transition to a more liberal testing policy amid efforts to safely reopen their economies, another potential use for our model is to prioritize patients in need of testing, allocating supplies to those with a predisposition to poor outcomes. Currently, criteria for testing is highly varied, unstandardized, and rapidly evolving, as availability of testing supplies fluctuates [36]. A predictive model that prioritizes high-risk patients may allow for more appropriate allocation of resources.
While this proof-of-concept study does not have the sample size or regional and institutional diversity to justify changes to management recommendations, we believe that there is value in utilizing larger emerging datasets to generate more robust predictive models based on variables that may be collected remotely. For example, the VIRUS study from the Society of Critical Care Medicine has collected many of the same variables as in our study on a larger scale [37]. A multivariable model based on these larger datasets may have sufficient validation to change clinical decision making and improve our approach to remote risk-stratification. Furthermore, in EMR systems where presenting symptoms and past medical history are reliability recorded as structured data, validation of our model or similar model may be performed rapidly.
4.2. Limitations
Limitations of our study include small sample size, which increases the potential for type-II error. Owing to insufficient event occurrence, conditions of clinical interest such as interstitial lung disease and neuromuscular disease could not be included in our model but likely possess prognostic utility. Our study also has the limitations of a single institution retrospective study. As there were 17 patients who were still in the hospital at the end of our data collection period, our mortality and mechanical ventilation numbers may be underestimated.
Due to the nature of retrospective chart-review, symptom data has the potential for bias based on history taking and documentation. We found that critically ill patients tended to have less thorough histories and reviews of systems. In addition, history taking may have been limited in patients with altered mental status or dementia. To gather the most accurate representation of each patient's presentation, we took a composite of multiple provider notes, which may represent a level of information that would be difficult to ascertain remotely during a brief phone or electronic interaction. Furthermore, although our chart review process utilized two senior medical students, we did not perform double data entry and thus were unable to assess interrater reliability. However, this is partially mitigated by the fact that only the presence of symptoms and past medical history were determined by chart review. Outcomes were extracted from the EMR by automated methods. While our study used a differential follow up time to assess the primary outcome, only 1/164 patients meeting the primary outcome did so after our minimum follow up time of 21 days. This patient died on hospital day 24. Finally, our cohort was obtained during the first Covid-19 surge with peak prevalence within the region. It is possible that, as we reach a lower prevalence steady state, the threshold for patients to reach outcomes such as ICU admission will differ.
Finally, as a tertiary care center in which complex patients present for care, our data may have reduced generalizability. Our cohort had higher rates of ICU admission than previous reports from New York, though we had similar rates of mechanical ventilation [38]. Thus, rates of and indications for ICU level care may differ by patient population and institution, limiting generalizability. To overcome these limitations, we encourage the application of larger multi-institutional datasets to develop predictive models based on the general workflow we have presented here.
5. Conclusions
In our single institution cohort, a model using only demographic data, comorbidities and the presenting symptom of dyspnea can be utilized to predict severity of Covid-19 infection. We encourage other groups with larger, multi-institutional datasets to develop predictive models to risk stratify patients by their risk of severe disease resulting from Covid-19 infection based on information that can be collected without direct patient contact.
Funding sources/disclosures
CF has received research support unrelated to this work from the National Heart, Lung, and Blood Institute (1K12HL133304). CR, JC, AB, AD, BN and SA received support from the MIT Covid-19 Challenge. No authors have reported relevant disclosures.
Author contributions
CR, AM, and JC have contributed equally to this work which include all phases of this study from concept, design, data collection, statistical analysis and manuscript preparation. All listed authors contributed substantially to the concept and design of this study. CR, AM, JC, AB, AD, BN and SA initially proposed the concept and goals of this study. CR, AM and JC performed data collection, organization, primary and adjudication of chart review. CF performed electronic data abstraction and validation. CR, AM, and JC performed the primary statistical design and analysis with consultation from FS and CF. CR, AM, and JC drafted the manuscript and figures and all authors contributed substantially to its revisions. CF takes responsibility for this manuscript as a whole and has supervised all aspects of study design, data collection, analysis and manuscript preparation.
Declaration of Competing Interest
CR, AM, JC, AB, AD, BN, FS, SS, CF report no conflicts of interest.
Acknowledgements
The authors acknowledge and thank Chiu-Mei Chen, Caryn Boyd, Dr. Richard Medlin for their roles in assembling the data upon which this work is based. We also thank the MIT Covid-19 Challenge who initially supported the conception of this idea.
References
- 1.Johns Hopkins University . Johns Hopkins Coronavirus Resource Center; 2020. COVID-19 dashboard by the center for system science and engineering (CSSE) at Johns Hopkins University (JHU)https://coronavirus.jhu.edu/map.html Accessed August 28th, 2020. [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López L., Rodó X. The end of social confinement and COVID-19 re-emergence risk. Nat Hum Behav. 2020 doi: 10.1038/s41562-020-0908-8. Published online June 22. [DOI] [PubMed] [Google Scholar]
- 4.Vaid S., McAdie A., Kremer R., Khanduja V., Bhandari M. Risk of a second wave of Covid-19 infections: using artificial intelligence to investigate stringency of physical distancing policies in North America. Int Orthop. 2020 doi: 10.1007/s00264-020-04653-3. Published online June 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bialek S., Bowen V., Chow N., Curns A., Gierke R., Hall A., et al. Geographic Differences in COVID-19 Cases, Deaths, and Incidence — United States, February 12–April 7, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(15):465–471. doi: 10.15585/mmwr.mm6915e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johns Hopkins University . Johns Hopkins Coronavirus Resource Center; 2020. COVID-19 United States cases by county.https://coronavirus.jhu.edu/us-map Accessed August 28th, 2020. [Google Scholar]
- 7.Ivorra B., Ferrández M.R., Vela-Pérez M., Ramos A.M. Mathematical modeling of the spread of the coronavirus disease 2019 (COVID-19) taking into account the undetected infections. The case of China. Commun Nonlinear Sci Numer Simul. 2020;88:105303. doi: 10.1016/j.cnsns.2020.105303. Published online April 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.COVIDView Weekly Summary Centers for Disease Control and Preventio. Published May 8. 2020. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html#virus Accessed May 8, 2020.
- 9.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yezli S., Otter J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ Virol. 2011;3(1):1–30. doi: 10.1007/s12560-011-9056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroter R.C. Social distancing for covid-19: is 2 metres far enough? BMJ. May 21, 2020:m2010. doi: 10.1136/bmj.m2010. Published online. [DOI] [PubMed] [Google Scholar]
- 12.Wilson N.M., Norton A., Young F.P., Collins D.W. Airborne transmission of severe acute respiratory syndrome coronavirus-2 to healthcare workers: a narrative review. Anaesthesia. 2020;75(8):1086–1095. doi: 10.1111/anae.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warnes S.L., Little Z.R., Keevil C.W. Human coronavirus 229E remains infectious on common touch surface materials. mBio. 2015;6(6):e01697. doi: 10.1128/mBio.01697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evaluating and Testing Persons for Coronavirus Disease 2019 (COVID-19) 2020. Centers for Disease Control and Prevention. Published. https://www.cdc.gov/coronavirus/2019- nCoV/hcp/clinical-criteria.html. [Google Scholar]
- 15.Xie J., Tong Z., Guan X., Du B., Qiu H. Clinical characteristics of patients who died of coronavirus disease 2019 in China. JAMA Netw Open. 2020;3(4):e205619. doi: 10.1001/jamanetworkopen.2020.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Center for Immunization and Respiratory Diseases (NCIRD) DoVD . Center for Disease Control and Prevention. Center for Disease Control and Prevention; Published 2020. Coronavirus self-checker.https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html Accessed May 8, 2020. [Google Scholar]
- 18.Judson T.J., Odisho A.Y., Neinstein A.B., Chao J., Williams A., Miller C., et al. Rapid Design and Implementation of an Integrated Patient Self-Triage and Self-Scheduling Tool for COVID-19. J Am Med Inform Assoc. April 8, 2020:ocaa051. doi: 10.1093/jamia/ocaa051. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isakov A.W.D., Quay Yaffee A., Schrager J. Emory University School of Medicine; 2020. Coronavirus checker. [Google Scholar]
- 20.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;12 doi: 10.1001/jamainternmed.2020.2033. Published online May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong J., Ou J., Qiu X., Jie Y., Chen Y., Yuan L., et al. A Tool to early predict severe 2019-novel coronavirus pneumonia (COVID-19): A Multicenter Study Using the Risk Nomogram in Wuhan and Guangdong, China. Public Global Health. 2020 doi: 10.1101/2020.03.17.20037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y., Yu X., Zhao H., Wang H., Zhao R., Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24(1):108. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., et al. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. Epidemiology. 2020 doi: 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- 24.Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020;11 doi: 10.1038/s41591-020-0916-2. Published online May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanauer D.A., Mei Q., Law J., Khanna R., Zheng K. Supporting information retrieval from electronic health records: a report of University of Michigan’s nine-year experience in developing and using the electronic medical record search engine (EMERSE) J Biomed Inform. 2015;55:290–300. doi: 10.1016/j.jbi.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du R.-H., Liang L.-R., Yang C.-Q., Wang W., Cao T.-Z., Li M., et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020;55(5):2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta S., Hayek S.S., Wang W., Chan L., Mathews K.S., Melamed M.L., et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;15 doi: 10.1001/jamainternmed.2020.3596. Published online July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fung M., Babik J.M. COVID-19 in immunocompromised hosts: what we know so far. Clin Infect Dis. 2020:ciaa863. doi: 10.1093/cid/ciaa863. Published online June 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry B.M., Lippi G. Chronic kidney disease is associated with severe coronavirus disease 2019 (mCOVID-19) infection. Int Urol Nephrol. 2020 doi: 10.1007/s11255-020-02451-9. Published online March 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues-Diez R.R., Tejera-Muñoz A., Marquez-Exposito L., Rayego-Mateos S., Sanchez L.S., Marchant V., et al. Statins: Could an old friend help the fight against COVID-19? Br J Pharmacol. 2020 doi: 10.1111/bph.15166. Published online June 19. bph.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radenkovic D., Chawla S., Pirro M., Sahebkar A., Banach M. Cholesterol in relation to COVID-19: should we care about it? JCM. 2020;9(6):1909. doi: 10.3390/jcm9061909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb Hooper M., Nápoles A.M., Pérez-Stable E.J. COVID-19 and racial/ethnic disparities. JAMA. 2020;323(24):2466. doi: 10.1001/jama.2020.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babyak M.A. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med. 2004;66(3):411–421. doi: 10.1097/01.psy.0000127692.23278.a9. [DOI] [PubMed] [Google Scholar]
- 35.Wynants L., Van Calster B., Bonten M.M.J., Collins G.S., Debray T.P.A., De Vos M., et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020:m1328. doi: 10.1136/bmj.m1328. Published online April 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitzer V.E., Chitwood M., Havumaki J., Menzies N.A., Perniciaro S., Warren J.L., et al. The impact of changes in diagnostic testing practices on estimates of COVID-19 transmission in the United States. Epidemiology. 2020 doi: 10.1101/2020.04.20.20073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walkey A.J., Kumar V.K., Harhay M.O., Bolesta S., Bansal V., Gajic O., et al. The viral infection and respiratory illness universal study (VIRUS): an international registry of coronavirus 2019-related critical illness. Critic Care Explor. 2020;2(4) doi: 10.1097/CCE.0000000000000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020 doi: 10.1001/jama.2020.6775. Published online April 22. [DOI] [PMC free article] [PubMed] [Google Scholar]