ABSTRACT
Tuberculosis (TB) is a preventable and curable disease, but increased mortality and morbidity associated with TB is one of the leading causes of deaths worldwide. MicroRNAs (miRNAs) are small, non-coding RNAs known to regulate the host immune response against TB. We investigated the expression profile of candidate circulating miRNAs, which could be used as a blood biomarker for the effective diagnosis of pediatric tuberculosis. A cross-sectional comparative study was conducted, including 30 children with active-TB and 30 healthy controls (HC) in a tertiary care hospital in Puducherry. We used the SYBR green-based miScript qRT-PCR assay to analyze the expression levels of miRNAs in plasma. Further, we used the receiver operating characteristic curve (ROC) to evaluate the diagnostic value of miRNAs. Active-TB included 25 (83.3%) pulmonary TB and 5 (16.7%) extrapulmonary TB cases. We found a significant upregulation of miR-21, miR-29a, miR-31, miR-155, and downregulation of miR-146a in children with active-TB compared to HC. The ROC analysis showed an excellent diagnostic value of miRNAs as follows: miR‑31> miR‑155> miR‑146a with AUC of (95% CI) miRNAs 0.978, 0.953, and 0.903, respectively. Altered circulating miRNA expression levels could be involved in the dysregulation of the host immune response to TB. The ROC analysis indicated that miRNAs miR-31, miR-155 and miR-146a could be effective diagnostic biomarkers for the detection of active-TB in children.
KEYWORDS: Tuberculosis, mycobacterium tuberculosis, microrna, TB diagnosis, pediatric TB, biomarker
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb), is a severe threat to humans, and increased morbidity and mortality reported annually due to TB puts it among the top three fatal infectious diseases [1,2]. In around one third of the world, the population presents a latent form of TB infection (LTBI). However, only 10–15% of infected people tend to develop active-TB during their lifetime [1,3]. Globally, an estimated total of 230,000 children died of TB in 2018, in which 80% were aged less than five years old [1]. Pediatric TB (less than 15 years old) is one of the ten major mortalities in children globally. In India, an estimated total of 132,711 pediatric TB cases (only 59% of estimated pediatric TB cases occur every year) were reported, including new and relapsed pediatric TB patients [1,3,4].
Numerous studies have reported that mycobacterial virulence, together with the host genetics determine the immune status of the infected individual [5]. The immune response to Mtb is highly complex and multifaceted, and the initial recognition initiates the release of proinflammatory cytokines that leads to the activation of the adaptive immune response. In general, children and elderly people are more prone to develop active-TB due to their weakened immune system. One such infection-induced host-genetic factor is the RNA-mediated immune regulation, which has been studied widely in respect to TB infection [5,6].
Several regulatory RNA molecules such as microRNA (miRNA), antisense RNA transcripts, non-coding RNA regulate a myriad of gene products [5,7–9]. The process of RNA-associated gene silencing introduces local heterochromatin formation by controlling the epigenetic machinery (DNA methylation and histone modifications) [9,10].
The discovery of miRNAs and their interaction with the target genes have been reported for more than two decades [5]. Currently, more than 2,000 miRNAs have been identified [11], which target around 60% of human genes [6]. In general, mature miRNA binds at 3ʹ-UTR (untranslated region) of target mRNA and inhibits the translation process [12,13]. Further, the RNA-induced silencing complex (RISC) degrades the target mRNA [11]. Recently, the miRNAs are mainly considered as gatekeepers to determine the host immune response to the infection and are involved in many cellular processes, including the activation and regulation of immune response and apoptosis processes [13,14].
Importantly, many cytokines are regulated by miRNAs, which are involved in modulating the host immune responses in various diseases [11]. The miRNAs are a highly conserved regulatory system in our genome that maintains the fluid intercellular communication by circulating through plasma, serum, urine and saliva to exhibit its translational inhibition processes [13,15]. Differential expression patterns of miRNAs and its association with various communicable and non-communicable disease conditions [10,11], including TB [6,9,12,13] was reported elsewhere. There are several circulating miRNAs found to be differentially regulated and have the ability to distinguish significantly active-TB from healthy individuals. Particularly, miR-29a [16], miR-144 [2], miR-146a [17], miR-155 [18], miR-155* [19], miR-292, miR-361-5p [20], miR-576-3p and miR-889 [21] were reported to be used as a biomarker for TB [21].
Given the importance of miRNA-based transcriptional regulation during TB disease conditions, we aimed to study the differential expression of selected circulatory miRNAs in children with active-TB compared with healthy controls.
Patients and methods
Ethical statements
The Scientific Advisory Committee and Institute Ethical Committees (Human Studies) approved the protocol of the study of tertiary care hospital JIPMER, Puducherry (JIP/IEC/2014/10/491), from December 2014 to December 2018. Blood samples from 30 children with active-TB and 30 disease-free healthy control children (HC) were collected after obtaining a signed informed consent form.
Study population
We performed a cross-sectional comparative study based on a convenience sampling strategy, including 30 children with the active-TB disease and 30 disease-free healthy control children (HC). Children were recruited according to stringent inclusion and exclusion criteria as follows: Children with active pulmonary TB (PTB) and extra-pulmonary TB (EPTB) (≤14 years, both genders) were confirmed with sputum smear-positive – AFB positive, MTBC complex culture and/or GeneXpert MTB/RIF positive (Cepheid, Sunnyvale, CA) and other clinical confirmations of TB. Children with HIV co-infection were not recruited in this study. Children with active-TB disease status and at least one positive result for one inclusion criteria (confirmed with both physician and laboratory tests) were recruited at the time of diagnosis before the start of TB treatment.
Healthy control children (HC) (≤14 years, both genders) were recruited without any known risk factors for TB and other associated infectious disease conditions and without any clinically relevant terms or surgical conditions.
Selection of candidate miRNAs
The candidate miRNAs that play a significant role in TB – associated inflammatory pathways were identified through a literature search, and bioinformatics tools include micro T-CDS v5.0, miRTarBase v6.0 [22], miRDB [23] and TargetScanHuman v6.2. The selection of target miRNAs was based on only those experimentally verified with miRNA – target interaction studies (Table 1).
Table 1.
Nucleotide sequence of microRNAs studied.
| miRNA | miRBase ID | Sequence | bp |
|---|---|---|---|
| miR-21 | MIMAT0000076 | UAGCUUAUCAGACUGAUGUUGA | 22 |
| miR-29a | MIMAT0000086 | UAGCACCAUCUGAAAUCGGUUA | 22 |
| miR-31 | MIMAT0000089 | AGGCAAGAUGCUGGCAUAGCU | 21 |
| miR-146a | MIMAT0000449 | UGAGAACUGAAUUCCAUGGGUU | 22 |
| miR-155 | MIMAT0000646 | UUAAUGCUAAUCGUGAUAGGGGUU | 24 |
miRNA – micro ribonucleic acid, bp – base pair.
Plasma collection and isolation of circulating miRNAs
Peripheral blood (0.5 mL) was collected in a tube containing an anticoagulant (ethylenediaminetetraacetic acid (EDTA) K2) from both groups. Blood was centrifuged at 3,000 rpm for 10 minutes at 4°C, and clear plasma was separated and stored at −80° C until further use.
The miRNeasy Serum/Plasma extraction kit (Qiagen, Hilden, Germany), according to the manufacturer’s recommendation, was used for the isolation of circulating miRNAs from plasma. The procedure given in the kit is suitable for the separation of total RNA, including small RNAs, in a low volume of samples like the serum, plasma, and other body fluids. Briefly, 200 µL of plasma was used for the miRNA isolation. The isolated miRNAs from both groups were stored at −80° C until further use.
NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific, MA, USA) with absorbance measurements at 260 and 280 nm were used to determine the RNA concentration and purity. The absorbance ratio of 260/280 (1.8–2.0) was used as an indicator for the sufficient RNA purity for the further qRT-PCR experiments.
Synthesis of miRNA complementary DNA (cDNA)
A total of 100 ng of isolated miRNAs was further used for cDNA synthesis. The synthesis of reverse-transcribed cDNA was followed according to the miScript II RT Kit (Qiagen, Hilden, Germany) manufacturer’s recommendation. We used 5x miScript HiSpec buffer in this study to estimate the expression levels of only circulating mature miRNAs from both study groups. Synthesized cDNAs were stored at −20° C until further use.
The expression level of candidate miRNAs
Stored, undiluted 20 µL of cDNA was further diluted with at least 200 µL of RNase-free water (Qiagen, Hilden, Germany) to quantify mature miRNAs according to miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) manufacturer’s procedure. A total of 20 µL of the reaction mixture was prepared as follows: 2× QuantiTect SYBR green PCR master mix, 10 µL; 10× miScript universal primer, 2 µL; 10× miScript primer assay, 2 µL; RNA sample, 2 µL; and nuclease-free water, 4 µL. The quantification of each candidate miRNA was performed in the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) with separate miScript specific primer assay (Table 1). The miScript small nucleolar RNA (SNORD61) primer assay was used as the endogenous housekeeping control. Each miRNA expression level was normalized with the endogenous miRNA expression level.
The quantitative real-time PCR (qRT-PCR) was run with the following cycling conditions as initial denaturation at 95°C for 15 minutes, followed by 3-step cycling of totally 40 cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 30 seconds and extension at 70°C for 30 seconds. Melt curve analysis was performed from 60°C to 95°C with a gradual increment of 0.2°C/min. The relative expression levels of the candidate miRNAs were determined using the comparative Ct method, also known as 2-delta (Ct) method [24].
Melt curve analysis was performed to examine the specificity of the miR-specific qRT-PCR experiment. The Ct values for all the miRNAs are approximately between 22 and 25 cycles, and the qRT-PCR was in linear association with specific target miRNA amplification. The qRT-PCR experiment produced a single peak in melting curve analysis, and no secondary peaks were observed in respective miRNAs dissociation curves. This indicating the specificity and the presence of a single PCR amplified product in the qRT-PCR experiment. The melting temperature of miRNAs falls in the range of 76.5°C ± 1°C for miR-21, 74.5°C ± 1°C for miR-29a, 77.5°C ± 1°C for miR-31, 76.5°C ± 1°C for miR-146a and 77°C ± 1°C for miR-155 in both groups of children enrolled in this study.
Statistical analysis
Normality was assessed using the Shapiro–Wilk test, and non-parametric tests were applied throughout this study as sample numbers did not agree with normal distributions. Categorical data were expressed as numbers and percentages. Results of all the parameters were presented as median with interquartile range (IQR). Mann–Whitney U test was used to compare the expression levels of candidate miRNAs between groups. The Kruskal–Wallis test was used to analyze the difference in miRNAs expression levels between groups and the comparison between different baseline characteristics. The receiver operative characteristic curve (ROC) was performed to investigate the diagnostic power of the miRNAs expression in study children. All statistical analyzes were carried out in SPSS 17 (SPSS, Chicago, IL), GraphPad Prism 6.0 (GraphPad Software Inc., CA), and MS Excel. A 95% confidence interval with p < 0.05 was considered as significant.
Results
Clinical characteristics of the study population
In total, 30 children with TB (60% males and 40% females) and 30 (70% males and 30% females) HC were recruited for this study based on the inclusion and exclusion criteria. The median (IQR) age of the enrolled TB cases was 8 (3.8–11), with 36.7% of children under five years old. In the HC group, the median age was 10 (7–12). A significant difference was observed in the distribution of baseline parameters such as age, height, and weight between the study groups. Baseline characteristics and clinical features of the enrolled children are represented in Table 2.
Table 2.
Baseline characteristics of the enrolled children.
| S. No | Variables | Cases (n = 30) | Controls (n = 30) | p value |
|---|---|---|---|---|
| 1. | Age (years) | 8 (3.8–11) | 10 (7–12) | 0.009 |
| 2. | Gender (n) Male Female |
18 (60%) 12 (40%) |
21 (70%) 09 (30%) |
|
| 3. | Height (cm) | 118 (95.2–135) | 136.5 (128–149.3) | 0.0004 |
| 4. | Weight (kg) | 17.4 (9.5–26.2) | 31 (23.7–34.2) | <0.0001 |
| Fever | 5 (4–6.25) | ND | ||
| 5. | Cough | 12 (10–14) | ND | |
| 6. | Chest pain | 4 (3–5) | ND | |
| 7. | Loss of Appetite (LOA) | 5.5 (4–10) | ND |
In table the median value of corresponding baseline clinical feature is represented. Data are median with IQR. n – Number; % – Percentage; cm – Centimeter; kg – kilogram; ND, No Data/Not Determined; IQR – Interquartile range; p-value, <0.5.
TB disease status
Among 30 TB cases, 25 (83.3%) were PTB and 5 (16.7%) were TB meningitis (TBM) cases. Out of 25 PTB cases, 11 (44%) were AFB smear positive and 14 (56%) were AFB scanty grade-positive or smear negative PTB cases. Smear-negative PTB cases were included in this study based on other diagnostic criteria such as CBNAAT positive (Cartridge based nucleic acid amplification test), chest X-ray positive and based on clinical findings with TB symptoms. Among 25 PTB cases, 8 (32%) were CBNAAT positive, 15 (60%) were chest X-ray positive and 3 (12%) were TST-positive cases. Children unable to expectorate sputum, underwent induced sputum and bronchoscopy procedure to get BAL fluid and gastric-aspirate, nasopharyngeal-aspirate were examined using smear microscopy.
Circulating plasma miRNAs expression level in TB patients
The housekeeping and target microRNA genes were amplified using the SYBR green-based qRT-PCR. Melt curve analysis was performed to confirm a single amplified product in qRT-PCR. Five candidate miRNAs, miR-21, miR-29, miR-31, miR-146a, and miR-155, were selected for this study based on their significant association with TB [17–19].
Plasma miRNA levels were normalized to small nucleolar RNA (SNORD61) that demonstrated significant differences between study groups. The expression level of each selected circulating miRNAs was determined with normalized control RNA in both TB and HC. The RT-qPCR expression analysis was performed in plasma samples alone to avoid the influence of other confounding factors.
The expression levels of miRNAs miR-21, miR-29a, miR-31, miR-146a, and miR-155 were analyzed in children with active-TB disease status and HC. Figure 1 shows the distribution of the median expression level of candidate miRNAs in plasma from the study groups. The RT-qPCR expression analysis revealed that miRNAs, miR-21 (Figure 1(a)), miR-29a (Figure 1(b)), miR-31 (Figure 1(c)), and miR-155 (Figure 1(e)) expression levels were upregulated. In contrast, the miR-146a (Figure 1(d)) expression level was downregulated in children with active-TB compared to HC. Expression levels of miRNAs, miR-21, miR-31, miR-146a and miR-155, showed a statistically significant difference (p = 0.0003, p < 0.0001, p < 0.0001 and p < 0.0001, respectively), while miR-29a did not show a significant difference (p = 0.16) between study groups.
Figure 1.
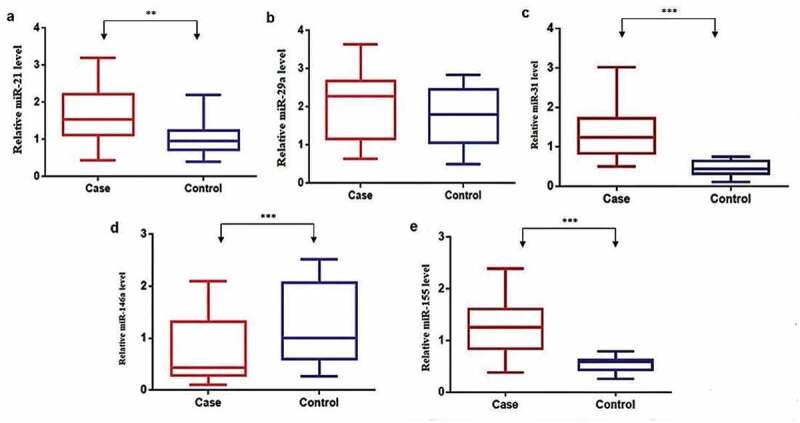
Candidate miRNAs’ expression in plasma from children with active-TB and HC.
We further compared the expression levels of candidate miRNAs among smear positive PTB cases and other baseline parameters of the enrolled children. Though the expression levels of miRNAs were comparable between sub-groups, we did not find a statistically significant difference. This observation might be attributable to the smaller sample size or that there was not a significant difference in inter-individual variables among the sub-groups analyzed. The EPTB cases were not included in the statistical analysis owing to the smaller size of the samples enrolled in this study.
The expression levels of miRNAs were compared with baseline and clinical features such as age, gender, and loss of appetite (LOA) among the enrolled children. We analyzed the different categories of age (<5, 6 to 10 and >10 in years), gender (male vs. female), and loss of appetite (<5 days and > 5 days) against miRNA expression level in TB cases. We found no statistically significant difference among them (Table 3).
Table 3.
Comparison miRNA expression and baseline characteristics of TB patients.
| Features | No of Cases (%) | Fold
change |
||||
|---|---|---|---|---|---|---|
| miR21 | miR29a | miR31 | miR146a | miR155 | ||
| Age | ||||||
| <5 y | 11 (36.7) | 1.53 (0.72–2.35) | 2.36 (1.16–2.67) | 1.04 (0.78–2.58) | 0.46 (0.37–0.67) | 0.81 (0.71–1.53) |
| 6 to 10 y | 11 (36.7) | 1.57 (1–2.36) | 2.27 (1.03–2.67) | 1.05 (0.84–1.48) | 0.42 (0.33–0.63) | 0.97 (0.89–1.76) |
| >10 y | 8 (26.7) | 1.54 (1.45–2.18) | 2.29 (1.15–2.87) | 1.28 (1.03–1.76) | 0.45 (0.4–0.78) | 1.29 (1.22–1.98) |
| p value | 0.93 | 0.93 | 0.66 | 0.47 | 0.18 | |
| Gender | ||||||
| Male | 17 (56.7) | 1.51 (0.8–2.23) | 2.36 (0.9–2.65) | 0.94 (0.8–1.96) | 0.51 (0.35–0.65) | 1.29 (0.85–1.69) |
| Female | 13 (43.3) | 1.6 (1.3–2.3) | 1.95 (1.2–2.73) | 1.37 (1.08–1.67) | 0.42 (0.4–0.7) | 1.18 (0.76–1.50) |
| p value | 0.2 | 0.97 | 0.34 | 0.87 | 0.57 | |
| LOA | ||||||
| < 5 days | 15 (50) | 1.53 (1.17–2.36) | 2.34 (1.03–2.64) | 1.04 (0.82–1.87) | 0.55 (0.4–0.66) | 1.31 (0.75–1.76) |
| >5 days | 15 (50) | 1.57 (1–2.11) | 2.27 (1.16–3.01) | 1.36 (0.92–1.58) | 0.41 (0.33–0.45) | 1.22 (0.92–1.37) |
| p value | 0.64 | 0.67 | 0.81 | 0.09 | 0.79 | |
Mann–Whitney U test was used to compare the expression levels of candidate miRNAs between the gender and loss of appetite. The Kruskal–Wallis test was used to analyze the difference in miRNAs expression levels between the different categories of age and Mann–Whitney U test was used to compare the expression levels of candidate miRNAs between the gender and loss of appetite of enrolled children. Data are median with IQR. TB, Tuberculosis; miRNA, Micro Ribonucleic Acid; IQR – Interquartile range; LOA – Loss of appetite; p-value, <0.5.
Diagnostic value of miRNAs
ROC curve and the area under the ROC curve (AUC) were generated for each miRNA to assess the diagnostic value of miRNAs included in this study. The cutoff point of each candidate miRNAs for the differentiation of active-TB from HC was analyzed further. Among the miRNAs, miR-31, miR-155, and miR-146a showed excellent diagnostic performance to determine significantly the active-TB cases from HC. ROC curves were constructed for the selected miRNAs, and the AUC was studied respectively. The AUC of miRNAs such as miR-31, miR-155 and miR-146a was 0.978 (0.945–1.000), 0.953 (0.894–1.000) and 0.903 (0.827–0.979), respectively (Figure 2). The complete details of the diagnostic values of three miRNAs are listed in Table 4.
Figure 2.
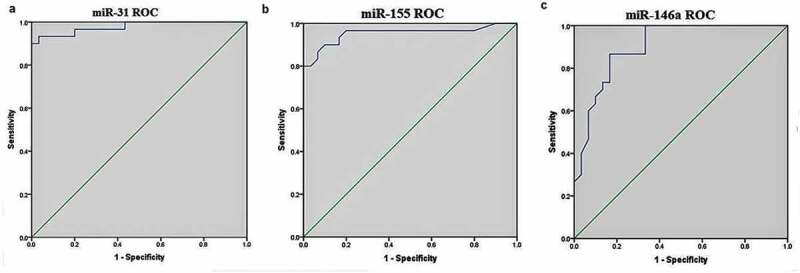
Receiver operating characteristic (ROC) curve analysis.
Table 4.
Receiver operating characteristic (ROC) analysis of miRNA in pediatric TB.
| miRNAs | AUC | 95% CI | Cutoff | Sensitivity | Specificity | p value |
|---|---|---|---|---|---|---|
| miR-31 | 0.978 | 0.945–1.000 | 0.73 | 93% | 97% | <0.0001 |
| miR-155 | 0.953 | 0.894–1.000 | 0.70 | 90% | 90% | <0.0001 |
| miR-146a | 0.903 | 0.827–0.979 | 0.69 | 83.3% | 86.7% | <0.0001 |
TB, Tuberculosis; miRNA, Micro Ribonucleic Acid; AUC, Area under the curve; CI, Confidence Interval 95%; p-value, <0.5.
Discussion
Earlier studies have focused on the role of miRNAs and their influence on gene expression in several immune cell types, including macrophages, dendritic cells, B cells, T cells, and natural killer cells (NK) from active and latent TB [12–14,16]. Several studies reported the importance of miRNA and its interaction with the host immune response during Mtb infection [14,20]. Some miRNAs are said to activate and others to suppress the immune response against TB such as miR-146a, miR-21, miR-142-3p, miR-155, and miR-26a [5,6,11–20]. This interaction between miRNAs and its target genes are principally to inhibit the translation process of target immune response genes in the host during Mtb infection. Therefore, miRNAs are considered a critical player in determining both innate and adaptive immunity.
Recent studies have focused on the concept of considering circulating miRNAs as a potent biomarker for diagnosis and therapeutic targets for TB, and emerging evidence suggests them as promising biomarkers for the early diagnosis of TB [14]. The miRNAs are capable of immune activation and are required for the activation of host innate immune cells such as miR-9, miR-21, miR-146a, and miR-155 [25]. However, tubercle bacilli can effectively regulate the host miRNA profile to subvert the immune response and to survive inside the host cells [15,17,21].
This study supports the existing evidence that differential expression pattern of miRNAs was observed and that they are involved in regulating the immune response against TB in children [15,17,21]. The candidate miRNAs selected in this study setting showed significant upregulation of miRNAs, miR-21, miR-29a miR-31, and miR-155 and downregulation of miR-146a in plasma from TB cases when compared to HC. Our results support the previous findings that significant upregulation of hsa-miR-29a-3p, hsa-miR-155-5p, and hsa-miR-361-5p in active-TB patients [16].
In contrast, downregulation of miR-26, miR-29a, and miR142-3p from whole blood was associated with decreased immune response in children against TB [26]. The miR-21, which is involved in the inhibition of pro-inflammatory cytokines secretion (TNF-α and IL-6) [27] and upregulation of the anti-inflammatory cytokine production (IL-10) [28] in TB patients. Besides, miR-21 suppressed host Th1 cell response to Mtb by inhibiting IL-12 expression [29], and its interaction with Bcl-2 is shown to induce DC apoptosis [30]. Our observation is similar to the earlier reports [27–31] suggesting that Mtb targeted the expression of miR-21 to subvert the host immune response in children.
Ma et al. [31] reported an increased level of miR-29 with the decreased expression level of IFN-γ gene, and Schaale et al. [32] described that overexpression of miR-29 deregulates Wnt signaling pathway by targeting its components that play an essential role in the regulation of pro-inflammatory cytokines to TB disease. A significant increase in the expression of the miR-29a mRNA level in our study follows a similar trend with previous reports describing that Mtb evades macrophage-mediated killing process by inhibiting the expression of the IFN-γ gene [14,31,32]. It is evident from our study that the expression level of miRNAs such as miR-21 and miR-29a was found to be significantly increased following previous reports that markedly reported a decrease in the immune response against TB.
The miR-31 was reported to be upregulated in children with TB [33] that inhibits MyD88, an adaptor molecule in TLR2-mediated immune response, and thus suppresses the TLR2 mediated immune response [34]. The miR-31 facilitates an immune evasive mechanism to autophagy response in post-TB infection, especially to Mtb by targeting WNT5A and SHH pathways [35]. Wang et al. [36] reported the negative regulation of miR-31 in TB patients. The authors observed a significantly lower level of miR-31 in children with TB associated with significantly higher levels of innate immunity cytokines such as IL-6, TNF-α, NF-κB, and IFN-γ. However, our study supports the findings of Zhou et al. [33], Ghorpade et al. [34] and Holla et al. [35] and suggest that the trend of upregulation of miR-31 was found to be responsible for the decreased expression level of TLR-2, MyD88, IL-6, TNF-α and IFN-γ genes in children with active-TB in comparison to HC.
Previous reports suggest that a significant increase in the miRNA-146a level negatively regulates the immune response to TB by downregulating the activation of proinflammatory cytokines (TNF-α, IL-1β, IL-6, and MCP-1) to facilitate intracellular survival of Mtb in macrophages [6,7,17]. Innate immune cells require the activation of miR146a to support protection against TB [17,21]. However, we found that in active-TB cases, miRNA-146a was downregulated and supported the notion that miRNA-146a is involved in downregulating an excessive immune response and restricting the expression level of pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6 in TB cases [15]. Besides, we have also observed a decrease in the expression level of IFN-γ, TNF-α, IL-1β, and IL-6 genes in our study settings (data not shown).
The miR-155 expression plays the primary role in the activation of immune cells include B cells, T cells, macrophages, dendritic cells [18] as and FOXP3+ regulatory T cells [21,37]. However, the expression level of miR-155 in TB disease condition is still not absolutely understood, and contradictory findings were reported [2,19,21,38,39]. This ambiguity might be dependent on the type of strain and the host model organism used in the study.
Virulent Mtb strain was shown to downregulate miR-155 in human macrophages, whereas the upregulation of miR-155 was reported in murine macrophages [21]. But upregulation of miR-155 with increased TNF level was seen in the case of an avirulent Mtb strain used in THP-1 cell line [40], and other studies also have found that an increased level of miR-155 was associated with TNF level in TB disease condition [41]. Active-TB patients showed that an increased level of miR-155 interlinked with a decreased level of FOXO3, thus inhibiting the apoptosis process [2].
Stahl et al. [18] reported that modulation of miR-155 level could play the leading role in Treg cell-mediated suppression in TB, and increased miR-155 level acted as an inhibitor of ATG3 expression in dendritic cells, thus silencing the autophagy process [42]. Thai et al. [38] and Wu et al. [19] have demonstrated that the level of miR-155 is interconnected with IFN-γ gene expression and reduced immune responses.
Also, Zhang et al. [39] reported that miR-155 was shown to suppress the activation of NK cells and was negatively associated with TNF-α in serum from TB patients. On the contrary, Wagh et al. [43] reported a decrease in the level of miR-155 in active-TB and MDR-TB patients. Thus, it is appealing to speculate that an increased level of miR-155 in TB cases could result in a decrease in TNF level. This finding is in accord with Fu et al. [44], Wu et al. [19] and Draz et al. [20] who respectively reported that miR-29a-3p (serum), miR-155-5p (PBMC) and miR-361-5p (serum) were significantly overexpressed in active-TB patients compared to HC.
Recently, several studies have reported the use of circulating miRNAs as a potential biomarker for TB diagnosis and treatment [16,19,20,27,33,36,39,43–45]. The previous report suggests that a single miRNA or combination of miRNAs can be used as a biomarker to distinguish active-TB from HC. Zhou et al. [33] studied the diagnostic values of selected miRNAs, including miR-1, miR-10a, miR-125b, miR-146a, miR-150, miR-155, and miR-31. They reported that combined miRNAs showed a promisingly increased diagnostic value compared to the use of a single miRNA. Indeed, the combination of miRNAs, miR-150, miR-21, miR-29 c, and miR-194 was tested for diagnostic value and showed that combined miRNAs could be suitable for biomarker studies [45].
Several other reports support our findings of the use of miRNAs as a potential biomarker for TB diagnosis in the pediatric population. These include Wang et al. [36] for miR-31, Wu et al. [18] for miR-155 and Zhou et al. [33] for miR-146a. We observed a favorable diagnostic value for the candidate miRNAs miR-31, miR-155, and miR-146a (AUC = 0.978, 0.953, and 0.903, respectively) to distinguish active-TB from HC (Table 3). At the same time, miR-21 and miR-29a did not show robust diagnostic ability in this study. Further, our data demonstrated that tested miRNAs showed accurate sensitivity and specificity as follows: miR-31> miR-155> miR-146a.
The limitations of our study need to be addressed. The number of patients included in our study to analyze the expression pattern and to assess the biomarker value is limited due to the small sample size. Further studies with a larger sample size (A cohort, case-control) are required to identify the potential use of miRNAs for the diagnosis of pediatric tuberculosis. Second, our study is limited to the recruitment of PTB cases alone, and this might influence the diagnostic value of the candidate miRNAs. Further analysis with an equal proportion of PTB vs. EPTB cases would give a better picture of the diagnostic performance of the candidate miRNAs. However, our study results are consistent with previous reports which strengthen the use of miRNAs as biomarkers for active-TB diagnosis. In this study, the healthy control children were recruited with no known disease conditions or symptoms for any infection (disease-free status) and did not test for the negative TST (or) negative IFN-γ assay for latent TB infection.
Conclusions
The study demonstrates the alteration in miRNA expression levels and the potential role of biomarker analysis for the effective identification of active-TB in children. The up- and downregulation of candidate miRNAs, miR-21, miR-29a, miR-31, miR-155, and miR-146a respectively suggest the importance of miRNAs’ role in determining the immune response in children with active-TB. The altered level of miRNAs is predominantly involved in determining the balance between the innate and adaptive immune response, thus impairing the host immune response to TB. The ROC analysis indicates that miRNAs such as miR-31, miR-146, and miR-155 could be suitable for the potential biomarker for the early and effective diagnosis of children with active-TB.
Acknowledgments
The authors much acknowledge the Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER) – Intramural Research Grant for financial support. The authors sincerely thank all the patients, their families, and the healthy volunteers for their participation.
Author contributions
All authors are equally contributed to study conception and design. Material preparation, data collection, analysis, and experiments were performed by Mr. Kathirvel M and Mrs. Saranya S. The first draft of the manuscript was written by Mr. Kathirvel M. Manuscript review, editing, and correction were done by Dr. S. Mahadevan. All authors have read and approved the final manuscript.
Disclosure statement
The authors declare that they have no conflict of interest.
Ethical conduct of research
The study was reviewed by the institute ethics committee of the tertiary care hospital (JIP/IEC/2014/10/491). All the children and their parents were signed in an informed consent form to participate in the study.
References
- [1].WHO . Global tuberculosis report. Geneva: World Health Organization; 2018. [cited 2018 December17]. Available from: https://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- [2].Huang J, Jiao J, Xu W, et al. miR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep. 2015;12(5):7102–7108. [DOI] [PubMed] [Google Scholar]
- [3].Ananya M. Tuberculosis Transmission. News-Medical. 2019. [cited 2019 June19]. Available from: <https://www.news-medical.net/health/Tuberculosis-Transmission.aspx>
- [4].India TB Report . 2019. Published on 2019 June6 [Cited 2020 Feb 21]. Available from: https://tbcindia.gov.in/WriteReadData/India%20TB%20Report%202019.pdf
- [5].Esterhuyse MM, Linhart HG, Kaufmann SHE.. Can the battle against tuberculosis gain from epigenetic research? Trends Microbiol. 2012;20(5):220–226. [DOI] [PubMed] [Google Scholar]
- [6].Spinelli SV, Diaz A, D’Attilio L, et al. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol Immunol. 2013;53(3):265–269. [DOI] [PubMed] [Google Scholar]
- [7].Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29(7):343–351. [DOI] [PubMed] [Google Scholar]
- [8].Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tollefsbol T. Handbook of Epigenetics: the new molecular and medical genetics. Oxford, UK: Academic Press, Elsevier Inc; 2011. [Google Scholar]
- [10].Simmons D. Epigenetic influences and disease. Nat Educ. 2008;1(1):6. [Google Scholar]
- [11].Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Furci L, Schena E, Miotto P, et al. Alteration of human macrophages microRNA expression profile upon infection with Mycobacterium tuberculosis. Int J Mycobacteriol. 2013;2(3):128–134. [DOI] [PubMed] [Google Scholar]
- [13].Wu LS, Lee SW, Huang KY, et al. Systematic expression profiling analysis identifies specific microRNA-gene interactions that may differentiate between active and latent tuberculosis infection. BioMed Res Int. 2014;895179. DOI:doi. 10.1155/2014/895179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yi Z, Fu Y, Ji R, et al. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS ONE. 2012;7(8):e43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Miotto P, Mwangoka G, Valente IC, et al. miRNA signatures in sera of patients with active pulmonary tuberculosis. PLoS ONE. 2013;8(11):e80149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ndzia EN, Nkenfou CN, Mekue LM, et al. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis. 2019;14:69–76. [DOI] [PubMed] [Google Scholar]
- [17].Li S, Yue Y, Xu W, et al. MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS ONE. 2013;8:e81438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stahl HF, Fauti T, Ullrich N, et al. miR-155 inhibition sensitizes CD4+ Th Cells for TREG mediated suppression. PLoS ONE. 2009;4(9):e7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu J, Lu C, Diao N, et al. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Human Immunol. 2012;73(1):31–37. [DOI] [PubMed] [Google Scholar]
- [20].Draz NI, Soha AR, Marwa SE, et al. Serum microRNA-29a and microRNA-361-5p as potential diagnostic biomarkers for active pulmonary tuberculosis. Egypt J Med Microbiol. 2014;3(4):27–35. [Google Scholar]
- [21].Harapan H, Fitra F, Ichsan I, et al. The roles of microRNAs on tuberculosis infection: meaning or myth? Tuberculosis. 2013;93:596e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vergoulis T, Vlachos I, Alexiou P, et al. Tarbase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucl Acids Res. 2012;40(D1):D222–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acids Res. 2015;43(D1):D146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2005;30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sabir N, Hussain T, Shah SZA, et al. miRNAs in Tuberculosis: new avenues for diagnosis and host-directed therapy. Front Microbiol. 2018;9:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kleinsteuber K, Heesch K, Schattling S, et al. Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4⁺ T cells and peripheral blood from tuberculosis patients. PLoS ONE. 2013;8(4):e61609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu Z, Lu H, Sheng J, et al. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586(16):2459–2467. [DOI] [PubMed] [Google Scholar]
- [28].Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11(2):141–147. [DOI] [PubMed] [Google Scholar]
- [29].Kumar R, Halder P, Sahu SK, et al. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012;14(10):1620–1631. [DOI] [PubMed] [Google Scholar]
- [30].Riendeau CJ, Kornfeld H. THP-1 cell apoptosis in response to mycobacterial infection. Infect Immun. 2003;71(1):254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ma F, Xu S, Liu X, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12(9):861–869. [DOI] [PubMed] [Google Scholar]
- [32].Schaale K, Neumann J, Schneider D, et al. Wnt signaling in macrophages: augmenting and inhibiting mycobacteria-induced inflammatory responses. Eur J Cell Biol. 2011;90(6–7):553–559. [DOI] [PubMed] [Google Scholar]
- [33].Zhou M, Yu G, Yang X, et al. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol Med Rep. 2016;13(6):4620–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ghorpade DS, Holla S, Kaveri SV, et al. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signalling. Mol Cell Biol. 2013;33(3):543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holla S, Kurowska-Stolarska M, Bayry J, et al. Selective inhibition of IFN-γ induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signalling. Autophagy. 2014;10(2):311–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang JX, Xu J, Han YF, et al. Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for paediatric pulmonary tuberculosis in Chinese patients. Genet Mol Res. 2015;14(4):17235–17243. [DOI] [PubMed] [Google Scholar]
- [37].Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. [DOI] [PubMed] [Google Scholar]
- [39].Zhang C, Xi X, Wang Q, et al. The association between serum miR-155 and natural killer cells from tuberculosis patients. Int J Clin Exp Med. 2015;8(6):9168–9172. [PMC free article] [PubMed] [Google Scholar]
- [40].Brook M, Tchen CR, Santalucia T, et al. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26(6):2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bala S, Marcos M, Kodys K, et al. Upregulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2010;286(2):1436–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Etna MP, Sinigaglia A, Grassi A, et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PLoS Pathog. 2018;14(1):e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wagh V, Urhekar A, Modi D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis. (Edinb). 2017;102: 24–30. [DOI] [PubMed] [Google Scholar]
- [44].Fu Y, Yi Z, Wu X, et al. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49(12):4246–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Latorre I, Leidinger P, Backes C, et al. A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur Respir J. 2015;45(4):1173–1176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ananya M. Tuberculosis Transmission. News-Medical. 2019. [cited 2019 June19]. Available from: <https://www.news-medical.net/health/Tuberculosis-Transmission.aspx>
- India TB Report . 2019. Published on 2019 June6 [Cited 2020 Feb 21]. Available from: https://tbcindia.gov.in/WriteReadData/India%20TB%20Report%202019.pdf


