ABSTRACT
Grafting of neural stem cells (NSCs) derived from human induced pluripotent stem cells (hiPSCs) has shown promise for brain repair after injury or disease, but safety issues have hindered their clinical application. Employing nano-sized extracellular vesicles (EVs) derived from hiPSC-NSCs appears to be a safer alternative because they likely have similar neuroreparative properties as NSCs and are amenable for non-invasive administration as an autologous or allogeneic off-the-shelf product. However, reliable methods for isolation, characterization and testing the biological properties of EVs are critically needed for translation. We investigated signatures of miRNAs and proteins and the biological activity of EVs, isolated from hiPSC-NSCs through a combination of anion-exchange chromatography (AEC) and size-exclusion chromatography (SEC). AEC and SEC facilitated the isolation of EVs with intact ultrastructure and expressing CD9, CD63, CD81, ALIX and TSG 101. Small RNA sequencing, proteomic analysis, pathway analysis and validation of select miRNAs and proteins revealed that EVs were enriched with miRNAs and proteins involved in neuroprotective, anti-apoptotic, antioxidant, anti-inflammatory, blood-brain barrier repairing, neurogenic and Aβ reducing activities. Besides, EVs comprised miRNAs and/or proteins capable of promoting synaptogenesis, synaptic plasticity and better cognitive function. Investigations using an in vitro macrophage assay and a mouse model of status epilepticus confirmed the anti-inflammatory activity of EVs. Furthermore, the intranasal administration of EVs resulted in the incorporation of EVs by neurons, microglia and astrocytes in virtually all adult rat and mouse brain regions, and enhancement of hippocampal neurogenesis. Thus, biologically active EVs containing miRNAs and proteins relevant to brain repair could be isolated from hiPSC-NSC cultures, making them a suitable biologic for treating neurodegenerative disorders.
KEYWORDS: Anti-inflammatory effects, extracellular vesicles, human induced pluripotent stem cells, ion-exchange chromatography, microRNAs, neurogenic properties, proteomics
Introduction
Grafting of neural stem cells (NSCs) derived from multiple sources, including the human induced pluripotent stem cells (hiPSCs), has shown promise for brain repair after injury or disease [1–6]. Although the transplanted NSCs differentiate into neurons, astrocytes and oligodendrocytes, the functional recovery mediated by NSCs has been attributed mostly to the trophic support provided by the graft-derived cells. Such bystander effects are efficient for reducing the loss of neurons and glia, creating a permissive substrate for axon growth and plasticity resulting in reconstruction of the local circuitry, and increased host hippocampal neurogenesis in several models of neurological diseases [1,2,7–9]. While grafting of NSCs derived from hiPSCs is devoid of ethical concerns and also beneficial for improving brain function after injury or disease, their translation to the clinic has been hampered by safety issues, which include immunogenic risks, and the possible genetic instability leading to incomplete differentiation or teratoma formation [10–13].
Although several approaches to circumvent the safety issues associated with the clinical use of NSCs derived from hiPSCs are in development [8], employing nano-sized extracellular vesicles (EVs) derived from hiPSC-NSCs appears to be a much safer alternative because of several reasons. Several studies have implied that the therapeutic outcome of stem cells, including NSCs, was due to the paracrine actions of their secretome [9,14]. A vital component of this paracrine secretion has been identified as EVs as they facilitate the transfer of genetic information and proteins from stem cells into injured cells in the host [9,14]. Because of the ability of EVs to mimic stem cell properties, stem-derived EVs have been increasingly considered as apt therapeutic alternatives to stem cells in regenerative medicine [15,16]. Indeed, several studies have reported the therapeutic efficacy of EVs derived from various stem cells in CNS repair and regeneration after injury or disease [17–21]. Moreover, EVs are amenable for repeated, non-invasive dispensation as an autologous or allogeneic off-the-shelf product for treating neurological diseases because they can quickly permeate the entire brain following an intranasal administration [22].
The mechanisms underlying the beneficial effects of exogenously administered EVs in the CNS likely involves the release and transfer of their molecular constituents into the target neurons and glia in a paracrine fashion [9]. Therefore, a rigorous characterization of EVs derived from different cell sources, particularly for their purity, miRNA and protein composition, biological activity and neuroreparative properties will be necessary before their application for treating CNS disorders. Such characterization would require the development of reliable methods that are efficient for isolating and analysing the molecular composition as well as investigating the biological properties of EVs. Therefore, in this study, we generated EVs from hiPSC-derived NSCs using a combination of anion-exchange chromatography (AEC) and size-exclusion chromatography (SEC) and investigated their composition through small RNA sequencing and proteomics. We also performed experiments to determine their ability to incorporate into neural cells as well as mediate anti-inflammatory and neurogenic effects in the host brain following intranasal administration. Our results demonstrate that biologically active EVs containing many miRNAs and proteins relevant to brain repair after injury or disease, and exhibiting anti-inflammatory and neurogenic properties could be isolated from hiPSC-NSCs.
Materials and methods
A schematic displaying the various experiments and analyses performed to characterize the properties and biological effects of EVs isolated from hiPSC-derived NSC cultures is illustrated in Figure 1.
Figure 1.
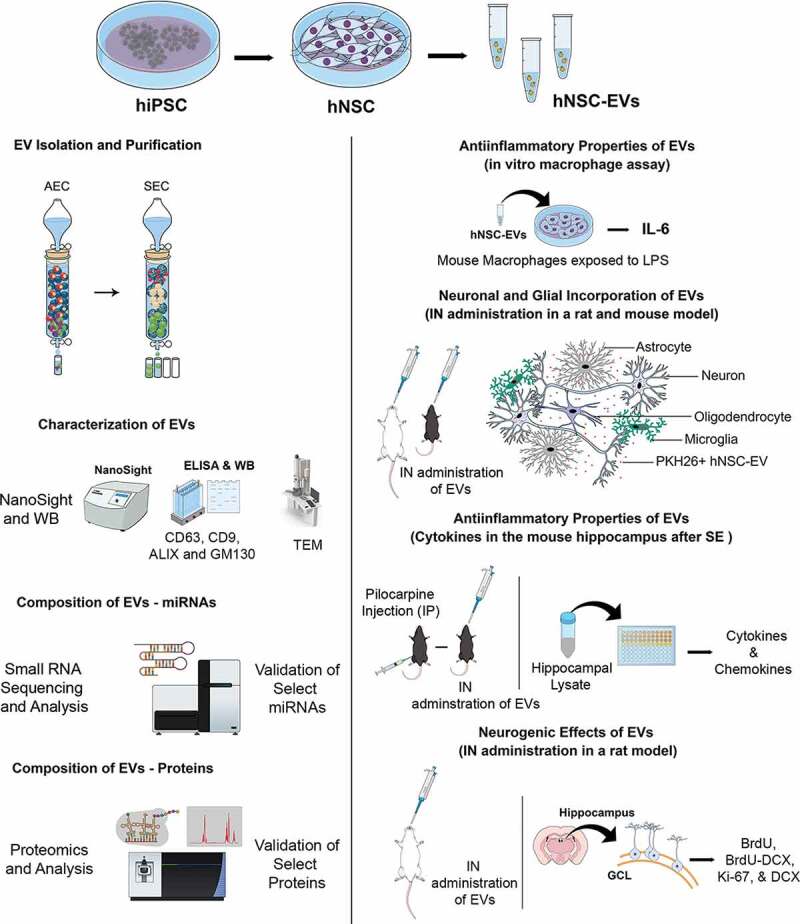
A schematic to show the various experiments and analyses performed to characterize the properties and biological effects of extracellular vesicles (EVs) derived from human induced pluripotent stem cells (hiPSCs). Human neural stem cells (hNSCs) were generated through the expansion of the hiPSCs in a chemically defined medium and expanded through serial passaging using an NSC proliferation medium (top panel). The various cartoons on the left illustrate the methods employed for the isolation and characterization of extracellular vesicles (EVs) from the spent media of passage 11 (P11) hNSC cultures. The methods include anion-exchange chromatography (AEC) and size exclusion chromatography (SEC) for EV isolation, ELISA and Western blotting (WB) studies on EV markers such as CD63, CD9 and ALIX and the deep cellular marker GM130, transmission electron microscopy (TEM) studies on EV morphology, evaluation of the size of EVs using NanoSight, and identification of small miRNAs and proteins using small RNA sequencing and proteomic analyses. The panels on the right show different experiments that examined the biological properties of EVs. First, the anti-inflammatory activity of EVs was measured through a macrophage assay in vitro with lipopolysaccharide exposure and quantification of interleukin-6 (IL-6) release. Next, the proficiency of EVs to target neurons, microglia and astrocytes in various regions of the brain in both rats and mice were examined 6 hours after an intranasal (IN) administration of PKH26-labelled EVs. Then, the anti-inflammatory activities of EVs in vivo was determined through quantification of various cytokine/chemokine in a mouse model of status epilepticus (SE). SE was induced through an intraperitoneal injection of pilocarpine. Finally, the neurogenic effects of IN administered EVs were evaluated via quantification of hippocampal neurogenesis in 6 months old rats. BrdU, 5ʹ-bromodeoxyuridine; DCX, doublecortin; GCL, granule cell layer.
Generation of NSCs from hiPSCs
The generation of NSCs from hiPSCs was done as described elsewhere [23]. Briefly, hiPSC colonies (IMR90-4; Wisconsin International Stem Cell Bank, Madison, WI, USA) were grown as cell clumps at a density of 2–2.5 × 104 cells/cm2 in six-well plates coated with matrigel (Corning, Tewksbury, MA, USA) using TeSR™-E8™ medium (STEMCELL Technologies, Vancouver, Canada). Twenty-four hours later, the culture medium was replaced with neural induction medium containing neurobasal (Gibco, Grand Island, New York, USA) and neural induction (Gibco) supplement. The medium was swapped every other day for seven days. Then primitive NSCs were dissociated with accutase (Gibco) and plated on matrigel-coated dishes with a density of 0.5–1.0 × 105 cells per cm2 in an NSC expansion medium containing 50% neurobasal, 50% advanced DMEM/F12, and 1X neural induction supplement. The culture medium was exchanged every other day until NSCs reached confluency on day 5 of plating. The NSC cultures were passaged every seven days, and NSCs from different passages were cryoprotected and stored in liquid nitrogen. The NSC status at different passages was confirmed through immunofluorescence staining for nestin (anti-nestin, 1:1000; EMD Millipore, Burlington, MA, USA) and Sox-2 (anti-Sox-2, 1:300; Santacruz Biotechnology, Dallas, TX, USA).
Collection of hiPSC-NSC culture media for harvesting EVs
For the isolation of NSC-derived EVs, frozen vials containing passage 11 NSCs were thawed at 37°C and plated on to a T-75 culture flask (Corning) and grown at 37°C in a CO2 incubator. Following 70% confluency, the cells were dislodged using 1 U/ml of dispase (Gibco), washed with NSC media (Gibco), and seeded at ~500 cells per cm2 into 150 × 20 mm diameter tissue culture plates (Corning) in NSC expansion medium. Once NSCs reached 90% confluency, the media was harvested and used for isolating EVs or stored at −80°C for further use.
EV isolation by Ion exchange and size exclusion chromatography
The conditioned media containing EVs were subjected to low-speed centrifugation at 1811 g for 10 minutes, which was followed by filtration through a 0.22 µm filter to remove the cell debris and larger suspending particles. The large volume of filtered media was then subjected to a 5–7 fold concentration using Amicon 10 kDa cut-off ultra-filtration device (EMD Millipore). Using a 1.5 × 12 cm chromatography column (Bio-Rad, Hercules, CA, USA), 10 ml of Q-Sepharose fast flow (GE Healthcare, Chicago, Illinois, USA) was equilibrated with 100 ml of equilibration buffer, and the chromatography was performed by adding the concentrated conditioned media. The EVs were selectively eluted using elution buffer containing 50 mM Tris and 1000 mM NaCl of pH 8.0. Fractions were collected at a flow rate of 1 ml/minute, and elution of EVs was continuously monitored by Nanoparticle tracking analysis (NanoSight LM10, Malvern Panalytical, Malvern, UK). After AEC, the fractions containing EVs were pooled and concentrated using an ultrafiltration device of 10 kDa cut-off and subjected to an SEC on a column made up of 25 ml of Sephacryl S-500 High Resolution (GE Healthcare). Using the mobile phase containing 50 mM phosphate buffer and 200 mM NaCl of pH 7.0, EVs were size-fractionated, and fractions were collected at a flow rate of 1 ml/minute. The elution of EVs was continuously monitored by quantifying the total protein content by BCA method and tracked by nanoparticle tracking analysis. Fractions containing a high number of EVs with less protein content were pooled, concentrated and stored at −20°C for further use.
Characterization of the number, size and markers of EVs
The final concentration and size distribution of particles was measured by nanoparticle tracking analysis, as described in our previous report [24].The expression of CD63 was investigated using ELISA [17]. The expression of other EV markers such as CD9 and ALIX were examined through Western blotting. For this, an aliquot of EVs in 100 µl volume was mixed with 300 µl of mammalian protein extractor reagent (Thermo Fischer Scientific, Waltham, MA, USA) and lysed, as detailed in our recent study [24]. The total protein in the lysate was quantified by the Pierce BCA protein assay kit (Thermo Fisher Scientific), and 40 µg of protein was loaded and separated by 4–12% NuPAGE Bis-Tris Gels (Thermo Fisher Scientific). Following the transfer of proteins onto a nitrocellulose membrane using the iBlot2 gel transfer device (Thermo Fisher scientific), the membrane was processed for protein detection using antibodies against CD9 (1:500; BD Biosciences, San Jose, CA, USA), or ALIX (1:500; Santa Cruz). Then, the signal was detected using the ECL detection kit (ThermoFisher) and visualized using a Versadoc Imaging System (Bio-Rad). In order to rule out the contamination of EV lysate with deep cellular proteins, the protein separated membrane was identified for GM130 (1:500; BD Biosciences) and compared with the NSC lysate [25,26].
Measurement of total protein, and protein-lipid ratio in EVs
We measured the amount of total protein in aliquots of EVs containing 5, 10, 20 and 50 × 109 EVs using the BCA protein assay kit (Thermo Fisher Scientific) and determined EV numbers per microgram of protein. The total lipid content in EV preparations was measured by the modified sulpho-phospho vanillin (SPV) method, as described elsewhere [27]. Briefly, both standard (1,2-Dioleoyl-sn-glycero-3-phosphocholine, DOPC; Sigma, St. Louis, MO, USA) and hiPSC-NSC-EV samples in SEC buffer (40 μl each) were sonicated at 35 amplitude (20 kHz) for 10 minutes, intensely vortexed for 2 minutes and mixed with 200 μl of 96% sulphuric acid. Following brief vortexing, the tubes were incubated at 90°C for 20 minutes, moved to 4°C for 5 minutes and mixed with 120 μl of phospho-vanillin reagent and vortexed. Next, 280 μl of each sample was transferred to a 96 well plate, and the colour reaction was allowed to progress for 10 minutes at room temperature. Absorbance at 540 nm was determined with a plate reader. The total lipid content in EV samples was calculated using the standard graph, and these values were used to determine the protein-lipid ratio in different samples.
Visualization of hiPSC-NSC EVs through transmission electron microscopy (TEM)
A suspension of EVs (~50 × 109/ml) was diluted in phosphate-buffered saline (1:10), and 1.5 μl of the suspension was placed on 300 mesh carbon-coated copper grids (Electron Microscopy Science, Hatfield, PA, USA) at room temperature. Five minutes later, excess liquid in grids was blotted with a filter paper, rinsed twice with drops of distilled water and the grids were stained by continuously dripping 150 μl of 0.5% uranyl acetate onto them while tilted at a 45° angle. The excess liquid was blotted, and the grids were air-dried at room temperature for 10 minutes. The images were collected using an FEI Morgagni 268 transmission electron microscope equipped with a MegaView III CCD camera. hiPSC NSC-EV diameters were determined using the “Analyze” tool in the ImageJ software as an average of the measured diameters along four different axes (x, y, x + 45°, y + 45°).
Small RNA-seq of hiPSC-NSC EVs and data analysis
System Biosciences (Palo Alto, CA, USA) performed small RNA-sequencing of hiPSC-NSC derived EVs (n = 3). The RNA was isolated using the mirVana miRNA Isolation kit (Thermo Fischer Scientific). Quantification of RNA comprised the use of 1:2 diluted RNA in a 20 µl RiboGreen Assay. The amount of small-RNA obtained (125.3–729.1 pg/µl) was sufficient for library preparation and sequencing. Small RNA libraries were created with the CleanTag Small RNA Library Preparation Kit (TriLink, San Diego, CA, USA) according to the manufacturer’s protocol. The final purified library was quantified with high sensitivity DNA reagents (Agilent Technologies, Santa Clara, CA, USA) and high sensitivity DNA chips (Agilent Technologies). The libraries were pooled, and the 140 to 300 bp region was size selected on an 8% TBE gel (Invitrogen, Carlsbad, CA, USA). The size selected library is quantified with high sensitivity DNA 1000 screen tape (Agilent Technologies), high sensitivity D1000 reagents (Agilent Technologies) and the TailorMix HT1 qPCR assay (SeqMatic, Fremont, CA, USA) followed by a NextSeq high output single-end sequencing run at SR75 using NextSeq 500/550 High Output v2 kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The small RNA-sequencing data analysis is performed using the Banana Slug analytics platform. In this sequencing platform, sequencing data are analysed for quality and contamination. The raw reads were trimmed to remove adapters, then trimmed and filtered based on quality scores. The sequence reads were mapped to the genome, and mapping statistics were generated. Next, the abundance levels for ncRNAs (miRNAs, tRNAs, rRNAs, lincRNAs, piRNAs, snoRNAs), antisense transcripts, coding genes and repeat elements (LTR, LINE, SINE and tandem repeats) were determined. Differences in expression of ncRNA, antisense transcripts and repeat elements between replicates were then calculated.
Validation of selected miRNA using quantitative real-time PCR
Among the most abundant 40 miRNAs from RNA-Seq data, eight miRNAs that are known to contribute to various aspects of brain function were chosen and validated through quantitative real-time PCR. The total RNA was isolated from two biological replicates of hiPSC-NSC-EVs (~25 × 109 each) using the SeraMir Exosome RNA amplification kit (System biosciences). miRCURY LNA RT Kit (Qiagen, Germantown, MD, USA) was employed for converting 5 ng/μl of total RNA into cDNA. miRCURY LNA miRNA SYBR Green PCR kit (Qiagen) and miRCURY LNA miRNA PCR assay primer mix (Qiagen) were used to measure the comparative expression of 8 different miRNAs (miRNAs- 320a, 103a-3p, 21–5p, 26a-5p, 320b, 30a-3p, 181a-5p, 191–5p).
miRNA pathway analysis
The enrichment pathway analysis was performed by using DIANA-279 miRPath v.3, a web tool that identifies enriched pathways targeted by selected miRNAs [28]. Since many predicted targets are false-positives, we have included only experimentally validated human target genes by using the Tarbase v7.0 database. The miR-ID was based on Mirbase version 21. For miRs with ambiguous IDs, we used both 3p and 5p arms (miRNA stem-loop). The functional annotation was performed according to the “Kyoto Encyclopedia” of Genes and Genomes (KEGG) databases. We used the “Pathways/Categories Union” algorithm to assess the combined action of selected miRNAs. Fisher´s exact test was chosen for enrichment analysis, with a microTthreshold of 0.8, false discovery rate (FDR) correction and p value threshold at ≤ 0.05. The heat map of microRNAs versus pathways was based on significance values, where darker colours represent lower p values.
Proteomic analysis of hiPSC-NSC EVs and pathway analysis using reactome
Extracellular vesicles were lysed in buffer containing 5% SDS/50 mM triethylammonium bicarbonate (TEAB) in the presence of protease and phosphatase inhibitors (Thermo Fisher Scientific) and nuclease (Thermo Fisher Scientific). Aliquots corresponding to 90 µg protein (EZQ™ Protein Quantitation Kit; Thermo Fisher Scientific) were reduced with tris(2-carboxyethyl) phosphine hydrochloride (TCEP), alkylated in the dark with iodoacetamide and applied to S-Traps mini columns (Protifi, Farmingdale, NY, USA) for tryptic digestion (Promega, Fitchburg, WI, USA) in 50 mM TEAB. Peptides were eluted from the S-Traps with 0.2% formic acid in 50% aqueous acetonitrile and quantified using Pierce Quantitative Fluorometric Peptide Assay (Thermo Fisher Scientific). Data-independent acquisition mass spectrometry was conducted on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). On-line HPLC separation was accomplished with an RSLC NANO HPLC system (Thermo Fisher Scientific/Dyonex): column, PicoFrit™ (New Objective, Woburn, MA, USA), 75 µm i.d, packed to 15 cm with C18 adsorbent (218MSB5 Vydac, Columbia, MD, USA); mobile phase A, 0.5% acetic acid (HAc)/0.005% trifluoroacetic acid in water; mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA/9.5% water; gradient 3 to 42% B in 120 minutes; flow rate, 0.4 μl/minute. A pool was made of all of the samples, and 2-µg peptide aliquots were analysed using gas-phase fractionation and 4-m/z windows (120 k resolution for precursor scans, 30 k for product ion scans, all in the orbitrap) to create a data independent acquisition (DIA) chromatogram library [29], by searching against a panhuman spectral library [30]. Experimental samples were blocked by replicate and randomized within each replicate. Injections of 2 µg of peptides were employed. MS data for experimental samples were acquired in the orbitrap using 12-m/z windows (staggered; 120 k resolution for precursor scans, 30 k for product ion scans) and searched against the chromatogram library. Scaffold DIA (v1.3.1; Proteome Software; Portland, OR, USA) was used for all DIA data processing. Only peptides that were exclusively assigned to a protein were used for quantitative analysis. The total proteins found in the two different preparations of EVs were first subjected to pathway analysis using Reactome version 71 (https://reactome.org/) [31,32]. The proteins were compared against the Vesiclepedia database (http://microvesicles.org/) and were mapped to Gene Ontology (GO) terms by using the Functional Enrichment analysis tool (FunRich 3.1.3) [33].
Validation of selected proteins using ELISA
Among the most abundant 100 proteins from proteomics data, we validated five proteins that are known to contribute to various brain functions and also perceived to be useful for treating neurodegenerative disorders, using ELISA. These include agrin, pentraxin 3 (PTX3), hemopexin, galectin-3 binding protein (Gal-3BP) and nidogen-1. Briefly, three biological replicates of hiPSC-NSC-EVs (~50 × 109 each) were lysed, and agrin (Abcam, Cambridge, UK), PTX3 (Aviscera Biosciences, Sunnyvale, CA, USA), hemopexin (Aviscera Biosciences), Gal-3BP (R&D systems, Minneapolis, MN, USA) and nidogen-1 (R&D systems) were measured using commercially available ELISA kits. The concentrations of these proteins were normalized with the total protein content in the EV lysate.
Analysis of the anti-inflammatory activity of EVs using lipopolysaccharide (LPS) stimulated mouse macrophages
The ability of the EVs generated from hiPSC-NSCs to suppress the expression of a pro-inflammatory cytokine was assayed using macrophages, as detailed in a previous report [34]. The assay used a line of transformed murine monocytes/macrophages (RAW 264.7; ATCC, Manassas, VA, USA) to measure the anti-inflammatory activity of EVs. Briefly, after an overnight culture of mouse macrophages, ~100,000 cells were seeded per well in 48 well plates, and the cultures were maintained overnight. The non-adhered cells were removed the following day, and the adhered cells were stimulated with a 10 ng/ml LPS alone, (Sigma), in combination with 1 µg/ml dexamethasone (DEX) (Sigma) or LPS in combination with various concentrations of hiPSC-NSC derived EVs for 4 hours. Then, the conditioned media were harvested, and the IL-6 in the medium was quantified via ELISA (R&D Systems).
In vivo studies using mice and rats
Two-month-old male mice (C57BL/6 J; Jackson Labs, Bar Harbor, ME, USA) and six-months-old male rats (F344; Harlan Sprague-Dawley, Indianapolis, IN, USA) were used for in vivo studies. All animal studies done were approved by the Animal Care and Use Committee (IACUC) of the Texas A&M University College of Medicine, and complied with all federal and state regulations for the purchase, transportation, housing and ethical use of animals for research. The euthanasia procedures were consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
Tracking of hiPSC-NSC EVs in the adult rat and mouse brain following intranasal administration
The EVs were labelled with PKH26 (Sigma) as per manufacturer’s instructions, and the free dye was separated from the bound dye by ultrafiltration using 10 kDa MWCO filter columns (Sartorius, Gottingen, Germany). The rats and mice (n = 2/species) were moderately anesthetized with a cocktail of ketamine, xylazine and acepromazine, and their nostrils were treated with 10 µl of hyaluronidase (Sigma-Aldrich, St. Louis, MO, USA) to enhance the permeability of the nasal mucous membrane. Thirty minutes later, labelled EVs were administered (150 × 109 [~75 µg]/rat and 25 × 109 [~12.5 µg]/mouse), as detailed in our recent report [22]. Six hours later, the animals were perfused, the brain tissues were processed for cryostat sectioning, and thirty-micrometre-thick coronal sections through the entire forebrain, midbrain and hindbrain, were collected serially. Serial sections through the entire brain were selected and processed for immunofluorescent staining using appropriate primary antibodies against NeuN (1:1000; Millipore), IBA-1 (1:1000; Abcam), GFAP (1:3000; Dako, Glostrup, Denmark), S-100β (1:1000; Sigma) and calbindin (1:500; Sigma), as described in our previous report [22]. The sections were examined using Z-section analysis in a confocal microscope for the distribution of EVs in different cell types (neurons, microglia and astrocytes) in multiple regions of the brain.
Analysis of anti-inflammatory activity of EVs in a mouse model of status epilepticus(SE)
Male C57BL/6J mice were randomly designated to three groups: naïve (n = 6), status epilepticus plus vehicle (SE + VEH, n = 10), and SE + EVs (n = 10). SE was induced using a protocol described in our previous report [18]. Briefly, the animals received a subcutaneous injection of scopolamine methyl nitrate (1 mg/kg; Sigma-Aldrich), which was followed 30 minutes later with an intraperitoneal injection of pilocarpine hydrochloride (Sigma-Aldrich) at a dose of 380 mg/kg [35], which induced SE. Animals were observed for the severity and duration of behavioural convulsions for two hours after SE onset [36],and the behavioural convulsions were then terminated with a subcutaneous injection of diazepam (10 mg/Kg). Only animals that displayed consistent stage 4 (i.e., bilateral forelimb myoclonus and rearing) or stage 5 (i.e., bilateral forelimb and hindlimb myoclonus and transient falling) convulsions were chosen for further experimentation. At 2 hours post-SE, animals in the SE+EVs group received IN administration of 50 × 109 EVs, whereas animals in the SE+VEH group received IN administration of the SEC buffer (50 mM phosphate buffer pH 7.0 containing 200 mM NaCl). Twenty-four hours later, animals were deeply anesthetized, the brains were removed, snap-frozen in dry ice and stored at −80°C. The hippocampi from both hemispheres were micro-dissected, and the samples were processed for measuring various cytokines using a cytokine array (Signosis, Santa Clara, CA, USA) [18]. The values for each cytokine/chemokine were compared between SE + VEH and SE + EVs groups (n = 6–7/group). The changes in TNF-α and IL-1β were also confirmed with individual ELISAs, as described in our previous report [18].
Characterization of neurogenic effects of hiPSC-NSC EVs in adult rats
Six-month-old male F344 rats acquired from Harlan Sprague-Dawley were employed for this experiment. Each animal received IN administration of either 100 × 109 EVs (EVs group, n = 6) or SEC buffer (VEH group, n = 6), as described in the previous section. Thirteen days later, the animals received three injections of 5ʹ-bromodeoxyuridine (BrdU, once every 8 hours at 75 mg/Kg). Eight hours later, the animals were euthanized through intracardiac perfusions with 4% paraformaldehyde, and the brain tissues were processed for cryostat sectioning as described in our previous reports [36,37]. Thirty-micrometre-thick serial sections (every 15th) through the entire septotemporal axis of the hippocampus were immunohistochemically processed for identifying BrdU (1:100; Sigma), doublecortin (DCX, 1:500; Santa Cruz) and Ki-67 (1:500; Millipore) [1,38]. The number of newly born cells (BrdU+ cells), newly born neurons (DCX+ cells) and proliferating cells (Ki-67+ cells) in the entire subgranular zone-granule cell layer (SGZ-GCL) were quantified using stereology [39,40]. Additional serial sections were also processed for BrdU-DCX dual immunofluorescence and Z-section analysis in a confocal microscope to measure the extent of neuronal differentiation of newly born cells and to calculate net hippocampal neurogenesis [38].
Statistical analyses
Statistical analyses were performed using Prism software 8.0. When comparisons involving three or more groups, statistical analyses were performed using one-way analyses of variance (one-way ANOVA) with Newman-Keuls multiple comparison post hoc tests. When comparisons involved only two groups, unpaired, two-tailed Student’s t test was employed. However, when standard deviations between the two groups were significantly different (p < 0.05), we employed the non-parametric Mann-Whitney U-test. Data were expressed as means ± SEM and a p value less than 0.05 was considered as statistically significant.
Results
All cells derived from P11-NSCs expressed nestin and Sox-2
Passaging and culturing of primitive NSCs with the NSC differentiation medium resulted in the transformation of virtually all hiPSC-derived cells into NSCs at passage 6. Such directed differentiation was evident from the expression of two NSC markers nestin and Sox-2. NSCs generated from passage 7–11 also showed consistent expression of nestin and Sox-2. EVs were collected from cultures of banked P11 NSCs in all studies. Dual immunofluorescence assays on P11-NSC sister cultures confirmed that all cells in these cultures expressed both nestin and Sox-2 (Figure 2(a1-a4)), and none of the cells in these cultures expressed the PSC marker Oct-4.
Figure 2.
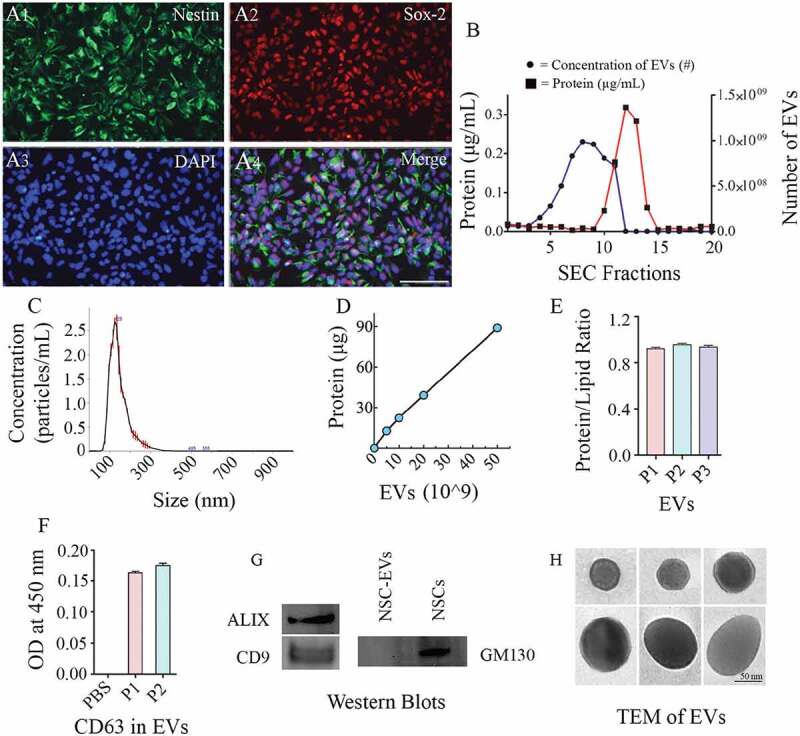
Human neural stem cells (hNSCs) express specific markers, and extracellular vesicles (EVs) isolated from hNSC cultures express multiple EV markers. (a1-a4) illustrate that all cells in the passage 11 hNSCs derived from human induced pluripotent stem cells (hiPSCs) express NSC markers nestin and Sox-2. Scale bar: 100 μm. (b) compares protein concentrations (the left Y-axis), and the concentration of EVs (the right Y-axis) in different EV fractions (X-axis) collected from size-exclusion chromatography (SEC). Note that fractions 5–9 contain most EVs with minimal protein content. (c). Representative graph from NanoSight analysis showing the size of EVs. (d, e and f) Illustrate a linear relationship between the number of EVs and the total protein (D), the protein-lipid ratio in EVs (E) and CD 63 protein content measured through ELISA (F). (g) shows the presence of ALIX and CD9 in EVs evaluated through Western blotting. The figure also indicates the absence of a deep cellular marker GM130 in hNSC-derived EVs, in contrast to its robust presence in the NSC lysate. Figure H shows the EVs of different size and shape visualized through transmission electron microscopy. Scale bar, 50 nm.
hiPSC-NSC-EVs exhibited CD63, CD9 and ALIX expression and characteristic ultrastructure, and lacked GM130
The EVs collected from AEC were subjected to SEC. The elutes obtained from SEC showed a different range of protein concentrations and the number of EV particles (Figure 2(b)). Fractions 5–9 contained considerably higher numbers of EV particles with a negligible amount of co-eluting proteins, implying that EVs in these fractions are highly concentrated with minimal protein contamination (Figure 2(b)). Therefore, only the fractions 5–9 were pooled and used for further analysis. Nanoparticle tracking analysis showed that the size of EVs ranged from 100 to 200 nm with a mean value of 145.3 ± 49.0 (Figure 2(c)). Measurement of the amount of total protein in different aliquots of EVs containing 0, 5, 10, 20 and 50 × 109 EVs (Figure 2(d)) revealed that the average amount of protein per 1 × 109 EVs was 2.14 ± 0.18 µg (Mean ± S.E.M), which amounted to ~0.5 × 109 EVs per 1 µg protein. Furthermore, the protein-lipid ratio measured from three different EV preparations showed comparable values (0.94 ± 0.009, Figure 2(e)) confirming the reproducibility of the EV isolation methods. Moreover, ELISA revealed the expression of CD63 in two different preparations of EVs isolated from SEC (Figure 2(f)). Also, Western blot analysis showed that EVs expressed several other EV markers, including CD9 and ALIX (Figure 2(g)). We also performed Western blotting of lysate from NSC-derived EVs and the lysate from NSCs for the expression of GM130, a cytoplasmic protein found to be tightly associated with Golgi membranes but not found in EVs. This analysis revealed robust expression of GM130 in NSCs but not in NSC-derived EVs (Figure 2(g)). Imaging with a TEM following negative staining demonstrated vesicles that are round or oval and ranged in diameter from 50 to 130 nm. Examples of individual EVs exhibiting different sizes and shapes found in our hiPSC-NSC-EV preparations are illustrated in Figure 2(h)). In some EVs, the lipid bilayer membrane could be observed. Thus, EV preparations made from the hiPSC-NSC culture medium through sequential AEC and SEC contained EVs displaying CD63, CD9 and ALIX expression, and characteristic morphology, but lacked deep cellular proteins.
Small RNA-seq revealed most abundant miRNAs having roles in different signalling pathways
We analysed three different preparations (biological replicates) of hiPSC-NSC derived EVs with small RNA sequencing to determine the consistency of our protocol in producing EVs with similar RNA composition. All 3 preparations contained similar fractions of miRNA (1%), piRNA (1.2%), tRNA (0.8%), rRNA (1.1%) and the other ncRNAs (4.5%) (Figure 3(a)). Differential expression data analysis also confirmed the negligible amount differentially expressed small RNAs among the three different EV preparations (Figure 3(b)). The number and types of small RNAs found in hiPSC-NSC-EVs are detailed in Supplementary Table 1. Furthermore, miRNAs showing ≥10 reads in all three preparations of hiPSC-NSC-EVs are detailed in Supplementary Table 2. From this list, eight miRNAs that are known to contribute to various aspects of brain function were chosen and evaluated through qPCR. All chosen miRNAs (miRNAs- 320a, 103a-3p, 21–5p, 26a-5p, 320b, 30a-3p, 181a-5p, 191–5p) showed comparable Ct values in two biological replicates (Figure 3(c)). Thus, miRNAs identified in hiPSC-NSC-EVs through RNA-seq could be validated through qPCR in additional EV samples. We next performed pathway analysis for the top 40 miRNAs using DIANA-mirPath v.3, followed by functional annotation based on KEGG databases. hiPSC-NSC derived EVs were enriched with miRNAs that are involved in different signalling pathways and diverse metabolic functions. These results are congruous with the role of EVs as mediators of cell-cell communication (Table 1). The hiPSC-NSC derived EVs also contained miRNAs that are involved in regulatory pathways in conditions such as cancers and prion diseases (Table 1). Notably, heatmaps on miRNAs versus functional categories showed that fatty acid metabolism, fatty acid biosynthesis, the extracellular matrix–receptor (ECM-receptor) interaction pathway, are targeted with higher significance (red squares) by most of miRs (Figure 4).
Figure 3.
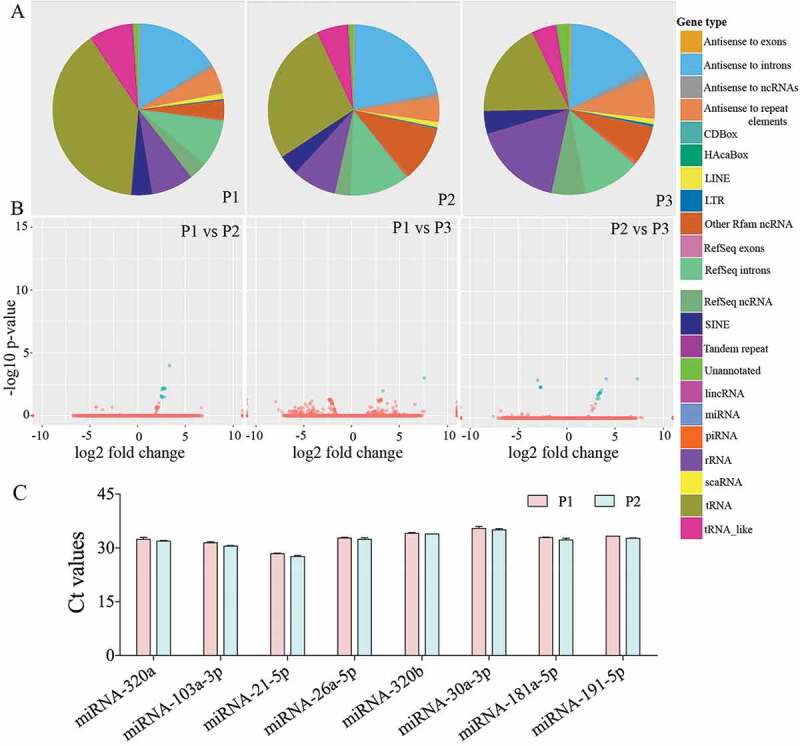
Small RNA-Sequencing showed a comparable composition of small RNAs in three different preparations of human neural stem cell-derived extracellular vesicles (hNSC-EVs). The pie charts in figure (a) depict fractions of various small RNAs in three different preparations of hNSC-EVs (P1-P3). The Volcano plots in (b) compare the composition of small RNAs between different EV preparations. Note that the composition of small RNAs is mostly similar in three different preparations of hNSC-EVs. The bar chart (c) shows Ct values of miRNAs (miRs- 320a, 103a-3p, 21–5p, 26a-5p, 320b, 30a-3p, 181a-5p, 191–5p) validated through real-time quantitative PCR.
Table 1.
Enriched KEGG pathways with functional categories. Relevant to the central nervous system.
| miRNA |
||||
|---|---|---|---|---|
| Pathway | p value | #genes | Number | Names |
| Hippo signalling pathway | 0 | 91 | 16 | miR-92a-3p, miR-92a-2-5p, miR-320a, miR-30a-3p, miR-30a-5p, miR-21-5p, miR-28-5p, miR-191-3p, miR-181a-5p, miR-10a-5p, miR-10a-3p, miR-320b, miR-26a-5p, miR-1291, miR-151a-3p miR-320 c |
| ECM-receptor interaction | 0 | 25 | 6 | miR-92a-1-5p, miR-92a-2-5p, miR-30a-3p, miR-28-5p, miR-181a-3p, miR-181a-2-3p |
| Fatty acid metabolism | 0 | 23 | 9 | miR-92b-5p, miR-30a-5p, miR-21-5p, miR-21-3p, miR-10a-5p, miR-423-5p, miR-151a-3p, miR-10b-5p, miR-103a-3p |
| Fatty acid biosynthesis | 0 | 5 | 7 | miR-30a-5p, miR-21-3p, miR-10a-5p, miR-423-5p, miR-10b-5p, miR-103a-3p, miR-148a-3p |
| Prion diseases | 0 | 5 | 4 | miR-30a-3p, miR-191-5p, miR-26a-1-3p miR-148a-3p |
| Glioma | 3.33E-16 | 45 | 11 | miR-320a, miR-30a-5p, miR-28-5p, miR-181a-5p, miR-181a-3p, miR-423-5p, miR-1291, miR-1246, miR-103a-3p, miR-320 c miR-148a-3p |
| Adherens junction | 1.94E-14 | 53 | 16 | miR-92a-3p, miR-92b-3p, miR-320a, miR-30a-5p, miR-21-3p, miR-28-3p, miR-28-5p, miR-191-5p, miR-181a-5p, miR-10a-3p, miR-423-5p, miR-320b, miR-26a-5p, miR-151a-5p, miR-103a-3p miR-320 c |
| Signalling pathways regulating pluripotency of stem cells | 2.16E-08 | 75 | 7 | miR-92a-3p, miR-92b-3p, miR-320a, miR-28-5p, miR-181a-5p, miR-26a-5p, miR-1291 |
| TGF-beta signalling pathway | 5.08E-08 | 45 | 9 | miR-92a-3p, miR-92b-3p, miR-320a, miR-30a-3p, miR-10a-3p, miR-26a-5p, miR-1291 miR-320 c miR-148a-3p |
| Cell cycle | 2.84E-06 | 72 | 7 | miR-92a-3p, miR-30a-3p, miR-21-5p, miR-26a-5p, miR-10b-5p, miR-103a-3p miR-148a-3p |
| FoxO signalling pathway | 3.75E-06 | 71 | 7 | miR-92a-3p, miR-92b-3p, miR-30a-3p, miR-21-5p, miR-181a-5p, miR-10a-3p miR-148a-3p |
| P53 signalling Pathway | 5.60E-06 | 47 | 11 | miR-30a-3p, miR-30a-5p, miR-21-5p, miR-181a-5p, miR-26a-5p, miR-26a-1-3p, miR-26a-2-3p, miR-10b-5p, miR-1246, miR-103a-3p miR-148a-3p |
| Thyroid hormone signalling pathway | 9.91E-04 | 57 | 6 | miR-92a-3p, miR-21-5p, miR-26a-5p, miR-1291, miR-1246, miR-103a-3p |
Figure 4.
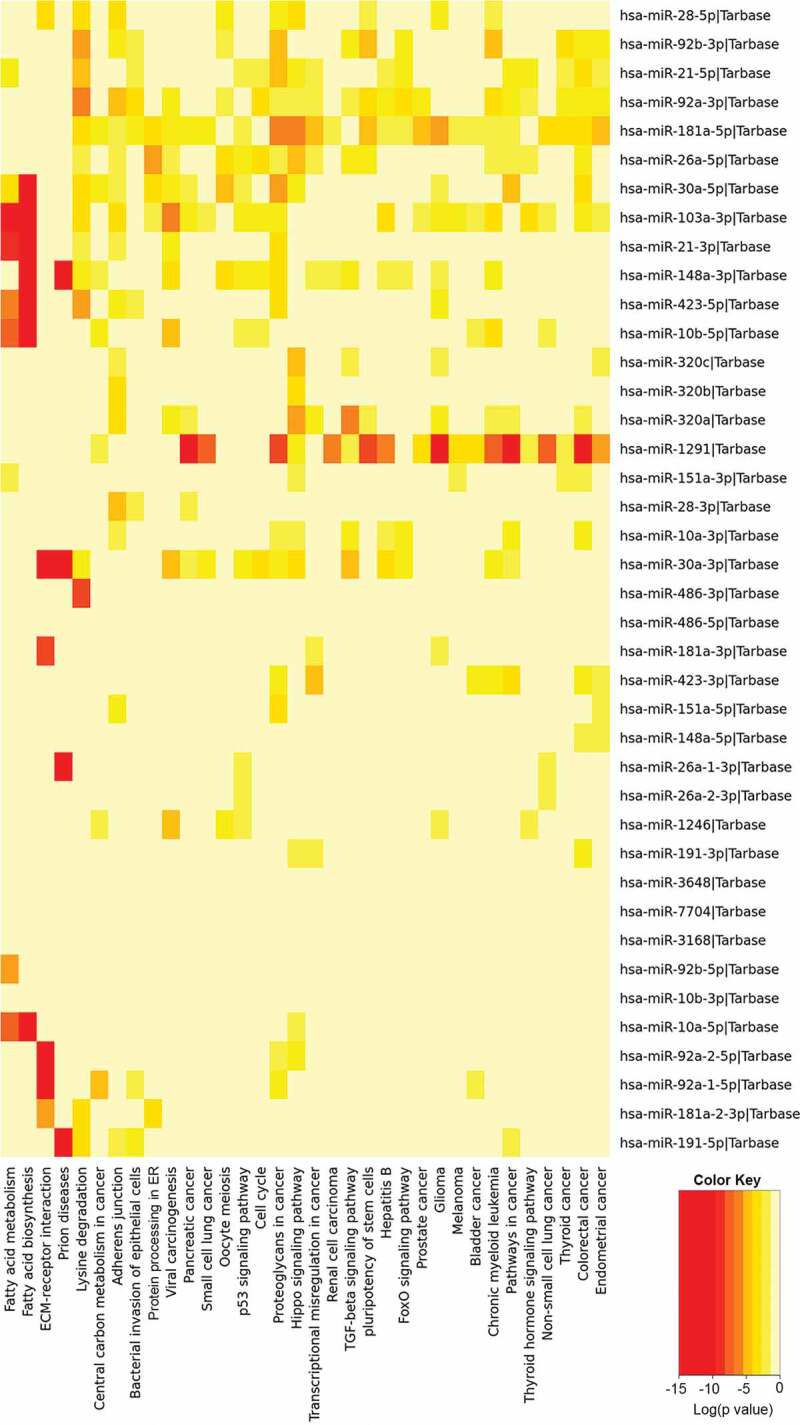
The results of pathway enrichment analysis on the top 40 most abundant miRNAs found in extracellular vesicles (EVs) derived from human neural stem cell-derived extracellular vesicles (hNSC-EVs). The figure shows the heatmap for the top 40 miRNAs in EVs versus KEGG pathways. The heatmap was calculated based on p values and the colour gradient depicts the significance of the interaction between miRNAs and pathways, where red squares mean the lowest p value and light-yellow squares imply a non-significant p value.
Proteomics and Reactome pathway analysis
Proteomic analysis identified a total of 1,086 proteins. Both EV preparations showed highly similar protein intensity values, which indicated the reproducibility of the AEC, and SEC procedures employed in the study for isolation of EVs. Among the proteins identified by mass spectrometry, there were multiple EV markers such as CD63, CD81, ALIX (PDC6I), CD9, TSG-101 (Table 2). The top 100 most abundantproteins found in the proteomic analysis along with associated biological functions and the values of protein intensity for two different EV preparations are listed in Supplementary Table 3. A complete list of identified/quantified proteins is presented in Supplementary Table 4.Pathway analysis was carried out using Reactome (https://reactome.org/); 960 out of 1086 identifiers in the sample were found, where 1478 pathways were hit by at least one of the proteins. The top 25 over-represented pathways from the submitted dataset are shown in Table 3. Many of these pathways are relevant to various molecular processes in the CNS in health and disease.
Table 2.
Common EV-specific proteins identified by mass spectrometry analysis.
| Peptides |
Intensitya |
|||||
|---|---|---|---|---|---|---|
| Name | Acc. No. | MW (kDa) | Identified | Quantifiedb | P1 | P2 |
| ALIX (PDC6I) | Q8WUM4-2 | 96 | 40 | 38 | 1.38E+09 | 1.38E+09 |
| TSG101 | Q99816 | 44 | 12 | 11 | 1.54E+08 | 1.63E+08 |
| CD81 | P60033 | 26 | 3 | 3 | 3.41E+08 | 3.70E+08 |
| CD9 | P21926 | 25 | 3 | 2 | 4.57E+07 | 3.34E+07 |
| CD63 | P08962-2 | 23 | 2 | 2 | 6.30E+07 | 7.38E+07 |
aSummed peptide intensities derived from tandem-MS peptide fragment ions. Only peptides exclusively assigned to a protein were used for determination of the intensity.
bNumber of peptides exclusively assigned to the protein used for quantitative analysis.
Table 3.
25 most relevant pathways over-represented in reactome analysis.
| Entities |
Reactions |
|||
|---|---|---|---|---|
| Pathway name | Found/total | p value | FDR | Found/total |
| Formation of the ternary complex, and subsequently, the 43S complex | 35/52 | 1.11E-16 | 4.11E-15 | 3/3 |
| Translation initiation complex formation | 39/59 | 1.11E-16 | 4.11E-15 | 2/2 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 39/60 | 1.11E-16 | 4.11E-15 | 6/6 |
| Ribosomal scanning and start codon recognition | 38/59 | 1.11E-16 | 4.11E-15 | 2/2 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | 36/56 | 1.11E-16 | 4.11E-15 | 4/4 |
| Peptide chain elongation | 57/90 | 1.11E-16 | 4.11E-15 | 5/5 |
| Eukaryotic Translation Elongation | 59/95 | 1.11E-16 | 4.11E-15 | 9/9 |
| Formation of a pool of free 40S subunits | 63/102 | 1.11E-16 | 4.11E-15 | 2/2 |
| Eukaryotic Translation Termination | 57/94 | 1.11E-16 | 4.11E-15 | 5/5 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 58/96 | 1.11E-16 | 4.11E-15 | 1/1 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | 68/113 | 1.11E-16 | 4.11E-15 | 3/3 |
| Cap-dependent Translation Initiation | 69/120 | 1.11E-16 | 4.11E-15 | 18/18 |
| Viral mRNA Translation | 55/101 | 1.11E-16 | 4.11E-15 | 2/2 |
| Nonsense-Mediated Decay (NMD) | 60/117 | 1.11E-16 | 4.11E-15 | 6/6 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 60/117 | 1.11E-16 | 4.11E-15 | 5/5 |
| SRP-dependent co-translational protein targeting to membrane | 55/113 | 1.11E-16 | 4.11E-15 | 5/5 |
| Neutrophil degranulation | 144/480 | 1.11E-16 | 4.11E-15 | 10/10 |
| Eukaryotic Translation Initiation | 69/120 | 1.11E-16 | 4.11E-15 | 20/21 |
| Membrane Trafficking | 141/635 | 1.11E-16 | 4.11E-15 | 169/218 |
| Vesicle-mediated transport | 177/761 | 1.11E-16 | 4.11E-15 | 191/251 |
| RHO GTPase Effectors | 83/295 | 1.11E-16 | 4.11E-15 | 85/112 |
| ER-Phagosome pathway | 60/153 | 1.11E-16 | 4.11E-15 | 7/10 |
| L13a-mediated translational silencing of Ceruloplasmin expression | 69/112 | 1.11E-16 | 4.11E-15 | 2/3 |
| Antigen processing-Cross presentation | 62/169 | 1.11E-16 | 4.11E-15 | 14/22 |
| Translation | 94/294 | 1.11E-16 | 4.11E-15 | 61/99 |
Out of 1,086 proteins detected in the hiPSC-NSC-EV preparations, 1,059 (97.5%) overlapped with the proteins in the Vesiclepedia database (Figure 5(a)), indicating that the procedures employed for isolation of EVs are reliable. A total of 27 proteins found in hiPSC-NSC-EVs were not reported in the database, which comprised immunoglobulins, Cell cycle and apoptosis regulator protein 2, mTOR-associated protein, Eak-7 homolog (MEAK7) and other proteins. A few proteins found in EVs (e.g., serum albumin and apolipoprotein A-1) are most likely contaminants from the media. Others that are not known to be components of EVs (such as histones and cytoskeletal proteins) may have been secreted from the cells and then subsequently became associated with the EVs during isolation. Several previous studies have validated such protein contaminants in EV preparations [41,42]. However, another study has reported the various histone subunits in EV preparations as exosomal proteins [43].
Figure 5.
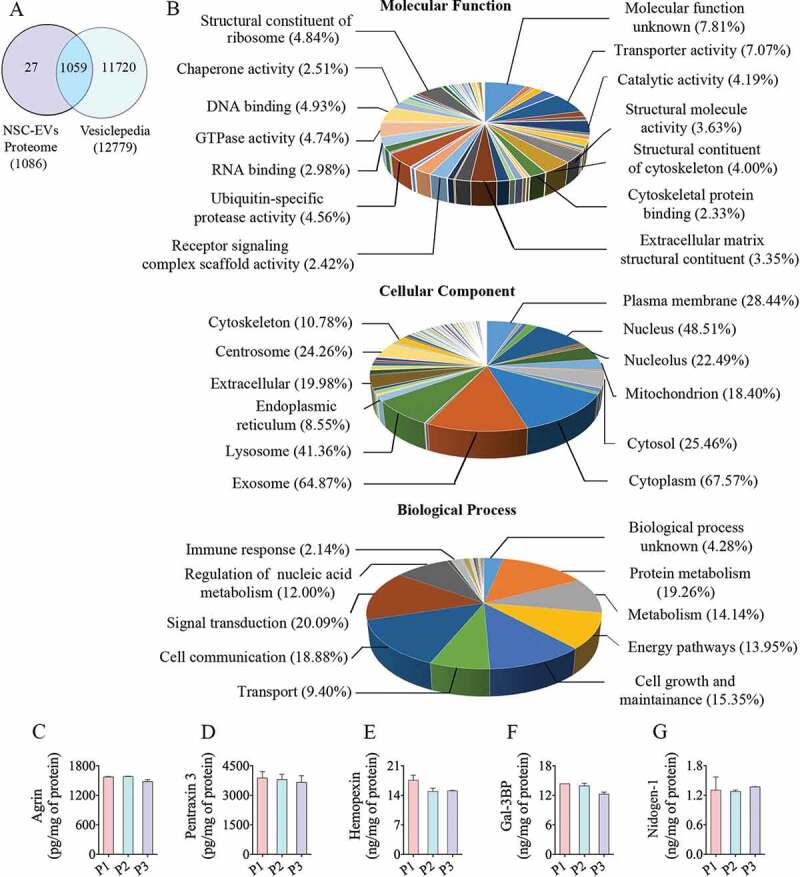
The results of Gene Ontology (GO) analysis of proteins found in extracellular vesicles (EVs) derived from human neural stem cell-derived extracellular vesicles (hNSC-EVs). Figure (a) is a Venn diagram showing that out of 1,086 proteins detected in the hiPSC-NSC-EV preparations, 1,059 overlapped with the proteins in Vesiclepedia database. Figure (b) shows the involvement of EV-associated proteins in molecular, cellular and biological processes. The bar charts in C-G illustrate EV-associated proteins agrin (c), pentraxin 3 (d), hemopexin (e), galectin-3 binding protein (Gal-3BP; f), and nidogen-1 (g), validated through quantitative ELISAs.
GO analysis and validation of proteomic data
The entire proteome dataset was subjected to GO analysis to predict the possible role of these proteins in molecular, cellular and biological processes (Figure 5(b)). The GO classification system revealed that a majority of proteins belonged to exosomes, the cytoplasm, lysosomes, plasma membrane and mitochondria (Figure 5(b)). GO analysis also revealed that these proteins are involved in protein metabolism, nucleic acid metabolism and other metabolic processes. Besides, these proteins are also involved in other biological processes such as signal transduction, cell communication, cell growth and maintenance, energy metabolism and transport (Figure 5(b)). The categorized protein dataset based on molecular functions revealed that these are involved in transport activity, catalytic activity, ubiquitin-specific protease activity, GTPase activity, DNA binding function, oxidoreductase activity, transcription regulation activity and calcium ion binding function (Figure 5(b)). Among the 100 most abundant proteins from proteomics data, we validated the presence of five proteins (agrin, PTX3, hemopexin, Gal-3BP and nidogen-1) that are known to participate in various brain functions and recognized to be useful for treating neurodegenerative disorders. All of these proteins showed nearly comparable concentrations in three preparations of hiPSC-NSC-EVs (Figure 5(c-g)), further confirming the reproducibility of the EV isolation procedure.
hiPSC-NSC-derived EVs suppressed the upregulation of IL-6 in LPS stimulated macrophages in a dose-dependent manner
LPS exposure alone greatly enhanced the release of IL-6 from macrophages (Figure 6(a)). When macrophages were simultaneously treated with LPS and DEX (a potent anti-inflammatory corticosteroid), IL-6 release was significantly inhibited. The addition of NSC-EVs to LPS treated macrophage cultures resulted in a dose-dependent anti-inflammatory effect. Significant suppression of IL-6 release was observed with doses of 4 × 109 EVs or higher. Maximal suppression was observed with a treatment of 64 × 109 EVs (Figure 6(a)). Dose-dependent suppression of IL-6 release revealed that hiPSC-NSC derived EVs mediate anti-inflammatory activity.
Figure 6.
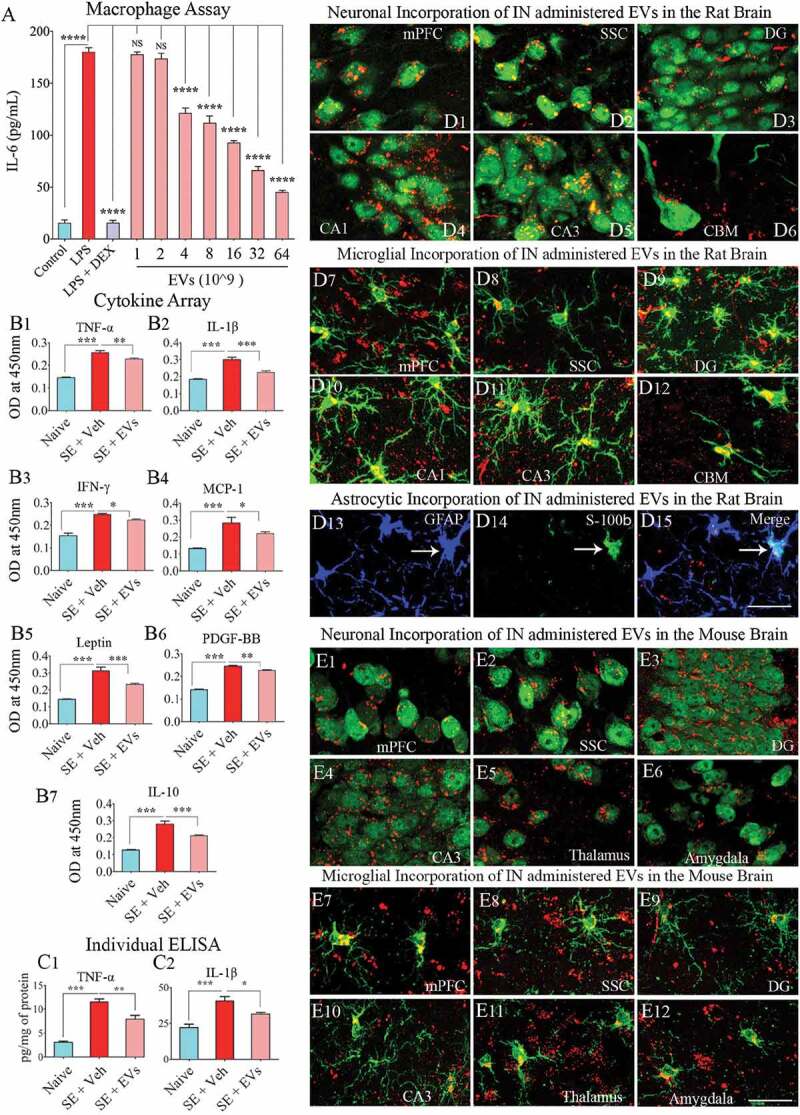
Extracellular vesicles (EVs) derived from human neural stem cells (hNSCs) display anti-inflammatory properties, and pervasively permeate the brain following an intranasal administration. Figure A shows data from the macrophage assay. LPS, lipopolysaccharide; DEX, dexamethasone; EVs, extracellular vesicles. Note that the addition of hNSC-EVs to LPS stimulated macrophage cultures resulted in a dose-dependent suppression of interleukin-6 (IL-6) release by macrophages, implying an anti-inflammatory effect. The effect was significant when 4 × 109 or more EVs were added to macrophage + LPS cultures (a). ****, p < 0.0001. Figures (b1–b6) show that an intranasal (IN) administration of hNSC-EVs after 2 hours of acute seizure activity (a mouse model of status epilepticus) is adequate for reducing the concentration of multiple cytokines in the hippocampus. The cytokines that showed significant reductions following hNSC-EV treatment include the tumour necrosis factor-alpha (TNF-α; B1), interleukin-1 beta (IL-1β; B2), interferon-gamma (IFN-γ; b3), monocyte chemoattractant protein −1 (MCP-1; b4), leptin (b5) and the platelet-derived growth factor (PDGF-BB; b6). hNSC-EV treatment also normalized the anti-inflammatory protein IL-10 after status epilepticus (b7). Figures c1 and c2 show that normalization of TNF-α and IL-1β found through cytokine array following EV treatment could also be confirmed with individual ELISA. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant. Figures d1-d15 illustrate the incorporation of PKH-26 labelled hNSC-EVs (red dots) by different cells in various regions of the rat brain at 6 hours after an IN administration. The figures illustrate NeuN+ neurons (d1-d5), calbindin+ Purkinje neuron (d6), IBA-1+ microglia (d7-d12) and astrocytes positive for GFAP (d13), S-100β (d14) or both GFAP and S-100β (d15). Figures e1-e12 illustrate the incorporation of PKH-26 labelled hNSC-EVs (red dots) by NeuN+ neurons (e1-e6) and IBA-1+ microglia (e7-e12) in various regions of the mouse brain at 6 hours after an IN administration. mPFC, medial prefrontal cortex (d1, d7, e1, e7); SSC, somatosensory cortex (d2, d8, e2, e8); DG, dentate gyrus (d3, d9, e3, e9); CA1 subfield (d4, d10); CA3 subfield (d5, d11, e4, e10); CBM, cerebellum (d6, d12); thalamus (e5, e11); and amygdala (e6, e12). Scale bar: 25 μm.
IN administration of hiPSC-NSC-derived EVs reduced status epilepticus-induced elevation of proinflammatory cytokines in the hippocampus
To further investigate the anti-inflammatory activity of hiPSC-NSCs derived EVs, we quantified the effects of their IN administration at 2 hours post-SE on the levels of proinflammatory cytokines at 24 hours post-SE. Cytokine/chemokine array results showed that induction of acute seizure activity greatly enhanced the concentration of multiple proinflammatory cytokines. Administration of NSC-EVs normalized the concentration of tumour necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β), interferon-gamma (IFN-γ), leptin, monocyte chemoattractant protein-1 (MCP-1 or CCL2), IL10 and platelet-derived growth factor – two B subunits (PDGF-BB) (Figure 6(b1-b7)). Among these cytokines, TNF-α and IL-1β were further validated through individual ELISA, as they are the primary mediators of acute and chronic inflammation after SE. The concentration of TNF-α and IL-1β were significantly elevated in SE animals receiving vehicle but reduced significantly in SE animals receiving NSC-EVs (Figure 6(c1-c2)). Thus, the anti-inflammatory activity of NSC-EVs observed in an in vitro macrophage assay could be replicated in an in vivo SE model.
IN administration of NSC-EVs resulted in their rapid incorporation by neurons, microglia astrocytes in all regions of the brain
We examined the ability of intranasally administered hiPSC-NSC derived EVs to rapidly get incorporated into the soma of neurons and glia in different regions of the rat and mouse brain. Examination of brain tissue sections using immunofluorescent markers of neurons, microglia and astrocytes through confocal microscopy at 6 hours after IN administration revealed the entry of EVs into virtually all regions of the forebrain, midbrain and hindbrain. This finding agrees with our previous study on mesenchymal stem cell-derived EVs [22]. PKH26+ EVs could be localized inside virtually all neurons and microglia in all regions of the brain examined. Significant fractions of astrocytes also incorporated EVs. Examples of neurons incorporating EVs in the rat medial prefrontal cortex (mPFC), the somatosensory cortex (SSC), the dentate gyrus (DG), and CA1 and CA3 subfields of the hippocampus, and the cerebellum are illustrated (Figure 6(d1-d6)). Likewise, microglia in all regions of the brain incorporated a more substantial amount of EVs (Figure 6(d7-d12)). Examples of mature astrocytes expressing GFAP and S-100β incorporating EVs inside their soma are also illustrated in Figure 6(d13-d15). A similar distribution of EVs was found in the adult mouse brain following an IN administration. Examples of neurons and microglia incorporating EVs in brain regions such as the mPFC, SSC, DG, CA3 subfield, thalamus and amygdala are illustrated in Figure 6(e1-e12). Thus, IN administration of NSC-EVs is an efficient approach to target EVs into neurons and glia in virtually all regions of the rat and mouse brain.
IN administration of hiPSC-NSC-derived EVs into adult rats enhanced hippocampal neurogenesis
We quantified neurogenesis in the hippocampus 13 days after an IN administration of hiPSC-NSC derived EVs in 6 months old F344 rats using BrdU labelling. Administration of EVs increased neurogenesis in the SGZ of the hippocampus, which was evident from an increased number of BrdU+ newly born cells and net hippocampal neurogenesis quantified from BrdU+ cells and the percentage of BrdU+ cells expressing DCX (Figure 7(a1-a10)). Next, we determined whether increased neurogenesis in EV-treated animals is linked to increased proliferation of NSCs through stereological counting of Ki-67+ proliferating cells (putative NSCs) in the SGZ. Indeed, animals receiving EVs contained a higher density of Ki-67+ cells in the SGZ (Figure 7(b1-b4)). In addition, these animals displayed more significant numbers of Ki-67+ clusters and a higher number of cells per cluster in the SGZ (Figure 7(b5-b6)). Thus, hiPSC-NSC derived EVs increased neurogenesis through increased proliferation of NSCs. We also quantified the number of newly born neurons positive for DCX (a marker of newly born neurons), which revealed no differences between naïve and EV-treated animals (Figure 7(c1-c4,c9)). This is not surprising because doublecortin expression of newly born neurons in mice lasts for up to 4 weeks, and the total number of DCX+ neurons represent all neurons that were born during the 4 weeks prior to euthanasia [44]. However, the examination of clusters of immature DCX+ neurons (presumably neurons born after EV administration) revealed an increased number of clusters as well as higher numbers of DCX+ neurons per cluster in animals receiving EVs (Figure 7(c5-c8,c10-c11)), implying that EV treatment-induced localized increase in the proliferation of NSCs leading to clusters of neuroblasts.
Figure 7.
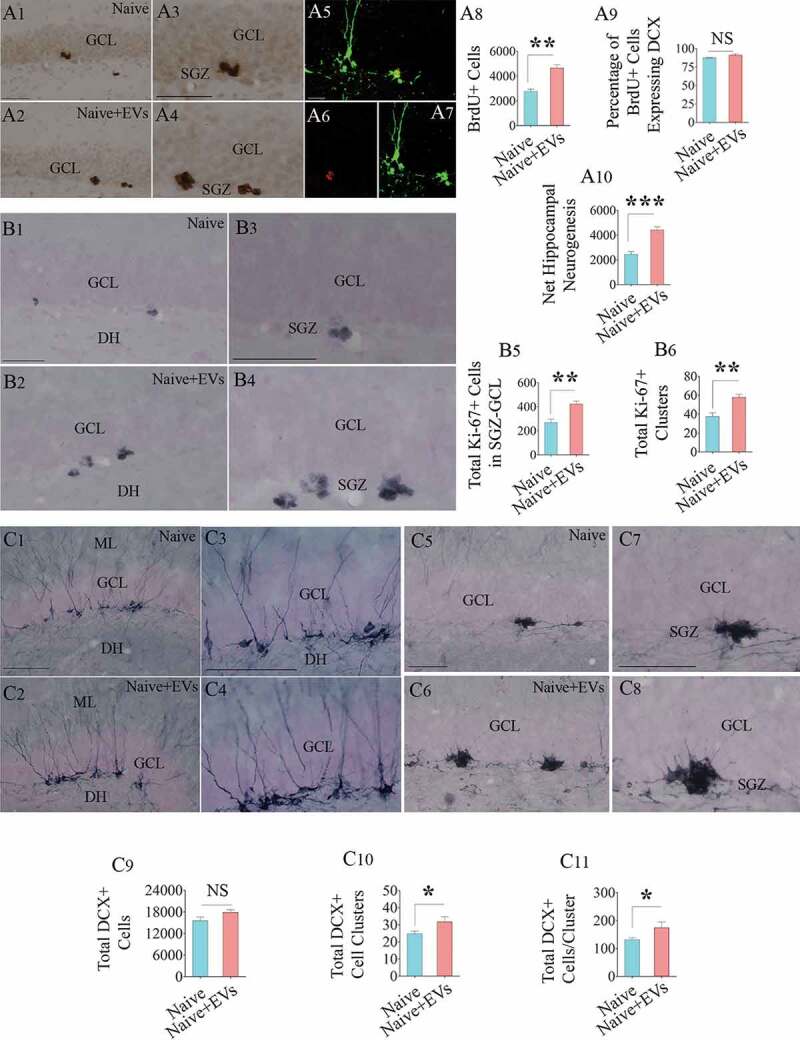
Intranasal administration of extracellular vesicles (EVs) derived from human neural stem cells (hNSCs) into adult rats increased the production of newly born neurons in the hippocampus. Figures a1-a4 show the distribution of 5ʹ-bromodeoxyuridine-positive (BrdU+) newly born cells from a naïve rat (a1) and a rat that received hNSC-EVs (a2). Figures a3 and a4 are magnified views from a1 and a2. Figures a5-a7 show examples of newly born neurons that express both BrdU (red, A5-A6) and doublecortin (DCX, green in A5 and A7). The bar charts in a8-a10 show that EV administration to adult rats increased the number of newly born cells (a8), had no effects on neuronal differentiation of newly born cells (a9) and enhanced net hippocampal neurogenesis (a10). Figures b1-b4 show the distribution of Ki-67+cells (i.e., proliferating cells) in the subgranular zone (SGZ) of a naïve rat (b1) and a rat that received hNSC-EVs (b2). Figures b3 and b4 are magnified views from b1 and b2. The bar charts in b5 and b6 show that EV administration to adult rats increased the number of proliferating cells in the SGZ (b5) with a higher number of Ki-67+ cell clusters (b6). Figures c1-c4 show the distribution of DCX+ neurons with vertical dendrites (relatively mature, newly born neurons) in the SGZ of a naïve rat (c1) and a rat that received hNSC-EVs (c2). Figures c3 and c4 are magnified views from c1 and c2. Figures c5-c8 show the distribution of clusters of DCX+ neurons with short dendrites (relatively immature, newly born neurons) in the SGZ of a naïve rat (c5) and a rat that received hNSC-EVs (c6). Figures c7 and c8 are magnified views from c5 and c6. The bar charts in c9-c11 show that EV administration to adult rats did not increase the number of total DCX+ neurons (c9) but increased the number of DCX+ immature neuron clusters (c10) and the number of DCX+ neurons/cluster (c11). *p < 0.05; **p < 0.01, ***p < 0.001; NS, not significant. Scale bar, a1-a4, b1-b4 and c1-c8 = 100 μm; a5-a7, 25 μm.
Discussion
Our study provides several novel findings. These include the development of a reproducible protocol that facilitates isolation of similar EVs from hiPSC-NSC cultures from batch to batch, and essential information on the miRNA and protein composition of EVs shed from hiPSC-NSCs. Besides, we demonstrated the ability of such EVs toquickly target a vast majority of neurons and microglia and some astrocytes in virtually all regions of the adult brain following an IN administration and mediate anti-inflammatory effects in the injured brain and enhanced neurogenesis in the intact adult brain. We employed a combination of AEC and SEC to obtain preparations of hiPSC-NSC-EVs with a consistent occurrence of 179 miRNAs and 1,086 proteins. The replicability of the methodology was evident from small RNA sequencing and proteomic studies showing negligible amounts of differentially expressed small RNAs and proteins in different EV preparations. Furthermore, ELISA, WB and proteomics studies confirmed that the EVs isolated through a combination of AEC and SEC consistently expressed many EV markers, which include CD63, CD81, CD9, ALIX and TSG-101. The preparations were also devoid of deep cellular proteins. The TEM analysis further confirmed that the procedure does not interfere with the morphological integrity of EVs.
The enrichment pathway analysis on the top 40 most abundant miRNAs demonstrated that the EVs from hiPSC-NSCs carry a cargo of miRNAs that are important for multiple cell signalling pathways. The pathways comprised fatty acid biosynthesis and metabolism, extracellular matrix-receptor interaction, adherens junction, protein processing in the endoplasmic reticulum, p53 signalling, cell cycle, hippo signalling, transforming growth factor-beta (TGF-β) signalling, the pluripotency of stem cells and FoxO and thyroid hormone signalling. The analysis also revealed that some miRNAs found in hiPSC-NSC-EVs display altered expression in conditions such as prion diseases, glioma and a variety of cancers. However, the presence of such miRNAs does not necessarily mean that they are involved in inducing prion disease or cancer.For example, miRNAs highly dysregulated in prion disease are not among the most abundant miRNAs present in hiPSC-NSC-EVs [45]. Furthermore, the manifestation of some miRNAs having a role as oncogenes or tumour suppressors in cancer in hiPSC-NSC-EVs is not particularly surprising, as there are some similarities between NSCs and cancer stem cells, which include their capacity for self-renewal, unlimited proliferation, extensive migration and generating progenitor or differentiated cells. Therefore, common miRNAs in EVs from NSCs and cancer stem cells may reflect their role in targeting genes and pathways supporting stemness properties [46].
We validated the occurrence of eight miRNAs in hiPSC-NSC-EVs. The known functions of these miRNAs (miRs-320a, 320b, 103a-3p, 21–5p, 26a-5p, 30a-3p, 181a-5p, 191–5p) and their potential application as therapeutics are briefly detailed here. The miRNAs, miR-320a and 320b, are involved in regulating the insulin-like growth factor-1 receptor, TGF-β signalling and neuron related target genes and acts as a tumour suppressor and metastasis inhibitor [47–51]. The miRNA-103a-3p plays a vital role in brain development and function through the regulation of p35 and cyclin-dependent kinase 5 (CDK5) pathway [52]. While miR-103a is downregulated in AD, it has been shown to reduce apoptosis and increase total neurite outgrowth in cellular AD models via inhibition of prostaglandin-endoperoxide synthase 2 (PTGS2) [52,53]. PTGS2 is involved in neuroinflammation and production of Aβ in AD [54,55], which alludes to the utility of miR-103a in treating AD. Dysregulation of miR21 has been linked to age-related cognitive impairment [56], and its activity promotes phosphatase and tensin homolog deleted on chromosome 10 (PTEN)-AKT (protein kinase B) signalling pathway, which promotes cell survival via PI3K/AKT pathway [57]. miR-21 has been shown to have neuroprotective and anti-inflammatory properties in models of traumatic brain injury (TBI) and stroke [58–60]. miR-21 mediates anti-inflammatory activity through the downregulation of NF-kB and TNF-α, and induction of the anti-inflammatory cytokine IL-10 [61–63].
The miRNA-26a-5p promotes angiogenesis from human brain microvascular endothelial cells [64], long-term potentiation and neurite growth by repressing PTEN [65,66]. Importantly, inhibition of miR-26a-5p in vivo caused synucleinopathy and loss of dopaminergic neurons, whereas its overexpression alleviated behavioural abnormalities in a mouse prototype of Parkinson’s disease (PD) [67].The miRNA-30a acts as a tumour suppressor in glioma [68], and vital for ubiquitin-proteasome mediated proteolysis (UPP), which has significance in AD and dementia [69]. In addition, overexpression of miR-30a results in the upregulation of the FOXO signalling pathway [70], which can mediate beneficial effects through the upregulation of the longevity protein silent mating type information regulation 2 homolog 1 (SIRT1) [71]. The miRNA-181-5p plays a role in hippocampus-dependent memory formation via inhibition of protein kinase AMP-activated catalytic subunit alpha 1 (PRKAA1), and overexpression of miR-181 enhances memory formation [72]. Also, the miR-181 family plays a significant role in mitochondrial function and homoeostasis [73]. However, inhibition of miR-181 was found to be beneficial for improving memory function in an AD model [74], and decreasing brain infarction is a prototype of stroke [75]. The miRNA-191-5p has a role in the maintenance of dendritic spines in neurons [76]. Thus, based on the known functions of validated miRNAs, hiPSC-NSC-EVs appear useful for treating different neurodegenerative conditions. However, as miRNAs function depends on the context or the type of disease, gain-of-function and loss-of-function studies on specific miRNAs in EVs using disease models are needed in the future for comprehending the net effect of EV-based therapies.
Nearly 98% of proteins we found in EVs through proteomics overlapped with the proteins in the Vesiclepedia database. Reactome analysis of proteomics data on EVs identified the presence of proteins involved in multiple cellular pathways relevant to health and disease. The known CNS-related functions of the top 100 proteins in these EVs are listed in Supplementary Table 3. GO analysis revealed that the majority of proteins found in EVs were components of EVs, plasma membrane, cytoplasm and lysosomes, and involved in energy metabolism, mitochondrial metabolism, signal transduction and several other biological processes. We also validated five proteins from the most abundant 100 proteins, including agrin, PTX3, hemopexin, Gal-3BP and nidogen-1. Agrin, widely expressed in neurons and microvascular basal lamina in the CNS, has a role in synaptogenesis [77,78], synaptic plasticity through phosphorylation of cAMP response element-binding protein (CREB) and facilitation of long-term potentiation (LTP) [79], reactive synaptogenesis after TBI [80], hippocampal neurogenesis [21] and blood-brain barrier integrity [81]. PTX3, on the other hand, is efficient for reducing the migration of neutrophils after brain injury [82], promoting neurogenesis, angiogenesis, BBB integrity and functional recovery after stroke [83,84]. PTX3 can also act as the first line of defence against pathogens [85], protect neurons against seizures [86], regulate complement activation during infection and inflammation [87,88]. Hemopexin has been shown to protect neurons against haem and reactive oxygen species (ROS) toxicity through haem oxygenase activity [89]. Hemopexin promotes new blood vessel formation, BBB integrity and synaptic plasticity and can provide neuroprotection in mouse models of stroke and intracerebral haemorrhage [90–92]. Hemopexin can also influence the transformation of the proinflammatory M1 microglia into an anti-inflammatory M2 microglia [93]. The protein Gal-3BP is associated with the immune system [94], as it functions as a suppressor of inflammatory responses by blocking the NF-kB signalling pathway. Remarkably, Gal-3BP can suppress Aβ production through inhibition of the amyloid precursor protein (APP) processing by β-secretase [95]. Thus, the delivery of Gal-3BP into neurons and microglia in the brain through IN administration of EVs might be beneficial for restraining neuroinflammation and reducing amyloid-beta (Aβ) plaques in AD. The other validated protein nidogen-1, a component of basement membranes, promotes cell adhesion and movement, BBB formation, network excitability and plasticity in the brain [96,97]. Disruption of nidogen-1 leads to seizure-like symptoms and loss of muscle control [98].
Furthermore, IN administration of EVs in this study resulted in their rapid incorporation by virtually all neurons and microglia, and some astrocytes in all regions of the rat and mouse brains examined in this study. These results are consistent with the findings observed with MSC-derived EVs recently [22]. Such rapid incorporation of EVs in neural cells within deep brain areas likely reflects the speedy entry of EVs into the subarachnoid space via perineurial spaces around olfactory nerves passing through the cribriform plate and the subsequent transportation through CSF flow into the interstitial fluid in the interstitial spaces of the brain [99]. The anti-inflammatory effects of hiPSC-NSCs-EVs could be determined from both in vitro and in vivo prototypes of inflammation. In the macrophage assay, EVs effectively inhibited the LPS-induced elevation of IL-6 by macrophages in a dose-dependent manner. Whereas, in a model of SE, EVs significantly inhibited the elevation of multiple proinflammatory cytokines, including TNF-α and IL-1β, the primary mediators of acute and chronic neuroinflammation after SE [100]. Such effects of EVs may be linked to specific miRNAs (e.g., miR-21-5p, miR-103a) and proteins (e.g., PTX3, hemopexin, Gal-3BP) in EVs having anti-inflammatory and antioxidant activities. The neurogenic effects of EVs were evident from their ability to increase hippocampal neurogenesis through the increased proliferation of NSCs in six-months-old rats. Enhanced neurogenesis following a dose of hiPSC-NSC EVs in the adult brain might be reflecting the activity of proteins in EVs promoting neurogenesis, including the migration and differentiation of newly born cells (e.g., agrin, PTX3).
In summary, this study presents an efficient protocol for isolating EVs from hiPSC-NSC cultures. The EVs isolated from different batches of cultures were comparable in terms of their miRNA and protein composition. Many miRNAs and proteins in EVs have neuroprotective, anti-apoptotic, antioxidant, anti-inflammatory, BBB repairing, neurogenic and Aβ reducing activities. Besides, some of them are known to promote synaptogenesis, synaptic plasticity and improve cognitive function. The study has also confirmed the anti-inflammatory and neurogenic properties of these EVs. All of these properties seem beneficial for the application of hiPSC-NSC-EVs in treating multiple neurodegenerative neuroinflammatory disorders.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institute of Neurological Disorders and Stroke (1R01NS106907-01 to A.K.S) and the State of Texas (Emerging Technology Fund to A.K.S.). DLGG was supportedby the Research Productivity Scholarship Program in Brazilian National Council for Scientific and Technological Development (CNPq). Gabriele Zanirati was supported by a doctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Governmentof Brazil. Mass spectrometry analyses were conducted in the Institutional Mass Spectrometry Laboratory of the University of Texas Health Science Center at San Antonio. The expert technical assistance of Sammy Pardo and Dana Molleur is greatly appreciated. Fund for purchase of the Lumos mass spectrometer were provided by the University of Texas System Proteomics Core Network (to S.T.W.).
Disclosure of interest
The authors report no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Hattiangady B, Shetty AK.. Neural stem cell grafting counteracts hippocampal injury-mediated impairments in mood, memory, and neurogenesis. Stem Cells Transl Med. 2012;1(9):696–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shetty AK. Hippocampal injury-induced cognitive and mood dysfunction, altered neurogenesis, and epilepsy: can early neural stem cell grafting intervention provide protection? Epilepsy Behav. 2014;38:117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen KS, Sakowski SA, Feldman EL. Intraspinal stem cell transplantation for amyotrophic lateral sclerosis. Ann Neurol. 2016;79:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Genc B, Bozan HR, Genc S, et al. Stem cell therapy for multiple sclerosis. Adv Exp Med Biol. 2019;1084:145–174. [DOI] [PubMed] [Google Scholar]
- [5].Boese AC, Hamblin MH, Lee JP. Neural stem cell therapy for neurovascular injury in Alzheimer’s disease. Exp Neurol. 2020;324:113112. [DOI] [PubMed] [Google Scholar]
- [6].Shetty AK. Neural stem cells in health and disease In: Shetty AK, editor. Neural stem cells in health and disease. Singapore: World Scientific Publishing Company; 2015. p. 1–20. [Google Scholar]
- [7].Eckert A, Huang L, Gonzalez R, et al. Bystander effect fuels human induced pluripotent stem cell-derived neural stem cells to quickly attenuate early stage neurological deficits after stroke. Stem Cells Transl Med. 2015;4:841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tsuji O, Sugai K, Yamaguchi R, et al Concise review: laying the groundwork for a first-in-human study of an induced pluripotent stem cell-based intervention for spinal cord injury. Stem Cells. 2019;37:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Willis CM, Nicaise AM, Peruzzotti-Jametti L, et al. The neural stem cell secretome and its role in brain repair. Brain Res. 2020;1729:146615. [DOI] [PubMed] [Google Scholar]
- [10].Martin U. Therapeutic application of pluripotent stem cells: challenges and risks. Front Med (Lausanne). 2017;4:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Attwood SW, Edel MJ. iPS-cell technology and the problem of genetic instability-can it ever be safe for clinical use? J Clin Med. 2019;8:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yoshihara M, Oguchi A, Murakawa Y. Genomic instability of iPSCs and challenges in their clinical applications. Adv Exp Med Biol. 2019;1201:23–47. [DOI] [PubMed] [Google Scholar]
- [13].Tucker BA, Mullins RF, Stone EM. Autologous cell replacement: a noninvasive AI approach to clinical release testing. J Clin Invest. 2020;130:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vogel A, Upadhya R, Shetty AK. Neural stem cell derived extracellular vesicles: attributes and prospects for treating neurodegenerative disorders. EBioMedicine. 2018;38:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wiklander OPB, Brennan MA, Lotvall J, et al. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med. 2019;11:eaav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Elahi FM, Farwell DG, Nolta JA, et al. Preclinical translation of exosomes derived from mesenchymal stem/stromal cells. Stem Cells. 2020;38:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim DK, Nishida H, An SY, et al. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc Natl Acad Sci U S A. 2016;113:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Long Q, Upadhya D, Hattiangady B, et al Intranasal MSC-derived A1-exosomes ease inflammation, and prevent abnormal neurogenesis and memory dysfunction after status epilepticus. Proc Natl Acad Sci U S A. 2017;114:E3536–E3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lombardi M, Parolisi R, Scaroni F, et al Detrimental and protective action of microglial extracellular vesicles on myelin lesions: astrocyte involvement in remyelination failure. Acta Neuropathol. 2019;138:987–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun MK, Passaro AP, Latchoumane CF, et al Extracellular vesicles mediate neuroprotection and functional recovery after traumatic brain injury. J Neurotrauma. 2019;37:1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang ZG, Buller B, Chopp M. Exosomes - beyond stem cells for restorative therapy in stroke and neurological injury. Nat Rev Neurol. 2019;15:193–203. [DOI] [PubMed] [Google Scholar]
- [22].Kodali M, Castro OW, Kim DK, et al Intranasally administered human MSC-derived extracellular vesicles pervasively incorporate into neurons and microglia in both intact and status epilepticus injured forebrain. Int J Mol Sci. 2019;21:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yan Y, Shin S, Jha BS, et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med. 2013;2:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Madhu LN, Attaluri S, Kodali M, et al Neuroinflammation in Gulf War Illness is linked with HMGB1 and complement activation, which can be discerned from brain-derived extracellular vesicles in the blood. Brain Behav Immun. 2019;81:430–443. [DOI] [PubMed] [Google Scholar]
- [25].Gitai DLG, Dos Santos YDR, Upadhya R, et al. Extracellular vesicles in the forebrain display reduced miR-346 and miR-331-3p in a rat model of chronic temporal lobe epilepsy. Mol Neurobiol. 2019;57:1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nakamura N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J Pharmacol Sci. 2010;112:255–264. [DOI] [PubMed] [Google Scholar]
- [27].Visnovitz T, Osteikoetxea X, Sodar BW, et al An improved 96 well plate format lipid quantification assay for standardisation of experiments with extracellular vesicles. J Extracell Vesicles. 2019;8:1565263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vlachos IS, Zagganas K, Paraskevopoulou MD, et al DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Searle BC, Pino LK, Egertson JD, et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun. 2018;9:5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rosenberger G, Koh CC, Guo T, et al A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fabregat A, Sidiropoulos K, Viteri G, et al Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jassal B, Matthews L, Viteri G, et al The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pathan M, Keerthikumar S, Ang C, et al FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. [DOI] [PubMed] [Google Scholar]
- [34].Bartosh TJ, Ylostalo JH. Macrophage inflammatory assay. Biol Protoc. 2014;4:e1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cunningham M, Cho JH, Leung A, et al hPSC-derived maturing GABAergic interneurons ameliorate seizures and abnormal behavior in epileptic mice. Cell Stem Cell. 2014;15:559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hattiangady B, Kuruba R, Shetty AK. Acute seizures in old age leads to a greater loss of CA1 pyramidal neurons, an increased propensity for developing chronic TLE and a severe cognitive dysfunction. Aging Dis. 2011;2:1–17. [PMC free article] [PubMed] [Google Scholar]
- [37].Shetty AK, Turner DA. Fetal hippocampal grafts containing CA3 cells restore host hippocampal glutamate decarboxylase-positive interneuron numbers in a rat model of temporal lobe epilepsy. J Neurosci. 2000;20:8788–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rao MS, Hattiangady B, Shetty AK. The window and mechanisms of major age-related decline in the production of new neurons within the dentate gyrus of the hippocampus. Aging Cell. 2006;5:545–558. [DOI] [PubMed] [Google Scholar]
- [39].Hattiangady B, Shetty AK. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol Aging. 2008;29:129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rao MS, Hattiangady B, Reddy DS, et al. Hippocampal neurodegeneration, spontaneous seizures, and mossy fiber sprouting in the F344 rat model of temporal lobe epilepsy. J Neurosci Res. 2006;83:1088–1105. [DOI] [PubMed] [Google Scholar]
- [41].Jeppesen DK, Fenix AM, Franklin JL, et al Reassessment of exosome composition. Cell. 2019;177:428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Thery C, Witwer KW, Aikawa E, et al Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Neerukonda SN, Tavlarides-Hontz P, McCarthy F, et al. Comparison of the transcriptomes and proteomes of serum exosomes from Marek’s disease virus-vaccinated and protected and lymphoma-bearing chickens. Genes (Basel). 2019;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kempermann G, Gast D, Kronenberg G, et al. Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development. 2003;130:391–399. [DOI] [PubMed] [Google Scholar]
- [45].Kanata E, Thüne K, Xanthopoulos K, et al MicroRNA alterations in the brain and body fluids of humans and animal prion disease models: current status and perspectives. Front Aging Neurosci. 2018;10:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Macharia LW, Wanjiru CM, Mureithi MW, et al. MicroRNAs, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front Genet. 2019;10:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guo T, Feng Y, Liu Q, et al MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumour Biol. 2014;35:11269–11275. [DOI] [PubMed] [Google Scholar]
- [48].Sun JY, Xiao WZ, Wang F, et al MicroRNA-320 inhibits cell proliferation in glioma by targeting E2F1. Mol Med Rep. 2015;12:2355–2359. [DOI] [PubMed] [Google Scholar]
- [49].Xiong W, Ran J, Jiang R, et al. miRNA-320a inhibits glioma cell invasion and migration by directly targeting aquaporin 4. Oncol Rep. 2018;39:1939–1947. [DOI] [PubMed] [Google Scholar]
- [50].Somel M, Liu X, Tang L, et al MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lv QL, Du H, Liu YL, et al Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol Rep. 2017;38:959–966. [DOI] [PubMed] [Google Scholar]
- [52].Yang H, Wang H, Shu Y, et al. miR-103 promotes neurite outgrowth and suppresses cells apoptosis by targeting prostaglandin-endoperoxide synthase 2 in cellular models of Alzheimer’s disease. Front Cell Neurosci. 2018;12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huynh RA, Mohan C. Alzheimer’s disease: biomarkers in the genome, blood, and cerebrospinal fluid. Front Neurol. 2017;8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang P, Guan PP, Wang T, et al. Aggravation of Alzheimer’s disease due to the COX-2-mediated reciprocal regulation of IL-1β and Aβ between glial and neuron cells. Aging Cell. 2014. August;13(4):605–615. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [55].Sawikr Y, Yarla NS, Peluso I, et al. Neuroinflammation in Alzheimer’s disease: the preventive and therapeutic potential of polyphenolic nutraceuticals. Adv Protein Chem Struct Biol. 2017;108:33–57. [DOI] [PubMed] [Google Scholar]
- [56].Sessa F, Maglietta F, Bertozzi G, et al Human brain injury and miRNAs: an experimental study. Int J Mol Sci. 2019;20:1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kriplani N, Hermida MA, Brown ER, et al. Class I PI 3-kinases: function and evolution. Adv Biol Regul. 2015;59:53–64. [DOI] [PubMed] [Google Scholar]
- [58].Han Z, Chen F, Ge X, et al. miR-21 alleviated apoptosis of cortical neurons through promoting PTEN-Akt signaling pathway in vitro after experimental traumatic brain injury. Brain Res. 2014;1582:12–20. [DOI] [PubMed] [Google Scholar]
- [59].Buller B, Liu X, Wang X, et al MicroRNA-21 protects neurons from ischemic death. Febs J. 2010;277:4299–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gaudet AD, Fonken LK, Watkins LR, et al. MicroRNAs: roles in regulating neuroinflammation. Neuroscientist. 2018;24:221–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Slota JA, Booth SA. MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Noncoding RNA. 2019;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. [DOI] [PubMed] [Google Scholar]
- [63].Barnett RE, Conklin DJ, Ryan L, et al Anti-inflammatory effects of miR-21 in the macrophage response to peritonitis. J Leukoc Biol. 2016;99:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang ZF, Liao F, Wu H, et al. Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. J Exp Clin Cancer Res. 2019;38:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gu QH, Yu D, Hu Z, et al miR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat Commun. 2015;6:6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li B, Sun H. MiR-26a promotes neurite outgrowth by repressing PTEN expression. Mol Med Rep. 2013;8:676–680. [DOI] [PubMed] [Google Scholar]
- [67].Su Y, Deng MF, Xiong W, et al. MicroRNA-26a/death-associated protein kinase 1 signaling induces synucleinopathy and dopaminergic neuron degeneration in Parkinson’s disease. Biol Psychiatry. 2019;85:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang Y, Wu Z, Li L, et al. miR-30a inhibits glioma progression and stem cell-like properties by repression of Wnt5a. Oncol Rep. 2017;38:1156–1162. [DOI] [PubMed] [Google Scholar]
- [69].Hegde AN, Smith SG, Duke LM, et al. Perturbations of ubiquitin-proteasome-mediated proteolysis in aging and Alzheimer’s disease. Front Aging Neurosci. 2019;11:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang T, Chen G, Ma X, et al MiR-30a regulates cancer cell response to chemotherapy through SNAI1/IRS1/AKT pathway. Cell Death Dis. 2019;10:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xiong S, Salazar G, Patrushev N, et al. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J Biol Chem. 2011;286:5289–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang SF, Chen JC, Zhang J, et al. miR-181a involves in the hippocampus-dependent memory formation via targeting PRKAA1. Sci Rep. 2017;7:8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Indrieri A, Carrella S, Romano A, et al miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol Med. 2019;11:e8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rodriguez-Ortiz CJ, Prieto GA, Martini AC, et al miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer’s disease. Aging Cell. 2020;19:e13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xu LJ, Ouyang YB, Xiong X, et al. Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp Neurol. 2015;264:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hu Z, Yu D, Gu QH, et al miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun. 2014;5:3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bose CM, Qiu D, Bergamaschi A, et al Agrin controls synaptic differentiation in hippocampal neurons. J Neurosci. 2000;20:9086–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].McCroskery S, Bailey A, Lin L, et al. Transmembrane agrin regulates dendritic filopodia and synapse formation in mature hippocampal neuron cultures. Neuroscience. 2009;163:168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ji RR, Bose CM, Lesuisse C, et al Specific agrin isoforms induce cAMP response element binding protein phosphorylation in hippocampal neurons. J Neurosci. 1998;18:9695–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Falo MC, Reeves TM, Phillips LL. Agrin expression during synaptogenesis induced by traumatic brain injury. J Neurotrauma. 2008;25:769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Barber AJ, Lieth E. Agrin accumulates in the brain microvascular basal lamina during development of the blood-brain barrier. Dev Dyn. 1997;208:62–74. [DOI] [PubMed] [Google Scholar]
- [82].Rajkovic I, Wong R, Lemarchand E, et al. Pentraxin 3 regulates neutrophil infiltration to the brain during neuroinflammation. AMRC Open Res. 2019;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rodriguez-Grande B, Varghese L, Molina-Holgado F, et al Pentraxin 3 mediates neurogenesis and angiogenesis after cerebral ischaemia. J Neuroinflammation. 2015;12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Shindo A, Maki T, Mandeville ET, et al Astrocyte-derived pentraxin 3 supports blood-brain barrier integrity under acute phase of stroke. Stroke. 2016;47:1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Doni A, Stravalaci M, Inforzato A, et al The long pentraxin PTX3 as a link between innate immunity, tissue remodeling, and cancer. Front Immunol. 2019;10:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ravizza T, Moneta D, Bottazzi B, et al Dynamic induction of the long pentraxin PTX3 in the CNS after limbic seizures: evidence for a protective role in seizure-induced neurodegeneration. Neuroscience. 2001;105:43–53. [DOI] [PubMed] [Google Scholar]
- [87].Ma YJ, Garred P. Pentraxins in complement activation and regulation. Front Immunol. 2018;9:3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lu J, Mold C, Du Clos TW, et al. Pentraxins and Fc receptor-mediated immune responses. Front Immunol. 2018;9:2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hahl P, Davis T, Washburn C, et al. Mechanisms of neuroprotection by hemopexin: modeling the control of heme and iron homeostasis in brain neurons in inflammatory states. J Neurochem. 2013;125:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Li RC, Saleem S, Zhen G, et al Heme-hemopexin complex attenuates neuronal cell death and stroke damage. J Cereb Blood Flow Metab. 2009;29:953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yang Y, Dong B, Lu J, et al. Hemopexin reduces blood–brain barrier injury and protects synaptic plasticity in cerebral ischemic rats by promoting EPCs through the HO-1 pathway. Brain Res. 2018;2018:177–185. [DOI] [PubMed] [Google Scholar]
- [92].Leclerc JL, Santiago-Moreno J, Dang A, et al Increased brain hemopexin levels improve outcomes after intracerebral hemorrhage. J Cereb Blood Flow Metab. 2018;38:1032–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Han D, Yu Z, Liu W, et al Plasma Hemopexin ameliorates murine spinal cord injury by switching microglia from the M1 state to the M2 state. Cell Death Dis. 2018;9:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hong CS, Park MR, Sun EG, et al Gal-3BP negatively regulates NF-kappaB signaling by inhibiting the activation of TAK1. Front Immunol. 2019;10:1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Seki T, Kanagawa M, Kobayashi K, et al Galectin 3-binding protein suppresses amyloid-beta production by modulating beta-cleavage of amyloid precursor protein. J Biol Chem. 2020;295:3678–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dong L, Chen Y, Lewis M, et al Neurologic defects and selective disruption of basement membranes in mice lacking entactin-1/nidogen-1. Lab Invest. 2002;82:1617–1630. [DOI] [PubMed] [Google Scholar]
- [97].Grimpe B, Probst JC, Hager G. Suppression of nidogen-1 translation by antisense targeting affects the adhesive properties of cultured astrocytes. Glia. 1999;28:138–149. [DOI] [PubMed] [Google Scholar]
- [98].Vasudevan A, Ho MS, Weiergraber M, et al Basement membrane protein nidogen-1 shapes hippocampal synaptic plasticity and excitability. Hippocampus. 2010;20:608–620. [DOI] [PubMed] [Google Scholar]
- [99].Shetty AK, Zanirati G. The interstitial system of the brain in health and disease. Aging Dis. 2020;11:200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Frigerio F, Pasqualini G, Craparotta I, et al n-3 Docosapentaenoic acid-derived protectin D1 promotes resolution of neuroinflammation and arrests epileptogenesis. Brain. 2018;141:3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


