Abstract
Genetic alterations affecting members of the Janus kinase (JAK) family have been discovered in a wide array of cancers and are particularly prominent in hematological malignancies. In this review, we focus on the role of such lesions in both myeloid and lymphoid tumors. Oncogenic JAK molecules can activate a myriad of canonical downstream signaling pathways as well as directly interact with chromatin in noncanonical processes, the interplay of which results in a plethora of diverse biological consequences. Deciphering these complexities is shedding unexpected light on fundamental cellular mechanisms and will also be important for improved diagnosis, identification of new therapeutic targets, and the development of stratified approaches to therapy.
Mature blood cells are predominantly short-lived, with more than 5 × 1011 cells turned over each day during steady state hematopoiesis. The continual replenishment of blood cells is the mandate of the hematopoietic stem cells (HSCs), which display both self-renewal capacity and the ability to differentiate into mature cells of the blood, such as erythrocytes, megakaryocytes, neutrophils, and lymphocytes. Hematopoiesis is a hierarchical process in which HSCs reside at the apex of a hierarchy of multilineage and unilineage progenitors of increasingly restricted potential that can ultimately develop into terminally differentiated, specialized blood cells. Homeostasis is ensured by intricate mechanisms that integrate differentiation, proliferation, and cell death, with disruption of these checks and balances leading to oncogenic consequences.
Cytokines play a critical role during hematopoietic ontogeny by initiating intracellular signals that govern cell fate choices such as proliferation and differentiation. Most cytokine receptors lack intrinsic kinase activity, and hence often employ Janus kinases (JAKs) as signaling intermediates to facilitate downstream signaling. JAKs are a family of four nonreceptor tyrosine kinases (JAK1, JAK2, JAK3, and TYK2), which are generally found constitutively bound to receptors and which are activated after cytokine receptor activation. JAK activation results in phosphorylation of the STAT transcription factors, of which seven members exist (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6). Upon activation, STAT complexes translocate into the nucleus, bind DNA, and initiate transcription.
The critical roles of JAK and STAT proteins in hematopoietic ontogeny have been clearly demonstrated via targeted gene disruption studies in mice (reviewed by Khwaja, 2006). Loss of Jak1, Jak3, or Tyk2 results in impaired lymphopoiesis whereas Jak2 deficiency results in embryonic lethality as a result of a failure of definitive erythropoiesis. These phenotypes largely reflect defects in specific cytokine signaling pathways within which JAK family members play key roles. Mice lacking Stat1, Stat2, Stat4, or Stat6 are viable but exhibit specific signaling abnormalities, predominantly in lymphoid cells. Stat3 deficiency results in early embryonic lethality as a result of severe developmental defects, and simultaneous loss of both Stat5a and Stat5b leads to perinatal lethality with anemia and leucopenia. Most studies have not distinguished between Stat5a and Stat5b and so the term “Stat5” will be used to represent their combined effects.
In many chronic and acute hematological malignancies, acquired genetic lesions cause aberrant JAK-STAT signaling. Neoplastic modulation of JAK-STAT pathways may be indirect and result from mutations that activate upstream receptors such as the Flt3-ITD mutation in acute myeloid leukemia (AML) or from activation of other signaling proteins such as the BCR-ABL protein in chronic myeloid leukemia (CML). Due to space limitations, these types of lesions will not be discussed in detail here. Instead, we will focus on genetic alterations that directly target components of the JAK-STAT pathway (Figure 1, Table 1).
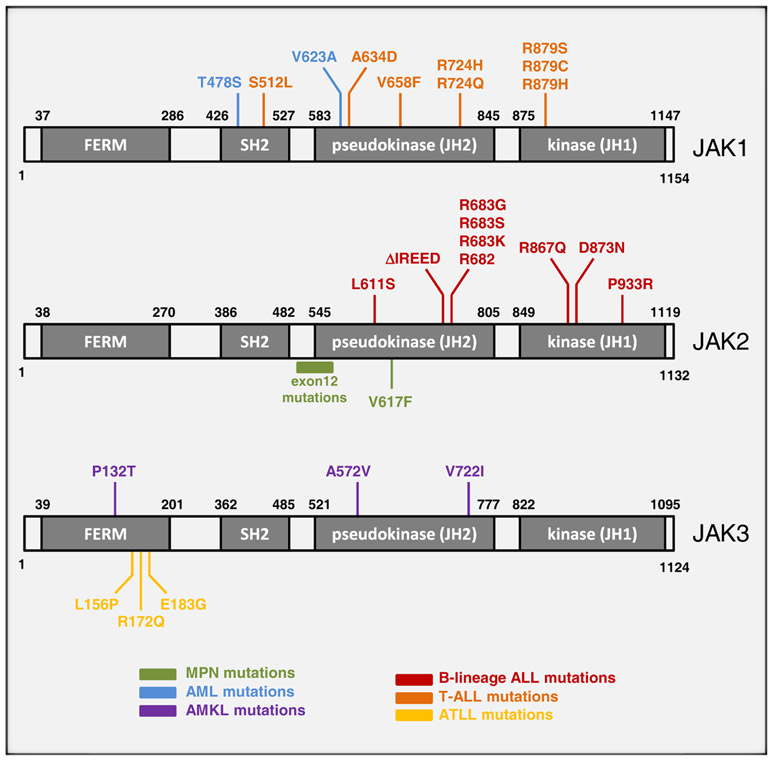
Figure 1. Summary of JAK Mutations Discovered in Hematological Malignancies.
Color-coded representation of the location of each mutant residue within the domain structure of each JAK protein. The majority of mutations in JAK proteins are found within the pseudokinase or kinase domain.
Table 1.
JAK-Related Mutations in Hematological Malignancies
| Type of Lesion | Mutation | Disease | Frequency | Reference |
|---|---|---|---|---|
| Fusion | TEL-JAK2 | T-ALL | rare | Lacronique et al. (1997) |
| BCR-JAK2 | atypical CML | rare | Griesinger et al. (2005) | |
| PCM1-JAK2 | AML, T-ALL | rare | Reiter et al. (2005) | |
| RPN1-JAK2 | PMF | rare | Mark et al. (2006) | |
| SSBP2-JAK2 | B-ALL | rare | Poitras et al. (2008) | |
| PAX5-JAK2 | B-ALL | rare | Nebral et al. (2009) | |
| Point Mutation: JAK2 | V617F | PV | ~95% | Baxter et al. (2005); James et al. (2005); Kralovics et al. (2005); Levine et al. (2005b) |
| ET | 50%–60% | see above | ||
| PMF | 50%–60% | see above | ||
| exon 12 variants | PV | ~3% | Scott et al. (2007) | |
| exon 12-15 variants | MPNs | rare | Ma et al. (2009) | |
| ΔIREED | B-ALL | rare | Malinge et al. (2007) | |
| L611S | B-ALL | rare | Kratz et al. (2006) | |
| R683 variants | DS-ALL | 18%–28% | Bercovich et al. (2008); Kearney et al. (2009) | |
| R683 variants | B-ALL | 7% | Mullighan et al. (2009b) | |
| Point Mutation: JAK1 | T478S, V623A | AML | rare | Xiang et al. (2008) |
| A634D | T-ALL | 4%–18% | Asnafi et al. (2010); Flex et al. (2008) | |
| Point Mutation: JAK3 | A572V, V722I, P132T | AMKL | rare | Walters et al. (2006) |
| L156P, R172Q, E183G | ATLL | rare | Elliott et al. (2011) | |
| Amplification | JAK2 | PMBL, HL | 30%–50% | Joos et al. (2000); Lenz et al. (2008); Rosenwald et al. (2003) |
Disease abbreviations are as follows: T-ALL, T cell acute lymphoblastic leukemia; CML, chronic myelogenous leukemia; AML, acute myelogenous leukemia; B-ALL, B lineage acute lymphoblastic leukemia; PV, polycythemia vera; ET, essential thrombocythemia; PMF, primary myelofibrosis; MPN, myeloproliferative neoplasms; DS-ALL, Down’s syndrome-associated acute lymphoblastic leukemias; AMKL, acute megakaryocytic leukemia; ATLL, adult T cell leukemia or lymphoma; PMBL, peripheral mediastinal B cell lymphoma; HL, Hodgkin’s lymphoma.
JAK2 Mutations in Chronic Myeloid Malignancies
Mutations of JAK family members recently came to prominence with the discovery of the V617F mutation in JAK2 in a substantial proportion of patients with chronic myeloproliferative neoplasms (MPNs) (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005b). MPNs encompass a spectrum of neoplastic disorders characterized by overproduction of terminally differentiated cells of the myelo-erythroid lineage and share a predisposition to the development of AML. The JAK2-V617F mutation was detected in ~95% of individuals with polycythemia vera (PV) and 50%-60% of those with essential thrombocythemia (ET) and primary myelofibrosis (PMF). Subclones homozygous for the JAK2-V617F mutation are readily detectable in most patients with PV but are undetectable or present at a low level in most patients with ET (Scott et al., 2006). The V617F mutation is also detectable at lower frequencies in other chronic myeloid malignancies including systemic mastocytosis (6%), chronic myelomonocytic leukemia (6%), Philadelphia-negative CML (19%), myelodysplastic syndrome (MDS) (3%), and refractory anemia with ringed sideroblasts associated with thrombocytosis (RARS-T) (~50%) (Jelinek et al., 2005; Jones et al., 2005; Levine et al., 2005a; Scott et al., 2005; Steensma et al., 2005). PV patients negative for JAK2-V617F often harbor one of several mutations in exon 12 of JAK2, a finding that revealed the existence of a previously unrecognized a PV subtype associated with a more isolated erythrocytosis (Scott et al., 2007). Additional rare variants in exons 12 to 15 have been identified (Ma et al., 2009).
Genetic predisposition to the development of an MPN is increasingly recognized. Large population studies of first-degree relatives of MPN patients have revealed a 5- to 7-fold increase in the risk of developing an MPN (Landgren et al., 2008). A haplotype block called “46/1” or “CCGG,” and which contains the JAK2 gene itself, has been reported by several groups to increase the odds of developing an MPN by 3- to 4-fold (Jones et al., 2009; Kilpivaara et al., 2009; Olcaydu et al., 2009). The 46/1 haplotype occurs predominantly in cis with the V617F mutation and is thought to be in linkage disequilibrium with a locus that either increases the occurrence of JAK2 mutations or provides a selective advantage for JAK2 mutations once they have arisen.
The V617F and exon 12 mutations are located within the JAK2 JH2 pseudokinase domain and normally inhibit the adjacent JH1 tyrosine kinase domain (Giordanetto and Kroemer, 2002). The mutations are predicted to be located at the JH1-JH2 interface and are thought to abrogate the JH2 inhibitory effect, thus resulting in dysregulated JAK2 kinase activity. A crystal structure of the JAK2 JH1 tyrosine kinase domain has been reported (Lucet et al., 2006) but none are available for the full-length protein or for a fragment containing both JH1 and JH2 domains, and so the details of the JH1-JH2 interaction remain unclear. However, recent data have provided some clues. It has been suggested that optimal activation of JAK2-V617F requires an aromatic amino acid at residue F595 to mediate a stacking interaction with the mutant F617 within the JH2 αC helix (Dusa et al., 2010). It has also been reported that the JH2 domain possesses low-level dual-specific kinase activity that is required for autophosphorylation at residues S523 and Y570, two modifications thought to be important for facilitating JH1-JH2 interaction and maintaining JAK2 in an inactive state (Ungureanu et al., 2011). According to this model, the V617F mutation abrogates the dual-specific kinase activity of the JH2 domain, resulting in diminished phosphorylation of S523 and Y570 and consequently dysregulated JH1 tyrosine kinase activity.
JAK Mutations in Acute Leukemias
In contrast to the morphologically normal hematopoietic differentiation that typifies chronic neoplasms, acute myeloid leukemias (AML) and acute lymphoblastic leukemias (ALL) are characterized by a differentiation block with accumulation of primitive blast cells in the bone marrow and/or peripheral blood. Acquired lesions involving JAK1, JAK2, and JAK3 (but not TYK2) have been reported in both AML and ALL.
Historically, acute leukemias were the first malignancies to be associated with a lesion in a JAK gene. The TEL-JAK2 fusion protein was originally observed as the gene product of a t(9;12)(p24;p13) translocation in a patient with T-ALL (Lacronique et al., 1997). The chimeric protein contains the oligomerization domain of the Ets protein TEL and the JH1 tyrosine kinase domain of JAK2. The TEL subunit facilitates homodimerization of TEL-JAK2 molecules, thus facilitating transphosphorylation and activation of the JAK2 kinase domains. Several analogous JAK2 fusion proteins have since been described in ALLs or AMLs, including PCM1-JAK2 (Reiter et al., 2005), BCR-JAK2 (Griesinger et al., 2005), RPN1-JAK2 (Mark et al., 2006), SSBP2-JAK2 (Poitras et al., 2008), and PAX5-JAK2 (Nebral et al., 2009). In all cases, the mechanism of JAK2 activation is thought to be similar, with the JAK2 fusion partner promoting dimerization and constitutive activation of the JAK2 tyrosine kinase component of the fusion protein. Detailed biochemical analyses of most of these fusion gene products have yet to be reported.
In recent years, point mutations that activate JAK family members have also been revealed to be a common in acute leukemias. Multiple different JAK2 mutations have been reported. The JAK2-V617F mutation is found in 3%–5% of de novo AMLs (Scott et al., 2005; Steensma et al., 2005), but at least some of these cases may represent transformation from an undiagnosed prior MPN. Several groups have also identified additional JAK2 mutations in 18%–28% of B progenitor ALLs that develop from Down’s syndrome patients (DS-ALL) (Bercovich et al., 2008; Kearney et al., 2009) and in 7% of high-risk pediatric B lineage ALL patients with or without Down’s syndrome (Mullighan et al., 2009b). Multiple mutations were found near the R683 residue, of which JAK2-R683G was the most common. In silico structure modeling of JAK2 predicts that the R683 residue resides within a deep and narrow binding pocket within the pseudokinse domain and forms a salt bridge with E685 (Bercovich et al., 2008). Mutation of R683 probably interferes with the formation of the salt bridge and changes the physicochemical properties of the binding pocket, potentially altering the capacity of the JH2 pseudokinase domain to undergo intramolecular or intermolecular interaction. At present, it is unclear whether the R683 mutations function to disrupt autoinhibition in the same way postulated for the archetypal V617F mutation. Whereas V617 is predicted to be situated at the interface between the pseudokinase and kinase domains, R683 is predicted to be located in a different region of the pseudokinase domain (Bercovich et al., 2008). Finally, three additional mutations within the catalytic tyrosine kinase domain of JAK2 were identified in B lineage ALL patients (Mullighan et al., 2009b), although functional characterization has yet to be performed.
JAK1 mutations have been reported in 4%–18% of T-ALLs (Asnafi et al., 2010; Flex et al., 2008), in 3% of poor-prognosis pediatric B lineage ALL patients (Mullighan et al., 2009b), and in 2% of de novo AMLs (Xiang et al., 2008). The majority of these mutations occurred within the pseudokinase domain. Indeed, the oncogenic potential of pseudokinase domain disruption within JAK1 had been predicted earlier by studies in which introduction of a V658F mutation in JAK1 (homologous to the V617F mutation in JAK2) led to its constitutive activation (Staerk et al., 2005). At least one of the JAK1 mutations may act in a similar manner as indicated by the fact that in silico structural modeling of the JAK1-A634D mutant predicted the mutated amino acid residue to reside on a surface of the pseudokinase domain that directly interacts with the kinase domain (Flex et al., 2008).
JAK3 mutations in ALL and AML are rare. However, several nonrecurrent JAK3 mutations were identified in the CMK megakaryoblastic cell line and patients with acute megakaryoblastic leukemia (AMKL) (Walters et al., 2006). All three mutations were found to constitutively activate the JAK3 protein, although only the A572V and V722I mutants are situated within the pseudokinase domain. A P132T mutation lies within the receptor binding region of JAK3 and the mechanism by which it activates JAK3 is not clear. Several different JAK3 mutations have since been reported in primary AMKL samples but are yet to be functionally characterized (Malinge et al., 2008; Sato et al., 2008).
JAK2 Amplifications in Lymphoma
Given the importance of JAK-STAT signaling in normal mature lymphoid subpopulations, it is not surprising that many lymphoid malignancies derived from these normal cells coopt these pathways to aid and abet their oncogenic missions. Although histologically very distinct, Hodgkin lymphoma (HL) and peripheral mediastinal B cell lymphoma (PMBL) share a common gene expression signature that distinguishes them from other aggressive lymphoma subtypes (Rosenwald et al., 2003; Savage et al., 2003). The PMBL and HL signatures stem from the action of JAK2 signaling, which is a feature of both lymphoma subtypes, and cell line models of PMBL and HL die when JAK2 is genetically or pharmacologically inhibited (Rui et al., 2010). The underlying genetic basis for these observations is the recurrent amplicon involving JAK2 on chromosome band 9p24 seen in 30%–50% of HL and PMBL cases (Joos et al., 2000; Lenz et al., 2008; Rosenwald et al., 2003). The minimally amplified region is ~3 Mb and includes not only JAK2 but also several other functionally interesting genes including JMJD2C and RANBP6. These three genes are coordinately overexpressed in a significant proportion of HL and PMBL where they function in concert to promote lymphomagenesis (discussed below).
Cell of Origin and Mutation Timing
MPNs represent a particularly attractive model in which to study cancer development. In the chronic phase, they are thought to represent a very early stage of tumor evolution inaccessible in most cancers, because many patients are diagnosed early after an incidental blood count, the neoplastic clone has not acquired lesions resulting in a differentiation block, and there is usually no overgrowth of a dominant clone as found in most acute malignancies. MPNs tend to evolve from a chronic phase to a more aggressive accelerated phase or to overt AML over a period of many years, thus allowing studies of disease evolution. Of particular note, clonal analysis can be performed by studying colonies grown in vitro from single progenitor cells.
Initial studies using X chromosome inactivation in MPN patients suggested the HSC as the cell of origin of MPNs (reviewed in Chen and Prchal, 2007). Subsequent to the discovery of the JAK2-V617F mutation, testing for its presence in different hematopoietic compartments was performed to determine the cell in which the mutation arose. The disease allele was detected in multipotent HSC subpopulations in PV and PMF patients (Jamieson et al., 2006), LTC-ICs derived from bone marrow of MPN patients (James et al., 2008), and long-term engrafted cells in NOD-SCID mice transplanted with CD34+ cells from MPN patients (Ishii et al., 2007). Cumulatively, the above data suggested that the JAK2-V617F mutation is present in a cell exhibiting HSC-like properties. However, it is also formally possible that the JAK2-V617F mutation arises in a myeloid progenitor cell that subsequently acquires properties of self-renewal and the capacity to undergo multipotential differentiation. Indeed, stochastic mathematical modeling studying the evolutionary dynamics of cancer-initiating cells predicts this path to MPN development to be the most likely (Haeno et al., 2009).
Whether it arises in the HSC or multipotent progenitor compartment, the JAK2-V617F mutation will be carried by differentiated progeny in multiple lineages and will therefore potentially influence the behavior of multiple different cell types. Additionally, terminally differentiated mature cells also have the ability to influence the behavior of stem and progenitor cells, thus further complicating the biological consequences of this single mutation. Studies of knockin mice engineered to express physiological levels of JAK2-V617F suggest that the mutant JAK2 imparts little or no selective advantage at the HSC level. Compared to wild-type HSCs, JAK2-V617F-positive HSCs show either no advantage (Mullally et al., 2010) or a disadvantage (Li et al., 2010b) in murine competitive transplantation assays. This is consistent with several additional lines of evidence suggesting that JAK2-V617F exerts little effect on HSC self-renewal capacity: (1) xenograft transplantation of MPN HSCs into SCID immunodeficient mice demonstrated successful engraftment of V617F-positive HSCs but no obvious selective advantage compared to normal HSCs (James et al., 2008); (2) in a case in which a patient with high-risk MDS was transplanted with V617F-positive allogeneic donor cells, the recipient remained in remission and exhibited no increase in V617F allele burden for at least 7 years posttransplant (Van Pelt et al., 2008); and (3) retroviral studies in mice have shown that expression of constitutively activated tyrosine kinases (e.g., BCR-ABL, FLT3-ITD) does not trigger self-renewal in recipient cells (Huntly et al., 2004).
In contrast, JAK2-V617F has been suggested to play key roles in later stages of myelopoiesis. Progenitor cells from MPN patients form erythroid or megakaryocytic colonies in the absence of exogenous growth factors, a phenomenon thought to be a direct consequence of mutant JAK activity. The increase in erythropoiesis observed in patients with PV is likely to reflect an effect of JAK2 activation on the late stages of erythroid differentiation, although the detailed cell biology has yet to be elucidated, and it also remains unclear why the consequences of mutant JAK2 are different in patients with ET. Consistent with this idea that mutant JAK2 acts at multiple levels within hematopoiesis, JAK2-V617F gene-targeted mice display expansion of lineage-restricted myeloid compartments, increased numbers of BFU-E, CFU-GM, and CFU-Megs, together with increased erythroid and megakaryocytic terminal differentiation (Li et al., 2010b; Mullally et al., 2010).
Whether JAK2-V617F represents the initiating lesion in MPNs remains a topic of intense investigation. Its involvement in an example of an early-stage cancer like the MPNs implies that it might have an initiating role. In addition, retroviral transduction-based and transgenic mouse studies showed that JAK2-V617F was sufficient to generate a myeloproliferative phenotype (reviewed in Li et al., 2011). This was corroborated by knockin mouse studies in which transplantable MPN phenotypes ranging from thombocytosis to polycythemia were also observed (Li et al., 2010b; Mullally et al., 2010). However, some caveats should be considered when evaluating these data. Retroviral and transgenic models often result in dysregulation of the pattern or level of expression of mutant JAK2, and even in knockin models, JAK2 is still activated simultaneously in large numbers of hematopoietic cells. This contrasts with the monoclonal origin of human malignancies. Indeed, several lines of evidence suggest that, in some patients, JAK2-V617F is not the initiating lesion. Secondary AMLs arising from a V617F-positive MPN are often wild-type for the JAK2 allele (Beer et al., 2010; Campbell et al., 2006; Theocharides et al., 2007), a finding that led to the concept that these individuals harbored (at least) two distinct clonal expansions, one with and one without the JAK2-V617F mutation. This notion of oligoclonality in some MPN patients is consistent with the observations that some patients possess granulocyte clonality in excess of the V617F allelic burden (Kralovics et al., 2006) and the existence of erythropoietin-independent erythroid colonies (EECs) that are negative for the JAK2 mutation in patients with a JAK2-V617F-positive MPN (Nussenzveig et al., 2007).
Definitive support for the presence of genetically distinct clones has come from studies of clonally derived hematopoietic colonies and the demonstration that individual colonies can carry mutually exclusive acquired genetic lesions. The presence of biclonal disease could reflect the existence of a shared founder clone, evidence for which has come from the demonstration that TET2 mutations (Beer et al., 2009; Delhommeau et al., 2009) or 20q deletion (Schaub et al., 2009) can precede acquisition of the JAK2 mutation. Of particular interest, X chromosome inactivation studies have demonstrated that biclonal disease can also reflect independent expansions arising from unrelated HSCs, a situation reported in two of three evaluable patients (Beer et al., 2009).
In summary, by exploiting the JAK2-V617F-positive preleukemic MPNs as an experimentally tractable system, significant insights have been gained into the underlying biology of blood cancer development at its earliest stages.
Mutant JAKs and Receptors
One of the distinguishing hallmarks of gain-of-function JAK mutations is their ability to confer cytokine-independent growth and survival when introduced into cytokine-dependent cell lines. For example, TEL-JAK2 homodimers in the cytosol leads to STAT5 activation without a requirement for cytokine stimulation (Lacronique et al., 2000) and expression of JAK2-V617F in BaF3 cells causes factor independent growth and survival, associated with increased JAK2 phosphorylation and STAT5 activation (James et al., 2005; Kralovics et al., 2005; Levine et al., 2005b). The ability of JAK2-V617F to produce factor-independent growth has been reported to require the presence of homodimeric type I receptors, which is thought to act as a scaffold that juxtaposes two JAK2 molecules and facilitates transphosphorylation (Lu et al., 2005).
The ALL-associated JAK2-R683G mutant also requires the presence of cytokine receptors. In 50%–60% of B lineage ALL patients, leukemic blasts exhibit increased cell surface expression of the cytokine receptor-like factor 2 (CRLF2) (Mullighan et al., 2009a; Russell et al., 2009). CRLF2 is a JAK2-binding cytokine receptor subunit that, together with IL-7Rα, forms the heterodimeric thymic stromal lymphopoietin (TSLP) receptor. Increased expression of CRLF2 in B lineage ALLs is achieved in two ways: (1) through an interstitial deletion within the X chromosome that brings the CRLF2 gene under the control of the P2RY8 promoter, or (2) chromosome translocations that place the CRLF2 gene under the control of the IGH promoter (Mullighan et al., 2009a). Illegitimate recombination involving a pseudoautosomal region adjacent to the CRLF2 gene is thought to be responsible for the formation of both lesions (Mullighan et al., 2009a). JAK2 mutations have been found in 34% of CRLF2-over-expressing DS-ALL patients (Mullighan et al., 2009b) and 69% of patients with high-risk pediatric B lineage ALL with CRLF2 rearrangements (Harvey et al., 2010a) but are uncommon in those lacking CRLF2 overexpression or rearrangements. This strong association suggests functional cooperativity and, consistent with this, JAK2-R683G and CRLF2 proteins interact and their coexpression is necessary for cytokine-independent growth of BaF3 cells (Mullighan et al., 2009a; Yoda et al., 2010).
JAK1 mutants seen in T-ALL and B lineage ALL have been reported to be capable of transforming Ba/F3 cells to factor independence without coexpression of a receptor (Flex et al., 2008). It is unclear whether this difference from JAK2 mutants is due to a true biological peculiarity of the JAK1 mutants or whether it reflects issues of experimental methodology such as the level of mutant JAK1 expression achieved. In addition, BaF3 cells are lymphoid and so may possess a repertoire of endogenous receptors that are able to provide scaffolding for mutant JAK1 molecules. It seems inherently unlikely that receptors do not play a role in mutant JAK1 signaling, and consistent with this, mutating the FERM domain, thus preventing receptor binding, abolished the activation and transforming ability of JAK1-V658F (Hornakova et al., 2009). A clue into the potential scaffold for mutant JAK1 molecules came with the recent identification of activating mutations in the α subunit of the IL-7 receptor (IL-7Rα) that stimulates a JAK1-STAT5 signaling axis to induce transformation in approximately 10% of T-ALLs (Shochat et al., 2011; Zenatti et al., 2011).
JAK2-V617F mutations are observed only in myeloid disorders, whereas JAK2-R683G and JAK1 mutations are seen in ALLs and not MPNs. These striking disease associations remain poorly understood, but it seems likely that distinct JAK family mutations require different cellular contexts to exert their neoplastic functions, with particular mutations requiring the presence of specific cytokine receptors.
Canonical Effects of Mutant JAK Signaling
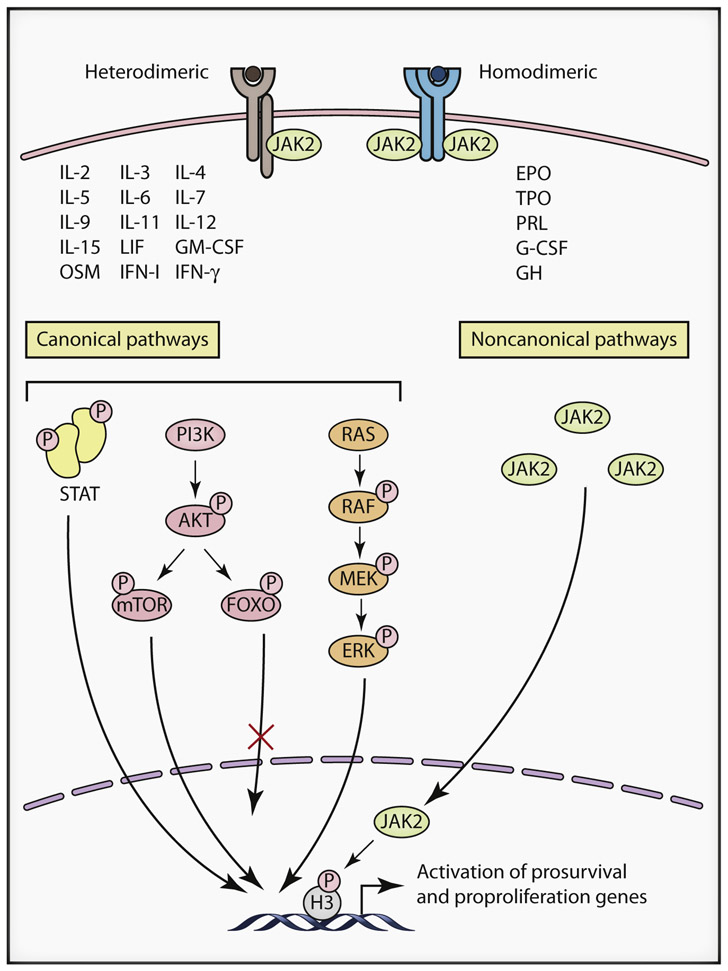
The canonical downstream signaling consequences of JAK mutations have been best studied in the context of JAK2 mutations. As discussed above, JAK2 is involved in signaling from multiple cytokine receptors. In addition, it is capable of activating multiple STATs, as well as the PI3K and the MAPK signaling pathways. The transcriptional programs activated by mutant JAK2 in a given cell type will therefore depend on the complement of receptors, STATs, and other signaling pathway components present in that cellular context (Figure 2). In the following sections, we will review the role of these canonical signaling pathways in the pathogenesis of various hematological disorders associated with mutant JAK signaling.
Figure 2. Signaling Complexities of Janus Kinases.
Janus kinases (JAKs) such as JAK2 can modulate signaling through multiple different cytokine receptor families, and can activate multiple downstream canonical pathways such as STATs, PI3K and ERK, as well as non-canonical pathways via direct nuclear targeting. The biological consequences elicited by oncogenic JAK signaling will therefore depend on the complement of receptors, STATs and other signaling pathway components present in that cellular context.
STAT5
Most is known about STAT5 and its role downstream of mutant JAK2. Neither TEL-JAK2 nor JAK2-V617F is capable of producing disease in a Stat5-deficient background (Funakoshi-Tago et al., 2010; Walz et al., 2012; Yan et al., 2011a), and mice reconstituted with bone marrow expressing a constitutively active Stat5a mutant develop a fatal myeloproliferative syndrome (Schwaller et al., 2000). Moreover, overexpression of Stat5 in the lymphoid compartment can also give rise to thymic T cell lymphoblastic lymphoma in mice (Kelly et al., 2003). Cumulatively, these data suggest that STAT5 is necessary for hematological disease downstream of mutant JAKs, and also that its activation is sufficient to generate a malignant phenotype when overexpressed. STAT5 activates large numbers of targets with at least three subcategories likely to relate to oncogenesis: (1) cell cycle regulators, such as the D-type cyclins, MYC, and PIM1 (Matsumura et al., 1999); (2) DNA repair proteins, such as RAD51 and DNA polymerase β (Slupianek et al., 2002)–this may contribute to the augmentation of error-prone DNA repair that has been reported in some MPN patients (Plo et al., 2008), and thus, could contribute to a degree of genomic instability; and (3) antiapoptotic proteins such as BCL-XL and BCL-2 (Dumon et al., 1999). BCL-XL overexpression is able to mimic some features of mutant JAK2 expression, including cytokine-independent growth and EEC formation (Garçon et al., 2006). In the presence of DNA damage, BCL-XL undergoes deamidation and, as a consequence, apoptosis is enhanced in normal cells. However, in primary cells from MPN patients, mutant JAK2 blocks DNA damaged-induced deamidation of BCL-XL and therefore promotes the accumulation of cells harboring DNA damage, thus providing a mechanism for disease evolution (Zhao et al., 2008).
STAT3
Although STAT3 is a well-known oncogene in the context of both solid and hematological malignancies, the role of JAK-STAT3 signaling in the pathogenesis of myeloid disorders remains unclear. Higher amounts of tyrosine-phosphorylated STAT3 have been reported in granulocytes of some JAK2-V617F-positive MPN patients and are associated with heightened resistance to apoptosis caused by cytokine withdrawal (Mesa et al., 2006), although others have failed to see this correlation (Teofili et al., 2007). Recent murine studies have shown Stat3 to be dispensable for myeloid expansion induced by JAK2-V617F (Yan et al., 2011a). Further studies will be required to clarify the role of STAT3 in myeloid malignancies.
In contrast, emerging evidence is implicating a prominent role for increased JAK-STAT3 signaling during lymphomagenesis. In the activated B cell-like (ABC) subtype of diffuse large B cell lymohoma (DLBCL), approximately half of all tumors exhibit a STAT3 gene expression signature (Figure 3, top; Ding et al., 2008; Lam et al., 2008). The underlying genetic basis for constitutive JAK-STAT3 signaling in ABC DLBCL was illuminated by the discovery of mutations in the TIR domain of MYD88, a key adaptor in Toll-like receptor signaling (Ngo et al., 2011). Overall, 39% of ABC DLBCL tumors have MYD88 mutations, including many recurrent point mutations indicative of selection. However, one mutant, termed L265P, is the clear evolutionary winner, occurring in 29% of ABC DLBCL cases but only rarely among other aggressive lymphoma subtypes. MYD88-L265P mutations are also frequent among primary central nervous system lymphomas, chronic lymphocytic leukemia, and marginal zone lymphomas (Ngo et al., 2011; Yan et al., 2011b).
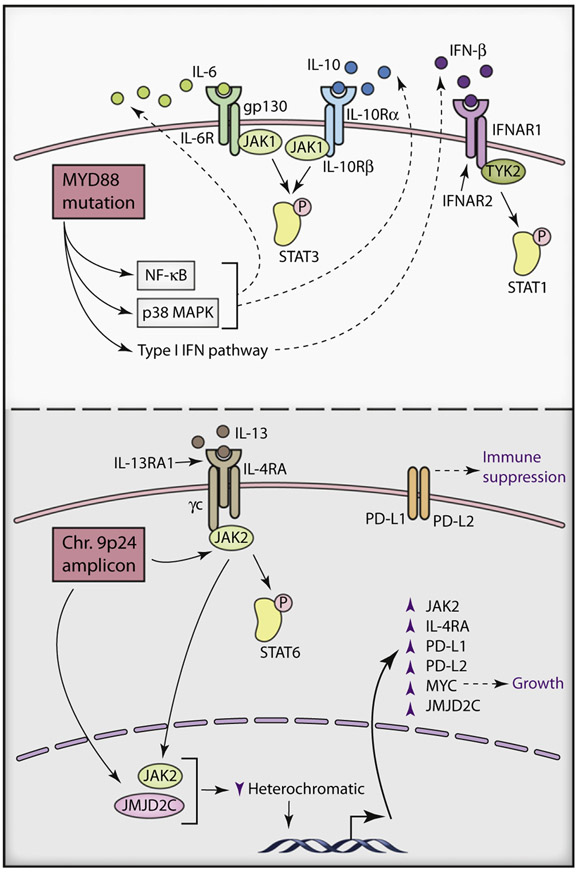
Figure 3. JAK-STAT Signaling in Lymphomas.
Top: Autocrine JAK-STAT3 signaling in activated B cell like diffuse large B cell lymphoma. MYD88 mutants activate NF-κB and p38 MAP kinase signaling pathways to increase production and secretion of IL-6 and IL-10, which in turn activate the JAK-STAT3 pathway by autocrine stimulation of their respective receptors. MYD88 mutants can also activate type I interferon pathways through increased production of IFN-β. Bottom, Molecular consequences of 9p24 amplification in Hodgkin lymphoma and primary mediastinal B cell lymphoma. Increased expression of JAK2 and JMJD2C following 9p24 amplification synergizes to destabilize heterochromatic state and positively regulate expression of multiple genes (such as those encoding MYC, JAK2, JMJD2C, IL4RA, PD-L1, and PD-L2) that promote increased growth factor signaling, immune suppression and proliferation.
These gain-of-function MYD88 mutations activate JAK-STAT3 signaling in an autocrine manner. The biochemical mechanism of action of MYD88 mutants involves the recruitment of the kinases IRAK4 and IRAK1 into a constitutive signaling complex in which IRAK1 is phosphorylated by IRAK4 (Ngo et al., 2011). Phosphorylation of IRAK1 engages the NF-κB and p38 MAP kinase signaling pathways, which coordinately induce the expression of the cytokines IL-6 and IL-10, which in turn activate the JAK-STAT3 pathway by autocrine stimulation of their respective receptors (Lam et al., 2008; Ngo et al., 2011). STAT3 also physically interacts with NF-κB p50-p65 heterodimers to potentiate transactivation of NF-κB target genes (Yang et al., 2007). A likely consequence of this regulatory cooperation is high relative expression of a set of NF-κB target genes in STAT3+ ABC DLBCL tumors (Lam et al., 2008). STAT3-NF-κB cooperation may also account for the fact that JAK-STAT3 signaling can promote secretion of IL-6 and IL-10, both of which are NF-κB targets (Lam et al., 2008).
The MYD88-dependent JAK-STAT3 signaling in ABC DLBCLs suggests several therapeutic strategies. A small molecule pan-JAK kinase inhibitor induces apoptosis in ABC DLBCL cell lines but has little if any effect on cell line models of the germinal center B cell-like (GCB) DLBCL subtype (Lam et al., 2008). Likewise, genetic or pharmacologic inhibition of STAT3 is toxic for ABC DLBCL cell lines (Scuto et al., 2011). Inhibitors of IκB kinase (IKK), which block NF-κB signaling, also induce apoptosis in ABC DLBCL cell lines (Lam et al., 2005). Combined treatment with JAK kinase and IKK inhibitors is synergistically toxic for ABC DLBCL cell lines, possibly as a result of the cooperation between STAT3 and NF-κB mentioned above (Lam et al., 2008). JAK inhibitors emerge from these studies as viable therapeutic candidates in ABC DLBCL that could be combined with other drugs that block constitutive NF-κB signaling. In addition, a small molecule inhibitor of IRAK4 kinase was selectively toxic for ABC DLBCL cell lines bearing the MYD88-L265P mutant (Ngo et al., 2011). IRAK4 kinase inhibitors, which are in clinical development for a variety of inflammatory and autoimmune diseases, could be repurposed to shut down MYD88 signaling in ABC DLBCL.
STAT1
JAK proteins also play pivotal roles in both type I and type II interferon signaling, and unsurprisingly, expression of constitutively active JAK molecules in cell line models can directly activate interferon signaling pathways via the direct phosphorylation of STAT1 (Chen et al., 2010; Xiang et al., 2008). Alternatively, lymphoma cell lines bearing the ABC DLBCL-associated oncogenic MYD88 signaling can also produce interferon β that signal in an autocrine fashion through the type I interferon receptor to activate classical interferon-response genes (Ngo et al., 2011). At present, the adaptive benefit of interferon signaling to malignant lymphoma cells is unclear because type I interferon is typically antiproliferative and/or proapoptotic. Conceivably, the immunomodulatory functions of interferon could help the tumor evade immune surveillance. However, it is also possible that interferon signaling is not a selected tumor phenotype but rather the “price of doing business” with constitutive JAK activity in lymphoma.
Recent data provide evidence that type II interferon signaling pathway may play a role in myeloid disorders and that modulation of its activity may be important in certain disease subtypes. Historically, studying signaling downstream of mutant JAK2 in primary cells from MPN patients has been challenging because of interindividual variability in signaling pathway activity combined with variable mixtures of mutant and normal cells. This is reflected by the highly variable levels of tyrosine phosphorylated STAT5 or STAT3 in MPN patients that do not always correlate with V617F allele burden (Aboudola et al., 2007; Mesa et al., 2006; Teofili et al., 2007). Comparison of clonally derived mutant and wild-type cells from each patient provides a powerful way to circumvent these confounding variables. Application of this strategy revealed unexpected cell-intrinsic differences between PV and ET patients in the signaling pathways activated by JAK2-V617F (Chen et al., 2010). STAT1 and its downstream transcriptional program were activated in response to JAK2-V617F in erythroblasts from patients with ET but not in those from PV patients. This observation is likely to reflect inherited or acquired genetic differences and may well contribute to the phenotypic differences between ET and PV. Extension of this approach to the study of other cancers is likely to reveal unexpected complexity in the signaling consequences of other oncogenic lesions.
PI3K-AKT Pathway
In addition to STAT proteins, JAKs can activate a variety of additional signaling pathways. JAK2 can activate the phosphoinositide 3-kinase (PI3K) by phosphorylating the erythropoietin receptor (EPOR) on Y479, which causes recruitment of the PI3K to the plasma membrane where it activates AKT. Activation of the PI3K-AKT signaling pathway is a feature of many neoplasms associated with aberrant JAK signaling. It appears to play a critical role in tumorigenesis, as shown by the fact that fibroblasts coexpressing JAK2-V617F and an EPOR scaffold unable to facilitate PI3K activation fail to form tumors when transplanted into mice (Kamishimoto et al., 2011). Increased PI3K-AKT signaling transforms cells by promoting increased cell survival and proliferative index. Inhibition of PI3K activity with pharmacological inhibitors reduces EEC formation from PV progenitors, increases apoptosis of cells expressing MPN- or ALL-associated JAK mutations, and reduces erythroid proliferation (Funakoshi-Tago et al., 2009; Ugo et al., 2004).
These effects probably relate to the >100 nonredundant substrates of AKT that have been identified. Upon phosphorylation, AKT targets are functionally inactivated by sequestration either from essential protein partners or from their correct subcellular compartments and many of these AKT targets are known to be disrupted in mutant JAK-associated hematological neoplasms: (1) inactivation of the proapoptotic protein BAD by serine phosphorylation is a feature of JAK2-V617F-positive cell lines (Gozgit et al., 2008); (2) inactivation of negative regulators of mTORC1 increases ribosome biogenesis, mRNA translation, and cell growth and proliferation, known features of JAK2-V617F-positive MPNs (Lelièvre et al., 2006)–indeed, the mTORC1 inhibitor, rapamycin, has been demonstrated to be cytotoxic for cell lines expressing many different oncogenic JAK molecules (Li et al., 2010a); (3) increased phosphorylation of glycogen synthase kinase 3β (GSK3β) promotes increased cycling of PV erythroid progenitors by relieving inhibition of cyclin D and E (Dai et al., 2005); and (4) inhibition of the forkhead box-containing transcription factors (FOXO) family leads to impaired activation of quiescence-associated genes and proapoptotic genes. Although inactivation of FOXO members has yet to be reported in malignancies associated with JAK mutations, it has been implicated in disorders associated with elevated JAK-STAT signaling such as CML and AML.
RAS-RAF1-MEK-ERK1 and -ERK2 Pathway
Oncogenic JAK molecules can also activate the RAS-RAF1-MEK-ERK1 and -ERK2 cascade, the predominant mitogen-activated kinase (MAPK) pathway initiated by cytokine receptors. In contrast to PI3K-AKT signaling, the contribution of ERK1 and ERK2 downstream of oncogenic JAK molecules seems to be largely restricted to increased proliferative drive. MEK inhibitors suppress S phase entry but not prosurvival signals in mutant JAK-expressing cells (Funakoshi-Tago et al., 2010; Ugo et al., 2004). Consistent with this, elevated expression of ERK1 and ERK2 targets JUNB and FOSB is seen in JAK2-V617F-positive MPNs, and JUNB overexpression stimulates growth of erythroid cells in vitro (da Costa Reis Monte-Mór et al., 2009; Puigdecanet et al., 2008). Additional insights into the hematological consequences of oncogenic RAS signaling have also been derived from studies of juvenile myelomoncytic leukemia (JMML) and chronic myelomonocytic leukemias (CMML). RAS or PTPN11 mutations account for ~60% of JMMLs and ~35% of CMMLs (Loh, 2011). Both classes of mutations lead to dysregulated signaling of the GM-CSF receptor, increased ERK1/2 activation, and excessive monocyte production. Mice expressing a JMML-associated mutation of K-Ras or N-Ras develop myeloproliferative disorders that closely resemble JMML or CMML with an isolated expansion of mature monocytes but no erythroid or lymphoid hyperplasia (Parikh et al., 2006). Although JAK2 is bound to the GM-CSF receptor and is required for its signaling, JAK2 mutations are rarely found in JMMLs. In addition, the few patients with JAK-V617F-positive CMMLs rarely harbor RAS mutations (Pèrez et al., 2010).
Noncanonical Effects of Mutant JAK Signaling
In recent years, there has been mounting evidence for functions of JAK molecules beyond their purview as adaptors in signal transduction pathways. An early clue that JAKs may possess additional functions was seen in Drosophila melanogaster where constitutive JAK activation is associated with global disruption of heterochromatic gene silencing in a fly leukemia model (Shi et al., 2006). The mechanism by which JAK signaling modulates heterochromatin stems from the unexpected observation that JAK2 exists in the nucleus, where it directly phosphorylates the histone H3 tail on tyrosine 41 (H3Y41) (Dawson et al., 2009). Heterochromatin formation is associated with recruitment of HP1α, which uses its carboxy-terminal chromo-shadow domain to bind to this portion of the histone H3 tail in its unphosphorylated state. HP1α separately uses its amino-terminal chromodomain to interact with the heterochromatin mark H3K9me3. H3Y41 phosphorylation leads to displacement of HP1α protein from chromatin and increased gene transcription at that locus (Dawson et al., 2009). The JAK2-H3Y41-HP1α signaling pathway specifically targets thousands of loci, many of which lack identifiable STAT binding elements (Rui et al., 2010), and are thus likely to activate an additional set of target genes distinct from those regulated by canonical JAK-STAT signaling pathways. These direct targets of nuclear JAK2 signaling are likely to have functional roles in disease biology, as indicated by the fact that phospho-Y41-mediated HP1α displacement can occur at loci of proto-oncogenes (Dawson et al., 2009). Recently, this noncanonical signaling pathway has been shown to be important in the biology of lymphoma (Figure 3, bottom).
In addition to JAK2, two additional genes–JMJD2C and RANBP6–situated within the HL/PMBL-associated 9p24 amplicon are known to be functionally important for tumor cell proliferation and survival (Rui et al., 2010). No insights into RANBP6 function have yet been provided in the scientific literature, although its inhibition potently reduced proliferation of many PMBL and HL cell lines. JMJD2C is a chromatin-modifying enzyme that demethylates trimethylated lysine 9 of the histone H3 tail (H3K9me3), thereby reducing heterochromatin formation (Loh et al., 2007; Whetstine et al., 2006). Thus, JAK2-mediated phosphorylation of H3Y41 and JMJD2C-mediated demethylation of H3K9me3 block both means of HP1α recruitment, potentially accounting for the synergism between JAK2 and JMJD2C in reducing heterochromatin formation in PMBL and HL. Consequently, combined inhibition of JAK2 and JMJD2C in HL and PMBL cells synergizes to increase heterochromatin and kill PMBL and HL cell lines, demonstrating the functional cooperation between these two amplicon genes (Rui et al., 2010).
Given that heterochromatin is typically associated with gene silencing, the effects of JAK2 and JMJD2C on HP1α recruitment positively regulate the expression of hundreds of genes in PMBL and HL, several of which are likely to be involved in lymphoma development: (1) MYC activation occurs through phosphorylation of the chromatin in the vicinity of the first intron of the MYC locus and is functionally important in promoting survival of PMBL and HL cells, although it is insufficient on its own to mediate all of the prosurvival effects of nuclear JAK2 signaling (Rui et al., 2010); (2) JAK2 and JMJD2C activation institutes a positive feedforward regulatory loop (Rui et al., 2010); (3) chromatin modification of IL4RA (which encodes an IL-13 receptor subunit) fosters autocrine IL-13 signaling (LaPorte et al., 2008); and (4) increased expression of immune modulators PD-L1 and PD-L2 blocks T cell receptor signaling–because PMBL and HL may arise amidst a sea of thymic T cells, such negative regulation may be essential for the malignant clone to escape immune surveillance (Rui et al., 2010).
These preclinical data provide a strong rationale for the clinical evaluation of JAK2 inhibitors in PMBL and HL. JMJD2C also emerges from these studies as an intriguing therapeutic target. JMJD2C inhibitors could have single agent activity in PMBL and HL and would be predicted to synergize with JAK2 inhibitors. A concern when considering epigenetic regulators such as JMJD2C as therapeutic targets is whether the side effects of inhibitors will be tolerable. In this regard, it is notable that reduced JMJD2C expression is not toxic for cell lines derived from lymphoma subtypes other than PMBL and HL (Rui et al., 2010). Thus, JMJD2C is not an essential gene in all cells, meaning that a therapeutic window may exist for the clinical development of JMJD2C inhibitors.
In summary, that JAK molecules have a noncanonical nuclear function has been an unexpected finding and is suggesting tantalizing links between the JAK-STAT pathway and heterochromatin regulation in the pathogenesis of myeloid and lymphoid malignancies. These observations may reflect a need for cells to tightly regulate the chromatin state of their genome, with dysregulation contributing to tumorigenesis. Further understanding of the role of these noncanonical pathways in disease processes will aid in devising strategies to exploit these new functions of JAK molecules for therapeutic intervention.
Clinical Implications
The discovery of JAK mutations has been rapidly translated into the clinic. Arguably the most significant effect has been the impact of JAK2 mutations on the diagnosis of MPNs, disorders of which can be difficult to distinguish from reactive causes of abnormal blood counts. Assays to detect the JAK2-V617F mutation have greatly simplified diagnostic algorithms and the identification of JAK2 exon 12 mutations have revealed the existence of a previously unrecognized subtype of PV (Scott et al., 2007). JAK2 mutation testing is already firmly embedded in national and international guidelines (Harrison et al., 2010; McMullin et al., 2007; Tefferi and Vardiman, 2008).
Detection of JAK mutations is also beginning to provide prognostic information. For example, in B precursor ALL, JAK mutations are associated with a “high-risk” subgroup that exhibits poor outcome (Mullighan et al., 2009b). Further deconvolution of this high-risk cohort by gene expression profiling shows the majority of JAK-mutated patients to be confined to a subgroup that has exceedingly poor outcome (21% 4-year relapse-free survival compared to 66% for the entire cohort) (Harvey et al., 2010b), suggesting that these patients may represent candidates for more aggressive treatment regimens. Within the MPNs, it has been suggested that the JAK2-V617F mutation is associated with increased risk of thrombosis and/or leukemic transformation (reviewed in Vannucchi et al., 2008).
Lastly, there is considerable interest in developing therapeutic JAK inhibitors. In 2010, the results of the first clinical trials with JAK kinase inhibitors for treatment of primary and post-PV and ET myelofibrosis were published (Santos et al., 2010; Verstovsek et al., 2010). As reviewed elsewhere in this issue, multiple selective JAK2 inhibitors are now in early-phase trials and showing promise in ameliorating symptoms in a subset of MPN patients. Moreover, there are strong grounds for exploring the efficacy of JAK inhibitors in multiple other hematological malignancies. For example, JAK3 inhibition has recently been shown to be effective in eradicating T cells from patients with adult T cell leukemia or lymphoma in preclinical studies (Ju et al., 2011). One point to emphasize is that clinical trials of JAK inhibitors need not be confined to cases with JAK mutations. For example, the gene expression signature of JAK2 activity is shared by most PMBL tumors, with or without 9p24 amplification (Rui et al., 2010), and MPN patients lacking a JAK2-V617F mutation respond to selective JAK2 inhibition (Verstovsek et al., 2010). However, these agents are limited by the toxic consequences of inhibiting normal JAK2 and, at the moment, there is little evidence that they alter the natural history of the MPNs. Therapeutic strategies that selectively target mutant proteins therefore remain attractive, albeit challenging. In the future, deciphering the complexities of mutant JAK signaling will facilitate identification of new therapeutic targets and is also likely to lead to improved approaches to stratified therapy.
ACKNOWLEDGMENTS
Research in the authors’ laboratories is supported by the Leukemia and Lymphoma Research, the Kay Kendall Leukaemia Fund, the NIHR Biomedical Cambridge Research Centre and the Leukemia and Lymphoma Society of America (A.R.G.), and the National Institutes of Health (L.M.S.).
REFERENCES
- Aboudola S, Murugesan G, Szpurka H, Ramsingh G, Zhao X, Prescott N, Tubbs RR, Maciejewski JP, and Hsi ED (2007). Bone marrow phospho-STAT5 expression in non-CML chronic myeloproliferative disorders correlates with JAK2 V617F mutation and provides evidence of in vivo JAK2 activation. Am. J. Surg. Pathol 31, 233–239. [DOI] [PubMed] [Google Scholar]
- Asnafi V, Le Noir S, Lhermitte L, Gardin C, Legrand F, Vallantin X, Malfuson JV, Ifrah N, Dombret H, and Macintyre E (2010). JAK1 mutations are not frequent events in adult T-ALL: a GRAALL study. Br. J. Haematol 148, 178–179. [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. ; Cancer Genome Project. (2005). Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061. [DOI] [PubMed] [Google Scholar]
- Beer PA, Jones AV, Bench AJ, Goday-Fernandez A, Boyd EM, Vaghela KJ, Erber WN, Odeh B, Wright C, McMullin MF, et al. (2009). Clonal diversity in the myeloproliferative neoplasms: independent origins of genetically distinct clones. Br. J. Haematol 144, 904–908. [DOI] [PubMed] [Google Scholar]
- Beer PA, Delhommeau F, LeCouédic JP, Dawson MA, Chen E, Bareford D, Kusec R, McMullin MF, Harrison CN, Vannucchi AM, et al. (2010). Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 115, 2891–2900. [DOI] [PubMed] [Google Scholar]
- Bercovich D, Ganmore I, Scott LM, Wainreb G, Birger Y, Elimelech A, Shochat C, Cazzaniga G, Biondi A, Basso G, et al. (2008). Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet 372, 1484–1492. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Baxter EJ, Beer PA, Scott LM, Bench AJ, Huntly BJ, Erber WN, Kusec R, Larsen TS, Giraudier S, et al. (2006). Mutation of JAK2 in the myeloproliferative disorders: timing, clonality studies, cytogenetic associations, and role in leukemic transformation. Blood 108, 3548–3555. [DOI] [PubMed] [Google Scholar]
- Chen GL, and Prchal JT (2007). X-linked clonality testing: interpretation and limitations. Blood 110, 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, Ingle CE, Dermitzakis ET, Campbell PJ, and Green AR (2010). Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell 18, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Reis Monte-Mór B, Plo I, da Cunha AF, Costa GG, de Albuquerque DM, Jedidi A, Villeval JL, Badaoui S, Lorand-Metze I, Pagnano KB, et al. (2009). Constitutive JunB expression, associated with the JAK2 V617F mutation, stimulates proliferation of the erythroid lineage. Leukemia 23, 144–152. [DOI] [PubMed] [Google Scholar]
- Dai C, Chung IJ, and Krantz SB (2005). Increased erythropoiesis in polycythemia vera is associated with increased erythroid progenitor proliferation and increased phosphorylation of Akt/PKB. Exp. Hematol 33, 152–158. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, and Kouzarides T (2009). JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 461, 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. (2009). Mutation in TET2 in myeloid cancers. N. Engl. J. Med 360, 2289–2301. [DOI] [PubMed] [Google Scholar]
- Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, Cattoretti G, and Ye BH (2008). Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 111, 1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumon S, Santos SC, Debierre-Grockiego F, Gouilleux-Gruart V, Cocault L, Boucheron C, Mollat P, Gisselbrecht S, and Gouilleux F (1999). IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene 18, 4191–4199. [DOI] [PubMed] [Google Scholar]
- Dusa A, Mouton C, Pecquet C, Herman M, and Constantinescu SN (2010). JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PLoS ONE 5, e11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott NE, Cleveland SM, Grann V, Janik J, Waldmann TA, and Davé UP (2011). FERM domain mutations induce gain of function in JAK3 in adult T-cell leukemia/lymphoma. Blood 118, 3911–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, Ariola C, Fodale V, Clappier E, Paoloni F, et al. (2008). Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J. Exp. Med 205, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi-Tago M, Tago K, Sumi K, Abe M, Aizu-Yokota E, Oshio T, Sonoda Y, and Kasahara T (2009). The acute lymphoblastic leukemia-associated JAK2 L611S mutant induces tumorigenesis in nude mice. J. Biol. Chem 284, 12680–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi-Tago M, Tago K, Abe M, Sonoda Y, and Kasahara T (2010).STAT5 activation is critical for the transformation mediated by myeloproliferative disorder-associated JAK2 V617F mutant. J. Biol. Chem 285, 5296–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garçon L, Rivat C, James C, Lacout C, Camara-Clayette V, Ugo V, Lecluse Y, Bennaceur-Griscelli A, and Vainchenker W (2006). Constitutive activation of STAT5 and Bcl-xL overexpression can induce endogenous erythroid colony formation in human primary cells. Blood 108, 1551–1554. [DOI] [PubMed] [Google Scholar]
- Giordanetto F, and Kroemer RT (2002). Prediction of the structure of human Janus kinase 2 (JAK2) comprising JAK homology domains 1 through 7. Protein Eng. 15, 727–737. [DOI] [PubMed] [Google Scholar]
- Gozgit JM, Bebernitz G, Patil P, Ye M, Parmentier J, Wu J, Su N, Wang T, Ioannidis S, Davies A, et al. (2008). Effects of the JAK2 inhibitor, AZ960, on Pim/BAD/BCL-xL survival signaling in the human JAK2 V617F cell line SET-2. J. Biol. Chem 283, 32334–32343. [DOI] [PubMed] [Google Scholar]
- Griesinger F, Hennig H, Hillmer F, Podleschny M, Steffens R, Pies A, Wormann B, Haase D, and Bohlander SK (2005). A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes Chromosomes Cancer 44, 329–333. [DOI] [PubMed] [Google Scholar]
- Haeno H, Levine RL, Gilliland DG, and Michor F (2009). A progenitor cell origin of myeloid malignancies. Proc. Natl. Acad. Sci. USA 106, 16616–16621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CN, Bareford D, Butt N, Campbell P, Conneally E, Drummond M, Erber W, Everington T, Green AR, Hall GW, et al. ; British Committee for Standards in Haematology. (2010). Guideline for investigation and management of adults and children presenting with a thrombocytosis. Br. J. Haematol 149, 352–375. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, et al. (2010a). Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 115, 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. (2010b). Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116, 4874–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornakova T, Staerk J, Royer Y, Flex E, Tartaglia M, Constantinescu SN, Knoops L, and Renauld JC (2009). Acute lymphoblastic leukemia-associated JAK1 mutants activate the Janus kinase/STAT pathway via interleukin-9 receptor alpha homodimers. J. Biol. Chem 284, 6773–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, et al. (2004). MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6, 587–596. [DOI] [PubMed] [Google Scholar]
- Ishii T, Zhao Y, Sozer S, Shi J, Zhang W, Hoffman R, and Xu M (2007). Behavior of CD34+ cells isolated from patients with polycythemia vera in NOD/SCID mice. Exp. Hematol 35, 1633–1640. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. (2005). A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148. [DOI] [PubMed] [Google Scholar]
- James C, Mazurier F, Dupont S, Chaligne R, Lamrissi-Garcia I, Tulliez M, Lippert E, Mahon FX, Pasquet JM, Etienne G, et al. (2008). The hematopoietic stem cell compartment of JAK2V617F-positive myeloproliferative disorders is a reflection of disease heterogeneity. Blood 112, 2429–2438. [DOI] [PubMed] [Google Scholar]
- Jamieson CH, Gotlib J, Durocher JA, Chao MP, Mariappan MR, Lay M, Jones C, Zehnder JL, Lilleberg SL, and Weissman IL (2006). The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc. Natl. Acad. Sci. USA 103, 6224–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek J, Oki Y, Gharibyan V, Bueso-Ramos C, Prchal JT, Verstovsek S, Beran M, Estey E, Kantarjian HM, and Issa JP (2005). JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood 106, 3370–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, et al. (2005). Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 106, 2162–2168. [DOI] [PubMed] [Google Scholar]
- Jones AV, Chase A, Silver RT, Oscier D, Zoi K, Wang YL, Cario H, Pahl HL, Collins A, Reiter A, et al. (2009). JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet 41, 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos S, Küpper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, Marynen P, Möller P, Pfreundschuh M, Trümper L, and Lichter P(2000). Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 60, 549–552. [PubMed] [Google Scholar]
- Ju W, Zhang M, Jiang JK, Thomas CJ, Oh U, Bryant BR, Chen J, Sato N, Tagaya Y, Morris JC, et al. (2011). CP-690,550, a therapeutic agent, inhibits cytokine-mediated Jak3 activation and proliferation of T cells from patients with ATL and HAM/TSP. Blood 117, 1938–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamishimoto J, Tago K, Kasahara T, and Funakoshi-Tago M (2011). Akt activation through the phosphorylation of erythropoietin receptor at tyrosine 479 is required for myeloproliferative disorder-associated JAK2 V617F mutant-induced cellular transformation. Cell. Signal 23, 849–856. [DOI] [PubMed] [Google Scholar]
- Kearney L, Gonzalez De Castro D, Yeung J, Procter J, Horsley SW, Eguchi-Ishimae M, Bateman CM, Anderson K, Chaplin T, Young BD, et al. (2009). Specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood 113, 646–648. [DOI] [PubMed] [Google Scholar]
- Kelly JA, Spolski R, Kovanen PE, Suzuki T, Bollenbacher J, Pise-Masison CA, Radonovich MF, Lee S, Jenkins NA, Copeland NG, et al. (2003). Stat5 synergizes with T cell receptor/antigen stimulation in the development of lymphoblastic lymphoma. J. Exp. Med 198, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A (2006). The role of Janus kinases in haemopoiesis and haematological malignancy. Br. J. Haematol 134, 366–384. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, Bass A, Marubayashi S, Heguy A, Garcia-Manero G, et al. (2009). A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet 41, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, and Skoda RC (2005). A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med 352, 1779–1790. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Teo SS, Li S, Theocharides A, Buser AS, Tichelli A, and Skoda RC (2006). Acquisition of the V617F mutation of JAK2 is a late genetic event in a subset of patients with myeloproliferative disorders. Blood 108, 1377–1380. [DOI] [PubMed] [Google Scholar]
- Kratz CP, Böll S, Kontny U, Schrappe M, Niemeyer CM, and Stanulla M (2006). Mutational screen reveals a novel JAK2 mutation, L611S, in a child with acute lymphoblastic leukemia. Leukemia 20, 381–383. [DOI] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffé M, Berthou C, Lessard M, Berger R, Ghysdael J, and Bernard OA (1997). A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Monni R, Dumon S, Mauchauffé M, Mayeux P, Gouilleux F, Berger R, Gisselbrecht S, Ghysdael J, and Bernard OA (2000). Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood 95, 2076–2083. [PubMed] [Google Scholar]
- Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, and Staudt LM (2005). Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin. Cancer Res 11, 28–40. [PubMed] [Google Scholar]
- Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, and Staudt LM (2008). Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-kappaB pathways in subtypes of diffuse large B-cell lymphoma. Blood 111, 3701–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren O, Goldin LR, Kristinsson SY, Helgadottir EA, Samuelsson J, and Björkholm M (2008). Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24,577 first-degree relatives of 11,039 patients with myeloproliferative neoplasms in Sweden. Blood 112, 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, and Garcia KC (2008). Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre H, Cervera N, Finetti P, Delhommeau F, Vainchenker W, Bertucci F, and Birnbaum D (2006). Oncogenic kinases of myeloproliferative disorders induce both protein synthesis and G1 activators. Leukemia 20, 1885–1888. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. (2008). Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA 105, 13520–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E, Berger R, Clark JJ, Willis SG, Nguyen KT, et al. (2005a). The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 106, 3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. (2005b). Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397. [DOI] [PubMed] [Google Scholar]
- Li G, Miskimen KL, Wang Z, Xie XY, Tse W, Gouilleux F, Moriggl R, and Bunting KD (2010a). Effective targeting of STAT5-mediated survival in myeloproliferative neoplasms using ABT-737 combined with rapamycin. Leukemia 24, 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Spensberger D, Ahn JS, Anand S, Beer PA, Ghevaert C, Chen E, Forrai A, Scott LM, Ferreira R, et al. (2010b). JAK2 V617F impairs hematopoietic stem cell function in a conditional knock-in mouse model of JAK2 V617F-positive essential thrombocythemia. Blood 116, 1528–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kent DG, Chen E, and Green AR (2011). Mouse models of myeloproliferative neoplasms: JAK of all grades. Dis. Model. Mech 4, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh ML (2011). Recent advances in the pathogenesis and treatment of juvenile myelomonocytic leukaemia. Br. J. Haematol 152, 677–687. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, and Ng HH (2007). Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Levine R, Tong W, Wernig G, Pikman Y, Zarnegar S, Gilliland DG, and Lodish H (2005). Expression of a homodimeric type I cytokine receptor is required for JAK2V617F-mediated transformation. Proc. Natl. Acad. Sci. USA 102, 18962–18967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, and Rossjohn J (2006). The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood 107, 176–183. [DOI] [PubMed] [Google Scholar]
- Ma W, Kantarjian H, Zhang X, Yeh CH, Zhang ZJ, Verstovsek S, and Albitar M (2009). Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias. J. Mol. Diagn 11, 49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S, Ben-Abdelali R, Settegrana C, Radford-Weiss I, Debre M, Beldjord K, Macintyre EA, Villeval JL, Vainchenker W, Berger R, et al. (2007). Novel activating JAK2 mutation in a patient with Down syndrome and B-cell precursor acute lymphoblastic leukemia. Blood 109, 2202–2204. [DOI] [PubMed] [Google Scholar]
- Malinge S, Ragu C, Della-Valle V, Pisani D, Constantinescu SN, Perez C, Villeval JL, Reinhardt D, Landman-Parker J, Michaux L, et al. (2008). Activating mutations in human acute megakaryoblastic leukemia. Blood 112, 4220–4226. [DOI] [PubMed] [Google Scholar]
- Mark HF, Sotomayor EA, Nelson M, Chaves F, Sanger WG, Kaleem Z, and Caughron SK (2006). Chronic idiopathic myelofibrosis (CIMF) resulting from a unique 3;9 translocation disrupting the janus kinase 2 (JAK2) gene. Exp. Mol. Pathol 81, 217–223. [DOI] [PubMed] [Google Scholar]
- Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell RG, and Kanakura Y (1999). Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin MF, Reilly O, Campbell P, Bareford D, Green AR, Harrison CN, Conneally E, and Ryan K National Cancer Research Institute, Myeloproliferative Disorder Subgroup; British Committee for Standards in Haematology. (2007). Amendment to the guideline for diagnosis and investigation of polycythaemia/erythrocytosis. Br. J. Haematol 138, 821–822. [DOI] [PubMed] [Google Scholar]
- Mesa RA, Tefferi A, Lasho TS, Loegering D, McClure RF, Powell HL, Dai NT, Steensma DP, and Kaufmann SH (2006). Janus kinase 2 (V617F) mutation status, signal transducer and activator of transcription-3 phosphorylation and impaired neutrophil apoptosis in myelofibrosis with myeloid metaplasia. Leukemia 20, 1800–1808. [DOI] [PubMed] [Google Scholar]
- Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, et al. (2010). Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 17, 584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, et al. (2009a). Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat. Genet 41, 1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, et al. (2009b). JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 106, 9414–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebral K, Denk D, Attarbaschi A, König M, Mann G, Haas OA, and Strehl S (2009). Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 23, 134–143. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. (2011). Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzveig RH, Swierczek SI, Jelinek J, Gaikwad A, Liu E, Verstovsek S, Prchal JF, and Prchal JT (2007). Polycythemia vera is not initiated by JAK2V617F mutation. Exp. Hematol 35, 32–38. [DOI] [PubMed] [Google Scholar]
- Olcaydu D, Harutyunyan A, Jäger R, Berg T, Gisslinger B, Pabinger I, Gisslinger H, and Kralovics R (2009). A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet 41, 450–454. [DOI] [PubMed] [Google Scholar]
- Parikh C, Subrahmanyam R, and Ren R (2006). Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood 108, 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez B, Kosmider O, Cassinat B, Renneville A, Lachenaud J, Kaltenbach S, Bertrand Y, Baruchel A, Chomienne C, Fontenay M, et al. (2010). Genetic typing of CBL, ASXL1, RUNX1, TET2 and JAK2 in juvenile myelomonocytic leukaemia reveals a genetic profile distinct from chronic myelomonocytic leukaemia. Br. J. Haematol 151, 460–468. [DOI] [PubMed] [Google Scholar]
- Plo I, Nakatake M, Malivert L, de Villartay JP, Giraudier S, Villeval JL, Wiesmuller L, and Vainchenker W (2008). JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood 112, 1402–1412. [DOI] [PubMed] [Google Scholar]
- Poitras JL, Dal Cin P, Aster JC, Deangelo DJ, and Morton CC (2008). Novel SSBP2-JAK2 fusion gene resulting from a t(5;9)(q14.1;p24.1) in pre-B acute lymphocytic leukemia. Genes Chromosomes Cancer 47, 884–889. [DOI] [PubMed] [Google Scholar]
- Puigdecanet E, Espinet B, Lozano JJ, Sumoy L, Bellosillo B, Arenillas L, Alvarez-Larrán A, Solé F, Serrano S, Besses C, and Florensa L (2008). Gene expression profiling distinguishes JAK2V617F-negative from JAK2V617F-positive patients in essential thrombocythemia. Leukemia 22, 1368–1376. [DOI] [PubMed] [Google Scholar]
- Reiter A, Walz C, Watmore A, Schoch C, Blau I, Schlegelberger B, Berger U, Telford N, Aruliah S, Yin JA, et al. (2005). The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer Res. 65, 2662–2667. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. (2003). Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favourable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med 198, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL, et al. (2010). Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 18, 590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, et al. (2009). Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood 114, 2688–2698. [DOI] [PubMed] [Google Scholar]
- Santos FP, Kantarjian HM, Jain N, Manshouri T, Thomas DA, Garcia-Manero G, Kennedy D, Estrov Z, Cortes J, Verstovsek S (2010). Phase 2 study of CEP-701, an orally available JAK2 inhibitor, in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. Blood 115, 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Toki T, Kanezaki R, Xu G, Terui K, Kanegane H, Miura M, Adachi S, Migita M, Morinaga S, et al. (2008). Functional analysis of JAK3 mutations in transient myeloproliferative disorder and acute megakaryoblastic leukaemia accompanying Down syndrome. Br. J. Haematol 141, 681–688. [DOI] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, et al. (2003). The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 102, 3871–3879. [DOI] [PubMed] [Google Scholar]
- Schaub FX, Jäger R, Looser R, Hao-Shen H, Hermouet S, Girodon F, Tichelli A, Gisslinger H, Kralovics R, Skoda RC (2009). Clonal analysis of deletions on chromosome 20q and JAK2-V617F in MPD suggests that del20q acts independently and is not one of the predisposing mutations for JAK2-V617F. Blood 113, 2022–2027. [DOI] [PubMed] [Google Scholar]
- Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, Lee CK, Gerthner R, Kitamura T, Frantsve, et al. (2000). Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 6, 693–704. [DOI] [PubMed] [Google Scholar]
- Scott LM, Campbell PJ, Baxter EJ, Todd T, Stephens P, Edkins S, Wooster R, Stratton MR, Futreal PA,and Green AR (2005). The V617F JAK2 mutation is uncommon in cancers and in myeloid malignancies other than the classic myeloproliferative disorders. Blood 106, 2920–2921. [DOI] [PubMed] [Google Scholar]
- Scott LM, Scott MA, Campbell PJ, Green AR (2006). Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood 108, 2435–2437. [DOI] [PubMed] [Google Scholar]
- Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, et al. (2007). JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med 356, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuto A, Kujawski M, Kowolik C, Krymskaya L, Wang L, Weiss LM, Digiusto D, Yu H, Forman S, Jove R (2011). STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 71, 3182–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Calhoun HC, Xia F, Li J, Le L, and Li WX (2006). JAK signalling globally counteracts heterochromatic gene silencing. Nat. Genet 38, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, Cario G, Cazzaniga G, Kulozik AE, Stanulla M, et al. (2011). Gain-of-function mutations in interleukin-7 receptor-α (IL7R) in childhood acute lymphoblastic leukemias. J. Exp. Med 208, 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, Fishel R, Skorski T (2002). Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol. Cell. Biol 22, 4189–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J, Kallin A, Demoulin JB, Vainchenker W, Constantinescu SN (2005). JAK1 and Tyk2 activation by the homologous polycythemia vera JAK2 V617F mutation: cross-talk with IGF1 receptor. J. Biol. Chem 280, 41893–41899. [DOI] [PubMed] [Google Scholar]
- Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A (2005). The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood 106, 1207–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, andVardiman JW (2008). Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia 22, 14–22. [DOI] [PubMed] [Google Scholar]
- Teofili L, Martini M, Cenci T, Petrucci G, Torti L, Storti S, Guidi F, Leone G, Larocca LM (2007). Different STAT-3 and STAT-5 phosphorylation discriminates among Ph-negative chronic myeloproliferative diseases and is independent of the V617F JAK-2 mutation. Blood 110, 354–359. [DOI] [PubMed] [Google Scholar]
- Theocharides A, Boissinot M, Girodon F, Garand R, Teo SS, Lippert E, Talmant P, Tichelli A, Hermouet S, Skoda RC (2007). Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood 110, 375–379. [DOI] [PubMed] [Google Scholar]
- Ugo V, Marzac C, Teyssandier I, Larbret F, Lécluse Y, Debili N, Vainchenker W, Casadevall N (2004). Multiple signaling pathways are involved in erythropoietin-independent differentiation of erythroid progenitors in polycythemia vera. Exp. Hematol 32, 179–187. [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O (2011). The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol 18, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pelt K, Nollet F, Selleslag D, Knoops L, Constantinescu SN, Criel A, andBilliet J (2008). The JAK2V617F mutation can occur in a hematopoietic stem cell that exhibits no proliferative advantage: a case of human allogeneic transplantation. Blood 112, 921–922. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, and Tefferi A (2008). Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia 22, 1299–1307. [DOI] [PubMed] [Google Scholar]
- Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, Estrov Z, Fridman JS, Bradley EC, Erickson-Viitanen S, et al. (2010). Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N. Engl. J. Med 363, 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M, Goss VL, Lee KA, Eide CA, Wong MJ, et al. (2006). Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 10, 65–75. [DOI] [PubMed] [Google Scholar]
- Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, Zaleskas VM, and Van Etten RA (2012). Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and Jak2V617F in mice. Blood, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, and Shi Y (2006). Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Zhao Y, Mitaksov V, Fremont DH, Kasai Y, Molitoris A, Ries RE, Miner TL, McLellan MD, DiPersio JF, et al. (2008). Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood 111, 4809–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Hutchison RE, and Mohi G (2011a). Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Huang Y, Watkins AJ, Kocialkowski S, Zeng N, Hamoudi RA, Isaacson PG, de Leval L, Wotherspoon A, and Du MQ (2011b). BCR and TLR signalling pathways are recurrently targeted by genetic changes in splenic marginal zone lymphomas. Haematologica, in press. Published online November 18, 2011 10.3324/haematol.2011.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, and Stark GR (2007). Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 21, 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, West N, Xiao Y, Brown JR, Mitsiades, et al. (2010). Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 107, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, Tritapoe J, Hixon JA, Silveira AB, Cardoso BA, et al. (2011). Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat. Genet 43, 932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Follows GA, Beer PA, Scott LM, Huntly BJ, Green AR, and Alexander DR (2008). Inhibition of the Bcl-xL deamidation pathway in myeloproliferative disorders. N. Engl. J. Med 359, 2778–2789. [DOI] [PubMed] [Google Scholar]