SUMMARY
Homeostatic control of core body temperature is essential for survival. Temperature is sensed by specific neurons, in turn eliciting both behavioral (i.e., locomotion) and physiologic (i.e., thermogenesis, vasodilatation) responses. Here, we report that a population of GABAergic (Vgat-expressing) neurons in the dorsolateral portion of the dorsal raphe nucleus (DRN), hereafter DRNVgat neurons, are activated by ambient heat and bidirectionally regulate energy expenditure through changes in both thermogenesis and locomotion. We find that DRNVgat neurons innervate brown fat via a descending projection to the raphe pallidus (RPa). These neurons also densely innervate ascending targets implicated in the central regulation of energy expenditure, including the hypothalamus and extended amygdala. Optogenetic stimulation of different projection targets reveals that DRNVgat neurons are capable of regulating thermogenesis through both a “direct” descending pathway through the RPa and multiple “indirect” ascending pathways. This work establishes a key regulatory role for DRNVgat neurons in controlling energy expenditure.
Graphical Abstract
In Brief
A population of heat-activated GABAergic neurons in the dorsal raphe nucleus (DRN) regulate energy expenditure through changes in thermogenesis and locomotion.
INTRODUCTION
Core body temperature is maintained with great precision in mammals. This is accomplished by coordinated thermoregulatory processes that include both behavioral and autonomic components, such as shivering, adaptive thermogenesis, and skin vasodilatation. In addition to regulating core temperature, these processes are also fundamental to maintaining energy balance, and concomitantly, body weight (Münzberg et al., 2016; Saltzman and Roberts, 1995). While several recent reports have identified neuronal populations that regulate thermogenesis, the neural mechanisms by which these pathways modulate the autonomic nervous system to control these processes is incompletely understood.
The anterior hypothalamus, particularly the preoptic area (POA), is a key node controlling thermoregulation (Hemingway et al., 1954; Magoun et al., 1938). Recent studies of the POA have identified cell types that are warm-sensitive and project broadly throughout the brain to regulate core body temperature. This is brought about in part by regulating the activity of interscapular brown adipose tissue (iBAT) and skin vasodilatation, as well as heat/cold seeking and avoidance behaviors (Tan et al., 2016; Zhao et al., 2017). While these data have established that the POA can sense and respond to changes in temperature, many other nuclei are likely to be directly capable of regulating thermogenesis.
In the current work, we use an unbiased whole-brain screen to identify brain regions that are activated in response to a thermal challenge. We find that GABAergic (Vgat-expressing) neurons within the DRN are activated by an increase in ambient temperature suggesting a possible role for these neurons in regulating body temperature. Indeed, converging lines of functional and anatomic evidence have implicated several brainstem nuclei, including the DRN, in the central regulation of thermogenesis. For example, pharmacological and electrophysiological manipulations of the DRN have been shown to lead to changes in core and iBAT temperature (Dib et al., 1994; Hillegaart, 1991), and neurons within the DRN have also been suggested to be sensitiveto local and ambient increases in temperature (Hori and Harada, 1976; Weiss and Aghajanian, 1971). The DRN and bordering ventrolateral periaqueductal gray (vlPAG) also receive inputs from warm-sensitive neurons within the medial preoptic area (MPOA) (Tan et al., 2016). Finally, the DRN potently regulates feeding, which is often linked to changes in thermogenesis as part of a general mechanism controlling energy homeostasis (Nectow et al., 2017b). However, the identity and functional properties of these brainstem populations in controlling thermogenesis has not been evaluated.
We thus set out to determine whether Vgat-expressing neurons within the DRN can regulate core temperature. We find that chemogenetic modulation of these neurons potently regulates energy expenditure through changes in both thermogenesis and locomotor activity. We also find that these neurons innervate iBAT through a descending projection to the raphe pallidus (RPa). These neurons also regulate thermogenesis via ascending projections to numerous loci previously implicated in the regulation of energy balance, such as the bed nucleus of the stria terminalis (BNST), dorsomedial hypothalamus (DMH), and MPOA. Together, these studies identify the DRN as an important node embedded within a distributed thermoregulatory circuit, and reveal a novel mechanism by which this brainstem nucleus regulates energy expenditure.
RESULTS
Whole-Brain Activity Mapping to Identify Neurons within a Heat-Sensing Circuit
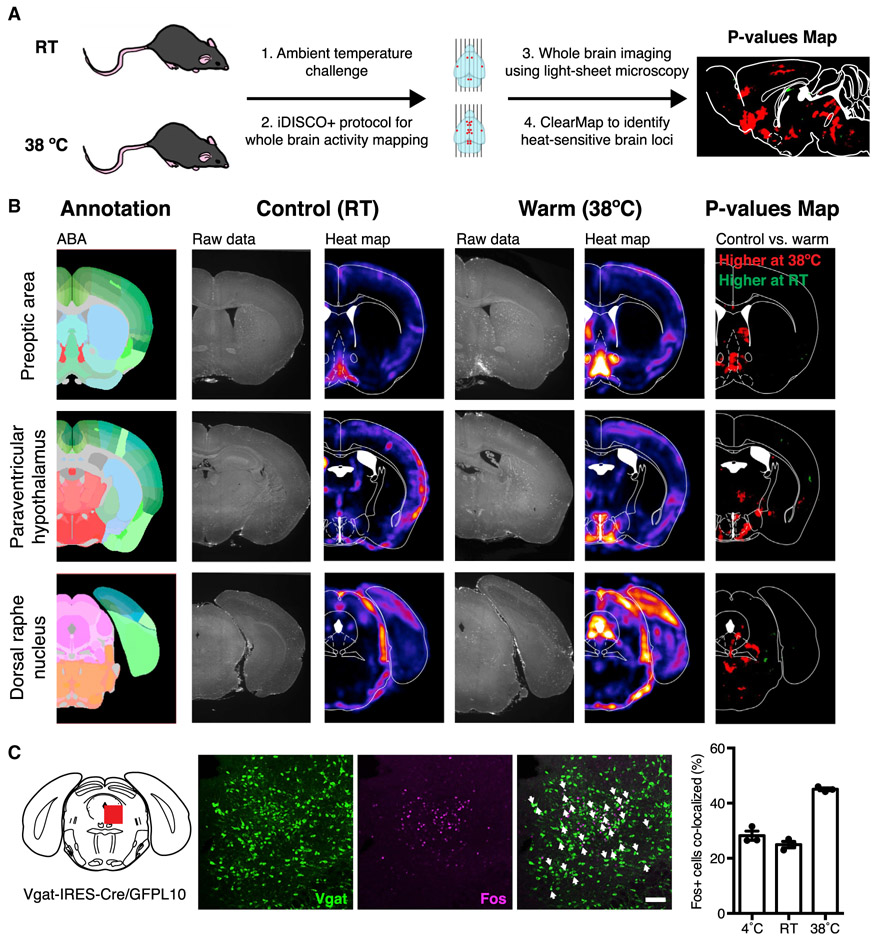
We set out to identify cell types within the brain that sense and respond to increases in ambient temperature using whole-brain activity mapping. Wild type mice were either housed at room temperature (RT) or exposed to a heat challenge of 38°C for 4 h. After brain isolation, we used whole-brain clearing technology in tandem with whole-mount immunostaining (iDISCO+) (Renier et al., 2016) to identify sites of immediate-early gene Fos expression (Figure 1A). ClearMap software was then used for unbiased cell detection and registration to the Allen Brain Atlas (Renier et al., 2016). In this unbiased manner, we identified a number of brain regions with significantly increased levels of Fos expression after heat exposure.
Figure 1. Whole-Brain Fos Mapping Identifies Heat-Sensitive Neurons.
(A) Schema for whole-brain activity mapping in response to thermal challenge.
(B) Whole-brain imaging and ClearMap results for the POA (top), hypothalamus (middle), and DRN (bottom). p values maps display differentially activated brain loci in 38°C versus RT (n = 5 mice per group).
(C) Vgat and Fos colocalization after 38°C thermal challenge. Significantly increased Fos and Vgat colocalization was observed at 38°C, compared to room temperature (p < 0.0001). Unpaired t test comparing treatments (n = 3 mice per group). White arrows designate double-labeled cells.
Scale bar, 100 mm. Data are presented as mean ± SEM.
Several of these identified brain regions have previously been implicated in thermoregulation, including the ventromedial preoptic area (VMPO), DMH, and paraventricular hypothalamus (PVH) (Figures 1B and S1A) (Morrison and Nakamura, 2011; Tan et al., 2016; Zhao et al., 2017). Indeed, the most significant activation was observed in the VMPO and the PVH, which are well-established sites of thermoregulation (Figure 1B; Table S1). After validating that this unbiased approach was capable of detecting known heat-sensitive neurons distributed throughout the brain, we next screened for additional brain regions that have not been directly implicated in the central regulation of thermogenesis.
One of the brain regions that showed significant neural activation in the warm (38°C) condition versus RT was the DRN. This region showed a time dependent, 3.8-fold increase in the number Fos+ neurons in mice treated at 38°C, as compared to RT controls (Figures 1B and S1B-S1D). Previous studies from our group and others have implicated the DRN in the control of food intake and locomotor activity, but a possible role for specific neurons in this region in regulating thermogenesis has not been tested (Dib et al., 1994; Nectow et al., 2017b; Veasey et al., 1997; Warden et al., 2012; Waterhouse et al., 2004). The pattern of neuronal activation we observed was predominantly in the dorsolateral aspects of the DRN, adjacent to, and in some cases, extending into the vlPAG. This sub-region of the DRN predominantly contains GABAergic neurons (Hioki et al., 2010), which led us to hypothesize that the heat-sensitive neurons in the DRN could be GABAergic.
To test this, we performed immunohistochemistry to co-localize Vgat, a marker of GABAergic neurons, and Fos in the DRN. Toward this end, we crossed Vgat-IRES-Cre knock-in mice to Cre-dependent GFPL10 reporter mice followed by exposure of animals to RT or 38°C. We found a significant increase in Fos and Vgat co-localization after a thermal challenge of 38°C (45% of Fos+ neurons were Vgat+ at 38°C, as compared to 20% at RT) (Figure 1C), corresponding to an ~7-fold increase in Vgat-positive neurons expressing Fos (Figure S1E). These results demonstrate that GABAergic neurons within the DRN (hereafter, DRNVgat neurons) are activated by increased ambient temperature.
DRNVgat Neuron Activation Suppresses Energy Expenditure through Multiple Mechanisms, Including iBAT Thermogenesis
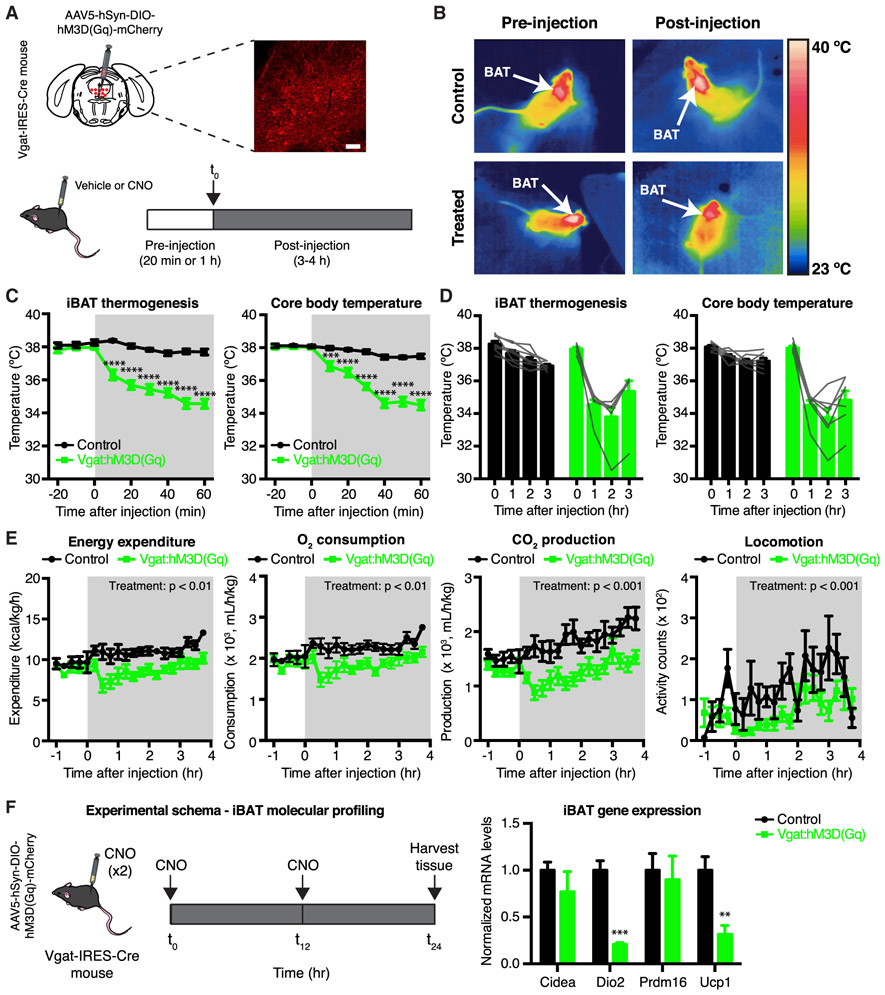
To assess whether DRNVgat neurons regulate thermogenesis, we used chemogenetics to activate these neurons and monitored changes in iBAT, core, and tail temperature. The activating designer receptor exclusively activated by designer drug (DREADD) hM3D(Gq) was targeted to Vgat-expressing neurons in the DRN and bordering ventral portion of vlPAG (for simplicity, referred to as DRNVgat neurons) by injecting a Cre-dependent AAV5-hSyn-DIO-hM3D(Gq)-mCherry virus (or control virus) into the DRN of Vgat-IRES-Cre mice (Figure 2A, top). After at least 3 weeks, mice received a set of sham injections to acclimate them to the procedure, after which they were given intraperitoneal (i.p.) injections of either vehicle (saline) or the DREADD ligand clozapine N-oxide (CNO) (Figure 2A, bottom). Changes in iBAT, core, and tail temperature were measured between 20 min pre-injection, through 3 h post-injection. iBAT temperature was measured initially using an infrared thermography camera for thermal imaging of the shaved interscapular region (Figure 2B) and was later measured using implantable temperature transponders implanted in iBAT; the data from these two methods were equivalent. Core body temperature was measured using an anal probe, while tail temperature was also assessed using thermal imaging.
Figure 2. Chemogenetic Activation of DRNVgat Neurons Suppresses Energy Expenditure.
(A) Experimental schema and representative IHC for DRNVgat neurons infected with excitatory DREADD hM3D(Gq).
(B) Representative thermal images of iBAT (control or Vgat:hM3D(Gq)) before and after injection of CNO. Chemogenetic activation of DRNVgat neurons suppresses iBAT thermogenesis.
(C and D) Short- and long-term changes in iBAT and core temperature after chemogenetic activation of DRNVgat neurons. (C) Activation of DRNVgat neurons significantly suppresses iBAT (treatment: p < 0.0001) and core temperature (treatment: p < 0.0001). Two-way repeated measures (RM) ANOVA comparing control and treated groups (n = 7–8 mice per group). (D) Average iBAT and core temperature at 0–3 h post-CNO injection. iBAT (p < 0.0001) and core (p < 0.0001) temperatures remain significantly suppressed 3 h post-CNO injection. Two-way RM ANOVA comparing t = 0 and t = 3 h post-CNO injection (n = 7–8 mice per group).
(E) Chemogenetic activation of DRNVgat neurons suppresses locomotor activity (treatment: p < 0.001), oxygen consumption (treatment: p < 0.01), carbon dioxide production (treatment: p < 0.001), and total energy expenditure (treatment: p < 0.01). Two-way RM ANOVA comparing control and treated groups after CNO injection (n = 7–8 mice per group).
(F) Molecular profiling of iBAT after chemogenetic activation of DRNVgat neurons. Genes tested: Cidea (p > 0.05), Dio2 (p < 0.001), Prdm16 (p > 0.05), Ucp1 (p < 0.01). Unpaired t tests comparing treatments (n = 4–5 samples per group).
Scale bar, 100 μm. **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are presented as mean ± SEM.
See also Figure S2.
We found that chemogenetically activating DRNVgat neurons significantly suppressed iBAT thermogenesis and concomitantly resulted in a significant decrease in core body temperature, effects which lasted as long as 3 h post-injection of CNO (Figures 2B-2D). No changes were observed in tail temperature at any of the time points that were tested (Figure S2D, left).
These studies were performed using a 2 × 2 design, where DREADD-expressing (Vgat:hM3D(Gq)) mice were compared to non-DREADD expressing (control) mice after either CNO or saline (vehicle) injections (on separate days). Critically, DREADD-expressing mice injected with vehicle (saline) instead of CNO failed to show changes in iBAT, core, or tail temperature within or between groups (Figures S2A and S2D). Similarly, control mice (not expressing DREADDs) given either CNO or saline also failed to show an effect on temperature (Figures S2A and S2D). Together, these data confirm that the observed suppression of iBAT and core body temperature was due specifically to DRNVgat neuron activation.
We next tested whether activation of DRNVgat neurons could suppress energy expenditure through mechanisms other than iBAT thermogenesis. Toward this end, a new cohort of Vgat-IRES-Cre mice were injected with AAV5-hSyn-DIO-hM3D(Gq)-mCherry and were then placed in metabolic cages, enabling automated phenotyping of whole animal metabolic activity. Activation of DRNVgat neurons by injection of CNO led to a significant decrease in locomotor activity, oxygen consumption, carbon dioxide production, and total energy expenditure, with no effect of CNO in animals that did not express the DREADD receptor (Figure 2E). We again used a 2 × 2 study design, confirming that none of these effects were seen after injections of CNO into control mice or saline into either treatment group (Figure S2B).
Given that activating DRNVgat neurons suppresses locomotor activity (Figure 2E, right), we next asked whether this effect accounted for some or all of the effects on thermogenesis. To address this question, we repeated the above studies in lightly sedated mice with suppressed locomotor activity. We found that chemogenetic activation of DRNVgat neurons in sedated animals was still able to significantly suppress both iBAT and core temperatures, with no effect on tail temperature (Figures S2C, left, and S2E, left). Here again, DREADD-expressing animals receiving saline (vehicle) injections and control animals receiving saline or CNO failed to show a similar effect (Figures S2C, right, and S2E, right). Consistent with an effect of DRNVgat neurons suppressing iBAT thermogenesis, we found that DRNVgat activation led to a significant decrease in mRNA levels of markers of iBAT thermogenesis, including Ucp1 and Dio2, but not Cidea or Prdm16 (Figure 2F). In aggregate, these data demonstrate that (1) activation of DRNVgat neurons reduces whole body energy expenditure by suppressing both thermogenesis and locomotor activity, (2) the effect on thermogenesis is observed even with the experimental suppression of locomotor activity, and (3) this is associated with a decreased expression of genes that are part of the molecular program regulating brown fat thermogenesis.
Inhibition of DRNVgat Neurons Augments Energy Expenditure Independent of Changes in iBAT Thermogenesis
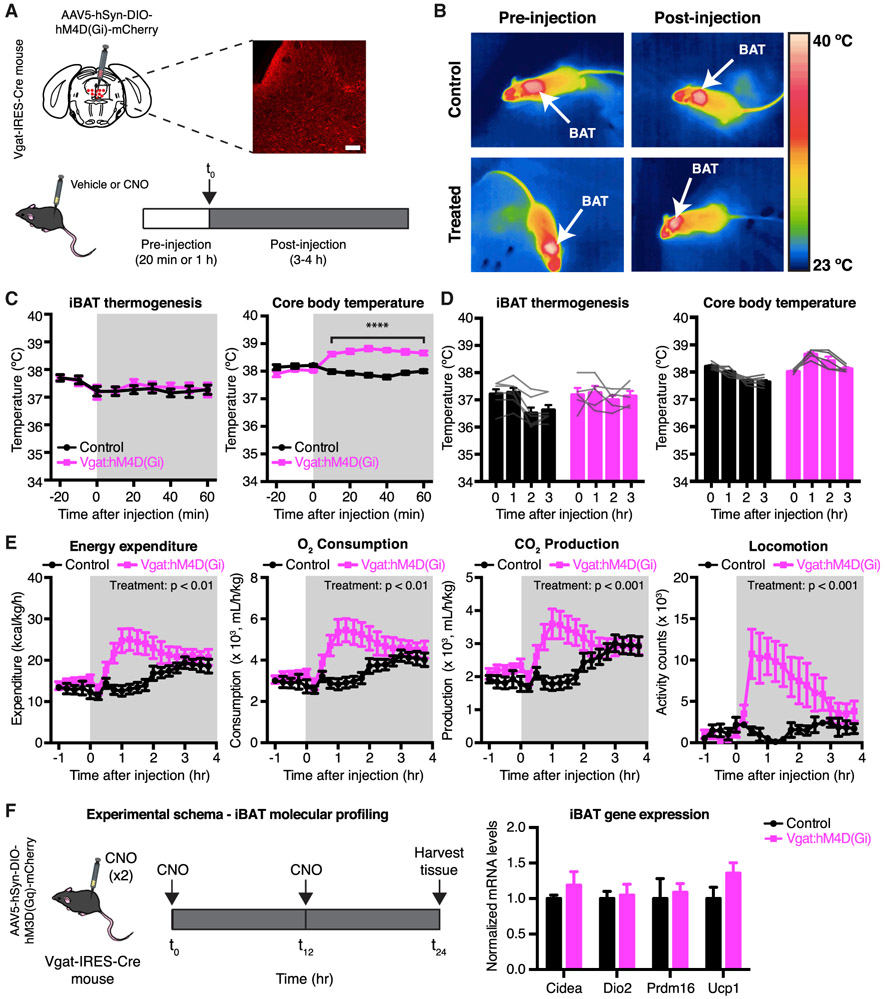
We next investigated whether inhibition of DRNVgat neurons could augment energy expenditure. We transduced DRNVgat neurons with the inhibitory DREADD hM4D(Gi) by injecting the DRN of Vgat-IRES-Cre mice with Cre-dependent AAV5-hSyn-DIO-hM4D(Gi)-mCherry (Figure 3A, top). Following viral incubation and a series of sham injections, control and treated mice were then injected i.p. with either vehicle or CNO (vehicle and CNO studies were performed using the same 2 × 2 experimental design described above) (Figure 3A, bottom). In contrast to activation of DRNVgat neurons, chemogenetic inhibition of these neurons did not affect iBAT temperature, as assessed by infrared thermography (Figure 3B). To ensure that the lack of thermogenic effects was not an artifact of the limited sensitivity of infrared thermography, we repeated these studies using implantable temperature transponders, while also monitoring for core and tail temperature, as in the chemogenetic activation studies. We again found that chemogenetic inhibition of DRNVgat neurons had no effect on iBAT temperature (Figures 3C and 3D). However, inhibiting these neurons nonetheless resulted in a small but significant increase in core body temperature (Figures 3C and 3D). No effect was observed on tail temperature, suggesting that this increase in core body temperature was not due to a change in vascular tone (Figure S3D). As expected, we did not observe any effect on iBAT, core, or tail temperature in control animals receiving CNO (Figures 3C and 3D), or after injection of vehicle into treated or control animals (Figure S3A), demonstrating that the observed effects are specific to inhibition of DRNVgat neurons.
Figure 3. Chemogenetic Inhibition of DRNVgat Neurons Augments Energy Expenditure.
(A) Experimental schema and representative IHC for DRNVgat neurons infected with inhibitory DREADD hM4D(Gi).
(B) Representative thermal images of iBAT (control or Vgat:hM3D(Gq)) before and after injection of CNO. Chemogenetic inhibition of DRNVgat neurons has no effect on iBAT thermogenesis.
(C and D) iBAT and core temperature after chemogenetic inhibition of DRNVgat neurons. (C) Inhibition of DRNVgat neurons has no effect on iBAT temperature (treatment: p > 0.05), but augments core temperature (treatment: p < 0.0001). Two-way RM ANOVA comparing control and treated groups (n = 5–7 mice per group). (D) Average iBAT (p > 0.05) and core temperature (p > 0.05) at 0 and 3 h post-CNO injection. Two-way RM ANOVA comparing t = 0 and t = 3 h post-CNO injection (n = 5–7 mice per group).
(E) Chemogenetic inhibition of DRNVgat neurons increases locomotor activity (treatment: p < 0.05), oxygen consumption (treatment: p < 0.05), carbon dioxide production (treatment: p = 0.087), and total energy expenditure (treatment: p < 0.05). Two-way RM ANOVA comparing control and treated groups after CNO injection (n = 7 mice per group).
(F) Molecular profiling of iBAT after chemogenetic inhibition of DRNVgat neurons. Genes tested: Cidea, Dio2, Prdm16, Ucp1 (all n.s.). Unpaired t tests comparing treatments (n = 6 samples per group).
Scale bar, 100 μm. ****p < 0.0001, n.s., not significant. Data are presented as mean ± SEM.
See also Figure S3.
The observation that DRNVgat neuron inhibition increases core temperature in the absence of an increased iBAT temperature (and produces no changes in peripheral vasodilatation) raised the possibility that this effect was mediated by an increase in locomotor activity. To address this, a separate cohort of Vgat-IRES-Cre mice was injected with AAV5-hSyn-DIO-hM4D(Gi)-mCherry, before being placed in metabolic cages. Inhibition of DRNVgat neurons resulted in a striking and significant increase in locomotor activity, as well as oxygen consumption and carbon dioxide production, with a concomitant increase in total energy expenditure, with no effects observed in CNO or saline injected control mice (Figures 3E and S3B). These results suggested that chemogenetic inhibition of DRNVgat neurons augments core body temperature secondary to an increase in locomotor activity.
To confirm that the increased temperature was a result of increased locomotor activity, we repeated the chemogenetic inhibition of DRNVgat neurons under a light sedating plane of anesthesia (as described earlier). In contrast to the earlier studies of neural activation, we found that light anesthesia completely blocked the effect of chemogenetically inhibiting DRNVgat neurons to increase core body temperature (Figures S3C, left). Similarly, inhibition of DRNVgat neurons under anesthesia also had no effect on iBAT thermogenesis or tail temperature, and no effects were observed on any of these parameters in saline or CNO treated mice (Figures S3C and S3E). These data show that inhibition of DRNVgat neurons raises core temperature by increasing locomotion without a discernible effect on iBAT thermogenesis. In line with these findings, we failed to see any changes in the expression level of thermogenic genes in iBAT (Figure 3F).
Overall, these results demonstrate that DRNVgat neurons can robustly and bidirectionally modulate overall energy expenditure, albeit via distinct mechanisms: activation of DRNVgat neurons suppresses energy expenditure through a reduction of both iBAT thermogenesis and locomotor activity, while inhibition of these neurons augments energy expenditure through an increase in locomotor activity, independent of iBAT thermogenesis.
The Neural Circuit Innervating iBAT
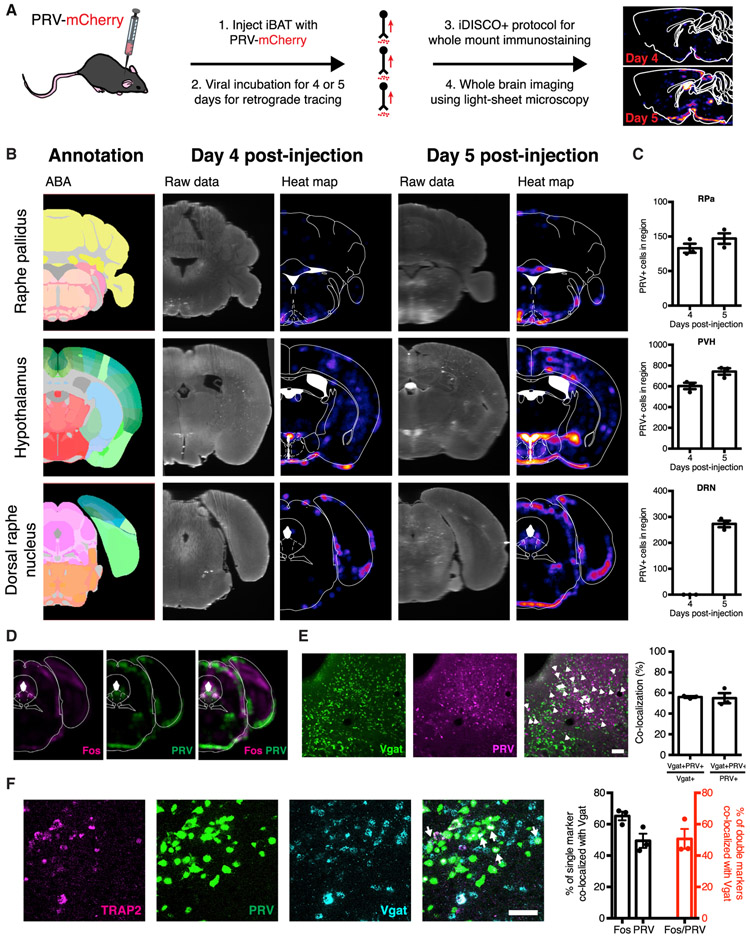
We next initiated studies to map the neural circuit through which DRNVgat neurons innervate brown fat by infecting iBAT with a recombinant PRV that enables retrograde tracing (and mCherry labeling) of neural circuits, in combination with whole-mount immunolabeling and clearing technology iDISCO+ (Figure 4A). The PRV strain we used is replication competent and constitutively expresses mCherry (PRV-lp298, hereafter PRV-mCherry) (Pomeranz et al., 2017). When injected into a neuron’s terminal field, PRV is taken up by local axon terminals and retrogradely transported to the cell’s soma, where it replicates and then “hops” to neurons directly upstream. Replicating PRV variants, such as PRV-mCherry, continue to “hop” synapses retrogradely until the animal succumbs to the infection (Figure 4A). The time course of the appearance of mCherry enables one to trace the neural pathways from iBAT to brain with the most proximate nodes in the circuit (with respect to iBAT) labeled at the earliest time points and more distal nodes being labeled at successively later time points.
Figure 4. DRNVgat Neurons Polysynaptically Project to Brown Fat.
(A) Schema for whole-brain mapping of neurons projecting polysynaptically to iBAT.
(B) iDISCO+ whole-brain imaging results (raw data and heatmaps) alongside the ABA annotation for the RPa (top), hypothalamus (middle), and DRN (bottom).
(C) Cell counts (manual) from brain slices for PRV-infected cellsat 4- and 5-days post-infection of PRV in iBAT. No changes were observed from day 4 to day 5 for RPa (n.s.), while significant increases in cell counts were observed for PVH (p < 0.05) and DRN (p < 0.0001). Unpaired t test comparing treatments (n = 3 mice per group).
(D) Regional colocalization (co-registration) between warm-sensitive (Fos-positive at 38°C) and iBAT-projecting (PRV-positive, at day 5 post-injection) neurons within the dorsolateral DRN (left).
(E) Colocalization (and quantification) between Vgat and PRV retrogradely labeled from iBAT 5 days post-injection (n = 3 mice per group). White arrows designate double-labeled cells. Scale bar, 100 μm.
(F) Cellular colocalization between DRNVgat neurons, tdTomato (warm-sensitive neurons), and EGFP (neurons polysynaptically innervating iBAT).
Scale bar, 50 μm.
n.s., not significant. Data are presented as mean ± SEM.
At 4 days post-infection, we found sparse viral expression within the brain, with robust labeling in the RPa and the PVH, known sites of sympathetic outflow to iBAT (Figure 4B; An et al., 2015; Cannon and Nedergaard, 2004; Ryu et al., 2015). These data suggest that these sites are early (i.e., first- or second-order) nodes in the brain circuit that innervates iBAT (Figure 4C). While there was no PRV expression in the DRN at day 4 post-injection, by day 5 we observed strong labeling of the DRN, raising the possibility that DRNVgat neurons innervate brown fat via projections to the PVH and/or RPa (Figures 4B and 4C).
The PRV labeling at day 5 post-injection was located predominantly in the dorsolateral portion of the DRN (bordering along, and in some cases extending into, the vlPAG), an area containing a significant number of Vgat-positive neurons. Image co-registration from iDISCO+ studies of Fos mapping and PRV tracing also demonstrated marked regional overlap between the heat-activated and polysynaptically iBAT-projecting DRN neurons, further suggesting that these could be DRNVgat neurons (Figure 4D). To confirm that DRNVgat neurons are upstream of the RPa and/or PVH, we repeated the PRV tracing studies in transgenic mice where DRNVgat neurons are fluorescently labeled using the progeny of a cross between Vgat-IRES-Cre mice and Cre-dependent GFPL10 reporter mice. PRV-mCherry was injected into the iBAT of these mice, and after a 5-day viral incubation period (allowing for labeling of the dorsolateral DRN), we found significant colocalization between Vgat+ neurons in this region and PRV, with ~56% of DRNVgat neurons showing expression of mCherry. Similarly, ~55% of mCherry-expressing (PRV+) neurons expressed Vgat (Figure 4E). These data together show that Vgat neurons in the dorsolateral compartment of the DRN (bordering along the vlPAG) are activated by ambient warmth and indirectly project to brown fat (Figures 1 and 4).
To confirm overlap between the heat-activated DRNVgat neurons and the DRNVgat neurons that project to iBAT, we performed activity-dependent genetic labeling of warm-activated DRN neurons in tandem with polysynaptic PRV mapping, followed by multiplex fluorescence in situ hybridization (FISH) to label Vgat-positive neurons. We labeled warm-activated DRN neurons by crossing TRAP2 mice, which express an activity-dependent, inducible iCreER transgene (activated by injection of 4-hydroxytamoxifen [4-OHT]), to Cre-dependent tdTomato reporter (Ai14) mice (Allen et al., 2017; Madisen et al., 2010). To fluorescently label warm-sensitive DRN neurons, these mice received an injection of 4-OHT (to activate expressed Cre), immediately followed by exposure to 38°C for 4 h. Using this approach, we found significant numbers of neurons within the dorsolateral DRN expressing tdTomato. After allowing for sufficient tdTomato expression in heat-activated neurons, we then injected a retrograde-tracing, replication-competent EGFP-expressing PRV (PRV-152, hereafter PRV-EGFP) into these mice (Smith et al., 2000). The mice were then sacrificed 5 days after PRV injection (the time point at which PRV labels DRNVgat neurons), and multiplex FISH was performed to co-localize warm-sensitive (tdTomato-positive), iBAT-projecting (EGFP-positive), Vgat-positive neurons within the DRN. We found significant numbers of Vgat-positive, warm-sensitive, iBAT-projecting neurons within the dorsolateral aspect of the DRN confirming that the thermally regulated DRNVgat neurons indirectly innervate iBAT (Figure 4F).
DRNVgat Neurons Project to Brain Regions Regulating Energy Expenditure
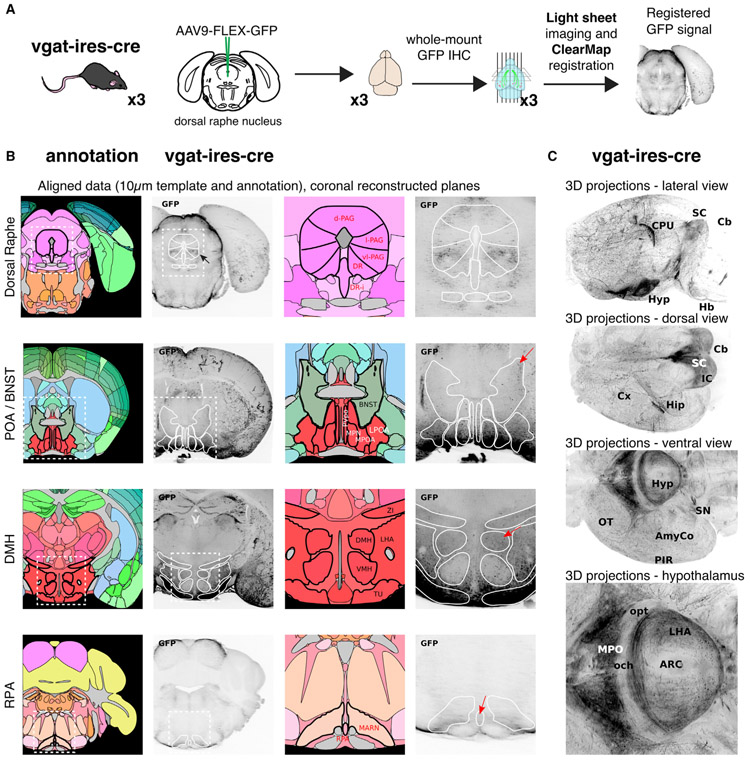
The DRN projects broadly throughout the brain; however, the precise downstream projection targets of DRNVgat neurons have not been established. Thus, while previous studies have mapped the ascending (Vertes, 1991) and brainstem (Vertes and Kocsis, 1994) projections of the DRN overall, cell-type-specific outputs have not been fully characterized. To systematically map these projections, we again used iDISCO+ whole-brain mapping combined with adeno-associated virus (AAV)-mediated tracing. To selectively label the projections of DRNVgat neurons, we injected AAV9-FLEX-GFP into the DRN of Vgat-IRES-Cre mice (Figure 5A). After 6 weeks, mice were sacrificed and their brains were immunostained for GFP and cleared using the iDISCO+ protocol. We then used light sheet imaging of the cleared whole brain to visualize the projection sites of DRNVgat neurons (Video S1).
Figure 5. DRNVgat Neurons Project to Brain Regions Implicated in Regulating Thermogenesis.
(A) Schema for whole-brain projection mapping of DRNVgat neurons.
(B) Ascending and descending projections from DRNVgat neurons are observed in numerous loci, such as the extended amygdala, hypothalamus, and RPa. Red arrows highlight (top to bottom) BNST, DMH, and RPa.
(C) Whole-brain, 3D images of DRNVgat projections throughout the brain from multiple viewpoints.
We observed dense innervation of the RPa, as well as several nuclei in the hypothalamus, notably the POA, DMH, and lateral hypothalamus (LH), but notably not in the PVH (Figure 5B; Table S2). We also noted significant projections to the amygdalar complex and extended amygdala (e.g., BNST), as well as the parabrachial nucleus (PBN), which have also been implicated in the central regulation of energy homeostasis (Figures 5B, 5C, and S4) (Waterson and Horvath, 2015; Yahiro et al., 2017). There were only a few sparse projections froms DRNVgat neurons to cortex, principally in motor cortex (Figure 5C). These results demonstrate that DRNVgat neurons send both ascending and descending projections to a number of brain regions known to regulate thermogenesis and/or energy balance.
DRNVgat Neurons Indirectly Innervate iBAT via a Functional Projection to the RPa
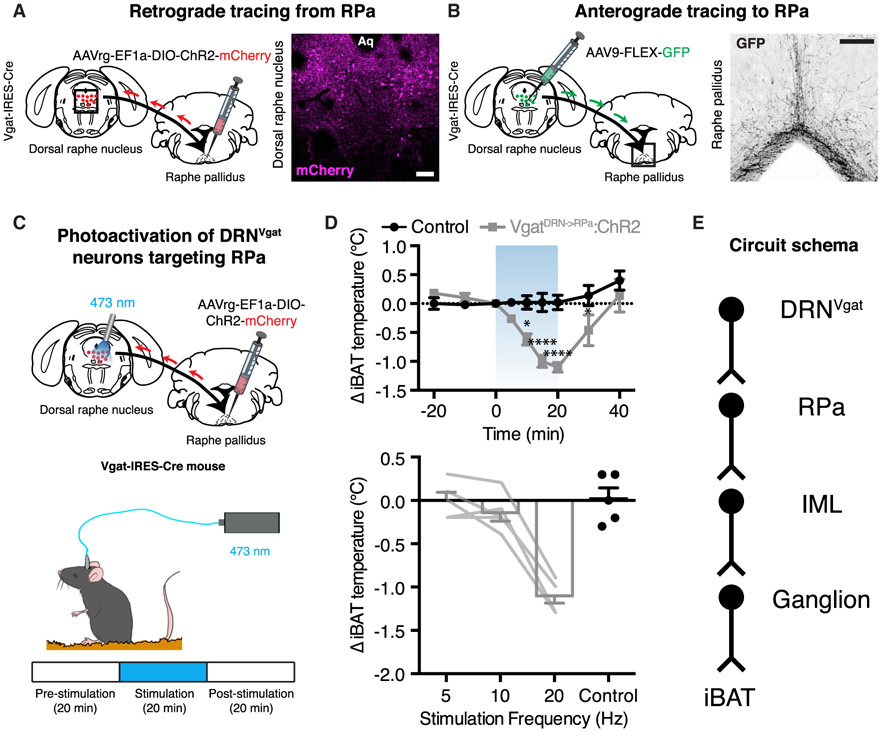
Retrograde tracing from brown fat revealed that DRNVgat neurons were labeled 1 day after labeling of PVH and RPa (Figure 4). However, the projection mapping studies shown above demonstrate that DRNVgat neurons appear to innervate the RPa, but not the PVH (Figure 5; Table S2). We thus set out to confirm that DRNVgat neurons project to the RPa and then test their function. We undertook two parallel approaches: retrograde tracing from the RPa to DRNVgat neurons and anterograde tracing from DRNVgat neurons to the RPa. For the retrograde tracing studies, we injected the retrograde-tracing, Cre-dependent AAVrg-EF1a-DIO-ChR2-mCherry into the RPa of Vgat-IRES-Cre mice. After 5 weeks, we performed immunohistochemical staining for mCherry within the DRN, observing significant labeling of neurons within the dorsolateral DRN (Figure 6A). In a separate cohort of mice, we injected the DRN of Vgat-IRES-Cre mice with the anterograde tracing AAV9-FLEX-GFP. After allowing for sufficient time for expression of GFP, we performed immunolabeling in the RPa against GFP, finding significant projections to this region (Figure 6B). These two anatomic studies together demonstrate that DRNVgat neurons project to the RPa.
Figure 6. DRNVgat Neurons Communicate with iBAT through the RPa to Regulate Thermogenesis.
(A) Retrograde labeling of DRNVgat neurons from the RPa.
(B) Anterograde labeling of RPa from DRNVgat neurons.
(C) Schematic for photoactivation of DRNVgat neurons innervating the RPa.
(D) Activation of DRNVgat neurons projecting to the RPa significantly suppresses iBAT temperature (top, treatment: p < 0.001) in a scalable fashion (bottom). Two-way RM ANOVA comparing control and treated groups (n = 5 mice per group). Blue-shaded region highlights Laser On epoch.
(E) Circuit schema of DRNVgat neurons polysynaptically innervating iBAT through the RPa.
Scale bars, 100 μm. *p < 0.05, ****p < 0.0001. Data are presented as mean ± SEM.
Having established that DRNVgat neurons project to the RPa (but not the PVH), we next asked whether this projection could suppress iBAT thermogenesis. We first injected the retrograde-tracing AAVrg-EF1a-DIO-ChR2-mCherry into the RPa of Vgat-IRES-Cre mice and placed an optical fiber into the DRN to photoactivate DRNVgat neurons projecting to the RPa (Figure 6C). We then tested the effect of stimulating the RPa-projecting DRNVgat neurons on iBAT thermogenesis. The assay was performed over the course of 1 h, and comprised of three, 20-min epochs: pre-stimulation, stimulation, and post-stimulation. Photoactivation of DRNVgat neurons projecting to the RPa at 20 Hz resulted in a significant suppression in iBAT temperature, an effect that rapidly diminished after cessation of photostimulation (Figure 6D). Lower frequency stimulation at 5 and 10 Hz had no effect on iBAT thermogenesis (Figure 6D).
Collectively, these data confirm that stimulation of DRNVgat neurons projecting to RPa is capable of suppressing thermogenesis (Figure 6E).
Ascending GABAergic DRN Projections to BNST, DMH, and MPOA Also Regulate iBAT Thermogenesis
In our projection mapping studies, we found that in addition to projecting to brainstem, DRNVgat neurons also send ascending projections into the forebrain. We thus set out to test whether these projection sites can also regulate thermogenesis, focusing on three nodes implicated in the central regulation of energy homeostasis: the BNST, DMH, and MPOA. If there are functional connections between DRNVgat neurons and these sites, it would suggest the intriguing possibility that DRNVgat neurons are an important new node within an established thermoregulatory network.
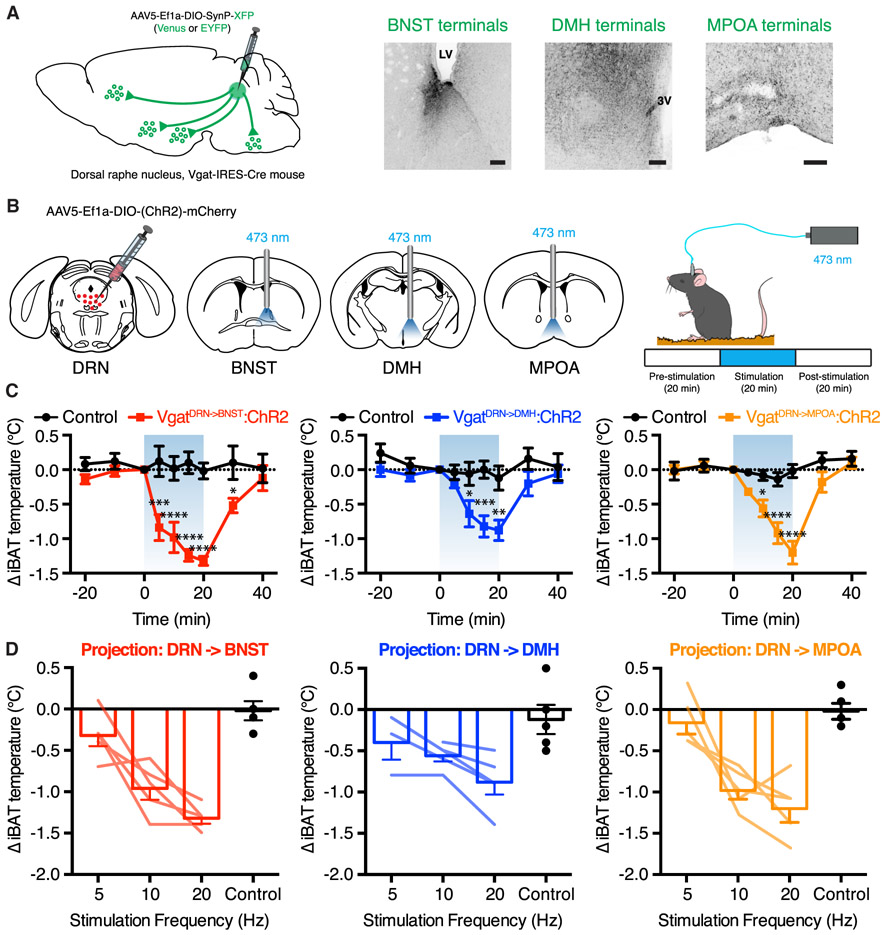
Before testing the functional effect of activating DRNVgat terminals, we reconfirmed that the DRNVgat neurons project to these regions using synaptophysin-XFP fusion constructs, which direct fluorophore expression selectively to nerve terminals. AAV expressing synaptophysin-XFP was injected into the DRN of Vgat-IRES-Cre mice, followed by an incubation period of 4 weeks to allow for anterograde transport of the synaptophysin-XFP to nerve terminals. Mapping of fluorophore expression in nerve terminals confirmed that DRNVgat neurons send projections to the BNST, DMH, and MPOA (Figure 7A).
Figure 7. DRNVgat Neurons Regulate iBAT Thermogenesis through Multiple Ascending Projections.
(A) Schema and representative IHC for validation of DRNVgat projections to BNST, DMH, and MPOA.
(B) Schema for DRNVgat neuron terminal stimulation.
(C and D) Activation of DRNVgat terminals in either the BNST (treatment: p < 0.001), DMH (treatment: p < 0.0001), or MPOA (treatment: p < 0.01) significantly suppresses iBAT temperature (C) in a scalable fashion (D). Two-way RM ANOVA comparing control and treated groups (n = 5 mice per group). Blue-shaded region highlights Laser On epoch.
Scale bars, 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are presented as mean ± SEM.
See also Figures S5 and S6.
We next asked whether activation of projections to these regions was capable of eliciting a decrease in iBAT thermogenesis. Toward this end, we injected AAV5-EF1a-DIO-ChR2-EYFP into the DRN of Vgat-IRES-Cre mice, followed by placement of an optical fiber above the BNST, DMH, or MPOA (Figure 7B, left). We then tested the effect of activating these projections by monitoring iBAT temperature during three 20-min epochs: pre-stimulation, stimulation, and post-stimulation (Figure 7B, right). Photoactivation (20 Hz) of projections to the BNST, DMH, and MPOA all led to a significant suppression of iBAT temperature, with temperature recovering to baseline levels 20 min after photostimulation ceased (Figure 7C). Photostimulation of DRNVgat terminals in each of the three loci at a lower frequency of 10 Hz (but not 5 Hz) also suppressed iBAT thermogenesis, with a maximal effect observed at 20 Hz (Figure 7D). There were no evident differences in the magnitude of the observed reduction in brown fat temperature among these three projection sites (Figures S5C and S5D).
In previous work, we demonstrated that DRNVgat neurons are capable of regulating food intake (Nectow et al., 2017b). The BNST and DMH are also known to regulate food intake, as well as thermogenesis (Betley et al., 2013; Garfield et al., 2016; Jennings et al., 2013). We thus tested whether any of these projection sites could also regulate food intake and found that DRNVgat projections to the BNST and DMH, but importantly not the MPOA or RPa, increase food intake with a maximal response at 20 Hz, and lesser effects at 5 and 10 Hz (Figures S5A, S5B, and S5D). These two projections (to BNST and DMH) also show minor collateralization (Figure S6A), which may partially influence how they regulate behavioral and physiologic processes (Figures S6B and S6C).
DISCUSSION
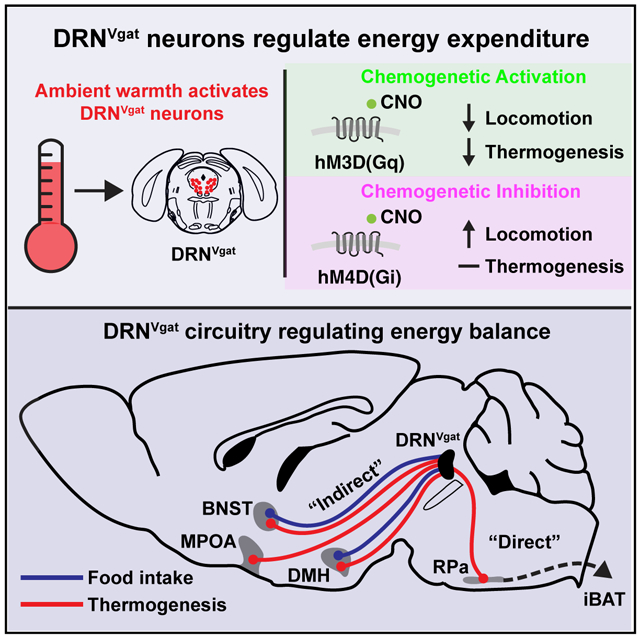
In mammals, deviations in core body temperature can have profound effects on many fundamental biological processes. The precise homeostatic control of core temperature is thus essential for survival. Thermogenesis is also a component of the homeostatic system that regulates energy balance, and altered energy homeostasis can lead to obesity, an important risk factor for cardiovascular disease and type 2 diabetes. However, while significant progress has been made in delineating CNS pathways that control energy intake, the circuit elements regulating energy expenditure, particularly thermogenesis, have only recently been identified (Piñol et al., 2018; Song et al., 2016; Tan et al., 2016; Tan and Knight, 2018; Zhao et al., 2017). In this report, we took an unbiased approach to identify populations of neurons that can regulate adaptive thermogenesis, a critical aspect of energy expenditure, using whole-brain activity mapping. We found that DRNVgat neurons within the dorsolateral aspect of the DRN were activated by increased ambient temperature (Figure 1). Consistent with this observation, we found that activation of DRNVgat neurons potently suppressed energy expenditure through changes in iBAT thermogenesis and locomotor activity, but had no effect on peripheral vascular tone (Figure 2). In contrast, we found that inhibition of these neurons augmented energy expenditure via increased locomotor activity, independent of thermogenesis (Figure 3).
A role for the DRN in controlling thermogenesis is consistent with numerous prior reports. Earlier electrophysiological studies demonstrated that subpopulations of DRN neurons are sensitive to ambient warmth as well as to direct (local) heating (Hori and Harada, 1976; Weiss and Aghajanian, 1971). Later studies using pharmacological and electrical modulation within the DRN also found that the DRN regulates some of the behavioral and physiologic components of energy expenditure, including locomotion and thermogenesis (Dib et al., 1994; Waterhouse et al., 2004). However, these studies did not establish the responsible cell types or the neural circuits in which they are embedded.
Building on our earlier findings, we set out to map the downstream circuit elements through which DRNVgat neurons regulate thermogenesis. Using anatomic and functional approaches, we found that DRNVgat neurons regulate thermogenesis through both a “direct” descending pathway to the brainstem (ultimately converging on brown fat), as well as multiple “indirect” pathways ascending into the forebrain (Figures 4, 5, and 6). The descending circuit terminates within brainstem premotor neurons of the RPa, but importantly not premotor neurons of the PVH; the ascending circuits terminate on the BNST, DMH, and MPOA, among other loci. These projection targets are key nodes embedded within hunger and/or thermoregulatory circuits. Consistent with these findings, we found that optical activation of these projection targets significantly suppressed thermogenesis, suggesting that DRNVgat neurons have multiple redundant or collateralized circuit mechanisms through which they can regulate energy expenditure (Figure 7).
In prior work, we also found that DRNVgat neurons are sensitive to changes in metabolic state and related hormonal signals. These neurons, in turn, drive potent short- and long-term changes in food intake, resulting in significant shifts in overall energy balance (Nectow et al., 2017b). In the current work, we demonstrated that DRNVgat neurons are also capable of promoting feeding through a subset of the projection targets (BNST and DMH), which we found could regulate thermogenesis. Importantly, we observed no such effect from stimulating DRNVgat projections to the MPOA or RPa, consistent with their hypothesized roles in regulating energy expenditure, but not energy intake (Morrison et al., 1999; Tan and Knight, 2018). This work demonstrates that different projection sets can regulate different aspects of energy balance, raising the important question of how these effects are achieved. In the future, it will thus be critical to parse out whether DRNVgat neurons exhibit substantial collateralization across projection targets, as this would enable a better understanding of how different DRNVgat sub-circuits coordinate energy balance.
While DRNVgat neurons potently and bidirectionally regulate energy homeostasis, the nature of the internal and external signals that regulate the activity of these neurons is largely unknown. Retrograde tracing studies from GABAergic neurons within the DRN have identified inputs from numerous nuclei implicated in the central regulation of energy balance, such as the hypothalamus, brainstem, and extended amygdala (Weissbourd et al., 2014). These are nuclei that often appear to have reciprocal interactions with the DRN, suggesting that it is an important node in the extended circuitry regulating energy balance. In further support of this notion, the DRN and bordering vlPAG receive inputs from arcuate nucleus “hunger neurons” expressing AgRP (Steculorum et al., 2016), as well as from VMPO “warm-sensitive” neurons expressing BDNF/PACAP (Tan et al., 2016). These anatomic data suggest that the DRN integrates multiple signals related to energy balance to effect changes in the animal’s behavior and physiology.
DRNVgat neurons mediate a number of diverse functions; however, the extent of this population’s heterogeneity is unclear. While we have demonstrated that different subsets of DRNVgat projections mediate different effects on energy balance, it is unclear whether further heterogeneity exists in terms of their molecular composition (i.e., neurotransmitter and neuromodulator profile). Molecular profiling of these neurons using viral TRAP revealed that they exhibit marked molecular heterogeneity (Nectow et al., 2017a, 2017b), and it is still unknown whether the DRNVgat neurons regulating thermogenesis are the same as those regulating hunger. Our photostimulation studies demonstrating simultaneous effects on food intake and thermogenesis (via projections to BNST and DMH) suggest that there may, indeed, be significant overlap or collateralization between these subpopulations. However, the functional contribution of this putative overlap is incompletely understood. Similarly, while we have shown that these neurons regulate energy balance through both behavioral and autonomic mechanisms, it is as yet unclear which DRNVgat cell subtypes are responsible for each of these effects. It will thus be essential in future studies to define the role of specific subpopulations that regulate the different functions known to be controlled by DRNVgat neurons, including the possible use of projection-specific and activity-dependent profiling approaches (Ekstrand et al., 2014; Knight et al., 2012; Nectow et al., 2015).
This work demonstrates that DRNVgat neurons potently regulate energy expenditure, and together with our previous work, suggests that the DRN is a key node in the extended circuit regulating energy homeostasis.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Alexander Nectow (arn2136@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were approved by Princeton University and The Rockefeller University Institutional Animal Care and Use Committees and were in accordance with the National Institutes of Health guidelines. Male and female mice were used for all studies, and mice were at least 6 weeks old at the time of surgery. Mice were housed in a 12 hr light-dark cycle with ad libitum access to food and water, except for fasting assays. All mouse lines are in a wild-type (C57BL/6J) background. Genotypes/sources for mice used in the above studies are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Abcam | ab13970; RRID: AB_300798 |
| Rabbit polyclonal anti-c-Fos | Synaptic Systems | 226003; RRID: AB_2231974 |
| Rabbit monoclonal anti-c-Fos | Cell Signaling | 2250S; RRID: AB_2247211 |
| Rabbit polyclonal anti-RFP | Rockland | 600-401-379; RRID: AB_2209751 |
| Rat monoclonal anti-mCherry (16D7) | ThermoFisher | M11217; RRID: AB_2536611 |
| Chicken polyclonal anti-mCherry | Abcam | ab205402; RRID: AB_2722769 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Clozapine N-oxide | Sigma-Aldrich | C0832-5MG |
| Critical Commercial Assays | ||
| RNAscope Multiplex Fluorescent Assay V2 | Advanced Cell Diagnostics (ACD Bio) | 323100 |
| RNAscope Probe- Mm-Slc32a1 | ACD Bio | 319191-C1 |
| RNAscope Probe- mCherry-C2 | ACD Bio | 431201-C2 |
| RNAscope Probe- EGFP | ACD Bio | 400281-C1 |
| RNAscope Probe- tdTomato-C2 | ACD Bio | 317041-C2 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Vgat-IRES-Cre: Slc32a1tm2(cre)Lowl/J | The Jackson Laboratory | 016962; RRID: ISMR_JAX:016962 |
| Mouse: Rosa26-LSL-EGFPL10a: B6.129S4-Gt(ROSA)26Sortm1(CAG-EGFP/Rp/10a,birA)Wtp/J | The Jackson Laboratory | 022367; RRID: IMSR_JAX:022367 |
| Mouse: C57BL/6J | The Jackson Laboratory | 000664; RRID: IMSR_JAX:000664 |
| Mouse: TRAP2: Fostm2.1(icre/ERT2)Luo/J | The Jackson Laboratory | 030323; RRID: IMSR_JAX:030323 |
| Mouse: Ai14: B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)/Hze/J | The Jackson Laboratory | 007908; RRID: IMSR_JAX:007908 |
| Recombinant DNA | ||
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | UNC Vector Core | N/A |
| AAV5-EF1a-DIO-EYFP | UNC Vector Core | N/A |
| AAV5-hSyn-DIO-hM3D(Gq)-mCherry | Addgene | 44361 |
| AAV5-hSyn-DIO-hM4D(Gi)-mCherry | Addgene | 44362 |
| AAV5-EF1a-fDIO-HA-NLS-Cre | This paper; available at Addgene | N/A |
| AAV9-CAG-FLEX-EGFP; AI854 | Penn Vector Core | N/A |
| AAV9-DIO-SynP-mCherry | MGH Vector Core | AAV-RN1 |
| AAV9-DIO-SynP-EYFP | MGH Vector Core | AAV-RN2 |
| PRV-lp298 (PRV-mCherry) | This paper | N/A |
| PRV-152 (PRV-GFP) | Smith et al., 2000 | N/A |
| Software and Algorithms | ||
| GraphPad Prism 6.0 | GraphPad | N/A |
| ClearMap | Renier et al., 2016 | N/A |
| Imaris 9.0.1 | Bitplane | N/A |
| Terastitcher | University Campus Bio-Medico of Rome | N/A |
METHOD DETAILS
Viral Vectors
All viruses used in these studies were obtained from UNC Vector Core, Penn Vector Core, MGH Vector Core, Addgene, or produced in-house by the PNI Vector Core (see Key Resources Table for further information). For optogenetic studies, AAV5-EF1a-DIO-hChR2(H134R)-EYFP (activation), AAV5-EF1a-DIO-eArch3.0-EYFP (inhibition), or AAV5-EF1a-DIO-EYFP (control virus) were used. For chemogenetic studies, AAV5-hSyn-DIO-hM3D(Gq)-mCherry (activation), AAV5-hSyn-DIO-hM4D(Gi)-mCherry (inhibition), or AAV5-hSyn-DIO-mCherry (control virus) were used. For whole-brain projection mapping studies, AAV9-FLEX-GFP was used. For terminal labeling studies, AAV9-DIO-SynP-XFP (mCherry, EYFP, or Venus) was used. For iBAT projection mapping, PRV-lp298 (PRV-mCherry) or PRV-152 (PRV-EGFP) were used. PRV-lp298 is a variant of PRV Bartha, engineered to express an mCherry-HA fusion by plaque-purification of PRV-Introvert-mCherry after passage through Cre-expressing cells (Pomeranz et al., 2017).
Stereotaxic Surgery
Mice were anesthetized using isoflurane anesthesia (induction: 3%–4%, maintenance: 1.5%–2%). Coordinates were identified using the Paxinos mouse brain atlas. For chemogenetic studies, Vgat-IRES-Cre mice were injected with 1.0 μL virus in the DRN (coordinates – from lambda: −2.8 mm DV or +0.8 mm ML, 0 mm AP, −3.0 mm DV:15° angle). For ascending projection-specific optogenetic studies, Vgat-IRES-Cre-driver mice were injected with 1.0 μL AAV5-EF1a-DIO-hChR2(H134R)-EYFP in the DRN (coordinates: −2.8 mm DV from lambda) followed by implantation of a fiber optic ferrule (Thor Labs) above the BNST (coordinates, relative to bregma: +1.0 mm ML, −0.15 mm AP, −3.1 mm DV), DMH (coordinates, relative to bregma: +0.5 mm ML, −1.5 mm AP, −5.15 mm DV), or MPOA (coordinates, relative to bregma: +0.25 mm ML, +0.45 mm AP, −4.60 mm DV). For descending projection-specific optogenetic studies, Vgat-IRES-Cre-driver mice were injected with 1.0 μL AAVrg-EF1a-DIO-hChR2(H134R)-EYFP in the RPa (coordinates, relative to bregma: 0 mm ML, −6.15 mm AP, −5.8 mm DV) followed by implantation of a fiber optic ferrule above the DRN (coordinates – from lambda: +0.8 mm ML, 0 mm AP, −2.4 mm DV:15° angle). Skin was closed using sutures, glue, or a surgical clip. All listed DV coordinates are relative to pia.
Immunohistochemistry
Mice were transcardially perfused with PBS, followed by 4% PFA. Brains were then post-fixed for 12-24 hr, followed by floating vibratome sectioning to a thickness of 40-50 μm. Primary antibodies used for immunohistochemistry can be found in the Key Resources Table; secondary antibodies were Alexa Fluor conjugated (Life Technologies). All images were captured using confocal microscopy (Zeiss or Leica).
Multiplex Fluorescence In Situ Hybridization (FISH)
Mice were transcardially perfused with RNase-free PBS, followed by 4% PFA. Brains were then harvested and post-fixed in 4% PFA for 12-24 hr. Brains were then incubated in increasing concentrations of sucrose solution (10% to 30%) overnight at 4°C, sliced with a temperature-controlled cryostat, and processed for FISH. Multiplex FISH was then performed using the RNAscope system (ACDBio). Probes for the following mRNAs were used: Vgat (Slc32a1), EGFP, tdTomato, and mCherry. Briefly, RNAscope (Advanced Cell Diagnostics) was used as per the manufacturer’s protocol. The target probe sets used included eGFP-C1, mCherry-C2, tdTomato-C2, and Slc32a1-C3. Formalin-fixed frozen brain tissue was sectioned using a cryostat. The sections obtained were attached on Superfrost Plus Adhesion Slides (Thermo Fisher), and a hydrophobic barrier created using Immedge Hydrophobic Barrier Pen (Vector Laboratories Inc.). Pre-treatment was done by serial submersion of the slides in 1X PBS, nuclease-free water, and 100% EtOH for two minutes each, at RT. Probe hybridization was achieved by incubation of 35 μL mRNA target probes for 2 h at 40°C using a HyBez oven. The signal was amplified by subsequent incubation of Amp-1, Amp-2, and Amp-3 one drop each for 30, 30, and 15 min respectively at 40°C using a HyBez oven. Each incubation step was followed by two 2 min washes using RNAscope washing buffer. Nucleic acids were stained using manufacturer’s supplied DAPI for 30 s followed by two washes with 1X PBS. The slides were mounted using Prolong Gold Antifade Mountant (Thermo Fisher).
Thermal Challenges
For thermal challenges, wild-type mice were kept in their home cages for 4 hours at either 4°C, RT, or 38°C using a temperature-controlled incubator. Immediately following thermal challenge, mice were transcardially perfused and processed for immunohistochemistry (above) or whole-brain clearing (below). For co-localization studies between Fos and DRNVgat neurons, the same immunohistochemistry studies were performed, except Vgat-IRES-Cre/GFPL10 reporter mice were used. Ambient warmth challenge validation was additionally performed at intervals of 0.5 hr, 1 hr, 2 hr, and 3 hr, followed by immunohistochemistry.
Genetic Labeling of Warm-Activated Neurons Using the TRAP2 System
To genetically label warm-activated neurons, TRAP2 mice (tamoxifen-inducible Cre expression placed downstream of an immediate early gene promoter) were crossed to (Cre-dependent) tdTomato reporter mice (Ai14). Mice were singly housed for one week prior to experimentation to enable habituation to social isolation, during which they were also sham injected once daily for habituation to injection. Mice were then injected with 40 mg/kg 4-hydroxytamoxifen (4-OHT, Sigma Aldrich), and moved from room temperature to a 38°C temperature-controlled incubator, and kept there for 4 hr post-injection. Mice were then moved back to their respective home cage at room temperature; 7 days were allowed to elapse post-injection for optimal expression of tdTomato.
Polysynaptic Circuit Tracing from iBAT
For polysynaptic, retrograde circuit mapping from iBAT, mice were injected with PRV-mCherry (PRV constitutively expressing mCherry) into iBAT, followed by an incubation period of 4 or 5 days. Mice were then transcardially perfused and processed for immunohistochemistry (above) or whole-brain clearing (below). For co-localization studies between PRV and DRNVgat neurons, the same immunohistochemistry studies were performed, except Vgat-IRES-Cre/GFPL10 reporter mice were used (at 5 days post-injection).
Triple-Labeling of Warm-Activated, iBAT-Projection DRNVgat Neurons
TRAP2 mice were crossed to tdTomato reporter mice and processed to label warm-activated neurons (as above). These mice were then injected with PRV-152 (polysynaptic PRV constitutively expressing EGFP), followed by an incubation period of 5 days (to label DRNVgat neurons). Mice were then perfused with RNase-free PBS, followed by 4% PFA, and processed following the multiplex FISH protocol, as above. Triple labeling was performed for Vgat, EGFP, and tdTomato RNA transcripts to label warm-activated, iBAT-projecting DRNVgat neurons.
Anterograde Projection Mapping from DRNVgat Neurons
Two approaches were taken for anterograde projection mapping: soluble GFP expression and synaptophysin-conjugated fluorophore to selectively label neuron terminals. For GFP expression, AAV9-FLEX-GFP was injected into the DRN of Vgat-IRES-Cre mice. After an incubation period of at least 5-6 weeks, mice were sacrificed and processed via either standard IHC methods (described above) or using iDISCO+ for whole-brain projection mapping. For synaptophysin studies, an AAV Cre-dependently expressing a synaptophysin-XFP (EYFP or Venus) fusion construct was injected into the DRN of Vgat-IRES-Cre mice, followed by an incubation period of at least 4-6 weeks. Following this incubation period, mice were sacrificed and brains were processed via standard IHC methods described above.
iDISCO+ Whole-Brain Clearing and Imaging
Mice were transcardially perfused with PBS, followed by 4% PFA. Brains were then put through a 24 hr post-fixing period, after which immunolabeling and whole-brain clearing were performed according to previously established protocols (Renier et al., 2016). Antibodies used for Fos and PRV (RFP) labeling can be found in the Key Resources Table. For Fos and PRV labeling studies, analysis was performed using ClearMap (Renier et al., 2016). For acquisition, cleared samples were imaged in a sagittal orientation (left lateral side up) on a light-sheet microscope (LaVision Biotec) equipped with a sCMOS camera and LVMI-Fluor 4x objective lens equipped with a 6-mm working distance dipping cap. Version v144 and v210 of Inspector Microscope controller software was used. Samples were scanned in the 640 nm channel. Images were taken every 6 mm and reconstructed with ClearMap software for quantification or Imaris 9.1 software for visualization. For autofluorescence, the 480 nm channel was used with a 1.3x objective lens.
ClearMap Analysis
All analyses for whole-brain studies were performed using ClearMap software (latest version available from https://www.idisco.info/) (see also Renier et al., 2016).
Molecular Profiling of iBAT Tissue
DREADD-expressing and control mice, as above, were sham-injected prior to the study. At t0 (at the beginning of the light cycle), mice were injected with 3 mg/kg CNO. Twelve hours later (t12), mice were again treated with a second 3 mg/kg dose of CNO. At t24, mice were sacrificed, and iBAT tissue was harvested. Total RNA was isolated from iBAT tissue using the TRI reagent (Sigma Aldrich), followed by purification using the RNeasy Mini Kit (QIAGEN). RNA quantity was measured with a Nanodrop 1000 spectrophotometer and quality was assayed on an Agilent 2100 Bioanalyzer. Only samples with RNA integrity values > 8.0 were used for qRT-PCR analyses. Following isolation and purification of RNA, reverse transcription was performed using the QIAGEN QuantiTect kit. Quantitative, real-time PCR (qPCR) was performed using SYBR green. Primers were used at a final concentration of 1 μM. Gene expression was normalized to housekeeping gene Tbp.
Awake Chemogenetic Studies to Measure Thermogenesis
Mice were injected with DREADD (hM3D(Gq) or hM4D(Gi)) or control virus in the DRN, followed by a recovery period of at least 3 weeks (see above for viruses injected). Mice were habituated with sham injections at least 5 days prior to the assay. Thermogenesis assays were performed in the home cage during the animal’s light phase. Mice were given ad libitum access to food prior to, during, and after the assay. Initial measurements of iBAT temperature were performed using a thermal camera (FLIR Systems), using an average of ~3 thermal images per time point. Measurements of core and iBAT temperature were obtained using, respectively, an anal probe (Braintree Scientific) and wireless implantable temperature probes (IPTT-300, Bio Medic Data Systems). For tail temperature monitoring, a thermal camera (FLIR Systems) was used, taking an average of ~2 thermal images per time point. All temperature measurements were monitored at intervals noted in the text (ranging from 10 min to 1 hr). Control studies were performed by injecting vehicle (saline) instead of CNO. All CNO injections were at a concentration of 1 mg/kg, unless stated otherwise.
Anesthetized Chemogenetic Studies to Measure Thermogenesis
As in the case of the awake preparation, DREADD-expressing or control mice were injected with virus in the DRN, followed by a recovery period of at least 3 weeks. Thermogenesis assays were performed during the animal’s light phase under a light plane of anesthesia (~1%–1.25% isoflurane); at this level of anesthesia, a toe pinch reflex could be elicited throughout the duration of the assay. Mice were given ad libitum access to food prior to and after the assay. Measurements of core, iBAT, and tail temperature were made using, respectively, an anal probe (Braintree Scientific), an implantable temperature probe in tandem with a portable reader (Bio Medic Data Systems), and a thermal camera (FLIR Systems). For tail temperature monitoring, an average of ~2 thermal images were used per time point. Both core and iBAT temperatures were monitored at intervals noted in the text. Studies controlling for CNO effect were performed by injecting vehicle (saline) instead of CNO. All CNO injections were at a concentration of 1 mg/kg.
Chemogenetic Studies using Metabolic Home Cages
Mice were injected with DREADD (hM3D(Gq) or hM4D(Gi)) or control virus in the DRN, followed by a recovery period of at least weeks (see above for viruses injected). Metabolic phenotyping of mice was performed using an automated home cage phenotyping system (TSE-Systems). Briefly, the system allows for phenotyping of mice in a home cage environment, which periodically measures a number of metabolic parameters in an automated fashion. In particular, we used a climate-controlled chamber (temperature: 22°C; humidity: 55%; 12 hr light-dark cycle). Locomotor activity is recorded as beam breaks, measuring activity in 3-D. Beam breaks are analyzed by custom software from the metabolic cage, which enables conversion of beam breaks into distance/velocity. Additionally, an indirect gas calorimetry module determines through an open circuit oxygen consumption and carbon dioxide production. TSE software calculates respiratory exchange ratio (RER) and energy expenditure from these values. All parameters can be measured continuously and simultaneously in up to 16 animals. In our paradigm, mice were placed in the metabolic home cage and left single-housed for one week, to enable habituation to social isolation. After adaptation, data were collected over the subsequent days. On day 1, basal measurements were evaluated to control for normal mouse behavior. On day 2, saline injection was performed and data were collected. On day 3, CNO injection (1mg/kg) was performed and data collected. Day 4 was collected to observe recovery of metabolic parameters to basal levels post-CNO injection. Data of the 1 hr pre-injection and 3 hr post-injection were collected and processed with PhenoMaster software from TSE Systems.
In Vivo Terminal Photostimulation
For terminal photostimulation studies, mice were either injected with virus in the DRN, followed by implantation of a fiber optic cannula over either the BNST, DMH, or MPOA or injected with virus in the RPa followed by implantation of a fiberoptic cannula over the DRN (see above for viruses and coordinates). Studies were performed at least 5-6 weeks post-surgery for recovery and sufficient expression of opsin in the terminals (for BNST, DMH, and MPOA) or retrogradely in the DRN (from the RPa). Mice received blue light laser stimulation at frequencies (described in the main text) of 1, 5, 10, and 20 Hz (473 nm, OEM Lasers/OptoEngine) with a 10-ms pulse width. Laser stimulation intensity was approximately 10 mW at the tip. Stimulation paradigms were programmed into an arbitrary waveform generator (Agilent), and mice were handled at least seven days prior to performing behavior. Core temperature was measured with an anal probe (ThermaWorks), and iBAT temperature was measured with an implantable temperature transponder in tandem with a portable wireless reader (Bio Medic Data Systems). Tail temperature was measured using infrared thermography (FLIR Systems). An average of ~2 thermal images were used per time point. Temperatures were monitored at times (in min) −20, 0, 5, 10, 15, 20, and 40 (thus, temperature was monitored 20 min before and after stimulation, as well as at 5 min intervals during stimulation).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters reported are: sample size (n = number of animals or samples per group), definition of center, dispersion, precision measures, statistical test used, and statistical significance. All data are displayed as mean ± SEM. Significance was defined as p < 0.05. Significance annotations are: *p < 0.05, **p <0.01, ***p < 0.001, ****p < 0.0001. Mice were randomized into control or treatment groups. Control mice were age-matched littermate controls where possible. All statistics and data analysis were performed using GraphPad Prism, ClearMap, R, or Python.
Supplementary Material
Highlights.
Ambient warmth activates DRNVgat neurons
DRNVgat neurons regulate energy expenditure through locomotion and thermogenesis
DRNVgat neurons exhibit vast projections and polysynaptically innervate brown fat
DRNVgat projections differentially regulate food intake and energy expenditure
ACKNOWLEDGMENTS
We thank the Princeton Neuroscience Institute’s Bezos Center for Neural Circuit Dynamics, and Rockefeller University Bio-Imaging Resource Center. We also thank Lisa Pomeranz for contributing PRV-lp298 for polysynaptic circuit mapping and Ferzin Sethna for assistance in creating AAV5-EF1a-fDIO-HA-NLS-Cre. A.R.N. acknowledges support from the Princeton Neuroscience Institute’s Innovation Award and CV Starr Fellowship, the Brain and Behavior Research Foundation’s NARSAD Young Investigator Award, the Foundation for Prader-Willi Research, the American Diabetes Association Pathway to Stop Diabetes Program, and the American Diabetes Association Core Program. J.M.F. acknowledges support from the JPB Foundation. M.S. is a Kavli NSI Postdoctoral Fellow. M.S. and J.M.F. acknowledge support from the Robertson Therapeutic Development Fund.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.05.048.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allen WE, DeNardo LA, Chen MZ, Liu CD, Loh KM, Fenno LE, Ramakrishnan C, Deisseroth K, and Luo L (2017). Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Liao GY, Kinney CE, Sahibzada N, and Xu B (2015). Discrete BDNF Neurons in the Paraventricular Hypothalamus Control Feeding and Energy Expenditure. Cell Metab. 22, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Cao ZF, Ritola KD, and Sternson SM (2013). Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell 155, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, and Nedergaard J (2004). Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359. [DOI] [PubMed] [Google Scholar]
- Dib B, Rompre PP, Amir S, and Shizgal P (1994). Thermogenesis in brown adipose tissue is activated by electrical stimulation of the rat dorsal raphe nucleus. Brain Res. 650, 149–152. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, and Friedman JM (2014). Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfield AS, Shah BP, Burgess CR, Li MM, Li C, Steger JS, Madara JC, Campbell JN, Kroeger D, Scammell TE, et al. (2016). Dynamic GABAergic afferent modulation of AgRP neurons. Nat. Neurosci 19, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway A, Forgrave P, and Birzis L (1954). Shivering suppression by hypothalamic stimulation. J. Neurophysiol 17, 375–386. [DOI] [PubMed] [Google Scholar]
- Hillegaart V (1991). Effects of local application of 5-HT and 8-OH-DPAT into the dorsal and median raphe nuclei on core temperature in the rat. Psychopharmacology (Berl.) 103, 291–296. [DOI] [PubMed] [Google Scholar]
- Hioki H, Nakamura H, Ma YF, Konno M, Hayakawa T, Nakamura KC, Fujiyama F, and Kaneko T (2010). Vesicular glutamate transporter 3-expressing nonserotonergic projection neurons constitute a subregion in the rat midbrain raphe nuclei. J. Comp. Neurol 518, 668–686. [DOI] [PubMed] [Google Scholar]
- Hori T, and Harada Y (1976). Responses of Midbrain raphe neurons to local temperature. Pflugers Arch. 364, 205–207. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Rizzi G, Stamatakis AM, Ung RL, and Stuber GD (2013). The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science 341, 1517–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, and Friedman JM (2012). Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoun HW, Harrison F, Brobeck JR, and Ranson SW (1938). Activation of heat loss mechanisms by local heating of the brain. J. Neurophysiol 1, 101–114. [Google Scholar]
- Morrison SF, and Nakamura K (2011). Central neural pathways for thermoregulation. Front. Biosci 16, 74–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SF, Sved AF, and Passerin AM (1999). GABA-mediated inhibition of raphe pallidus neurons regulates sympathetic outflow to brown adipose tissue. Am. J. Physiol 276, R290–R297. [DOI] [PubMed] [Google Scholar]
- Munzberg H, Qualls-Creekmore E, Berthoud HR, Morrison CD, and Yu S (2016). Neural Control of Energy Expenditure. Handb. Exp. Pharmacol 233, 173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nectow AR, Ekstrand MI, and Friedman JM (2015). Molecular characterization of neuronal cell types based on patterns of projection with Retro-TRAP. Nat. Protoc 10, 1319–1327. [DOI] [PubMed] [Google Scholar]
- Nectow AR, Moya MV, Ekstrand MI, Mousa A, McGuire KL, Sferrazza CE, Field BC, Rabinowitz GS, Sawicka K, Liang Y, et al. (2017a). Rapid Molecular Profiling of Defined Cell Types Using Viral TRAP. Cell Rep. 19, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nectow AR, Schneeberger M, Zhang H, Field BC, Renier N, Azevedo E, Patel B, Liang Y, Mitra S, Tessier-Lavigne M, et al. (2017b). Identification of a Brainstem Circuit Controlling Feeding. Cell 170, 429–442. [DOI] [PubMed] [Google Scholar]
- Pihol RA, Zahler SH, Li C, Saha A, Tan BK, Škop V, Gavrilova O, Xiao C, Krashes MJ, and Reitman ML (2018). Brs3 neurons in the mouse dorsomedial hypothalamus regulate body temperature, energy expenditure, and heart rate, but not food intake. Nat. Neurosci 21, 1530–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Ekstrand MI, Latcha KN, Smith GA, Enquist LW, and Friedman JM (2017). Gene expression profiling with Cre-conditional pseudorabies virus reveals a subset of midbrain neurons that participate in reward circuitry. J. Neurosci 37, 4128–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu V, Garretson JT, Liu Y, Vaughan CH, and Bartness TJ (2015). Brown adipose tissue has sympathetic-sensory feedback circuits. J. Neurosci 35, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman E, and Roberts SB (1995). The role of energy expenditure in energy regulation: findings from a decade of research. Nutr. Rev 53, 209–220. [DOI] [PubMed] [Google Scholar]
- Smith BN, Banfield BW, Smeraski CA, Wilcox CL, Dudek FE, Enquist LW, and Pickard GE (2000). Pseudorabies virus expressing enhanced green fluorescent protein: A tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. USA 97, 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Wang H, Kamm GB, Pohle J, Reis FC, Heppenstall P, Wende H, and Siemens J (2016). The TRPM2 channel is a hypothalamic heat sensor that limits fever and can drive hypothermia. Science 353, 1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steculorum SM, Ruud J, Karakasilioti I, Backes H, Engström Ruud L, Timper K, Hess ME, Tsaousidou E, Mauer J, Vogt MC, et al. (2016). AgRP Neurons Control Systemic Insulin Sensitivity via Myostatin Expression in Brown Adipose Tissue. Cell 165, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CL, and Knight ZA (2018). Regulation of Body Temperature by the Nervous System. Neuron 98, 31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CL, Cooke EK, Leib DE, Lin YC, Daly GE, Zimmerman CA, and Knight ZA (2016). Warm-Sensitive Neurons that Control Body Temperature. Cell 167, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veasey SC, Fornal CA, Metzler CW, and Jacobs BL (1997). Single-unit responses of serotonergic dorsal raphe neurons to specific motor challenges in freely moving cats. Neuroscience 79, 161–169. [DOI] [PubMed] [Google Scholar]
- Vertes RP (1991). A PHA-L analysis of ascending projections of the dorsal raphe nucleus in the rat. J. Comp. Neurol 313, 643–668. [DOI] [PubMed] [Google Scholar]
- Vertes RP, and Kocsis B (1994). Projections of the dorsal raphe nucleus to the brainstem: PHA-L analysis in the rat. J. Comp. Neurol 340, 11–26. [DOI] [PubMed] [Google Scholar]
- Warden MR, Selimbeyoglu A, Mirzabekov JJ, Lo M, Thompson KR, Kim SY, Adhikari A, Tye KM, Frank LM, and Deisseroth K (2012). A prefrontal cortex-brainstem neuronal projection that controls response to behavioural challenge. Nature 492, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse BD, Devilbiss D, Seiple S, and Markowitz R (2004). Sensorimotor-related discharge of simultaneously recorded, single neurons in the dorsal raphe nucleus of the awake, unrestrained rat. Brain Res. 1000, 183–191. [DOI] [PubMed] [Google Scholar]
- Waterson MJ, and Horvath TL (2015). Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metab. 22, 962–970. [DOI] [PubMed] [Google Scholar]
- Weiss BL, and Aghajanian GK (1971). Activation of Brain Serotonin Metabolism by Heat - Role of Midbrain Raphe Neurons. Brain Res. 26, 37–48. [Google Scholar]
- Weissbourd B, Ren J, DeLoach KE, Guenthner CJ, Miyamichi K, and Luo L (2014). Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron 83, 645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro T, Kataoka N, Nakamura Y, and Nakamura K (2017). The lateral parabrachial nucleus, but not the thalamus, mediates thermosensory pathways for behavioural thermoregulation. Sci. Rep 7, 5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZD, Yang WZ, Gao C, Fu X, Zhang W, Zhou Q, Chen W, Ni X, Lin JK, Yang J, et al. (2017). A hypothalamic circuit that controls body temperature. Proc. Natl. Acad. Sci. USA 114, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.