Figure 4.
SREBP1-Mediated Upregulation of mRNAs in the Liver of Agpat3LKO Mice
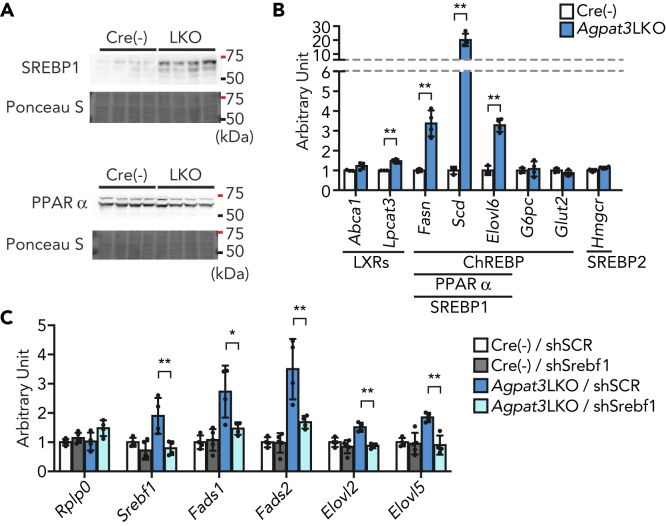
(A–C) Liver samples were prepared from 2-week-old Agpat3 LKO and Cre(−) mice. (A) Immunoblot analysis of the amount of SREBP1 and PPARα in the nucleus (n = 4 for each group). Ponceau S staining was as a loading control. (B) Relative mRNA expression of representative LXRs-, SREBP1-, SREBP2-, ChREBP-, and PPARα-target genes in the liver (n = 3 for Cre(−) and n = 4 for Agpat3 LKO mice). Significance is based on unpaired t test (∗p < 0.05, ∗∗p < 0.01). (C) The effect of short hairpin RNA (shRNA)-mediated Srebf1 knockdown on the expression of mRNAs encoding the PUFA biosynthetic enzymes in the liver (n = 3 for each group). Rplp0 is used as an internal control. shSCR, scrambled shRNA (control); shSrebf1, Srebf1 shRNA. Significance is based on two-way ANOVA followed by Bonferroni's post-hoc test (∗p < 0.05, ∗∗p < 0.01).
(B and C) Data are shown as means ± SD. See also Figure S4.