Abstract
Risk prediction of de novo donor specific antibody (DSA) would be very important for long term graft outcome after organ transplantation. The purpose of this study was to elucidate the association of eplet mismatches and predicted indirectly recognizable HLA epitopes (PIRCHE) scores with de novo DSA production. Our retrospective cohort study enrolled 691 living donor kidney transplantations. HLA-A, B, DRB and DQB eplet mismatches and PIRCHE scores (4 digit of HLA-A, B, DR, and DQ) were determined by HLA matchmaker (ver 2.1) and PIRCHE-II Matching Service, respectively. Weak correlation between eplet mismatches and PIRCHE scores was identified, although both measurements were associated with classical HLA mismatches. Class II (DRB+DQB) eplet mismatches were significantly correlated with the incidence of de novo class II (DR/DQ) DSA production [8/235 (3.4%) in eplet mismatch ≤ 13 vs. 92/456 (20.2%) in eplet mismatch ≥ 14, p < 0.001]. PIRCHE scores were also significantly correlated with de novo class II DSA production [26/318 (8.2%) in PIRCHE ≤ 175 vs. 74/373 (19.8%) in PIRCHE ≥ 176, p < 0.001]. Patients with low levels of both class II eplet mismatches and PIRCHE scores developed de novo class II DSA only in 4/179 (2.2%). Analysis of T cell and B cell epitopes can provide a beneficial information on the design of individualized immunosuppression regimens for prevention of de novo DSA production after kidney transplantation.
Keywords: kidney transplantation, eplet mismatch, PIRCHE-II, donor specific antibody, epitope analysis
Introduction
Chronic antibody-mediated rejection (ABMR) caused by de novo donor specific antibody (DSA) is a major cause of graft failure in solid organ transplantation (1). Randomized clinical trials have been undertaken in order to explore the efficacies of various treatments for ABMR (2). Although intravenous immunoglobulin (IVIG) and plasmapheresis have been advocated as standard of care, particularly in cases of acute ABMR, there are no effective treatments for chronic ABMR that would prevent the gradual deterioration of graft function (3). A means to prevent chronic ABMR is likely to be far superior than any available cure (4). While not all DSAs promote ABMR (5–8), the development of de novo DSAs remains among the most definitive of the known risk factors that promote this adverse event. Therefore, risk prediction of de novo DSA would be important for long term graft outcome.
Recently, a rigorous analysis of B cell epitopes was conducted in order to assess the immunogenicity of HLA mismatch in greater detail (9). The HLAMatchmaker algorithm was developed based on the concept of the HLA molecule as a linear sequence of amino acid triplets and via evaluation of the eplets, which are the small three-dimensional structure of amino acid residues that are the essential components of immunogenicity. Results from HLA epitope matching based on this concept have been reported to be superior to those obtained from more conventional HLA matching modalities. This new methodology provides greater insight into the risk of developing de novo DSAs as well as the possibility of reorganizing the organ allocation system (10). Many research groups have explored this issue, and reported that the degree of epitope mismatches recognized by B cell receptors as defined by an eplet, amino acid sequence and electrostatic mismatch would have a significant correlation with DSA production, ABMR and graft outcome in organ transplantation (11–19).
In parallel with B cell epitopes, attention has also been focused on T cell epitopes, specifically, those associated with donor-derived HLA molecules presented by HLA class II on recipient antigen presenting cells (20). T cell epitopes are recognized by the T cell receptor of CD4+ T cells at the first step toward DSA production via T-dependent B cell activation (Supplementary Figure 1). The number of potential T cell epitopes has been correctly assessed by the PIRCHE (Predicted indirectly recognizable HLA epitopes)-II algorithm (21, 22).
The purpose of this study was to examine the association of the eplet mismatch level and PIRCHE scores with de novo DSA production after kidney transplantation. Our goal was to elucidate the clinical significance of both T cell and B cell epitope prediction as a risk factor for de novo DSA production.
Materials and Methods
Study Design and Subjects
We conducted a retrospective cohort study of adult patients (n = 793) who underwent living donor kidney transplantation at Aichi Medical University or the Nagoya Daini Red Cross Hospital between 2008 and 2015. We excluded recipients with pre-existing DSAs (n = 66) and those who were lost to follow-up within 1 year due to death (n = 3), graft failure (n = 5) or transfer of care to a remote hospital (n = 28). The remaining 691 patients were enrolled in the retrospective cohort study. The final date for the analysis of graft survival was April 30, 2019; the mean follow-up period after transplantation was 78.7 ± 27.7 months.
HLA Typing, Eplet Mismatch and PIRCHE Score
HLA (-A, -B, -DRB1, -DQA1, and -DQB1) typing of donors and recipients was performed by xMAP® Technology of Luminex Corp. using PCR-sequence specific oligonucleotide (SSO) probes (Wakunaga Pharmaceutical Co. Ltd., Hiroshima, Japan or One Lambda, Canoga Park, CA, USA) at high resolution. Some low-resolution typing or missing data on HLA-A, B and DQA1 were extrapolated to second field HLA typing using the HLA-haplotype frequencies in Japanese population (23). Eplet mismatches and PIRCHE scores were determined using HLA types at four-digit levels. Eplet mismatch levels for HLA class I (A, B) and class II (DRB1, 3, 4, 5, and DQB1) were determined by HLAMatchmaker software v2.1. PIRCHE scores were calculated as a sum of mismatched HLA-A, HLA-B, HLA-DRB1, 3, 4, 5, HLA-DQA1, and HLA-DQB1-derived peptide counts presented with respect to the recipients' HLA-DRB1, 3, 4, 5, HLA-DQA1, and HLA-DQB1 using PIRCHE-II algorithm via the matching service.
Anti-HLA Antibody Detection and Identification of DSAs
For all recipients, anti-HLA antibodies were analyzed before transplantation and monitored annually after transplantation. Serum samples collected from 2009 to 2019 were examined for IgG antibodies against HLA class I or II using methodologies including Flow PRA, LABScreen Mixed and LABScreen PRA (One Lambda). Any positive evaluations were re-screened and the DSA was identified using LABScreen Single Antigen and Supplement (One Lambda). Mean fluorescence intensity (MFI) values above 1,000 for DSAs against HLA-A, -B, -DR, and -DQ at the 4-digit level were scored as positive.
Immunosuppressive Agents
All the patients received 500 mg of intravenous (IV) methylprednisolone prior to graft reperfusion and 20 mg of IV basiliximab as induction therapy on days 0 and 4. Maintenance immunosuppressive therapy consisted of a calcineurin inhibitor (cyclosporine or tacrolimus), steroid (prednisolone), and antimetabolites (mycophenolate mofetil or mizoribine) or an mTOR inhibitor (everolimus). Dosages of all oral immunosuppressive medications except for prednisolone were strictly adjusted according to pharmacokinetics, including area under the curve (AUC) or trough levels (24). Recipients of ABO-incompatible transplants were additionally pre-treated with mycophenolate mofetil from day−14 as well as double-filtration plasmapheresis and either splenectomy, rituximab (200 mg/body) on day−14 and/or day−1 or neither (due to low anti-A/B antibody titers).
Statistical Analysis
Statistical analyses were performed using JMP software v13.2. Nominal variables were examined using Fisher's exact test or chi-square test. Continuous variables were presented as mean ± SD and analyzed by Student's t-test or Mann-Whitney U-test. Spearman's rank correlation and simple linear regression analysis were conducted for quantifying the association. Receiver operating characteristic (ROC) curve analyses were performed to obtain the best predictive value of eplet mismatches and PIRCHE scores. Logistic regression model for univariate and multivariate analysis was used to assess the valuables associated with DSA production. DSA-free graft survival was defined as the time between kidney transplantation and the date of final follow-up without DSA detection. DSA-free survival rates were estimated using Kaplan-Meier survival curves and Wilcoxon tests. Cox proportional hazards regression model for univariate and multivariate analysis was used to find variables that impacted DSA-free survival. P < 0.05 were considered to be statistically significant.
Results
De novo DSA Production
De novo DSAs were detected in 114 (16.5%) of the 691 recipients enrolled in this study, including antibodies targeting HLA-class I (n = 14), class I + DR (n = 1), class I+ DQ (n = 2), DR (n = 19), DQ (n = 69) and DR + DQ (n = 9). DSAs detected were predominantly those directed against class II (n = 100), most notably HLA-DQ followed by HLA-DR. The incidence of HLA class I DSA was comparatively low; most of the HLA class I DSAs presented with low MFIs that fluctuated around cutoff level. MFI levels of class I DSA and class II DSA were 2,481 +/- 2,073, and 11,404 +/- 8,389, respectively. Chronic ABMR was reported to be mainly associated with HLA class II DSA (5, 6, 11, 12, 19, 25, 26). For these reasons, only class II (DR and/or DQ) DSAs were considered in the risk assessment.
Patient Data
Donor gender, classical HLA-A and B mismatches, eplet mismatches of HLA-A and B, and DRB showed statistically significant difference between de novo HLA-class I DSA-positive (n = 17) and negative (n = 674) patients, although positive number might be too small for precise analysis (Table 1A). There were no significant differences with respect to observation period, recipient/donor age, gender, relationship, use of basic immunosuppressive agents, use of desensitization therapy, or incidence of CMV infection between the de novo DR/DQ DSA-positive (n = 100) and negative patients (n = 591; Table 1B). Furthermore, no significant differences in levels of classical HLA mismatch were detected. A history of acute T cell-mediated rejection and blood group ABO compatibility were both identified as significant risk factors.
TABLE 1A.
Patient Characteristics by de novo class I DSA status.
| Factors | De novo A/B | De novo A/B | P-value |
|---|---|---|---|
| DSA (+) (n = 17) | DSA (−) (n = 674) | ||
| Observation period | 85.7 +/– 28.4 | 78.5 +/– 27.6 | |
| Recipient age | 46.3 +/– 15.4 | 16.3 +/– 15.9 | |
| Recipient M/F | 14/3 | 437/237 | |
| Donor age | 56.9 +/– 11.1 | 58.0 +/– 10.1 | |
| Donor M/F | 2/15 | 240/434 | 0.0422 |
| Parent/sibling/spouse/others | 7/1/9/0 | 282/60/305/27 | |
| CSA/TAC | 12/5 | 340/434 | |
| MMF/MZR/EVR | 13/4/0 | 565/34/75 | |
| RIT/SPX/NONE | 4/2/11 | 170/8/496 | |
| HLA-A+B MM | 2.6 +/– 0.8 | 2.1 +/– 1.0 | 0.0446 |
| HLA-DRB1 MM | 1.2+/– 0.8 | 1.2 +/– 0.6 | |
| HLA-DQB1 MM | 1.2 +/– 0.7 | 1.1 +/– 0.6 | |
| HLA-DRB1+DQB1 MM | 2.4 +/– 1.5 | 2.4 +/– 1.2 | |
| HLA-A+B+DRB1+DQB1 MM | 5.1 +/– 2.0 | 4.5 +/– 2.0 | |
| ABO-I/ABO-Id/C | 6/11 | 231/443 | |
| Eplet MM (A+B) | 15.7 +/– 5.2 | 11.1 +/– 6.4 | 0.0033 |
| Eplet MM (DRB) | 7.1 +/– 7.0 | 11.4 +/– 8.8 | 0.0426 |
| Eplet MM (DQB) | 9.8 +/– 7.2 | 8.3 +/– 6.5 | |
| Eplet MM (DRB+DQB) | 16.9 +/– 12.6 | 19.7 +/– 13.3 | |
| PIRCHE score | 211.9 +/– 136.7 | 214.0 +/– 139.7 | |
| Acute TCMR | 0 (0%) | 56 (8.3%) | |
| CMV infection | 2 (26.0%) | 196 (29.1%) |
TABLE 1B.
Patient Characteristics by de novo class II DSA status.
| Factors | De novo DR/DQ | De novo DR/DQ | P-value |
|---|---|---|---|
| DSA (+) (n = 100) | DSA (−) (n = 591) | ||
| Observation period | 80.8 +/– 28.7 | 78.3 +/– 27.5 | |
| Recipient age | 44.7 +/– 17.4 | 46.6 +/– 15.6 | |
| Recipient M/F | 73/27 | 378/213 | 0.0886 |
| Donor age | 56.9 +/– 11.1 | 58.0 +/– 10.1 | |
| Donor M/F | 29/71 | 213/378 | |
| Parent/sibling/spouse/others | 45/8/38/9 | 244/53/276/18 | |
| CSA/TAC | 60/40 | 292/299 | 0.0522 |
| MMF/MZR/EVR | 77/8/15 | 501/30/60 | |
| RIT/SPX/NONE | 20/2/78 | 154/8/429 | |
| HLA-A+B MM | 2.2 +/– 0.9 | 2.2 +/– 1.0 | |
| HLA-DRB1 MM | 1.3 +/– 0.5 | 1.2 +/– 0.6 | |
| HLA-DQB1 MM | 1.3 +/– 0.5 | 1.1 +/– 0.7 | |
| HLA-DRB1+DQB1 MM | 2.6 +/– 0.9 | 2.3 +/– 1.3 | |
| HLA-A+B+DRB1+DQB1 MM | 4.8 +/– 1.6 | 4.5 +/– 2.0 | |
| ABO-I/ABO-Id/C | 24/76 | 213/378 | 0.0223 |
| Eplet MM (A+B) | 11.0 +/– 6.8 | 11.2 +/– 6.4 | |
| Eplet MM (DRB) | 15.0 +/– 7.9 | 10.7 +/– 8.8 | <0.0001 |
| Eplet MM (DQB) | 11.0 +/– 5.7 | 7.9 +/– 6.5 | <0.0001 |
| Eplet MM (DRB+DQB) | 25.9 +/– 10.9 | 18.6 +/– 13.3 | <0.0001 |
| PIRCHE score | 265.5 +/– 139.1 | 205.3 +/– 137.8 | <0.0001 |
| Acute TCMR | 20 (20.0%) | 36 (6.1%) | <0.0001 |
| CMV infection | 26 (26.0%) | 172 (29.1%) |
DSA, donor specific antibody; CSA, cyclosporine; TAC, tacrolimus; MMF, mycophenolate mofetil; MZR, mizoribine; EVR, everolimus; RIT, rituximab; SPX, splenectomy; MM, mismatch; ABO-I, ABO-incompatible transplantation; ABO-Id/C, ABO-identical/compatible transplantation; TCMR, T cell mediated rejection.
Association Between Eplet Mismatch and PIRCHE Score
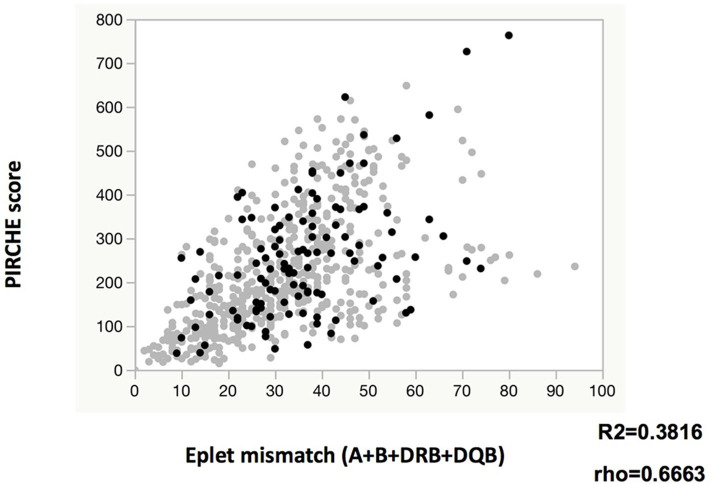
The eplet mismatches of class I (A+B) and class II (DRB, DQB, or both) were significantly higher among the de novo class I and class II DSA-positive patients than among those who were de novo DSA-negative, respectively (Tables 1A,B). PIRCHE scores were significantly higher in the de novo class II DSA-positive patients than in those were DSA-negative. Both eplet mismatch levels and PIRCHE scores were associated with classical HLA mismatches (Supplementary Figures 2A–C), there were weak positive correlations between eplet mismatches (A, B, DRB, and DQB) and PIRCHE scores (Figure 1). Compared to DSA-negative patients, DSA-positive patients tended to have a higher degree of eplet mismatches and higher PIRCHE scores. However, DSA-positive patients showed positive correlation between eplet mismatches and PIRCHE score to a lesser extent (DSA-positive; R2 = 0.2340, rho = 0.4621 vs. DSA-negative; R2 = 0.3940, rho = 0.6968).
Figure 1.
Relationship between eplet mismatch and PIRCHE score. Gray circles and black circles indicate de novo DR/DQ DSA negative and positive, respectively.
De novo DR/DQ DSA Production Associated With Eplet Mismatch and PIRCHE Score
ROC curve revealed that the number of eplet mismatches = 14 and PIRCHE score = 176 provide the best predictive values (Supplementary Figure 2). Eplet mismatches were significantly associated with the incidence of de novo DSAs, observed in 8/235 (3.4%) of those with eplet mismatches ≤ 13 vs. 92/456 (20.2%) of those with eplet mismatches ≥ 14 (14–67 vs. 0–13: OR 7.172, 95% CI 3.418–15.050, p < 0.0001; Table 2A). PIRCHE scores were also significantly associated with de novo DSA production, identified in 26/318 (8.2%) of those with PIRCHE scores ≤ 175 and 74/373 (19.8%) among those with PIRCHE scores ≥ 176 (176–763 vs. 0–175: OR 2.780, 95% CI 1.728–4.470, p < 0.0001; Table 2B). Patients were divided into groups, including those with eplet mismatches of 0, 1–13, and 14–67, and PIRCHE scores of 0, 1–175, 176–763 as per the optimized predictive value based on ROC curve analysis. Only 4/99 (4.0%) of the patients with low levels of both eplet mismatches (1–13) and PIRCHE scores (1–175) produced de novo DSAs, whereas, de novo DSAs were detected in 70 (22.1%) of 317 patients with high levels of both parameters (Table 2C).
TABLE 2.
Incidence of de novo DR/DQ DSA by (A) eplet mismatch and (B) PIRCHE score, (C) de novo DSA positive rate by both of eplet mismatch and PIRCHE score.
| De novo DR/DQ DSA | ||||
|---|---|---|---|---|
| (+) | (–) | Positive rate | ||
| (A) | ||||
| Eplet MM (DRB+DQB) | 0–13 | 8 | 227 | 3.4% |
| 14–67 | 92 | 364 | 20.2% | |
| P < 0.0001 | ||||
| (B) | ||||
| PIRCHE score | 0–175 | 26 | 292 | 8.2% |
| 176–763 | 74 | 299 | 19.8% | |
| P < 0.0001 | ||||
| (C) | ||||
| Eplet MM (DRB+DQB) | ||||
| 0 | 1–13 | 14–67 | ||
| PIRCHE score | 0 | 0/28 (0%) | 0/0 (0%) | 0/0 (0%) |
| 1–175 | 0/52 (0%) | 4/99 (4.0%) | 22/139 (15.8%) | |
| 176–763 | 0/2 (0%) | 4/54 (7.4%) | 70/317 (22.1%) | |
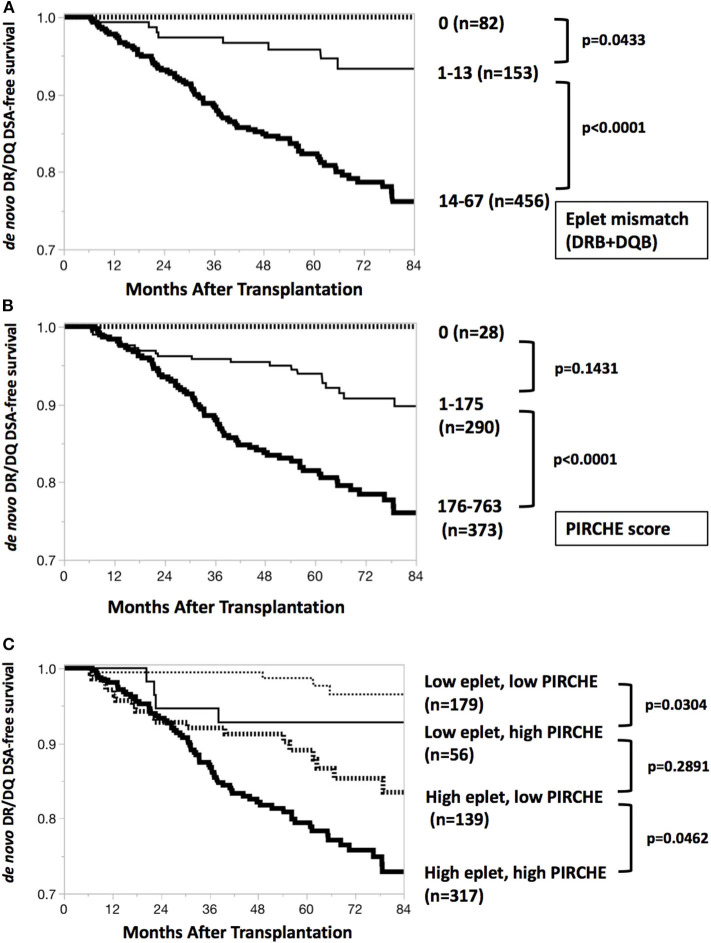
DSA-Free Graft Survival Predicted by Eplet Mismatch and PIRCHE Score
De novo DR/DQ DSA-free graft survivals by eplet mismatch and PIRCHE score were depicted in Figures 2A,B. Patients with eplet mismatches from 14 to 67 or PIRCHE scores from 176 to 763 responded with a significantly higher incidence of de novo DSAs. Subgroup comparisons revealed that higher PIRCHE scores were also associated with a significantly higher incidence of de novo DSAs than were low PIRCHE scores; this was the case among patients grouped in either the low or high eplet mismatch group (Figure 2C).
Figure 2.
De novo DR/DQ DSA-free survival curves. (A) De novo DR/DQ DSA-free survival by eplet mismatch. Kaplan-Meier de novo DR/DQ-free survival curves and Wilcoxon test show significant difference (p < 0.0001) between epitope mismatches 1–13 and 14–67. Thick dotted line, thin black line and thick black line indicate DSA-free survival (months after transplantation) of groups with eplet mismatches 0, 1–13, and 14–67, respectively. (B) De novo DR/DQ DSA-free survival by PIRCHE score. Kaplan-Meier de novo DR/DQ-free survival curves and Wilcoxon test show significant difference (p < 0.0001) between PIRCHE scores 1–175 and 176–763. Thick dotted line, thin black line and thick black line indicate DSA-free survival (months after transplantation) of groups with PIRCHE scores 0, 1–175, and 176–763, respectively. (C) De novo DR/DQ DSA-free survival by both eplet mismatch and PIRCHE score. Subgroup analysis by Kaplan-Meier de novo DR/DQ-free survival curves and Wilcoxon test shows significant differences between low and high PIRCHE scores among patients with low eplet mismatches (p = 0.0304) and high eplet mismatches (p = 0.0462). Thin dotted line, thin black line, thick dotted line and thick black line indicate DSA-free survival (months after transplantation) of groups including low eplet + low PIRCHE, low eplet + high PIRCHE, high eplet + low PIRCHE, and high eplet + high PIRCHE, respectively. Low eplet mismatches = 0–13; high eplet mismatches = 14–67; low PIRCHE scores = 0–175; high PIRCHE scores = 176–763.
Multivariate analysis of a Cox hazard regression model revealed that ABO-compatibility, eplet mismatch, PIRCHE score and history of acute T cell mediated rejection were all significantly associated with de novo DSA production (Table 3).
TABLE 3.
Risk factors associated with de novo DR/DQ DSA production.
| Cox proportional hazards regression model (DR, DQ) | ||||
|---|---|---|---|---|
| Variables | Univariate analysis | Multivariate analysis | ||
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| CSA vs. TAC | 1.314 (0.883–1.977) | 0.179 | ||
| ABO-I vs. ABO-Id/C | 0.613 (0.379–0.954) | 0.0295 | 0.465 (0.283–0.736) | 0.0009 |
| Eplet MM (DRB+DQB) | 1.035 (1.021–1.049)* | <0.0001 | 1.026 (1.009–1.043)* | 0.0028 |
| PIRCHE score | 1.026 (1.013–1.039)** | 0.0001 | 1.016 (1.001–1.032)** | 0.0347 |
| Acute TCMR | 3.052 (1.819–4.880) | <0.0001 | 3.309 (1.937–5.409) | <0.0001 |
Hazard ratio for one unit increase is expressed.
Hazard ratio for 10 unit increase is expressed.
HR, Hazard ratio; CI, confidence interval; CSA, cyclosporine; TAC, tacrolimus; ABO-I, ABO-incompatible transplantation; ABO-Id/C, ABO-identical/compatible transplantation; MM, mismatch; TCMR, T cell mediated rejection.
Discussion
Early diagnosis for chronic ABMR may be essential, because most treatment would be ineffective once there is evidence of graft dysfunction including elevated levels of serum creatinine and overt proteinuria (6). Effective treatment under subclinical condition of ABMR has been recently reported (27). However, early detection of ABMR is somewhat difficult due to consensus guidelines that suggest that routine monitoring of DSA monitoring is overall not cost-effective (28, 29). Furthermore, even if annual monitoring for all transplant patients was implemented, this interval might be too prolonged for meaningful detection of DSAs in some patients. The issue on the frequency of HLA antibody monitoring remains to be clarified. At the same time, innovative methods including high throughput technology and bioinformatics are currently in use in an effort to identify a biomarker for early detection of ABMR (30–32). Preventive measures are currently considered to be more likely to provide superior patient care when compared to the impact of therapeutic or preemptive strategies.
Our finding that the correlation between eplet mismatches and PIRCHE scores was weaker in DSA-positive patients than in DSA-negative patients corresponds to the previous report (33), although eplet mismatches for class I DSA and PIRCHE scores for donor-derived HLA class I were analyzed at that time. Two approaches, eplet mismatches and PIRCHE scores, seemed to be complementary to each other for predicting the risk of DSA production.
We found that analyses of both B cell and T cell epitopes (i.e., eplet mismatches and PIRCHE scores, respectively), had positive predictive capabilities with respect to de novo DSA production. Recent studies reported strong associations of eplet mismatch and/or PIRCHE-II scores with de novo DSA production or graft outcomes (25, 26, 34, 35), whereas it was also reported that allelic and epitope mismatch analysis presented no additional value with respect to risk management (36). Differences in immunogenicity might be dependent on the nature of the eplet and the precise mismatch position (37, 38). The immunological impact of PIRCHE-II is determined by the interactions between T cell receptor and the donor-derived peptides presented by HLA class II (22). Complete development of these algorithms in order to take into account both T and B cell epitopes remains a substantial challenge.
Our study has several limitations. This retrospective study features a relatively small sample size and brief follow-up period. There was substantial heterogeneity with respect to the immunosuppressive protocols used; this was to some extent related to the multi-center nature of this study. Our eplet mismatch data (primarily associated with HLA-DRB1,3,4,5, and DQB1) were comparable to findings from previous reports (12, 16, 17, 19, 25, 26, 34), although class I DSAs were not considered in this analysis. We calculated PIRCHE score based on typing information of recipient HLA-DRB1,3,4,5, DQA1, and DQB1 as the presenting HLA class II molecules; the addition of DRB3,4,5, and DQA1/DQB1 explains why our PIRCHE scores were higher in range than those included in previous reports that were based on DRB1 alone (21, 34, 35), even though we did not consider HLA-C among the donor-derived peptides. Patients with preformed DSAs were excluded so that our study could focus on primary immune responses rather than memory responses associated with long-term graft outcome (39). The primary endpoint in this study was de novo DSA production, not chronic ABMR or graft failure. Further analysis that included these data could potentially provide more convincing evidence of clinical benefit of the analyses of T cell and B cell epitopes; this would require significantly longer follow-up times. Furthermore, we need to consider the fact that de novo DSA production does not necessarily result in ABMR; this incidence of ABMR may relate to the amount and specificity of the DSAs, their ability to bind complement as well as graft accommodation resulting from ABO-incompatibility (7, 8, 40, 41). It was recently reported that imputed HLA alleles could lead to false findings particularly in multi-ethnic non-Caucasian individuals (42). Although we cannot deny such a possibility, we expect that single ethnic subjects used in this study would reduce the risk of estimation error.
Despite the above-mentioned limitations, the currently available data have clearly revealed the potential value of epitope analysis. These modalities offer predictive factors that are more reliable with respect to de novo DSA production than conventional HLA matching. This enhanced reliability suggests that these methodologies are likely to provide important contributions toward the development of individualized immunosuppression strategies in the not too distant future.
In conclusion, our findings revealed significant associations of eplet mismatches and PIRCHE scores with the prevalence of de novo DSAs. Further analysis of T cell and B cell epitopes is likely to provide critical information for the development of individualized immunosuppression strategies for the prevention graft rejection. Computer-based algorithms that predict T cell and B cell epitopes are undergoing rapid development. Further study will be needed in order to draw definitive conclusions regarding the clinical value of these predictive algorithms with respect to ABMR after organ transplantation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by The Institutional Ethics Committees of Aichi Medical University Hospital and the Institutional Review Board of Nagoya Daini Red Cross Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SS, KI, TT, and TK designed the research. SS and TK wrote the manuscript. SS, TT, KH, AT, NG, SN, YW, and TK performed the research. SS, TT, MN, ES, and TK participated in data analysis. ES and TK reviewed/edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
MN is an employee of PIRCHE AG. The UMC Utrecht has filed a patent application on the prediction of an alloimmune response against mismatched HLA. ES is listed as inventor on this patent. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Harue Fukami and Kohei Nishida of the Histocompatibility Laboratory at Nagoya Red Cross Hospital for their excellent technical assistance.
Footnotes
Funding. This work was supported by a Grant-in-Aid for Scientific Research (Grant Numbers JP16H05465, JP18K19593, and JP20H03818) from the Japan Society for the Promotion of Science (KAKENHI).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.02000/full#supplementary-material
References
- 1.Terasaki PI, Cai J. Human leukocyte antigen antibodies and chronic rejection: from association to causation. Transplantation. (2008) 86:377–83. 10.1097/TP.0b013e31817c4cb8 [DOI] [PubMed] [Google Scholar]
- 2.Wan SS, Ying TD, Wyburn K, Roberts DM, Wyld M, Chadban SJ. The treatment of antibody-mediated rejection in kidney transplantation: an updated systematic review and meta-analysis. Transplantation. (2018) 102:557–68. 10.1097/TP.0000000000002049 [DOI] [PubMed] [Google Scholar]
- 3.Böhmig GA, Eskandary F, Doberer K, Halloran PF. The therapeutic challenge of late antibody-mediated kidney allograft rejection. Transpl Int. (2019) 32:775–88. 10.1111/tri.13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley JA, Baldwin WM, Bingaman A, Ellenrieder C, Gebel HM, Glotz D, et al. Antibody-mediated rejection–an ounce of prevention is worth a pound of cure. Am J Transplant. (2011) 11:1131–9. 10.1111/j.1600-6143.2011.03581.x [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, Maruya E, Niwa M, Saji H, Kohara S, Katayama A, et al. Significant association between chronic antibody-mediated rejection and donor-specific antibodies against HLA-DRB rather than DQB in renal transplantation. Hum Immunol. (2011) 72:11–7. 10.1016/j.humimm.2010.10.018 [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto T, Watarai Y, Takeda A, Tsujita M, Hiramitsu T, Goto N, et al. De novo anti-HLA DSA characteristics and subclinical antibody-mediated kidney allograft injury. Transplantation. (2016) 100:2194–202. 10.1097/TP.0000000000001012 [DOI] [PubMed] [Google Scholar]
- 7.Schmitz R, Fitch ZW, Schroder PM, Choi AY, Jackson AM, Knechtle SJ, et al. B cells in transplant tolerance and rejection: friends or foes? Transpl Int. (2020) 33:30–40. 10.1111/tri.13549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan JH, Tinckam K. Clinical utility of complement dependent assays in kidney transplantation. Transplantation. (2018) 102(Suppl 1):S14–22. 10.1097/TP.0000000000001819 [DOI] [PubMed] [Google Scholar]
- 9.Tambur AR, Claas FH. HLA epitopes as viewed by antibodies: what is it all about? Am J Transplant. (2015) 15:1148–54. 10.1111/ajt.13192 [DOI] [PubMed] [Google Scholar]
- 10.Kramer CSM, Israeli M, Mulder A, Doxiadis IIN, Haasnoot GW, Heidt S, et al. The long and winding road towards epitope matching in clinical transplantation. Transpl Int. (2019) 32:16–24. 10.1111/tri.13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PI, Karpinski M, et al. Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant. (2013) 13:3114–22. 10.1111/ajt.12478 [DOI] [PubMed] [Google Scholar]
- 12.Wiebe C, Rush DN, Nevins TE, Birk PE, Blydt-Hansen T, Gibson IW, et al. Class II eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol. (2017) 28:3353–62. 10.1681/ASN.2017030287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sapir-Pichhadze R, Tinckam K, Quach K, Logan AG, Laupacis A, John R, et al. HLA-DR and -DQ eplet mismatches and transplant glomerulopathy: a nested case-control study. Am J Transplant. (2015) 15:137–48. 10.1111/ajt.12968 [DOI] [PubMed] [Google Scholar]
- 14.Sullivan PM, Warner P, Kemna MS, Albers EL, Law SP, Weiss NS, et al. HLA molecular epitope mismatching and long-term graft loss in pediatric heart transplant recipients. J Heart Lung Transplant. (2015) 34:950–7. 10.1016/j.healun.2014.12.017 [DOI] [PubMed] [Google Scholar]
- 15.Walton DC, Cantwell L, Hiho S, Ta J, Wright S, Sullivan LC, et al. HLA class II eplet mismatch predicts de novo DSA formation post lung transplant. Transpl Immunol. (2018) 51:73–5. 10.1016/j.trim.2018.10.002 [DOI] [PubMed] [Google Scholar]
- 16.Do guyen NHT, Wong G, Chapman JR, McDonald SP, Coates PT, Watson N, et al. The association between broad antigen HLA mismatches, Eplet HLA mismatches and acute rejection after kidney transplantation. Transplant Direct. (2016) 2:e120. 10.1097/TXD.0000000000000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosmoliaptsis V, Mallon DH, Chen Y, Bolton EM, Bradley JA, Taylor CJ. Alloantibody responses after renal transplant failure can be better predicted by donor-recipient HLA amino acid sequence and physicochemical disparities than conventional hla matching. Am J Transplant. (2016) 16:2139–47. 10.1111/ajt.13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiebe C, Kosmoliaptsis V, Pochinco D, Taylor CJ, Nickerson P. A comparison of HLA molecular mismatch methods to determine HLA immunogenicity. Transplantation. (2018) 102:1338–43. 10.1097/TP.0000000000002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiebe C, Kosmoliaptsis V, Pochinco D, Gibson IW, Ho J, Birk PE, et al. HLA-DR/DQ molecular mismatch: a prognostic biomarker for primary alloimmunity. Am J Transplant. (2019) 19:1708–19. 10.1111/ajt.15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geneugelijk K, Spierings E. Matching donor and recipient based on predicted indirectly recognizable human leucocyte antigen epitopes. Int J Immunogenet. (2018) 45:41–53. 10.1111/iji.12359 [DOI] [PubMed] [Google Scholar]
- 21.Lachmann N, Niemann M, Reinke P, Budde K, Schmidt D, Halleck F, et al. Donor-recipient matching based on predicted indirectly recognizable HLA epitopes independently predicts the incidence of de novo donor-specific HLA antibodies following renal transplantation. Am J Transplant. (2017) 17:3076–86. 10.1111/ajt.14393 [DOI] [PubMed] [Google Scholar]
- 22.Geneugelijk K, Spierings E. PIRCHE-II: an algorithm to predict indirectly recognizable HLA epitopes in solid organ transplantation. Immunogenetics. (2020) 72:119–29. 10.1007/s00251-019-01140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima F, Nakamura J, Yokota T. Analysis of HLA haplotypes in Japanese, using high resolution allele typing. MHC. (2001) 8:1–32. 10.12667/mhc.8.1 [DOI] [Google Scholar]
- 24.Okada M, Watarai Y, Iwasaki K, Murotani K, Futamura K, Yamamoto Y, et al. Favorable results in ABO-incompatible renal transplantation without B cell-targeted therapy: advantages and disadvantages of rituximab pretreatment. Clin Transplant. (2017) 31:e13071. 10.1111/ctr.13071 [DOI] [PubMed] [Google Scholar]
- 25.Tafulo S, Malheiro J, Santos S, Dias L, Almeida M, Martins LS, et al. Degree of HLA class II eplet mismatch load improves prediction of antibody-mediated rejection in living donor kidney transplantation. Hum Immunol. (2019) 80:966–75. 10.1016/j.humimm.2019.09.010 [DOI] [PubMed] [Google Scholar]
- 26.Sapir-Pichhadze R, Zhang X, Ferradji A, Madbouly A, Tinckam KJ, Gebel HM, et al. Epitopes as characterized by antibody-verified eplet mismatches determine risk of kidney transplant loss. Kidney Int. (2020) 97:778–85. 10.1016/j.kint.2019.10.028 [DOI] [PubMed] [Google Scholar]
- 27.Parajuli S, Joachim E, Alagusundaramoorthy S, Blazel J, Aziz F, Garg N, et al. Subclinical antibody-mediated rejection after kidney transplantation: treatment outcomes. Transplantation. (2019) 103:1722–9. 10.1097/TP.0000000000002566 [DOI] [PubMed] [Google Scholar]
- 28.Tait BD, Susal C, Gebel HM, Nickerson PW, Zachary AA, Claas FHJ, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. (2013) 95:19–47. 10.1097/TP.0b013e31827a19cc [DOI] [PubMed] [Google Scholar]
- 29.Tambur AR, Campbell P, Claas FH. Sensitization in transplantation: assessment of risk (STAR) 2017 working group meeting report. Am J Transplant. (2018) 18:1604–14. 10.1111/ajt.14752 [DOI] [PubMed] [Google Scholar]
- 30.Van Loon E, Gazut S, Yazdani S, Lerut E, de Loor H, Coemans M, et al. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: A multicentre, prospective study. EBioMedicine. (2019) 46:463–72. 10.1016/j.ebiom.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matz M, Heinrich F, Lorkowski C, Wu K, Klotsche J, Zhang Q, et al. MicroRNA regulation in blood cells of renal transplanted patients with interstitial fibrosis/tubular atrophy and antibody-mediated rejection. PLoS ONE. (2018) 13:e0201925. 10.1371/journal.pone.0201925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon HJ, Lee JG, Kim K, Jang JY, Han SW, Choi J, et al. Peripheral blood transcriptome analysis and development of classification model for diagnosing antibody-mediated rejection vs accommodation in ABO-incompatible kidney transplant. Am J Transplant. (2020) 20:112–24. 10.1111/ajt.15553 [DOI] [PubMed] [Google Scholar]
- 33.Otten HG, Calis JJ, Keşmir C, van Zuilen AD, Spierings E. Predicted indirectly recognizable HLA epitopes presented by HLA-DR correlate with the de novo development of donor-specific HLA IgG antibodies after kidney transplantation. Hum Immunol. (2013) 74:290–6. 10.1016/j.humimm.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 34.Daniëls L, Naesens M, Bosmans JL, Abramowicz D, Nagler E, Van Laecke S, et al. The clinical significance of epitope mismatch load in kidney transplantation: a multicentre study. Transpl Immunol. (2018) 50:55–9. 10.1016/j.trim.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 35.Geneugelijk K, Niemann M, Drylewicz J, van Zuilen AD, Joosten I, Allebes WA, et al. PIRCHE-II Is related to graft failure after kidney transplantation. Front Immunol. (2018) 9:321. 10.3389/fimmu.2018.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delion A, Girerd S, Duarte K, Girerd N, Schikowski J, Kessler M, et al. Which is the best predictor of de novo donor-specific antibodies in a cohort of non-sensitized first kidney transplantation: antigenic, allelic, epitope, or physiochemical HLA mismatches? Clin Transplant. (2019) 33:e13508 10.1111/ctr.13508 [DOI] [PubMed] [Google Scholar]
- 37.McCaughan JA, Battle RK, Singh SKS, Tikkanen JM, Moayedi Y, Ross HJ, et al. Identification of risk epitope mismatches associated with de novo donor-specific HLA antibody development in cardiothoracic transplantation. Am J Transplant. (2018) 18:2924–33. 10.1111/ajt.14951 [DOI] [PubMed] [Google Scholar]
- 38.Tambur AR. HLA-epitope matching or Eplet risk stratification: the devil is in the details. Front Immunol. (2018) 9:2010. 10.3389/fimmu.2018.02010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aubert O, Loupy A, Hidalgo L, Duong van Huyen JP, Higgins S, Viglietti D, et al. Antibody-mediated rejection due to preexisting versus de novo donor-specific antibodies in kidney allograft recipients. J Am Soc Nephrol. (2017) 28:1912–23. 10.1681/ASN.2016070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki K, Miwa Y, Haneda M, Kuzuya T, Ogawa H, Onishi A, et al. AMP-activated protein kinase as a promoting factor, but complement and thrombin as limiting factors for acquisition of cytoprotection: implications for induction of accommodation. Transpl Int. (2013) 26:1138–48. 10.1111/tri.12186 [DOI] [PubMed] [Google Scholar]
- 41.Okada M, Watarai Y, Iwasaki K, Futamura K, Yamamoto T, Hiramitsu T, et al. Lower incidence of de novo donor-specific antibodies against HLA-DR in ABO-incompatible renal transplantation. Hum Immunol. (2019) 80:169–75. 10.1016/j.humimm.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 42.Engen RM, Jedraszko AM, Conciatori MA, Tambur AR. Substituting imputation of HLA antigens for high resolution HLA typing: Evaluation of a multiethnic population and implications for clinical decision making in transplantation. Am J Transplant. (2020). 10.1111/ajt.16070. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.