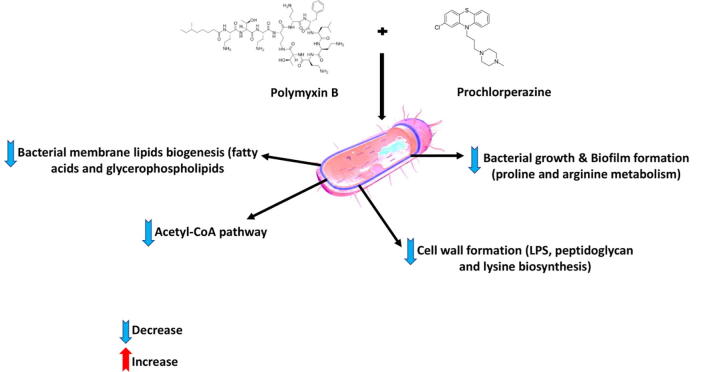
Graphical abstract
Keywords: Antimicrobial resistance, Antimicrobial peptides, Phenothiazines, Metabolomics, MDR, Gram-negative
Abstract
The status quo for combating uprising antibacterial resistance is to employ synergistic combinations of antibiotics. Nevertheless, the currently available combination therapies are fast becoming untenable. Combining antibiotics with various FDA-approved non-antibiotic drugs has emerged as a novel strategy against otherwise untreatable multi-drug resistant (MDR) pathogens. The apex of this study was to investigate the mechanisms of antibacterial synergy of the combination of polymyxin B with the phenothiazines against the MDR Gram-negative pathogens Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa. The synergistic antibacterial effects were tested using checkerboard and static time-kill assays. Electron microscopy (EM) and untargeted metabolomics were used to ascertain the mechanism(s) of the antibacterial synergy. The combination of polymyxin B and the phenothiazines showed synergistic antibacterial activity in checkerboard and static time-kill assays at clinically relevant concentrations against both polymyxin-susceptible and polymyxin-resistant isolates. EM revealed that the polymyxin B-prochlorperazine combination resulted in greater damage to the bacterial cell compared to each drug monotherapy. In metabolomics, at 0.5 h, polymyxin B monotherapy and the combination (to a greatest extent) disorganised the bacterial cell envelope as manifested by a major perturbation in bacterial membrane lipids (glycerophospholipids and fatty acids), peptidoglycan and lipopolysaccharide (LPS) biosynthesis. At the late time exposure (4 h), the aforementioned effects (except LPS biosynthesis) perpetuated mainly with the combination therapy, indicating the disorganising bacterial membrane biogenesis is potentially behind the mechanisms of antibacterial synergy. In conclusion, the study highlights the potential usefulness of the combination of polymyxin B with phenothiazines for the treatment of polymyxin-resistant Gram-negative infections (e.g. CNS infections).
1. Introduction
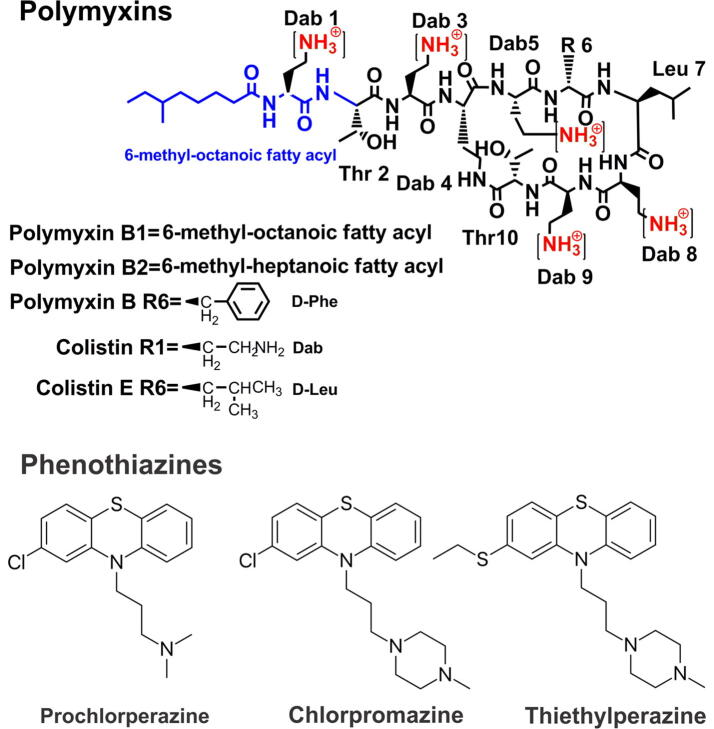
Antibiotic resistance has evolved as a serious global threat to mankind inevitably leading to increasing social and economic cost. The World Health Organisation (WHO) warned of the possibility of a post-antibiotic era where simple bacterial infections become untreatable [1]. According to a recent report of the World Health Organisation (WHO), multi-drug resistant (MDR) A. baumannii are amongst of the top priority pathogens that desperately require new antibiotic treatments [2]. These MDR pathogens often cause life-threatening nosocomial infections particularly in immuno-compromised and critically-ill patients, such as pneumonia and sepsis;[3], [4] and They can readily develop resistance to all available antibiotics including the last-resort polymyxins (i.e. polymyxin B and colistin, also known as polymyxin E) [5], [6], [7]. Colistin and polymyxin B are the cyclic lipopeptides which are comprised of an intra-molecular heptapeptide loop and a linear tripeptide side chain with a hydrophobic N-terminal fatty acyl group (Fig. 1). The intra-molecular heptapeptide ring is formed by the connection between the carboxyl group of the C-terminal threonine (L-Thr) residue at position 10 and amino group side chain of L-α,γ-diaminobutyric acid (Dab) residue at the position 4 [8], [9]. It is postulated that polymyxins exert their antimicrobial action via direct interaction between the positively charges Dab residues and negatively charges lipid A component of the LPS which then enables the insertion of the hydrophobic tail and the hydrophobic moieties of amino acid 6 and 7 into the outer membrane to induce membrane expansion [10]. This event is thought to be followed by polymyxin-mediated fusion of the inner leaflets of the outer membrane and the outer leaflet of the cytoplasmic membrane surrounding the periplasmic space. The fusion causes phospholipid exchange and then, osmotic imbalance that eventually leads to cell death [10], [11], [12]. The most common mode of polymyxins resistance observed in A. baumannii is developed through at least two distinct mechanisms, namely, complete LPS loss or modification of lipid A via the introduction of phosphoethanolamine (PEtN) or aminoarabinose (Ara4N) moieties onto the phosphates of its lipid A component [13], [14], [15].
Fig. 1.
The chemical structures for polymyxins and the phenothiazines drugs (prochlorperazine, chlorpromazine and thiethylperazine) employed in this study.
Phenothiazines were initially used in histochemistry for staining plasmodia and latter explored as an antimalaria in the late 1880s, and were used as antiparasitic and antibacterial drugs in livestock in the 1930s [16]. Latter derivatives such prochlorperazine, thiethylperazine and chlorpromazine were developed as antipsychotics to treat mania, psychotic depression, agitation, schizophrenia or as antiemetic drugs [17]. Phenothiazine itself was sprayed on orchards due to its high potency against apple maggot, corn borer, worms [16], [18]. In addition to the insecticidal activity of phenothiazine there was also a delay in the rotting of fruits, which indicated the potential antibacterial activity of the phenothiazine core [19]. This antibacterial activity was later utilised in the clinic to treat urinary tract infections [20]. Studies have shown antibacterial activity of different phenothiazine derivatives against Staphylococcus aureus, P. aeruginosa, A. baumannii, K. pneumoniae and Enterococcus spp. [21], [22], [23], [24]. Moreover, many phenothiazine derivatives such as thioridazine, chlorpromazine and prochlorperazine were found to have the ability to re-sensitize antibiotic-resistant bacteria (i.e. methicillin-resistant Staphylococcus aureus and erythromycin-resistant Streptococcus pyogenes) [25]. Notably, there was a study in 1960 showing that a patient with tuberculosis was treated successfully by chlorpromazine therapy [26]. Overall, it is clear that phenothiazines have great potential as anti-infective agents either per se or in combination with other compounds.
An emerging ‘off-the-shelf’ approach to combating polymyxin resistance is to repurpose FDA approved non-antibiotic drugs that present synergistic activity when combined with polymyxins [27], [28], [29], [30], [31]. Notably, the combination of chlorpromazine with the antifungal amphotericin B was synergistic against Cryptococcus neoformans [32]. Our laboratory previously demonstrated that the antibacterial effect of polymyxin B is synergistic in combination with non-antibiotic drugs such as closantel, ivacaftor, tamoxifen, raloxifene, toremifene, mitotane, and zidovudine [27], [28], [29], [30], [33], [34], [35]. The focus of the present study was to evaluate the antibacterial synergy of phenothiazine neuroleptic drugs in combination with polymyxin B against a range of polymyxin resistant and susceptible Gram-negative isolates and investigate potential antimicrobial mode of action using untargeted metabolomics and scanning and transmission electron microscopy. The presented findings highlight the effective synergy and clinical potential of this novel combination for the treatment of MDR polymyxin-resistant Gram-negative infections.
2. Results and discussion
2.1. Synergy testing of polymyxin B – phenothiazine combinations: determination of MICs and FICs
The synergistic antimicrobial activity of polymyxin B and the phenothiazines (prochlorperazine, chlorpromazine and thiethylperazine) monotherapy, and in combination was tested against a panel of clinical isolates of P. aeruginosa, A. baumannii, and K. pneumoniae (Table 1). Overall, the polymyxin B and phenothiazine combinations showed synergistic activity against more than half of the strains tested. The combination of polymyxin B and prochlorperazine displayed a synergistic effect against 13 out of 22 P. aeruginosa isolates tested (including 12 out of 13 polymyxin B resistant isolates); 4 out of 6 A. baumannii isolates tested, and 2 out of 4 K. pneumoniae isolates tested. Polymyxin B and prochlorperazine monotherapies were found to be ineffective (MICs of both drugs ranging from 8 to > 128 mg/L) against the polymyxin B resistant isolates. The polymyxin B and chlorpromazine combination demonstrated synergistic effect against 14 out of 22 P. aeruginosa isolates tested (including 12 out of 13 polymyxin B resistant strains); 1 out of 3 A. baumannii isolates tested, and 2 out of 3 K. pneumoniae isolates tested. Polymyxin B and chlorpromazine monotherapies were found to be ineffective (MICs of both drugs ranging from 8 to >128 mg/L) against the polymyxin B resistant isolates. Polymyxin B and thiethylperazine in combination showed a synergistic effect against 16 out of 22 P. aeruginosa isolates tested (including 12 out of 13 polymyxin B resistant strains); 2 out of 3 A. baumannii isolates tested, and 2 out of 3 K. pneumoniae isolates tested. Polymyxin B and thiethylperazine monotherapies were found to be ineffective (MICs of both drugs ranging from 8 to >128 mg/L) against all polymyxin B resistant isolates except for A. baumannii FADDI-AB065 wherein thiethylperazine monotherapy displayed low MIC 4 mg/L.
Table 1.
Antimicrobial activity of polymyxin B (PMB), prochlorperazine (PCH), chlorpromazine (CHP) and thiethylperazine (THY) monotherapy, and in combination.
|
MIC (mg/L) |
FIC indexd |
|||||||
|---|---|---|---|---|---|---|---|---|
| Isolates | PMB | PCH | CHP | THY | PMB-PCH | PMB-CHP | PMB-THY | |
| Polymyxin-resistant isolates | ||||||||
| P. aeruginosa | FADDI-PA006 muca | 8 | >128 | >128 | >128 | 0.27 | 0.27 | 0.26 |
| FADDI-PA092 | 16 | >128 | >128 | >128 | 0.28 | 0.19 | 0.28 | |
| FADDI-PA067 n/ma (s)b | 16 | >128 | >128 | >128 | 0.28 | 0.19 | 0.19 | |
| FADDI-PA093 | 32 | >128 | >128 | >128 | 0.25 | 0.19 | 0.19 | |
| FADDI-PA066 n/m | 32 | 128 | >128 | >128 | 0.25 | 0.27 | 0.13 | |
| FADDI-PA112 | 16 | >128 | >128 | >128 | 0.19 | 0.31 | 0.31 | |
| FADDI-PA070 n/m | 64 | 128 | >128 | >128 | 0.16 | 0.19 | 0.14 | |
| FADDI-PA071 n/m | >64 | >128 | >128 | >128 | 0.1 | 0.14 | 0.16 | |
| FADDI-PA016 n/m (L)b | >64 | >128 | >128 | >128 | 0.13 | 0.13 | 0.13 | |
| FADDI-PA068 n/m | >64 | >128 | >128 | >128 | 0.13 | 0.08 | 0.14 | |
| FADDI-PA063 muc | >64 | >128 | >128 | >128 | 0.13 | 0.08 | 0.06 | |
| FADDI-PA072 n/m | >64 | >128 | >128 | >128 | 0.16 | 0.27 | 0.28 | |
| FADDI-PA064 n/m | >64 | 64 | 128 | 32 | 0.63 | 1 | 1 | |
| Polymyxin-susceptible isolates | ||||||||
| FADDI-PA002 muc | 0.125 | 128 | >128 | >128 | 0.53 | 0.5 | 0.27 | |
| FADDI-PA005 n/m | 0.25 | 128 | >128 | >128 | 0.52 | 0.56 | 0.52 | |
| FADDI-PA021 muc | 0.25 | >128 | 128 | >128 | 0.5 | 1 | 0.53 | |
| FADDI-PA111 | 1 | >128 | >128 | >128 | 0.51 | 0.56 | 0.53 | |
| FADDI-PA117 | 0.5 | >128 | >128 | >128 | 0.5 | 0.28 | 0.52 | |
| FADDI-PA007 n/m | 0.5 | >128 | >128 | >128 | 0.53 | 0.56 | 1 | |
| FADDI-PA020 muc | 0.5 | >128 | >128 | >128 | 0.53 | 0.56 | 0.28 | |
| FADDI-PA024 | 1 | >128 | >128 | >128 | 0.26 | 0.56 | 0.27 | |
| FADDI-PA019 muc | 1 | 128 | >128 | >128 | 0.52 | 0.31 | 0.27 | |
| Polymyxin-resistant isolates | ||||||||
| A. baumannii | FADDI-AB065 | >64 | 8 | 8 | 4 | 1 | 1 | 1 |
| FADDI-AB148 | 8 | >128 | NAc | NA | 0.13 | NA | NA | |
| FADDI-AB144 | 8 | >128 | NA | NA | 0.13 | NA | NA | |
| Polymyxin-susceptible isolates | ||||||||
| ATCC 17,978 | 0.5 | 128 | NA | NA | 0.28 | NA | NA | |
| ATCC 19,606 | 1 | >128 | >128 | 32 | 0.27 | 0.27 | 0.31 | |
| FADDI-AB146 | 0.5 | 64 | 64 | 32 | 0.51 | 0.51 | 0.27 | |
| Polymyxin-susceptible isolates | ||||||||
| K. pneumoniae | ATCC 700,721 | 0.125 | >128 | >128 | >128 | 0.75 | 0.63 | 0.75 |
| Kp BM1 | 0.125 | >128 | NA | NA | 0.63 | NA | NA | |
| FADDI-KP001 | 1 | >128 | >128 | >128 | 0.38 | 0.25 | 0.19 | |
| FADDI-KP002 | 0.25 | >128 | >128 | >128 | 0.25 | 0.13 | 0.25 | |
muc, mucoid; n/m, nonmucoid.
S, small colony; L, large colony.
NA, not accessed.
FIC = FIC INDEX= (MIC polymyxin B in combination with phenothiazine/MIC polymyxin B monotherapy) + (MIC phenothiazine in combination with polymyxin B/MIC phenothiazine monotherapy); synergy FIC < 0.5 [green]; additivity FIC = 0.5–1.0 [yellow]; indifference FIC = 1–4 [not observed]; antagonism FIC ≥ 4 [not observed].
To investigate whether the synergy between polymyxin B and the phenothiazines is a result of the permeabilizing activity of polymyxin on the Gram-negative OM that enables the intracellular entry of phenothiazines to exert their action on their intracellular targets, polymyxin nonapeptide plus prochlorperazine treatment was utilised against one polymyxin-resistant P. aeruginosa strain (FADDI-PA070) and the polymyxin-susceptible strain P. aeruginosa ATCC 27853. Polymyxin nonapeptide is the deacylated amino derivative of polymyxin B that lacks the direct bactericidal activity but retains the OM permeabilizing activity. It is often employed as a sensitizer for hydrophobic antibiotics [36]. The polymyxin nonapeptide–prochlorperazine combination was ineffective against P. aeruginosa FADDI-PA070, whereas showed an excellent synergy (FIC = 0.15) against P. aeruginosa ATCC 27853.
2.2. Static time-kill studies
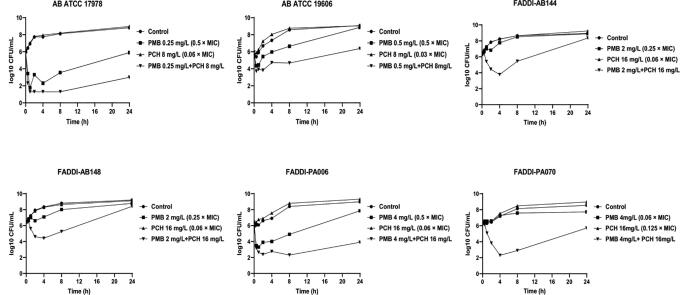
The antimicrobial activity of polymyxin B and prochlorperazine was further assessed in static time kill studies against polymyxin B-resistant P. aeruginosa strains, FADDI-PA070 (polymyxin B MIC = 128 mg/L, prochlorperazine MIC = 128 mg/L) and FADDI-PA006 (polymyxin B MIC = 8 mg/L, prochlorperazine MIC > 128 mg/L); polymyxin B-susceptible A. baumannii strains, A. baumannii ATCC 19606 (polymyxin B MIC = 1 mg/L, prochlorperazine MIC > 128 mg/L) and A. baumannii ATCC 17978 (polymyxin B MIC = 0.5 mg/L, prochlorperazine MIC = 128 mg/L); and against polymyxin B-resistant A. baumannii strains FADDI-AB148 (polymyxin B MIC = 8 mg/L, prochlorperazine MIC > 128 mg/L) and FADDI-AB144 (polymyxin B MIC = 8 mg/L, prochlorperazine MIC > 128 mg/L) (Fig. 2). Clinically relevant concentrations of polymyxin B and prochlorperazine were assessed [37], [38]. Polymyxin B monotherapy showed insignificant killing activity against P. aeruginosa FADDI-PA070 as manifested by a ≤ 1 log10 CFU/mL decrease in bacterial counts compared with untreated controls across all the time points. Although, a higher killing curve was observed after polymyxin B monotherapy against P. aeruginosa FADDI-PA006 at early time points with a ≥ 2.0- log10 CFU/ml decrease in the bacterial burden, inconsistent regrowth was also observed, with only a 1.0- log10 CFU/ml difference compared to the control after 24 h. Against A. baumannii strains, polymyxin B per se did not display significant bacterial killing except for A. baumannii ATCC 17978 wherein>2 log10 CFU/mL decrease in bacterial counts compared with control was seen after polymyxin B monotherapy (Fig. 2). Prochlorperazine monotherapy displayed no antimicrobial activity against all the strains assessed during 24 h treatment, which was demonstrated by its comparable time kill curves compared with untreated controls (Fig. 2). On the other hand, the combination therapy of polymyxin B and prochlorperazine demonstrated more effective bacterial killing activity against all tested strains peaking at 4 and 8 h with an 4.0–6.0-log10 CFU/ml decrease in the bacterial burden compared to the control, which was sustained to an ~5.0-log10 CFU/ml (P. aeruginosa FADDI-PA006 and A. baumannii ATCC 17978) and ~2.5-log10 CFU/ml (P. aeruginosa FADDI-PA070 and A. baumannii ATCC 19606) decline after 24 h compared to the control (Fig. 2).
Fig. 2.
Time kill curves for polymyxin B and prochlorperazine monotherapies and in combinations against polymyxin-resistant P. aeruginosa strains, FADDI-PA070 (polymyxin B MIC = 64 mg/L, prochlorperazine MIC = 128 mg/L) and FADDI-PA006 (polymyxin B MIC = 8 mg/L, prochlorperazine MIC > 128 mg/L); against polymyxin B-susceptible A. baumannii strains, ATCC 19606 (polymyxin B MIC = 1 mg/L, prochlorperazine MIC > 128 mg/L) and ATCC 17978 (polymyxin B MIC = 0.5 mg/L, prochlorperazine MIC = 128 mg/L); and against polymyxin B-resistant A. baumannii strains FADDI-AB148 and FADDI-AB144 (polymyxin B MIC = 8 mg/L, prochlorperazine MIC > 128 mg/L for both strains).
2.3. Scanning electron microscopy and transmission electron microscopy
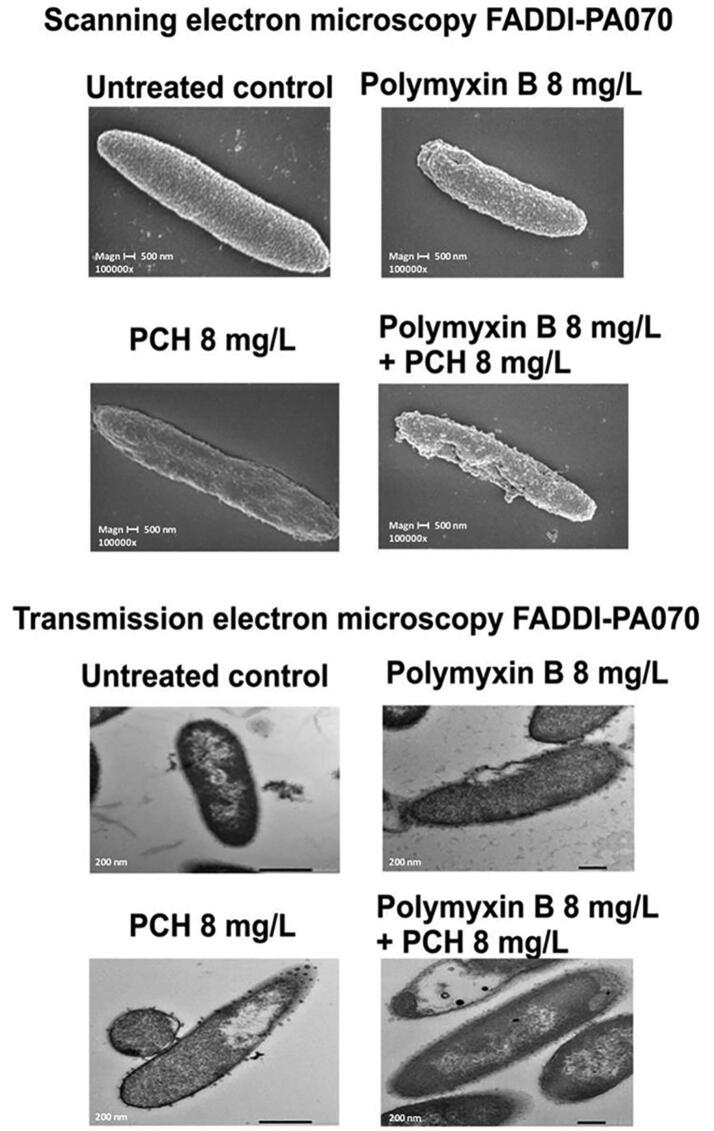
Among Gram-negative species, P. aeruginosa are notoriously difficult to treat due to its a formidable outer membrane barrier [39]. Therefore, scanning (SEM) and transmission electron microscopy (TEM) were conducted to examine the effect of polymyxin B and prochlorperazine monotherapy and in combination on the cellular morphology of the polymyxin B resistant P. aeruginosa strain FADDI-PA070 at 2 h (Fig. 3). SEM images showed blebbing, protrusions and minor damage of the bacterial cell membrane under polymyxin B monotherapy. The bacterial cells looked ‘dried out’ with shrinkage under prochlorperazine monotherapy. The combination therapy resulted in blebbing, protrusions and extensive damage to the bacterial cell membrane, which appeared to result in the leakage of cellular contents. In the TEM images, blebbing, protrusions and cell membrane damage are observed under polymyxin B monotherapy. Shrinkage and vesicle formation are observed under prochlorperazine monotherapy. The combination therapy resulted in greater shrinkage of the bacterial cell with vesicle formation.
Fig. 3.
Scanning and transmission electron microscopy images of polymyxin-resistant P. aeruginosa isolate FADDI-PA070 (polymyxin B MIC = 64 mg/L, prochlorperazine MIC = 128 mg/L) treated with polymyxin B or prochlorperazine monotherapy and in combination.
2.4. Metabolomics analysis of A. baumannii ATCC17978 treated with polymyxin B, prochlorperazine, and their combination
A. baumannii ATCC 17978 was specifically selected for the metabolomics studies as polymyxin B-prochlorperazine combination displayed superior killing kinetics against this strain in the time kill studies. Across all treatment conditions a total of 986 putatively identified metabolites were obtained, including: 39 metabolites in carbohydrate metabolism; 124 metabolites in amino acid metabolism; 231 metabolites in peptides metabolism; 46 metabolites in nucleotide metabolism and 206 in lipid metabolism. The Multivariate Data Analysis using one-way analysis of variance (ANOVA) was performed to determine the significant metabolites affected by the treatment conditions (≥0.58-log2-fold; p ≤ 0.05; FDR ≤ 0.05). The reproducibility of all sample groups was acceptable across both time points (0.5 and 4 h), where the median RSD across all time points was (16–19%) for untreated (control) groups and (17–26%) for treated samples, consistent with some baseline variability in the dynamics of ordinary bacterial metabolism with and without antibiotic treatment (Table S1). Notably, the PCA plots showed that the untreated control and combination treated samples were significantly separated across all the time points 0.5 and 4 h (Figure S1). Polymyxin B and combination treatments were overlapped across all exposure times which suggested the polymyxin B was the initial driving force behind the combination treatment; whereas prochlorperazine closely resembled the untreated control samples in particular at 0.5 h (Figure S1). Similarly, the heatmaps reflected the aforementioned differences between the treated groups and untreated (control) groups (Figure S2). The combination treatment perturbed 387 (0.5 h) and 221 (4 h) significant metabolites; whereas polymyxin B monotherapy induced far less perturbations in particular at 4 h, with only 53 significantly perturbed metabolites. Prochlorperazine monotherapy showed a latent effect, significantly perturbing only 12 significant metabolites at 4 h (Figure S3). The combination treatment induced 185 and 181 unique significant metabolites at 0.5 and 4 h, respectively (Figure S4). The classification analysis of the significantly impacted metabolites illustrated that amino acids, peptides, lipids, nucleotides and carbohydrates were largely perturbed (mainly decreased) compared to cofactor, vitamin and energy metabolism; which were less significantly impacted after combination treatment (Figure S5). This pattern was observed across both time points 0.5 and 4 h (Figure S5A and B). Similarly, but to a lesser extent polymyxin B monotherapy produced a similar scenario wherein lipids, amino acids and carbohydrates were the impacted metabolite classes (Figure S5A and B). In contrast, prochlorperazine monotherapy caused an elevation in the levels of amino acid and peptides intermediates at 4 h (Fig. 5B).
Fig. 5.
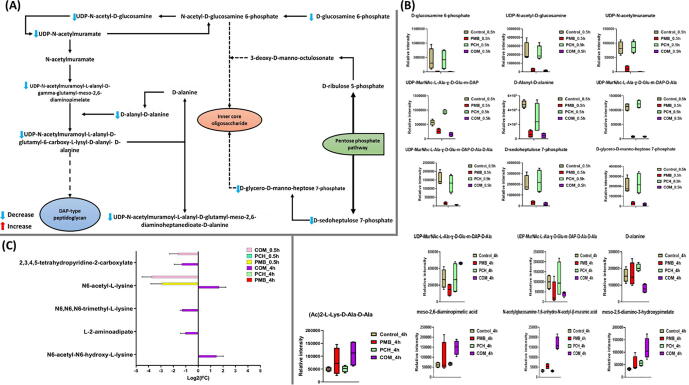
The schematic diagram depicting significantly impacted metabolites of amino-sugar and nucleotide-sugar metabolism, peptidoglycan biosynthesis, lipopolysaccharides biosynthesis and pentose phosphate pathway for A. baumannii ATCC 17978 treated with polymyxin B (PMB) or prochlorperazine (PCH) monotherapy and the combination (COM) after 0.5 h exposure (A). Bar charts for the significantly influenced metabolites of amino-sugar and nucleotide-sugar metabolism, peptidoglycan biosynthesis, lipopolysaccharides biosynthesis and pentose phosphate pathways at 0.5 h and 4 h (B), and (C) Lysine metabolism at 0.5 and 4 h (≥1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05).
2.5. Analyses of glycerophospholipid and fatty metabolism perturbations
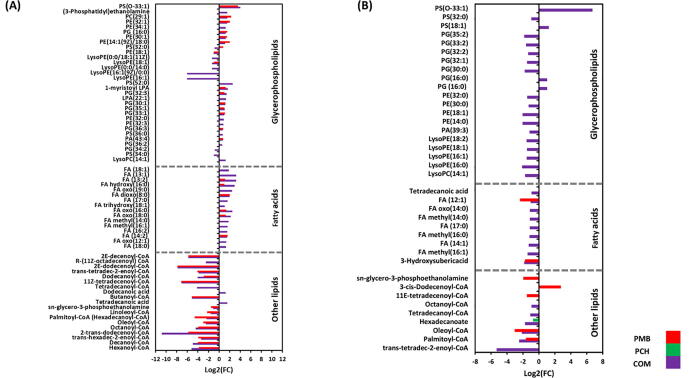
At 0.5 h the combination treatment induced significantly more perturbations across all lipid classes (i.e. fatty acids and glycerophospholipids [e.g. phosphatidylethanolamine, PE; phosphatidylserine, PS; phosphatidylglycerol, PG]) compared to the polymyxin B and prochlorperazine monotherapies (Fig. 4A). Notably, alterations in glycerophospholipid levels were overrepresented compared to other lipid classes such as fatty acids after combination treatment. Among the glycerophospholipids, the lysophospholipids (LPLs) e.g. lysophosphatidylethanolamines underwent a dramatic decline following combination treatment, notably lysoPE(16:1) (log2FC = -6.1), lysoPE(18:1) (log2FC = -1.3) and lysoPE(0:0/14:0) (log2FC = -1.3). Furthermore, essential precursors of bacterial membrane lipids such as sn-glycero-3-phosphoethanolamine were markedly decreased following combination treatment (log2FC = -1.1) (Fig. 4A). Likewise, fundamental fatty acids which are involved in fatty acid elongation and degradation underwent significant changes following combination treatment, notably trans-hexadec-2-enoyl-CoA (log2FC = -3.4), trans-tetradec-2-enoyl-CoA (log2FC = -3.9), dodecanoyl-CoA (log2FC = -4.1) and 2E-dodecenoyl-CoA (log2FC = -7.8) (Fig. 4A). The impact of polymyxin B monotherapy was less than the combination treatment at 0.5 h; and there were greater perturbations in glycerophospholipids compared to fatty acids (Fig. 4A). Fatty acid elongation and degradation metabolites were among the most significantly impacted lipids after polymyxin B monotherapy, including 2E-dodecenoyl-CoA (log2FC = -7.8), trans-tetradec-2-enoyl-CoA (log2FC = -4.1) and trans-hexadec-2-enoyl-CoA (log2FC = -3.9) (Fig. 4A). Prochlorperazine monotherapy did not induce any significant effect on lipid metabolism at 0.5 h.
Fig. 4.
Perturbations of bacterial lipids. Significantly perturbed lipids in A. baumannii ATCC17978 following treatment with polymyxin B (PMB, red), prochlorperazine (PCH, green) and the combination (COM, purple) for (A) 0.5 and (B) 4 h. Lipid names are putatively assigned based on accurate mass (≥0.58553-log2-fold, p ≤ 0.05; FDR ≤ 0.05). Control, untreated samples; PE, phosphoethanolamines; PG, glycerophosphoglycerols; PS, glycerophosphoserines; PC, glycerophosphocholines; PA, glycerophosphates; LysoPE, lysophosphatidylethanolamines; LysoPC, lysophosphatidylcholines; LPA, lysophosphatidic acid FA, fatty acids. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
On the other hand, at 4 h, the combination therapy produced a remarkable perturbation patterns on lipid intermediates particularly against glycerophospholipids (Fig. 4B). Notably, the levels of bacterial membrane lysophosphatidylethanolamines were decreased after combination treatment, including lysoPE(16:0) (log2FC = -2.1), lysoPE(16:1) (log2FC = -1.6), lysoPE(18:1) (log2FC = -1.5) and lysoPE(18:2) (log2FC = -1.5) (Fig. 4B). Furthermore, the impact on fatty acid elongation was sustained following the combination therapy as manifested by a significant decline in the abundance of palmitoyl-CoA, trans-tetradec-2-enoyl-CoA, hexadecanoate, tetradecanoyl-CoA and octanoyl-CoA (>-1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 4B). Similar pattern was seen on fatty acid intermediates which underwent a marked decrease in their abundance after the combination treatment, such as FA (14:1) (log2FC = -1.2), FA methyl(14:0) (log2FC = -1.1), FA methyl(16:0) (log2FC = -1.1) and tetradecanoic acid (myristic acid) (log2FC = -0.93) (Fig. 4B).
In comparison, polymyxin B monotherapy impact at 4 h has started to fade away with only slight perturbation on fatty acid elongation and degradation intermediates, namely 3-cis-dodecenoyl-CoA (log2FC = 2.7), 11E-tetradecenoyl-CoA (log2FC = -1.5), palmitoyl-CoA (log2FC = -1.6) and oleoyl-CoA (log2FC = -3.0) (Fig. 4B). Not unlike the effects at 0.5 h, no significant changes in lipid metabolism [except for hexadecanoate (log2FC = -0.74)] were observed following prochlorperazine monotherapy at 4 h (Fig. 4B).
2.6. Analyses of perturbations in peptidoglycan, lipopolysaccharides and lysine biosynthesis; amino-sugar and sugar-nucleotide metabolism; the pentose phosphate pathway
Several metabolites related to amino-sugar, sugar-nucleotide metabolism; the pentose phosphate pathway (PPP) and its downstream peptidoglycan, lipopolysaccharide (LPS) and lysine biosynthesis pathways were significantly perturbed at 0.5 and 4 h post treatment with the combination (Fig. 5A-C). At 0.5 h after combination treatment, three essential precursors of amino-sugar and nucleotide-sugar metabolism were significantly decreased, namely D-glucosamine-6-phosphate, UDP-N-acetyl-D-glucosamine and UDP-N-acetylmuramate (>-4-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5A&B). As a downstream consequence, the combination treatment also caused a marked decline in the abundance of fundamental peptidoglycan building blocks (i.e. which the amino-sugar and nucleotide-sugar pathways feed into), including UDP-N-acetylmuramoyl-L-alanyl-D-γ-glutamyl-meso-2,6-diaminopimelate, UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate-D-alanine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanyl-D-alanine and D-alanyl-D-alanine (>-1.5-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5B). A dramatic decline in the abundance of common intermediates of the LPS biosynthetic pathway (D-glycero-D-manno-heptose 7-phosphate) and pentose phosphate pathway (D-sedoheptulose-7-phosphate) were also seen following combination therapy at 0.5 h (>3.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5B). Simultaneously, the abundance of two precursors of lysine metabolism was also undergoing a significant decrease following the combination treatment at 0.5 h, namely N6-acetyl-L-lysine (log2FC = -3.6) and 2,3,4,5-tetrahydropyridine-2-carboxylate (log2FC = -1.5) (Fig. 5C).
At 0.5 h, although polymyxin B monotherapy significantly perturbed the main pathways involved in the bacterial cell envelope biogenesis, it was less potent than the combination therapy (Fig. 5B&C). The levels of three amino-sugar and sugar-nucleotide intermediates underwent a substantial decrease after polymyxin B monotherapy at 0.5 h, namely D-glucosamine-6-phosphate, UDP-N-acetyl-D-glucosamine and UDP-N-acetylmuramate (>-2.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5B). Peptidoglycan biosynthesis was also significantly disrupted after polymyxin B monotherapy wherein the levels of four essential intermediates experienced a significant decline, including UDP-N-acetylmuramoyl-L-alanyl-D-γ-glutamyl-meso-2,6-diaminopimelate, UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6 diaminoheptanedioate-D-alanine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanyl-D-alanine and D-alanyl-D-alanine (>-1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5B). Not unlike the combination treatment, albeit, to a lesser extent, polymyxin B monotherapy induced a marked decrease in the levels of PPP intermediate (D-sedoheptulose 7-phosphate) and the LPS biosynthesis precursor (D-glycero-D-manno-heptose-7-phosphate) (>-2.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5B). Only N6-acetyl-L-lysine of lysine metabolism experienced a significant reduction in its level after polymyxin B monotherapy (>-2.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 5C). Prochlorperazine monotherapy did not show any significant effects at 0.5 h.
Different patterns of perturbation were seen at 4 h wherein the combination therapy displayed more profound impact on peptidoglycan and lysine biosynthesis with unnoticeable impact on LPS biosynthesis (Fig. 5B&C). The levels of seven principle building blocks of peptidoglycan biosynthesis were significantly perturbed, namely UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate-D-alanine (log2FC = 0.75), UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanyl-D-alanine (log2FC = -1.32), D-alanine (log2FC = -0.98), N-acetylglucosamine-1,6-anhydro-N-acetyl-β-muramic acid (log2FC = 2.3), meso-2,6-diamino-3-hydroxypimelate (log2FC = 1.7), meso-2,6-diaminopimelic acid and (Ac)2-L-Lys-D-Ala-D-Ala (log2FC = 2.3) (Fig. 5B). Likewise, the combination treatment induced marked changes in the abundance of five essential lysine biosynthetic intermediates, including N6-acetyl-L-lysine (log2FC = 1.7), N6-acetyl-N6-hydroxy-L-lysine (log2FC = 1.5), 2,3,4,5-tetrahydropyridine-2-carboxylate (log2FC = -1.3), N6,N6,N6-trimethyl-L-lysine (log2FC = -1.3) and L-2-aminoadipate (log2FC = -1.0) (Fig. 5C). In contrast, at 4 h, polymyxin B monotherapy did not exhibit a noticeable influence on peptidoglycan and LPS biosynthesis; however, far less perturbation on lysine metabolism was observed wherein the level of N6-acetyl-L-lysine was declined after polymyxin B monotherapy (log2FC = -1.4) (Fig. 5C). Similarly, there was no significant impact on peptidoglycan, LPS and lysine biosynthesis following prochlorperazine monotherapy at 4 h.
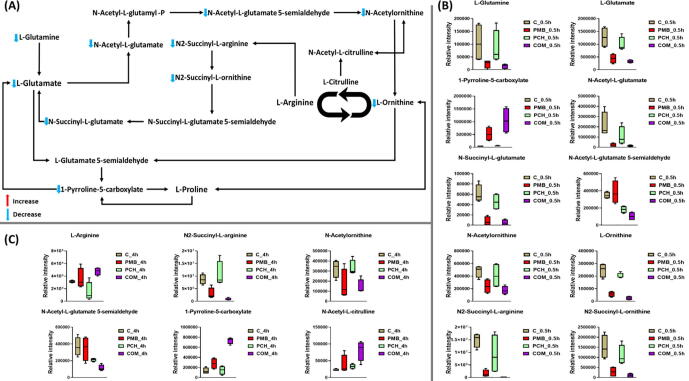
2.7. Analyses of perturbations in arginine and proline metabolism
The amino acids L-glutamate, L-ornithine, L-arginine and proline are key regulators of bacterial polyamine (i.e. putrescine and spermidine) metabolism as well as serving as a source of carbon and/or nitrogen [40], [41]. A high number of metabolites involved in arginine and proline metabolism were significantly altered following combination treatment at 0.5 and 4 h (Fig. 6A-C). At 0.5 h, the abundance of ten principle intermediates of arginine and proline metabolism were diminished [except (S)-1-pyrroline-5-carboxylate (log2FC = 4.5)] by the combination treatment, including L-glutamate, L-glutamine, L-ornithine, N-acetyl-L-glutamate, N-acetyl-L-glutamate 5-semialdehyde, N-succinyl-L-glutamate, N2-succinyl-L-ornithine and N2-succinyl-L-arginine (≥-1.5-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 6A&B). Similarly but to a lesser extent, polymyxin B monotherapy caused a significant inhibitory effect [ except (S)-1-pyrroline-5-carboxylate (log2FC = 3.5) ] on seven main precursors of arginine and proline metabolism at 0.5 h, namely L-glutamate, L-ornithine, N-acetyl-L-glutamate, N-succinyl-L-glutamate, N2-succinyl-L-ornithine and N2-succinyl-L-arginine (≥-1.5-log2-fold, p ≤ 0.05; FDR ≤ 0.05) (Fig. 6B). At 0.5 h, no significant effect was observed in the samples treated with prochlorperazine monotherapy except of N-acetyl-L-glutamate 5-semialdehyde which underwent a significant decrease in its abundance (log2FC = -0.92).
Fig. 6.
The graph shown the significantly impact metabolites of fatty acids metabolism for A. baumannii ATCC 17978 after 0.5 h treatment with polymyxin B (PMB), prochlorperazine (PCH) and the combination (COM) (A). Bar charts for the significantly impacted metabolites of fatty acid metabolism after treatment with polymyxin B (PMB) or prochlorperazine (PCH) monotherapy and the combination (COM) at 0.5 h (B) and 4 h (C) (≥1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05).
At 4 h, the aforementioned effects had continued as a result of the combination treatment which produced a significant perturbation in six essential precursors of arginine and proline metabolism, namely (S)-1-pyrroline-5-carboxylate (log2FC = 2.5), N2-succinyl-L-arginine (log2FC = -3.4), N-acetyl-L-glutamate-5-semialdehyde (log2FC = -1.7), L-arginine (log2FC = 0.61), N-acetylornithine (log2FC = -1.2) and N-acetyl-L-citrulline (log2FC = 1.8) (Figure C). Notably, polymyxin B monotherapy impact on arginine and proline metabolism had begun to subside at 4 h except for the inhibitory effect on the abundance of N-succinyl-L-glutamate (log2FC = -1.7) (Figure C). Likewise, prochlorperazine per se induced a considerable decline in the level of only one arginine and proline metabolism intermediate N-acetyl-L-glutamate 5-semialdehyde (log2FC = -0.8) (Figure C).
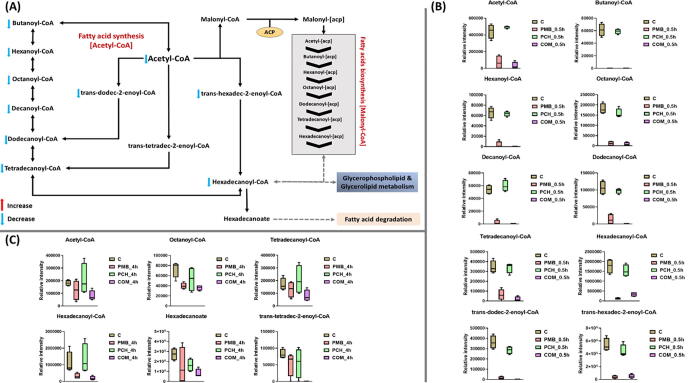
2.8. Analyses of perturbations of the acetyl-CoA pathway
The acetyl-CoA is an essential cofactor which acts as carrier of acyl groups that are critical for the elongation of bacterial membrane fatty acids and energy metabolism [42]. The combination treatment induced significant perturbations in the bacterial fatty acid biosynthesis across all time points 0.5 and 4 h (Fig. 7A-C). In different circumstances, marked changes in the fatty acid biosynthesis were observed following polymyxin B monotherapy mainly at the early time point (0.5 h) whilst its effects were far less pronounced compared to the combination treatment at 4 h (Fig. 7A-C). Unsurprisingly, no significant changes in fatty acid biosynthesis were seen after prochlorperazine monotherapy at 0.5 h; however, a very slight effect for prochlorperazine per se was seen at 4 h (Fig. 7A-C). The maximal impact of the combination occurred at 0.5 h, wherein the levels of ten key fatty acid precursors underwent a dramatic decline, namely acetyl-CoA (log2FC = -4.2), butanoyl-CoA (log2FC = -5.1), hexanoyl-CoA (log2FC = -5.2), octanoyl-CoA (log2FC = -4.4), dodecanoyl-CoA (log2FC = -16.7), tetradecanoyl-CoA (log2FC = -5.2), palmitoyl-CoA (log2FC = -2.4), trans-hexadec-2-enoyl-CoA (log2FC = -3.4), trans-dodec-2-enoyl-CoA (log2FC = -10.8) and decanoyl-CoA (log2FC = -4.9) (Fig. 7B). Although polymyxin B alone perturbed a similar number of fatty acid biosynthetic intermediates, it was relatively less potent in compared to the combination therapy at 0.5 h. This is manifested by a less intensity reduction in the levels of eight essential fatty acid intermediates due to polymyxin B monotherapy, including acetyl-CoA (log2FC = -2.6), butanoyl-CoA (log2FC = -5.0), hexanoyl-CoA (log2FC = -1.7), octanoyl-CoA (log2FC = -3.9), dodecanoyl-CoA (log2FC = -2.9), tetradecanoyl-CoA (log2FC = -2.4), trans-dodec-2-enoyl-CoA (log2FC = -5.8) and decanoyl-CoA (log2FC = -4.9) (Fig. 7B).
Fig. 7.
(A) Schematic diagram depicted the significantly impacted arginine and proline pathways of A. baumannii ATCC 17978 following treatment with polymyxin B (PMB), prochlorperazine (PCH) and the combination (COM) at 0.5 h (B) Bar charts for the significantly impacted of A. baumannii ATCC 17978 treated by polymyxin B (PMB), prochlorperazine (PCH) and the combination (COM) in arginine and proline pathways at 0.5 h and at 4 h (C) (≥1.0-log2-fold, p ≤ 0.05; FDR ≤ 0.05).
These effects were perpetuated at 4 h by the combination treatment wherein the abundance of seven essential intermediates of fatty acid metabolism underwent a significant decline, namely acetyl-CoA (log2FC = -1.1), palmitoyl-CoA (log2FC = -2.5), octanoyl-CoA (log2FC = -1.0), tetradecanoyl-CoA (log2FC = -1.0), trans-tetradec-2-enoyl-CoA (log2FC = -5.3), and hexadecanoic acid (log2FC = -1.7) (Fig. 7C). Whereas at 4 h polymyxin B monotherapy decreased the levels of only two intermediates palmitoyl-CoA (log2FC = -1.6) and octanoyl-CoA (log2FC = -0.88). Prochlorperazine alone also showed little influence as manifested by a significant decline in the level of only one intermediate hexadecanoic acid (log2FC = -0.74) (Fig. 7C).
2.9. Conclusions
This study is the first to demonstrate the synergistic activities of polymyxin B and phenothiazine combinations. Among the phenothiazines tested for polymyxin B combination therapy, prochlorperazine showed a superior antibacterial effect. EM studies revealed that the polymyxin B and prochlorperazine combination produced greater damage to the bacterial cell compared to the treatments with each drug per se. The metabolomics study showed that treatment of A. baumannii ATCC17978 with the combination of polymyxin B-prochlorperazine significantly affected the bacterial cells envelope biogenesis as reflected by the major perturbation of bacterial membrane glycerophospholipids and fatty acids, and acetyl-CoA pathway, as well as inhibiting the synthesis of lipopolysaccharide, peptidoglycan and lysine (Fig. 8). Notably, the late inhibitory impact on the bacterial cell envelope due to the combination therapy is mediated by non-LPS involvement, suggesting the unique mechanism of synergistic killing for polymyxin B-phenothiazines treatments. Polymyxin B monotherapy caused far fewer metabolome perturbations, whereas prochlorperazine monotherapy did not cause any significant metabolome perturbations. It is also noteworthy to mention that the metabolome perturbations induced by the combination were largely unique and did not entirely overlap with the effects seen with treatments of each drug per se particularly at 4 h. Pharmacokinetic studies of phenothiazines (e.g. prochlorperazine) in humans are sparse and have not been undertaken using contemporary analytical methodology[43]. The only data currently available is from a study conducted in 1980 s, which indicated that the plasma concentrations of the prochlorperazine in healthy volunteers vary between 0.04 and 0.025 µg/mL following single intravenous doses of 6.25–12.5 mg [44]. Prochlorperazine has been administered at higher tolerated doses with a maximum of 150 mg/day[45]. Notably, phenothiazine drugs concentrate in the central nervous system (CNS) at levels approximately 70-fold higher than in the plasma[43]. Our study highlights the potential for polymyxin B-phenothiazine combinations to be directly administered into the intra-cerebroventricular space via intracerebroventricular (ICV) device. This approach has long been used in the treatment of CNS infections[46], [47]. This direct route of delivery allows high drug bioavailability at the site of infection as well as minimises the unwanted effects[48]. Polymyxin B is among several antimicrobials that is already being given intraventricularly to treat deadly CNS infections caused by MDR Gram-negative bacteria[47], [49]. Therefore, we purport that polymyxin B-phenothiazines combination treatment would be potentially useful for treatment of CNS infections via intraventricular or intrathecal injections. Overall, this study provides a theoretical basis for the repurposing of the FDA approved prochlorperazine for polymyxin B combination therapy. Also, it provides valuable information for novel antibiotic target development.
Fig. 8.
Schematic summary for the main significantly impacted pathways in A. baumannii ATCC 17978 following treatment with polymyxin B (PMB) prochlorperazine (PCH) monotherapy and the combination (COM).
3. Methods
3.1. Bacterial isolates
22 different P. aeruginosa isolates, including 9 polymyxin B susceptible and 13 polymyxin B resistant; 6 different A. baumannii isolates, including 3 polymyxin B susceptible and 3 polymyxin B resistant; 4 different polymyxin B susceptible K. pneumoniae isolates were also employed in this study (Table 1).
3.2. Determination of MIC and FIC
The MICs of polymyxin B, prochlorperazine, thiethylperazine and chlorpromazine were determined for all bacterial isolates in three replicates on separate days using broth microdilution checkerboard assays. The MIC is defined as the lowest concentration of antimicrobial agent showing complete inhibition of bacterial growth as detected by the unaided eye. The stock solutions for polymyxin B and the nonantibiotics were prepared immediately before each experiment. For preparing stock solutions, polymyxin B powder (Betapharma, China) was dissolved in MilliQ water and sterilized by membrane filtration using 0.22 µm pore sized syringe filters (ThermoFisher Scientific, Australia). Prochlorperazine, thiethylperazine and chlorpromazine were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich, Australia) (preliminary studies showed serial concentrations of DMSO (0.25%, 0.5%, 1%, 2.5% v/v) to which the bacteria were exposed had no effect on their growth). The stock solutions were further serially (2-fold) diluted in cation-adjusted Mueller-Hinton broth (CAMHB; Oxoid, UK) to yield solutions with different concentrations. All experiments were performed in 96-well microtiter plates (Techno Plas, Australia) in CAMHB with a bacterial inoculum of approximately 106 CFU/mL. Plates were incubated at 37 °C for 18–20 h.
The FICs were determined using the same method as for MICs. The only difference was that combinations of drugs were used instead of single drugs. The FICs were calculated using the formula FIC= (MIC of Drug A in combination ÷ MIC of Drug A) + (MIC of Drug B in combination ÷ MIC of Drug B). The results were interpreted as: synergism FIC < 0.5 ; addition FIC = 0.5–1.0; indifference FIC = 1–4; antagonism FIC ≥ 4 [27].
3.3. Static time-kill studies
Static time-kill studies of polymyxin B monotherapy and in combination with phenothiazines (prochlorperazine, thiethylperazine and chlorpromazine) were conducted against selected Gram-negative bacterial isolates. The selection of those bacterial isolates was based on previously determined synergistic combination FIC results. Before each time-kill experiment, bacterial isolates to be studied were sub-cultured onto nutrient agar plates (Media Preparation Unit, University of Melbourne, Australia) and incubated at 37 °C for 18 ~ 24 h. To prepare the overnight culture, one colony of the bacterial isolate to be tested was added to 10 mL of CAMHB (Oxoid, UK) in a 50 mL Falcon tube (ThermoFisher Scientific, Australia) and incubated overnight (~16 h) in a shaking water bath at 37 °C with 150 rpm shaking speed. The overnight culture was then inoculated into another 50 mL Falcon tube containing 10 mL fresh CAMHB media and incubated in shaking water bath at 37 °C (shaking speed, 150 rpm) for 2 h to obtain an early log-phase. Aliquots (200 µL) of the bacterial suspension were inoculated into each 50 mL borosilicate glass treatment tube containing 20 mL of fresh CAMHB. Drugs (polymyxin B and prochlorperazine) per se or in combination were then added into the respective treatment tubes to yield the desired concentrations. At 0.5, 1, 2, 4, 8, 24 h, the samples were removed from each tube and serially diluted in 0.9% saline, and then 50 µL plated onto nutrient agar plates using Don Whitley automated spiral plater (. After 24 h incubation at 37 °C, the plates were read (the countable range is 20–300), and the time-kill curves were constructed as time versus log10 CFU/mL.
3.4. Bacterial samples preparation for electron microscopy
Scanning and transmission electron microscopy were performed as we have previously described [50].
3.5. Bacterial culture preparation for metabolomics
Acinetobacter baumannii ATCC 17978 (polymyxin B MIC = 0.5 mg/L) was sub-cultured from frozen stock onto a nutrient agar plate (Media Preparation Unit, University of Melbourne, Australia) and incubated at 37 °C for 18 ~ 24 h prior to the experiment. The overnight culture was prepared by adding one colony of the bacterial isolate to 10 mL of CAMHB (Oxoid, UK) in a 50 mL Falcon tube (ThermoFisher Scientific, Australia) and incubated overnight (~16 h) in shaking water bath at 37 °C (shaking speed, 150 rpm). A small amount from the overnight culture was added to each 500 mL conical flask containing a fresh CAMHB (100-fold dilution). The flasks were then incubated in Bioline incubator shaker at 37 °C with 180 rpm to reach log phase of ~ 108 CFU/mL (OD600 ~ 0.5). The flasks were then spiked with previously prepared antimicrobial stock solution to make the concentrations of interest (three treatment flasks with polymyxin B of 2 mg/L, prochlorperazine of 8 mg/L and the combination of both and one drug-free flask acted as control). The OD600 for each sample was measured at each time point (0.5 and 4 h) and normalized to ~ 0.5 with fresh CAMHB. The quenching and metabolite extraction were lastly performed. To reduce the bias from inherent random variation, four biological replicates were prepared for each treatment condition.
3.6. Metabolite extraction
Following bacterial culture preparation, extraction of metabolites was immediately performed to minimize further drug effects on bacterial metabolism. The samples were washed twice in 1 mL of 0.9% saline (centrifuged at 3220 × g at 4 °C for 3 min). The washed pellets were resuspended in cold chloroform:methanol:water (CMW; 1:3:1, v/v) extraction solvent containing 1 µM each of the internal standards (CHAPS, CAPS, PIPES and TRIS). The selected internal standards are physicochemically diverse small molecules not naturally occurring in any microorganism. Samples were then frozen in liquid nitrogen, thawed on ice and vortexed to release the intracellular metabolites. The samples were then centrifuged for 10 min at 3220 × g at 4 °C and 300 µL of the supernatant was transferred to 1.5 mL Eppendorf tubes for immediate storage at −80 °C. Before analysis, samples were thawed and centrifuged at 14,000 × g at 4 °C for 10 min. A 200 µL of sample was then transferred into the injection vial for LC-MS analysis. An equal volume of each sample was combined and used as a quality control since the combined sample contained all the analytes that will be encountered during the analysis [51].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
J. L. is an Australian National Health and Medical Research Council (NHMRC) Principal Research Fellow, and T. V. is an Australian NHMRC Industry Career Development Level 2 Research Fellow.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.08.008.
Contributor Information
Jian Li, Email: jian.li@unimelb.edu.au.
Tony Velkov, Email: tony.velkov@unimelb.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.WHO. Antimicrobial resistance: global report on surveillance 2014. 2014.
- 2.Organization W.H. WHO; Geneva: 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. [Google Scholar]
- 3.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Rice L.B. The University of Chicago Press; 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. [DOI] [PubMed] [Google Scholar]
- 5.Breidenstein E.B., de la Fuente-Nunez C., Hancock R.E. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Li J., Nation R.L., Turnidge J.D., Milne R.W., Coulthard K., Rayner C.R. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 7.Nation R.L., Li J., Cars O., Couet W., Dudley M.N., Kaye K.S. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis. 2015;15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 8.Velkov T., Thompson P.E., Nation R.L., Li J. Structure− activity relationships of polymyxin antibiotics. J Med Chem. 2009;53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogler K., Studer R. The chemistry of the polymyxin antibiotics. Experientia. 1966;22:345–354. doi: 10.1007/BF01901127. [DOI] [PubMed] [Google Scholar]
- 10.Clausell A., Garcia-Subirats M., Pujol M., Busquets M.A., Rabanal F., Cajal Y. Gram-negative outer and inner membrane models: insertion of cyclic cationic lipopeptides. J Phys Chem B. 2007;111:551–563. doi: 10.1021/jp064757+. [DOI] [PubMed] [Google Scholar]
- 11.Trimble M.J., Mlynárčik P., Kolář M., Hancock R.E. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harbor perspectives in medicine. 2016;6 doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velkov T., Roberts K.D., Nation R.L., Thompson P.E., Li J. Pharmacology of polymyxins: new insights into an ‘old’class of antibiotics. Future microbiology. 2013;8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beceiro A., Llobet E., Aranda J., Bengoechea J.A., Doumith M., Hornsey M. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arroyo L.A., Herrera C.M., Fernandez L., Hankins J.V., Trent M.S., Hancock R.E. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother. 2011;55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt J.H., Harper M., Harrison P., Hale J.D., Vinogradov E., Seemann T. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell S.C. Phenothiazine: the parent molecule. Curr Drug Targets. 2006;7:1181–1189. doi: 10.2174/138945006778226552. [DOI] [PubMed] [Google Scholar]
- 17.Australian Medicines Handbook 2014. Adelaide: Australian Medicines Handbook Pty Ltd; 2014.
- 18.Müller PaS M. Die Chemie der Insektizide, ihre Entwicklung und ihr heutiger Stand. Cell Mol Life Sci. 1954;10:91–131. doi: 10.1007/BF02158514. [DOI] [PubMed] [Google Scholar]
- 19.Ohlow M.J., Moosmann B. Phenothiazine: the seven lives of pharmacology's first lead structure. Drug Discovery Today. 2011;16:119–131. doi: 10.1016/j.drudis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Fea DeEds. Studies on phenothiazine VIII. Antiseptic value of phenothiazine in urinary tract infections. J Pharmacol Exp Ther. 1939;65:353–371. [Google Scholar]
- 21.Kristiansen J.E., Amaral L. The potential management of resistant infections with non-antibiotics. The Journal of antimicrobial chemotherapy. 1997;40:319–327. doi: 10.1093/jac/40.3.319. [DOI] [PubMed] [Google Scholar]
- 22.Kristiansen J.E., Vergmann B. The antibacterial effect of selected phenothiazines and thioxanthenes on slow-growing mycobacteria. Acta pathologica, microbiologica, et immunologica Scandinavica Section B, Microbiology. 1986;94:393–398. doi: 10.1111/j.1699-0463.1986.tb03073.x. [DOI] [PubMed] [Google Scholar]
- 23.Hendricks O., Butterworth T.S., Kristiansen J.E. The in-vitro antimicrobial effect of non-antibiotics and putative inhibitors of efflux pumps on Pseudomonas aeruginosa and Staphylococcus aureus. Int J Antimicrob Agents. 2003;22:262–264. doi: 10.1016/s0924-8579(03)00205-x. [DOI] [PubMed] [Google Scholar]
- 24.Nehme H, Saulnier P, Ramadan AA, Cassisa V, Guillet C, Eveillard M, et al. Antibacterial activity of antipsychotic agents, their association with lipid nanocapsules and its impact on the properties of the nanocarriers and on antibacterial activity. PLoS One. 2018;13:e0189950-e. [DOI] [PMC free article] [PubMed]
- 25.Kristiansen J.E., Hendricks O., Delvin T., Butterworth T.S., Aagaard L., Christensen J.B. Reversal of resistance in microorganisms by help of non-antibiotics. The Journal of antimicrobial chemotherapy. 2007;59:1271–1279. doi: 10.1093/jac/dkm071. [DOI] [PubMed] [Google Scholar]
- 26.Hollister L.E., Eikenberry D.T., Raffel S. Chlorpromazine in nonpsychotic patients with pulmonary tuberculosis. Am Rev Respir Dis. 1960;81:562–566. doi: 10.1164/arrd.1960.81.4.562. [DOI] [PubMed] [Google Scholar]
- 27.Hussein M.H., Schneider E.K., Elliott A.G., Han M., Reyes-Ortega F., Morris F. From Breast Cancer to Antimicrobial: Combating Extremely Resistant Gram-Negative “Superbugs” Using Novel Combinations of Polymyxin B with Selective Estrogen Receptor Modulators. Microb Drug Resist. 2017;23:640–650. doi: 10.1089/mdr.2016.0196. [DOI] [PubMed] [Google Scholar]
- 28.Schneider E.K., Azad M.A., Han M.L., Tony Zhou Q., Wang J., Huang J.X. An “Unlikely” Pair: The Antimicrobial Synergy of Polymyxin B in Combination with the Cystic Fibrosis Transmembrane Conductance Regulator Drugs KALYDECO and ORKAMBI. ACS Infect Dis. 2016;2:478–488. doi: 10.1021/acsinfecdis.6b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran T.B., Bergen P.J., Creek D.J., Velkov T., Li J. Synergistic Killing of Polymyxin B in Combination With the Antineoplastic Drug Mitotane Against Polymyxin-Susceptible and -Resistant Acinetobacter baumannii: A Metabolomic Study. Front Pharmacol. 2018;9:359. doi: 10.3389/fphar.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran T.B., Wang J., Doi Y., Velkov T., Bergen P.J., Li J. Novel Polymyxin Combination With Antineoplastic Mitotane Improved the Bacterial Killing Against Polymyxin-Resistant Multidrug-Resistant Gram-Negative Pathogens. Front Microbiol. 2018;9:721. doi: 10.3389/fmicb.2018.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider E.K., Reyes-Ortega F., Velkov T., Li J. Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative 'superbugs'. Essays Biochem. 2017;61:115–125. doi: 10.1042/EBC20160058. [DOI] [PubMed] [Google Scholar]
- 32.Rossato L., Loreto E.S., Zanette R.A., Chassot F., Santurio J.M., Alves S.H. In vitro synergistic effects of chlorpromazine and sertraline in combination with amphotericin B against Cryptococcus neoformans var. grubii. Folia Microbiol. 2016;61:399–403. doi: 10.1007/s12223-016-0449-8. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y.W., Abdul Rahim N., Zhao J., Han M.L., Yu H.H., Wickremasinghe H. Novel Polymyxin Combination with the Antiretroviral Zidovudine Exerts Synergistic Killing against NDM-Producing Multidrug-Resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.02176-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussein M., Han M.L., Zhu Y., Schneider-Futschik E.K., Hu X., Zhou Q.T. Mechanistic Insights From Global Metabolomics Studies into Synergistic Bactericidal Effect of a Polymyxin B Combination With Tamoxifen Against Cystic Fibrosis MDR Pseudomonas aeruginosa. Comput Struct Biotechnol J. 2018;16:587–599. doi: 10.1016/j.csbj.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran T.B., Cheah S.E., Yu H.H., Bergen P.J., Nation R.L., Creek D.J. Anthelmintic closantel enhances bacterial killing of polymyxin B against multidrug-resistant Acinetobacter baumannii. The Journal of antibiotics. 2016;69:415–421. doi: 10.1038/ja.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaara M., Vaara T. Polycations sensitize enteric bacteria to antibiotics. Antimicrob Agents Chemother. 1983;24:107–113. doi: 10.1128/aac.24.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Company F.A.D. F.A; Davis Company: 2015. prochlorperazine. [Google Scholar]
- 38.Isah A.O., Rawlins M.D., Bateman D.N. The pharmacokinetics and effects of prochlorperazine in elderly female volunteers. Age Ageing. 1992;21:27–31. doi: 10.1093/ageing/21.1.27. [DOI] [PubMed] [Google Scholar]
- 39.Zgurskaya H.I., Lopez C.A., Gnanakaran S. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis. 2015;1:512–522. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michael A.J. Polyamines in eukaryotes, bacteria, and archaea. J Biol Chem. 2016;291:14896–14903. doi: 10.1074/jbc.R116.734780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong L., Teng J.L., Botelho M.G., Lo R.C., Lau S.K., Woo P.C. Arginine metabolism in bacterial pathogenesis and cancer therapy. Int J Mol Sci. 2016;17:363. doi: 10.3390/ijms17030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cronan J.E., Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afeltra J., Verweij P. Antifungal activity of nonantifungal drugs. Eur J Clin Microbiol Infect Dis. 2003;22:397–407. doi: 10.1007/s10096-003-0947-x. [DOI] [PubMed] [Google Scholar]
- 44.Taylor W.B., Bateman D. Preliminary studies of the pharmacokinetics and pharmacodynamics of prochlorperazine in healthy volunteers. Br J Clin Pharmacol. 1987;23:137–142. doi: 10.1111/j.1365-2125.1987.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sweetman S.C., Martindale B.P. The complete drug reference. 2011;33 doi: 10.1136/bmj.331.7511.292-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziai W.C., Lewin J.J., III Improving the role of intraventricular antimicrobial agents in the management of meningitis. Curr Opin Neurol. 2009;22:277–282. doi: 10.1097/wco.0b013e32832c1396. [DOI] [PubMed] [Google Scholar]
- 47.Cook A.M., Mieure K.D., Owen R.D., Pesaturo A.B., Hatton J. Intracerebroventricular administration of drugs. Pharmacotherapy: The Journal of Human Pharmacology and Drug. Therapy. 2009;29:832–845. doi: 10.1592/phco.29.7.832. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q., Chen H., Zhu C., Chen F., Sun S., Liang N. Efficacy and safety of intrathecal meropenem and vancomycin in the treatment of postoperative intracranial infection in patients with severe traumatic brain injury. Experimental and Therapeutic Medicine. 2019;17:4605–4609. doi: 10.3892/etm.2019.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan S., Huang X., Wang Y., Li L., Zhao C., Yao Z. Efficacy of intravenous plus intrathecal/intracerebral ventricle injection of polymyxin B for post-neurosurgical intracranial infections due to MDR/XDR Acinectobacter baumannii: a retrospective cohort study. Antimicrobial Resistance & Infection Control. 2018;7:8. doi: 10.1186/s13756-018-0305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdul Rahim N., Cheah S.-E., Johnson M.D., Yu H., Sidjabat H.E., Boyce J. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’antibiotics—polymyxin B and chloramphenicol. J Antimicrob Chemother. 2015;70:2589–2597. doi: 10.1093/jac/dkv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gika H.G., Theodoridis G.A., Wingate J.E., Wilson I.D. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res. 2007;6:3291–3303. doi: 10.1021/pr070183p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.