Abstract
Increasing data show that intronic derived regulatory elements, such as transcription factor binding sites (TFBs), play key roles in gene regulation, and malfunction. Accordingly, characterizing the sequence context of the intronic regions of the human coagulation factor VIII (hFVIII) gene can be important. In this study, the intronic regions of the hFVIII gene were scrutinized based on in-silico methods. The results disclosed that these regions harbor a rich array of functional elements such as repetitive elements (REs), splicing sites, and transcription factor binding sites (TFBs). Among these elements, TFBs and REs showed a significant distribution and correlation to each other. This survey indicated that 31% of TFBs are localized in the intronic regions of the gene. Moreover, TFBs indicate a strong bias in the regions far from splice sites of introns with mapping to different REs. Accordingly, TFBs showed highly bias toward Short Interspersed Elements (SINEs), which in turn they covering about 12% of the total of REs. However, the distribution pattern of TFBs-REs showed different bias in the intronic regions, spatially into the Introns 13 and 25. The rich array of SINE-TFBs and CR1-TFBs were situated within 5′UTR of the gene that may be an important driving force for regulatory innovation of the hFVIII gene. Taken together, these data may lead to revealing intronic regions with the capacity to renewing gene regulatory networks of the hFVIII gene. On the other hand, these correlations might provide the novel idea for a new hypothesis of molecular evolution of the FVIII gene, and treatment of Hemophilia A which should be considered in future studies.
Keywords: Developmental biology, Molecular biology, Epigenetics, Developmental genetics, Health sciences, Human genetics, Genetic disorders, Molecular evolution, Regulatory motif, Repeat elements, TFBs, Gene regulation, Hemophilia A, Gene therapy
Developmental biology; Molecular biology; Epigenetics; Developmental genetics; Health sciences; Human genetics; Genetic disorders; Molecular evolution; Regulatory motif; Repeat elements; TFBs; Gene regulation; Hemophilia A; Gene therapy.
1. Introduction
Intronic sequence context and structures (ISCSs) and intronic derived regulatory elements (IDREs) play key roles in gene regulations and fine-tuning gene expressions [1, 2]. Among them, transposable elements (TEs) are the source of a variety of regulatory sequences such as transcription factor binding sites (TE-TFBs) which help to control the expression of the host genes. The precise expressions of the genes are dependent on the binding of transcription factors (TFs) to corresponding TFBs loci on the genomic regions [3, 4, 5]. TFBs are short, degenerate nucleotide sequences which are usually 6–20 bp long and may reach up to 200 bp in length. Although, TFs bind to promoters immediately upstream of the transcription start site (TSS) in single cell eukaryotes, in more complex multicellular organisms, additional area at distal sites of TSS are involved in regulatory networks. However, many of TFBs are lineage-specific and involved in the previously existing networks. Accordingly, the prediction and identification of these elements throughout the context of the genome as well as intronic regions is a crucial step towards understanding their evolutionary rates and patterns and mechanism of action in gene regulation and regulatory networks in details [6, 7]. Moreover, several studies have shown the improvement of gene expression by intronic sequences. These studies have revealed that the higher frequency of TFBs in used intron, the more enhancement in the gene expression [8, 9]. In this regard, several methods such as using DNase I hypersensitive sites, chromatin immunoprecipitation (ChIP) and SLIM-ChIP, next-generation sequencing, in-silico and genome-wide mapping of the TFBSs methods, have been applied extensively to map these regions and elements throughout the genomes and provided great tools for these purposes [5, 10, 11, 12].
The most basic elements used for identifying TFBs from the sequences are the characteristic binding properties for each TF, comprising the width of DNA binding site and the nucleotide preferences at each position which can be deduced from aligning a set of DNA sequences that are experimentally known to bind the TF [13]. However, the vast majority of these predicted sites are not functional in the cell. On the other hand, although high-throughput sequencing data are a powerful way to map regulatory relationships, they suffer from a high false-positive rate and therefore do not resolve TF-DNA binding footprints at base pair resolution [14]. Accordingly, our knowledge about the repertoire of regulatory regions of individual genes is very limited. Subsequently, their prediction using multiple genome information based statistical survey models is becoming a hot topic in bioinformatics to overcome these problems for revealing comprehensive catalogs of regulatory [11, 15]. Considering these challenges, in this study, we present a framework of in-silico methods to detect regulatory motifs throughout the intronic regions of the hFVIII gene in order to get more data about these important regions and try to scatter their functional elements in this individual gene.
The hFVIII gene with a large locus at the tip of the long arm of X chromosome consisting of 26 exons and 25 introns ranging in size from 69 to 3,106 bps and 207 to 32849 bps, respectively. Deficiencies in the regulation of the gene expression could lead to hemophilia A(HA), X-linked recessive bleeding disorder, affecting approximately 1–2 in every 10000 males worldwide [16, 17]. However, complications of its regulatory system considering intronic regions have not been paid proper attention so far. Therefore, it would be essential to mining potentially regulatory non-coding regions that might be involved in hFVIII gene regulation. Indeed, many contributing DNA variants have been reported to date in the hFVIII gene including its deep intronic variations, highlight the importance of variation studies in the nucleotide sequences of the hFVIII gene including non-coding regions as a cause of monogenic disorders [18, 19]. Moreover, it has been revealed that the genotypes and even genetic variants on multiple ethnic backgrounds of hFVIII gene have impact on bleeding, thrombosis and haemophilia outcomes [20, 21]. In this regard, mining the potential regulatory deep intronic regions of the hFVIII gene may be helpful for inferring a regulatory network of the hFVIII gene, which in turn may improve our understanding regarding the pathways in which the hFVIII is involved and would have potential to improve the existing strategies for treatment of hemophilia A and may led to new therapeutic horizons.
2. Materials and methods
2.1. Sequence extraction
The complete nucleotide sequence of the hFVIII gene, with accession number AY769950 and the full length of 192029 bp, including its intronic, 5′UTR (3284 bps) and 3′UTR (3770 bps) regions, retrieved from GenBank, from the National Center for Biotechnology Information database (NCBI, http://www.ncbi.nlm.nih.gov). This was involved complete cds of the gene including exonic, intronic as well as 5′UTR and 3′UTR. Based on the data that annotated in the page of this gene at Nucleotide databank, we can conclude that this sequence is a healthy individual.
2.2. Programs used for in-silico investigations
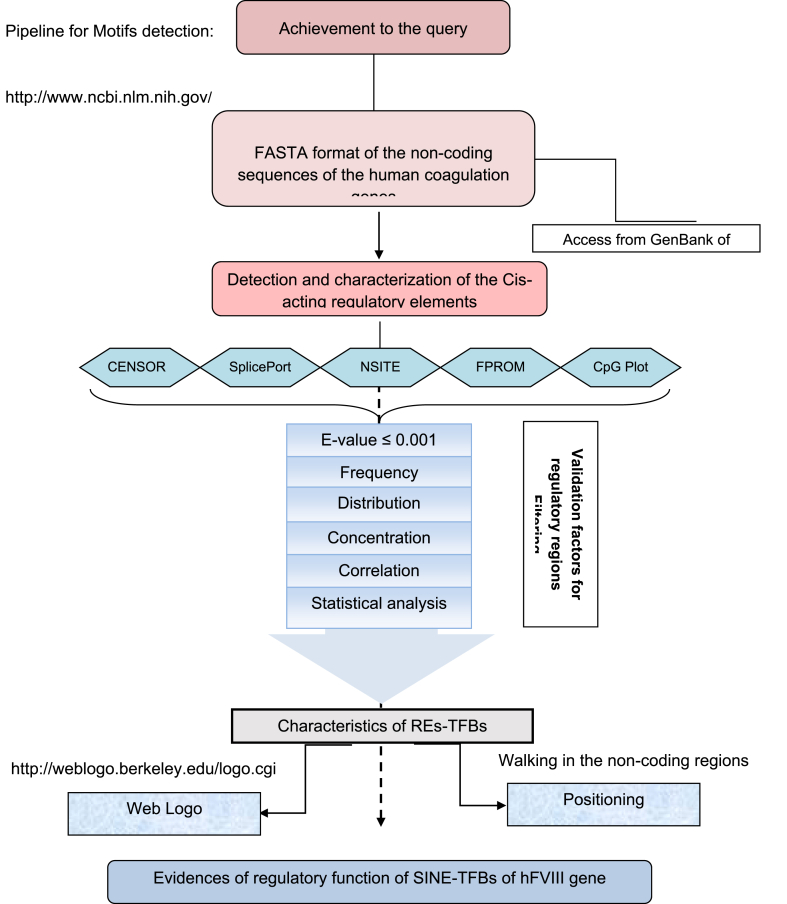
The non-coding regions of the hFVIII gene were scrutinized for cis-acting regulatory motifs with several programs which denote promoter and/or enhancer-like sequence, TFBs, CpG islands, and repetitive elements. In order to effectively decreased false positive results, the FASTA files of the non-coding regions of the hFVIII gene were given into each program based on the designed pipeline in Figure 1 for each analysis as follows:
Figure 1.
Flowchart describing the pipeline for determining correlation among TFBs and Repetitive elements throughout the human coagulation factor VIII gene.
2.2.1. CENSOR
In order to clarify the composition of non-coding regions of the hFVIII gene, CENSOR was employed. This software screens the query sequences against a reference collection of REs, as well as generating a report classifying all the found repeats [22].
2.2.2. FPROM/human promoter prediction
FPROM program was contacted to investigate the presence of any promoter areas throughout the gene based on potential transcription start positions by linear discriminant function, combining characteristics describing functional motifs and oligonucleotide composition of these sites [23].
2.2.3. CpG plot
In order to establish the association between the CpG islands and the promoters, any possible correlations between the CpG islands and intronic promoters were investigated using CpG Plot program [24].
2.2.4. NSITE/recognition of regulatory motifs
The potential TFBs were disclosed throughout the non-coding regions of the hFVIII gene utilizing NSITE tool in the Softberry software package, which is based on the statistical estimation of the expected number of a nucleotide consensus pattern in a given sequence [25].
2.2.5. SplicePort
The splice sites of the hFVIII gene were predicted by employing SplicePort tool. The splice site prediction scheme gives an accuracy of donor site recognition on the test set. For 86% accuracy poly-A region prediction (Sp = 50%), the algorithm has 8% false predictions (C = 0.62). We can choose between splice-site prediction and motif detection. After that the potential splice sites were forecasted and scored, the features on which those predictions are based can then be discovered. The method is a good alternative to the neural network approach [26].
2.2.6. Sequence logos
The Sequence Logos of detected regulatory regions were generated using the WebLogo server [27], by depicting stacks of letters. The height of each letter within a stack is proportional to the base frequency at that position, and the letters are sorted by size, with the tallest (i.e. most frequent) on top. The height of the stack is the sequence conservation measured in bits of information; 1 bit measures the choice between two equally likely possibilities.
2.3. Data validation
As shown in Figure 1, E-value, frequency, distribution, and correlation are confirming factors to discriminate between the positive and false positive results from each other. In this regard, in the first step, the data without references and E-values up to 10-3 were eliminated. Subsequently, the frequencies, distribution patterns as well as spatially connections of detected regulatory elements were investigated throughout the intronic regions of the hFVIII gene as well as its 5′UTR and 3′UTR. Furthermore, the occurrences frequencies of the TFBs throughout the REs of the hFVIII gene and the distribution pattern of them were assessed as evidence of the importance of these regions.
2.4. Statistical analysis
Correlations among the frequencies and distribution patterns of TFBs and REs throughout the regions were investigated by Pearson's correlation coefficient. All statistical analyses were carried out with SPSS 22.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Intronic regulatory motifs throughout the hFVIII gene
Our investigation led to disclose that the non-coding regions of the hFVIII gene harbor a rich array of functionally significant elements including promoter and enhancer-like elements, CpG islands, splicing donor and acceptor sites, polyadenylation signals, repetitive elements, and transcription factor binding sites. More inspection in the frequencies, situation, and correlation of these elements to each other showed that they are different in the frequency, positioning as well as their relationship.
3.2. TFBs and REs throughout the intronic regions of the hFVIII gene
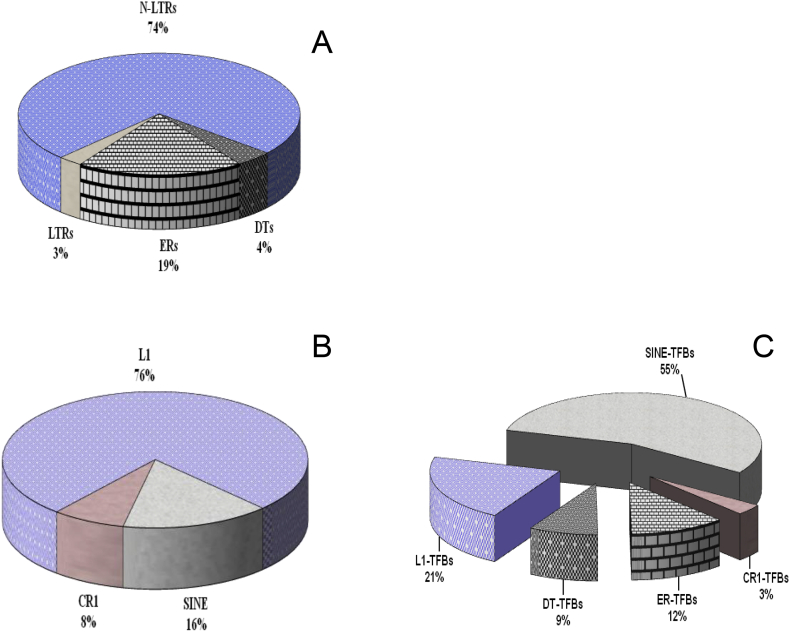
Surveying the sequence context of the hFVIII gene led to the detection 2852 motifs of different types of TFBs that are scattered in a length independent manner among the two strands of the intronic regions of the gene. So that they are condensed into the shorter introns, including; In-8, In-16, In-17, In-19, In-23, and In-24, compared with long introns such as introns In-1, In-22, and In-25 (Table 1). On the other hand, this survey revealed that more than 65% of the length of this gene consists of 377 repetitive elements falling into four classes, including Endogenous Retroviruses (ERs), Long Terminal Repeat retrotransposons (LTRs), Non-LTR retrotransposons (N-LTRs), and DNA transposons (DTs) (Figure 2-A). As shown in this Figure, N-LTRs with about 74% frequency are the most abundant ones, whereas LTRs with 3% frequency, meet the lowest frequencies in the gene.
Table 1.
Percentage of the density of REs-TFBs throughout the non-coding regions of the hFVIII gene (L. Kbp: length of different regions of the gene, T. TFBs: Total number of REs-TFBs in each region).
| Region | L.Kbp | T.TFBs | L1-TFBs |
SINE-TFBs |
CR1-TFBs |
ER-TFBs |
DT-TFBs |
Total TFBs |
|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | |||
| 5 UTR | 3.284 | 73 | 0 | 27.4 | 4.1 | 0 | 0 | 31.5 |
| In 1 | 22.808 | 247 | 2.02 | 23.88 | 0 | 4.45 | 0 | 30.35 |
| In 2 | 2.383 | 44 | 0 | 0 | 6.81 | 9.09 | 0 | 15.9 |
| In 3 | 3.824 | 89 | 1.12 | 20.22 | 6.74 | 3.37 | 0 | 31.45 |
| In 4 | 5.63 | 51 | 23.52 | 0 | 0 | 11.76 | 1.96 | 37.24 |
| In 5 | 2.433 | 84 | 0 | 35.7 | 1.19 | 0 | 0 | 36.89 |
| In 6 | 15.134 | 194 | 17.5 | 34.02 | 0.51 | 0 | 1.03 | 53.06 |
| In 7 | 2.643 | 46 | 0 | 0 | 0 | 6.52 | 0 | 6.52 |
| In 8 | 0.284 | 21 | 0 | 0 | 0 | 0 | 0 | 0 |
| In 9 | 4.801 | 119 | 5.88 | 5.04 | 0 | 3.36 | 5.88 | 20.16 |
| In 10 | 3.903 | 92 | 6.52 | 25 | 0 | 1.08 | 0 | 32.6 |
| In 11 | 2.914 | 90 | 0 | 12.22 | 2.2 | 18.88 | 0 | 33.3 |
| In 12 | 5.984 | 89 | 3.37 | 30.33 | 0 | 3.37 | 1.12 | 38.19 |
| In 13 | 16.021 | 161 | 32.92 | 13.04 | 0 | 0 | 0.62 | 46.58 |
| In 14 | 21.997 | 105 | 20 | 12.38 | 2.85 | 10.47 | 0 | 45.7 |
| In 15 | 1.396 | 29 | 6.9 | 0 | 0 | 0 | 3.44 | 10.34 |
| In 16 | 0.286 | 43 | 0 | 0 | 0 | 0 | 0 | 0 |
| In 17 | 0.207 | 36 | 0 | 0 | 0 | 0 | 0 | 0 |
| In 18 | 1.738 | 83 | 0 | 10.84 | 2.4 | 26.5 | 0 | 39.74 |
| In 19 | 0.608 | 52 | 0 | 19.23 | 0 | 0 | 0 | 19.23 |
| In 20 | 1.419 | 49 | 2.04 | 28.57 | 6.12 | 0 | 10.2 | 46.93 |
| In 21 | 3.653 | 142 | 0 | 0 | 4.22 | 0 | 0 | 4.22 |
| In 22 | 32.849 | 394 | 4.82 | 9.64 | 0 | 1.77 | 2.53 | 18.76 |
| In 23 | 1.216 | 77 | 15.58 | 20.77 | 0 | 0 | 3.89 | 40.24 |
| In 24 | 1.109 | 94 | 0 | 23.4 | 0 | 0 | 0 | 23.4 |
| In 25 | 22.679 | 285 | 2.45 | 22.1 | 0 | 2.1 | 14.7 | 41.35 |
| 3 UTR | 3.77 | 63 | 1.58 | 25.39 | 0 | 9.52 | 3.17 | 39.66 |
Figure 2.
A: Frequencies of repetitive elements throughout the hFVIII gene, including Endogenous Retroviruses (ERs), Long Terminal Repeat retrotransposons (LTRs), Non-LTR retrotransposons (N-LTRs), and DNA transposons (DTs). B: Frequencies of the families of N-LTRs elements throughout the hFVIII gene, and C: Distribution pattern of TFBs within repetitive elements of the hFVIII gene. L1, SINE and CR1 are the families of N-LTRs repetitive elements which are scattered throughout the hFVIII gene with various frequencies.
3.3. TFBs of the hFVIII gene has strong positioning bias to SINE
The distribution pattern of TFBs revealed that more than 69% of them are located out of the situation of REs throughout the hFVIII gene, although more than 65% of the length of this gene consists of these elements. However, the distribution pattern of these motifs among REs is consistent with their frequencies, so that the most of them, with about 74% TFBs, are situated in N-LTRs (Figure 2-A), while none of them were positioned in LTRs elements. Unlike this type of distribution of TFBs throughout the REs, their propagation within N-LTRs families was interesting. Although the most portion of N-LTRs belongs to LINE (L1) family (Figure 2-B), the most of TFBs are positioned within SINE (SINE-TFBs) family (Figure 2-C). Distribution pattern of TFBs within repetitive elements of the hFVIII gene revealed that L1, SINE and CR1, which are the families of N-LTRs repetitive elements, are scattered throughout the hFVIII gene with 21%, 55%, and 3% frequencies, respectively (Figure 2-C).
3.4. REs-TFBs throughout the non-coding regions of the hFVIII gene
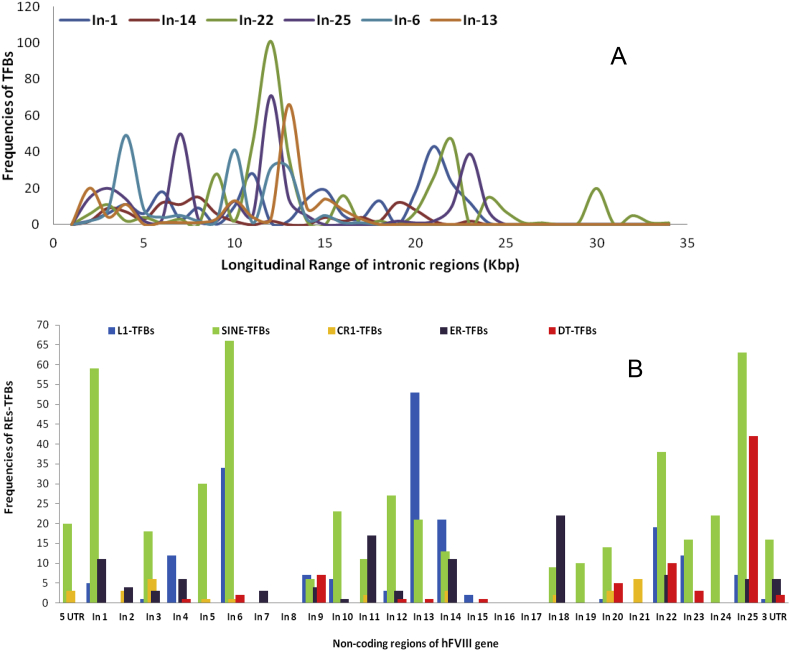
REs-TFBs are positioned in a length independent pattern in the non-coding regions of the gene(Table 1), so that the highest length intron of the gene, In-22, comprise 18.76% of total of REs-TFBs, while In-1, In-6, In-13, and In-25, with the shorter lengths, comprise 30.35%, 53.06%, 46.58% and 41.35% of these elements, respectively. On the other hand, the distribution pattern of TFBs in the long introns of the hFVIII gene is notable everywhere, so that they follow different patterns (Figure 3-A). As shown in Figure 3-B, the most elements of DT-TFBs are in In-25, while most of L1-TFBs are positioned within In-13. In this regard, SINE-TFBs are distributed in most of the non-coding regions, with the highest bias to the In-1, In-6, and In-25. Among these elements, SINE-TFBs and CR1-TFBs are the only elements which are situated within 5′UTR of the gene which is considerable.
Figure 3.
A: Distribution pattern of TFBs throughout the long introns of the hFVIII gene. Longitudinal range of each intron represented by different color, including blue: intron 1(In-1), Red: intron 14 (In-14), Green: Intron 22 (In-22), Violet: intron 25 (In-25), Cyan: intron 25 (In-25), and Orange: intron 13 (In-13). B: Frequencies and distribution pattern of REs-TFBs throughout the non-coding regions of the hFVIII gene.
3.5. Characteristics of the sequence context of REs-TFBs
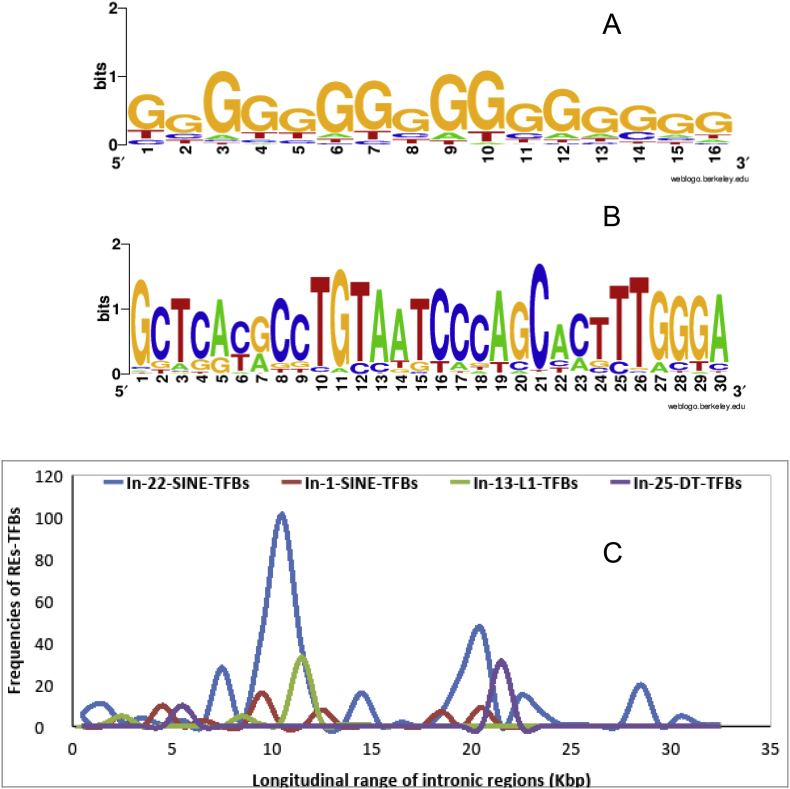
Assessment the features of sequence context of REs-TFBs throughout the hFVIII gene revealed that they have situated on both strands of intronic regions, with differences in the length and sequence context. This investigation revealed that these sites, with E-value equal to zero, have various lengths ranging in size from 7 up to 30 bps. On the other hand, a deeper inspection in the sequence context of the motifs showed that some of them have nucleotide mismatched in some position to the original sites. Among these motifs, DT-TFBs are G riche nucleotide and most of them have 16 nucleotides in length (Figure 4-A). While SINE-TFBs showed a composition of each of four nucleotides in context with 7 up to 30 bp in the length; however, most of them showed 30 bp lengths (Figure 4-B). Moreover, the sequence context of other REs-TFBs did not show a specific pattern.
Figure 4.
A: Sequence logos of DT-TFBs, B: SINE-TFBs throughout the non-coding regions of the hFVIII gene. C: Distribution pattern of REs-TFBs throughout the long introns of the hFVIII gene. Longitudinal range of each intron represented by the different color of the frequency of REs-TEBs, including Blue: SINE-TFBs throughout intron 22 (In-22-SINE-TFBs), Red: SINE-TFBs throughout intron 1 (In-1-SINE-TFBs), Green: L1-TFBs throughout intron 13 (In-13-L1-TFBs), and Purple: DT-TFBs throughout intron 25 (In-25-DT-TFBs).
3.6. TFBs distribution peak at regions far from splice sites
The distribution pattern of TFBs throughout the intronic regions of the hFVIII gene is indicating a strong positioning bias in the regions far from splice sites (Figure 4-C). As shown, they are peaked as significantly between 1 up to 7 restricted regions throughout the intronic area and they are restricted in seven regions within the In-22, and in three regions throughout the In-13. On the other hand, the most of TFBs which are condensed between 10 up to 15 Kbp and upstream of the donor splice site as well as the other introns, follow the similar pattern, but within the shorter area.
4. Discussion
Irrespective of the various effects of the introns on gene regulation, attention to the size of the intron sequences, from 31 to over 210,000 nucleotides [28, 29], as well as the number of them in a gene, from 1 up to 77 introns [30], aroused a serious debate, whether all introns and their sequences in a gene have effects with the same mechanism, or certain intron and definite sequences of them have critical effect with various mechanisms on gene regulation. Hence, it would be essential to understand which introns or intronic regions are potentially involved in gene regulation. Moreover, the importance of exploring genetic regulatory elements such as TFBS in the intronic regions is further highlighted when we consider that more than 90% of the identified single nucleotide polymorphisms (SNPs), located within regulatory or interoic regions, and not in coding regions of the genes. The functional regulatory SNPs (rSNPs) in TFBS may lead to multiple consequences such as variations in gene expression, phenotypes, and even susceptibility to environmental exposure (epigenetic trait) [31]. Accordingly, the complete non-coding regions of the hFVIII gene were surveyed in order to identify and describe the characteristics of its Cis-acting regulatory elements especially hFVIII-TFBs. Moreover, the varieties, frequencies as well as the distribution of the repetitive elements throughout the regions of the gene were investigated then were mapped to the hFVIII-TFBs. It was detected that 377 REs there are along with the two strands of the intronic regions (Figure 2-A), which are consistent with the corresponding frequency throughout the whole genome of human [32]. Among these elements, the frequency, distribution and spatial connection of TFBs with REs showed a remarkable pattern in the gene. They revealed an independent pattern of distribution with a strong positioning bias in some introns, spatially within the first and last introns, as well as in several restricted regions far from splice sites (Figure 3-A). It has been demonstrated that the first intron is enriched by highly conserved sequences such as TFBs, relative to other downstream introns, which is the further support that the first intron is likely involved in transcriptional regulation [33]. Remarkably, about 55% of TFBs are overlapped to the SINE elements of the hFVIII gene, while they consist 16% of N-LTR elements (Figure 2-C). This pattern may be a reason for the smart and targeted distribution of TFBs, through the non-coding regions of the hFVIII gene. These data are consistent with previous reports describing these regions carrying functional sequences [2, 15, 34].
On the other hand, the SINE elements in this study showed the binding peaks more than expected. In coordinating with our data, the previous reports have confirmed that the dependency of the peak positioning of TFBs to REs is imputed to the cell type as well as the type of species [1, 2]. Therefore, it is suggested that SINE-TFBs through the whole of the hFVIII gene might be an important driving force for the regulatory innovation of the hFVIII gene. Moreover, the density and distribution of REs-TFBs in the non-coding regions of the gene (Table 1, Figure 3-B), might be another reason for potential regulatory roles of these elements. This feature of the distribution of RE-TFBs is in agreement with several independent pieces of evidence, concerning the presence of regulatory elements in both first and last introns. Based on these data it was hypothesized that long first introns were involved in regulatory functions and their longer length was attributed to the presence of additional transcription factor binding sites [27, 28]. Among these elements, SINE-TFBs and CR1-TFBs are the only elements which are situated within 5′UTR of the hFVIII gene (Figure 3-B), which might be another reason of their regulatory function.
Moreover, the sequence context of REs-TFBs reveals that they are different in the length and context, ranging in size from 7 up to 30 bps and G rich sequence for DT-TFBs (Figure 4-A). However, most of SINE-TFBs showed 30 bps lengths with a composition of each of four nucleotides in context (Figure 4-B). In this regard, several TFBs that are harbored into the SINE elements have been determined [35, 36]. Meanwhile, REs-TFBs is following the distribution pattern of the whole TFBs in intronic regions (Figure 4-C), so that they are the peak at regions far from the splice site, which is inferring their correlation to each other. The distribution pattern of TFBs-REs showed different bias in the intronic regions of the hFVIII gene, spatially into the Introns 13 and 25. However, most of TFBs which are condensed upstream of the donor splice site as well as the other introns, follow the similar pattern, but within the shorter area. It suggests that the surveying the deep intronic variations which are overlapped to these regions might confirm the regulatory functions of them in the future studies. In this regard, the mutations which have reported within the introns of over 75 disease-associated genes, including the c.6429 + 14194T > C variant detected in patients carrying the intron 22 inversion of the hFVIII gene, highlight the importance of studying variation in deep intronic sequence as a cause of monogenic disorders [18, 19]. The structural variants (SVs) including hFVIII inversion variants in introns 1 and 22, have been explained in nearly one-half of patients with severe hemophilia A [37]. These data are consistent with previous reports describing these regions carrying functional sequences [2, 15, 26]. However, the functionality of these TFB sites may be influenced by other modes of gene regulation, e.g. epigenetic controls, and need to investigate and experience.
Indeed, epigenetic modifications, including CpG-site methylation of the promoter regions and DNA and histones, can affect epigenetic regulation of gene expression and disease development. DNA methylation close to transcription start sites (TSSs) represses directly transcription by interfering with binding of TF and indirectly by methyl-CpG-binding proteins and reducing chromatin remodeling activities. However, modeling the relationship of epigenetic modifications to transcription factor binding has revealed location- and cell type-specific relationships between epigenetic modifications and binding affinities of TFs [38, 39]. In this regard, a significant enrichment of epigenomic signals in first introns, relative to other introns, has been described for three cell lines - GM12878, H1-hESC, and K562 in human. Moreover, general regulatory signals (e.g., TFBSs, DNase I hypersensitivity sites (DHS)) and active regulatory chromatin marks (e.g., H3K4me1 and H3K4me3) have displayed a clear positive correlation between the regulatory signal ratio and the number of exons. This study showed that a conservation of enrichment of accessible chromatin and TFBS binding in conserved regions of the first intron is consistent with their role in active gene regulation [33].
On the other hand, it has been shown that the genotypes and even ethnicity have influence on haemophilia consequences [20, 21]. Pathogenicity describing for variants hFVIII gene is a critical and common clinical practice in etiology of the patient's haemophilia A. The study of hFVIII genetic variation has targeted in the "1000 Genomes Project" which showed the presence of the significant benign variations in the hFVIII gene across ethnic groups. This project has aimed to discover novel and rare hFVIII variants, and to characterize hFVIII variants in diverse population backgrounds. Indeed, 3030 single nucleotide variants, 31 short deletions/insertions and a large, 497 kb, deletion were recognized among 26 ethnic groups by analyzing 2535 subjects of F8 genetic variations, during 1000 Genomes Project, phase 3 dataset(http://1000genomes.org). Among them, 86.4% and 55.6% were rare and novel variants, respectively. While the most of these HA variants were ethnic-specific with low allele frequency, p.M2257V was current variant in 27% of African subjects. Additionally, the p. E132D, p. T281A, p. A303V and p. D422H were presented as sex-dependent HA variants. These results emphasized the complexity of hFVIII variants and the significance of questioning genetic variants on multiple ethnic backgrounds for associations with bleeding and thrombosis [20].
MyLifeOurFuture (MLOF) as the largest genetic program in haemophilia and nationwide U.S. initiative, has been provided genotyping for patients affected by haemophilia and their families to establish segregation and classification of hFVII and hFVIII variants, to support clinical care and reproductive planning to the haemophilia community and to inform a better understanding of haemophilia genotype interpretation. Moreover, the programe are constructing a valuable resource for research in haemophilia and associated disorders that combines omics data with wide phenotypic data for 6000 patients registered in the Research Repository [21]. In the first 3000 MLOF patients (2900 males and 100 females), structural variants (SVs) have been reported more common in hemophilia A, 43% of severe male cases, due to hFVIII gene intron 22 and intron 1 inversions. While, the incidence of other large SVs was 6% and 10% in severe male hemophilia A and B, respectively. Moreover, the complex intron 22 and intron 1 inversions, Alu insertions, and a complex partial exon 14 duplication were distinguished in hFVIII gene. These data emphasized the necessity for devoted assessments of structural variation in the genotyping of hemophilia patients, particularly the patients without any variant detected by other approaches [40].
Considering clinical aspects, it has shown that the high plasma level of hFVIII coagulant activity (FVIII:C) is a highly heritable quantitative trait and strongly associated with an increased risk for venous and arterial thrombosis within families [41] and the risk of regular venous thromboembolism. Lipoprotein receptor-related protein (LRP), is the only receptor of hFVIII identified, so far, which is assumed to be complicated in hFVIII degradation. Accordingly, the possible polymorphisms detected by molecular screening of FVIII:C and von Willebrand factor antigen (VWF:Ag) levels have been measured. A durable effect of VWF:Ag, the carrier molecule of hFVIII in the circulation, and ABO blood groups described more than 50% of FVIII:C variability in healthy population of nuclear families containing 200 parents and 224 offspring. Although, the N allele of the LRP/D2080N polymorphism has been accompanied with decreased levels of FVIII:C and VWF:Ag levels, no polymorphism has been detected in the LRP-binding domains of the hFVIII gene. This study strengthened the hypothesis of a genetic influence of hFVIII levels outside the influence of VWF:Ag and ABO blood groups. The D2080N polymorphism of the LRP gene weakly contributed to the variability of FVIII:C levels in this healthy population [42].
Bearing in mind the hypothesis that genetic variations in the three coding regions identified for the binding of hFVIII to LRP might influence the clearance of hFVIII, a molecular screening of the three regions in hFVIII gene, has been performed. Among a 137 unrelated nonhemophilic population, all known functional regions in a collection of 222 potentially distinct alleles was resequenced. Although, they failed to find any frequent sequence variation, it has been shown that the alleles of 56010G > A, a SNP within the 3′ splice junction of intron 7, are meaningfully accompanied with the 92714C > G, a SNP encoding the B-domain substitution D1241E of F8, which has suggested that significantly associated with increasing FVIII:C level. Nevertheless, supplementary studies was suggested mandatory to determine whether D1241E is itself a functional variant [43]. Accordingly, it can be suggested that more attention to intronic variations, along with exon or replicon changes, could shed more light on the dark corners of the gene's hFVIII gene functions, and might increase our understanding of some of its disorders.
On the other hand, there are several findings which confirmed the increased age-related plasma Factor VIII level through both enhanced gene expression and clearance mechanisms in normal individuals. Moreover, increasing data support the hypothesis that high levels of hFVIII in plasma may play a role in hypercoagulability as a prevalent, dose-dependent risk factor or a biomarker for venous thromboembolism (VTE), a significant health concern due to its high morbidity and mortality [44].
Recently two independent studies have demonstrated that introns play important roles in the regulation of cell growth [45, 46]. A study described 34 introns in Saccharomyces cerevisiae that appeared to be unusually stable and lingered around the spliceosome complex that inhibit its degradation under stresses conditions during the stationary phase [45]. Another study revealed that intronic deletions slow down cell growth and reducing energy consumption by promoting the cell's entry into the stationary phase, and thereby to survive for a longer time. The genetic and transcriptomic assessments shown that the introns enhance the repression of ribosomal protein genes (RPGs), which ake up about 90% of the spliced RNAs in yeast grown in nutrient-rich conditions, and promote resistance to starvation [46].
Accordingly, it is suggested that the mentioned repression of RPGs function which is correlated with the introns of the hFVIII gene might be disturbed during the aging process in normal individuals that influences gene regulatory networks of the hFVIII gene which in turn arise the plasma Factor VIII level. This idea can be evaluated by population studies. Recently, a population genomic study has revealed the positive selection of recent human TE insertions on polymorphic human TEs in 15 populations [47].
In overall, our data revealed the functional elements such as TFBs binding sites in the intronic regions of the hFVIII gene and their correlation with short interspersed elements, the density and distribution of TFBs-REs in each intron and the gene. This study presented the candidate introns for next studies regarding the gene regulation, evolution of the hFVIII genes and maybe invaluable resource for research in haemophilia and associated disorders with hFVIII as a functional protein and might have potential to improve the existing strategies for treatment of hemophilia A.
Declarations
Author contribution statement
Aliakbar Haddad-Mashadrizeh, Jafar Hemmat: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Muhammad Aslamkhan: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by the grant of Eastern Mediterranean Health Genomics & Biotechnology Network (EMGEN) (Grant No: 20145) and Deputy for Research and Technology of Ministry of Health and Medical Education of Iran.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to express our special thanks to Nazanin Gholampour, who helped us in data analyzing and revising the article. We are grateful to Masoomeh Ghobadnejad, Irfan-Masood M and Mahdi Nohtani for their valuable suggestions with respect to the manuscript.
Contributor Information
Aliakbar Haddad-Mashadrizeh, Email: a.haddad@um.ac.ir.
Jafar Hemmat, Email: j.hemmat@gmail.com.
References
- 1.Rebollo R., Romanish M.T., Mager D.L. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 2.Chuong E.B., Elde N.C., Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2017 Feb;18(2):71. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thornburg B.G., Gotea V., Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006 Jan 3;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 4.Li H., Chen D., Zhang J. Analysis of intron sequence features associated with transcriptional regulation in human genes. PloS One. 2012;7(10) doi: 10.1371/journal.pone.0046784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conley A.B., Jordan I.K. Identification of transcription factor binding sites derived from transposable element sequences using ChIP-seq. In: Ladunga I., editor. Computational Biology of Transcription Factor Binding. Humana Press; Totowa, NJ: 2010. pp. 225–240. [DOI] [PubMed] [Google Scholar]
- 6.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008 May;9(5):397–4057. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polavarapu N., Marino-Ramirez L., Landsman D., McDonald J.F., Jordan I.K. Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genom. 2008;9:226. doi: 10.1186/1471-2164-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad-Mashadrizeh A., Zomorodipour A., Izadpanah M., Sam M.R., Ataei F., Sabouni F. A systematic study of the function of the human beta-globin introns on the expression of the human coagulation factor IX in cultured Chinese hamster ovary cells. J. Gene Med. 2009 Oct;11(10):941–950. doi: 10.1002/jgm.1367. [DOI] [PubMed] [Google Scholar]
- 9.Zomorodipour A., Jahromi E.M., Ataei F., Valimehr S. Position dependence of enhancer activity of the human beta-globin intron-ii, within a heterologous gene. J. Mol. Med. Ther. 2017 Nov 10;1(1):19–24. [Google Scholar]
- 10.Gross D.S., Garrard W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 11.Chen H., Li H., Liu F., Zheng X., Wang S., Bo X., Shu W. An integrative analysis of TFBS-clustered regions reveals new transcriptional regulation models on the accessible chromatin landscape. Sci. Rep. 2015 Feb 16;5:8465. doi: 10.1038/srep08465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutin J., Sadeh R., Bodenheimer N., Joseph-Strauss D., Klein-Brill A., Alajem A., Ram O., Friedman N. Fine-resolution mapping of TF binding and chromatin interactions. Cell reports. 2018 Mar 6;22(10):2797–2807. doi: 10.1016/j.celrep.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000 Jan 1;16(1):16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 14.Whitfield T.W., Wang J., Collins P.J., Partridge E.C., Aldred S.F., Trinklein N.D., Myers RM, Weng Z. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012 Sep;13(9):R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nazanin G.F., Aliakbar H.M., Mahdi M., Hassan M., Safoora S.M., Reza B.A., Mohammadreza H., Maryam M.M., Alireza Z., Parisa D., Mahdi N. In-silico evidences of regulatory roles of WT1 transcription factor binding sites on the intervening Sequences of the human bcl-2 gene. Curr. Bioinf. 2018 Jun 1;13(3):260–272. [Google Scholar]
- 16.Antonarakis S.E., Kazazian H.H., Tuddenham E.G. Molecular etiology of factor VIII deficiency in hemophilia A. Hum. Mutat. 1995;5(1):1–22. doi: 10.1002/humu.1380050102. [DOI] [PubMed] [Google Scholar]
- 17.Pang J., Wu Y., Li Z., Hu Z., Wang X., Hu X., Wang X., Liu X., Zhou M., Liu B., Wang Y. Targeting of the human F8 at the multicopy rDNA locus in hemophilia A patient-derived iPSCs using TALENickases. Bioch. Bioph. Res. Co. 2016 Mar 25;472(1):144–149. doi: 10.1016/j.bbrc.2016.02.083. [DOI] [PubMed] [Google Scholar]
- 18.Vaz-Drago R., Custodio N., Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017 Sep;136(9):1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 19.Inaba H., Shinozawa K., Amano K., Fukutake K. Identification of deep intronic individual variants in patients with hemophilia A by next-generation sequencing of the whole factor VIII gene. Res. Pract. Thromb. Haemost. 2017 Oct;1(2):264–274. doi: 10.1002/rth2.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J.N., Carrero I.G., Dong J.F., Yu F.L. Complexity and diversity of F8 genetic variations in the 1000 genomes. J. Thromb. Haemostasis. 2015;13(11):2031–2040. doi: 10.1111/jth.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konkle B.A., Johnsen J.M., Wheeler M., Watson C., Skinner M., Pierce G.F. My Life Our Future programme. Genotypes, phenotypes and whole genome sequence: approaches from the My Life Our Future haemophilia project. Haemophilia. 2018;24:87–94. doi: 10.1111/hae.13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohany O., Gentles A.J., Hankus L., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinf. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solovyev V.V., Shahmuradov I.A., Salamov A.A. Identification of promoter regions and regulatory sites. Methods Mol. Biol. 2010;674:57–83. doi: 10.1007/978-1-60761-854-6_5. [DOI] [PubMed] [Google Scholar]
- 24.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015 Jul 1;43(W1):W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahmuradov I.A., Solovyev V.V. Nsite, NsiteH and NsiteM computer tools for studying transcription regulatory elements. Bioinformatics. 2015 Nov 01;31(21):3544–3545. doi: 10.1093/bioinformatics/btv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dogan R.I., Getoor L., Wilbur W.J., Mount S.M. SplicePort–an interactive splice-site analysis tool. Nucleic Acids Res. 2007 Jul;35:W285–W291. doi: 10.1093/nar/gkm407. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004 Jun;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss S.P., Joyce D.A., Humphries S., Tindall K.J., Lunt D.H. Comparative analysis of teleost genome sequences reveals an ancient intron size expansion in the zebrafish lineage. Genome Biol. Evolution. 2011 Jan 1;3:1187–1196. doi: 10.1093/gbe/evr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.JiaYan W., JingFa X., LingPing W., Jun Z., HongYan Y., ShuangXiu W., Zhang Z., Jun Y. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci. China Life Sci. 2013 Oct 1;56(10):968–974. doi: 10.1007/s11427-013-4540-y. [DOI] [PubMed] [Google Scholar]
- 30.Rose A.B. Introns as gene regulators: a brick on the accelerator. Front. Genet. 2019 Feb 7;9:672. doi: 10.3389/fgene.2018.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buroker N.E. Regulatory SNPs and transcriptional factor binding sites in ADRBK1, AKT3, ATF3, DIO2, TBXA2R and VEGFA. Transcription. 2014;5(4) doi: 10.4161/21541264.2014.964559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 33.Park S.G., Hannenhalli S., Choi S.S. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genom. 2014 Jun 26;15:526. doi: 10.1186/1471-2164-15-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sam M.R., Zomorodipour A., Shokrgozar M.A., Ataei F., Haddad-Mashadrizeh A., Amanzadeh A. Enhancement of the human factor IX expression, mediated by an intron derived fragment from the rat aldolase B gene in cultured hepatoma cells. Biotechnol Lett. 2010; Oct;32(10):1385–1392. doi: 10.1007/s10529-010-0321-x. [DOI] [PubMed] [Google Scholar]
- 35.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018 Feb 8;172(4):650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Spitz F., Furlong E.E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012 Sep;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 37.Bagnall R.D., Waseem N., Green P.M., Giannelli F. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood. 2002;99:168–174. doi: 10.1182/blood.v99.1.168. [DOI] [PubMed] [Google Scholar]
- 38.Curradi M., Izzo A., Badaracco G., Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell Biol. 2002 May 1;22(9):3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., Jin G., Zhou X. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 2015;43(8):3873–3885. doi: 10.1093/nar/gkv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnsen J.M., Fletcher S.N., Huston H., Roberge S., Martin B.K., Kircher M.…Morales J. Novel approach to genetic analysis and results in 3000 hemophilia patients enrolled in the My Life, Our Future initiative. Blood Adv. 2017;1(13):824–834. doi: 10.1182/bloodadvances.2016002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bank I., Libourel E.J., Middeldorp S., Hamulyák K., Van Pampus E.C., Koopman M.M., Prins M.H., Van Der Meer J., Büller H.R. Elevated levels of FVIII: C within families are associated with an increased risk for venous and arterial thrombosis. J. Thromb. Haemostasis. 2005 Jan;3(1):79–84. doi: 10.1111/j.1538-7836.2004.01033.x. [DOI] [PubMed] [Google Scholar]
- 42.Morange P.E., Tregouet D.A., Frere C., Saut N., Pellegrina L., Alessi M.C., Visvikis S., Tiret L., Juhan-Vague I. Biological and genetic factors influencing plasma factor VIII levels in a healthy family population: results from the Stanislas cohort. Br. J. Haematol. 2005 Jan;128(1):91–99. doi: 10.1111/j.1365-2141.2004.05275.x. [DOI] [PubMed] [Google Scholar]
- 43.Viel K.R., Machiah D.K., Warren D.M. A sequence variation scan of the coagulation factor VIII (FVIII) structural gene and associations with plasma FVIII activity levels. Blood. 2007;109:3713–3724. doi: 10.1182/blood-2006-06-026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albánez S., Ogiwara K., Michels A., Hopman W., Grabell J., James P., Lillicrap D. Aging and ABO blood type influence von Willebrand factor and factor VIII levels through interrelated mechanisms. J. Thromb. Haemostasis. 2016 May;14(5):953–963. doi: 10.1111/jth.13294. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan J.T., Fink G.R., Bartel D.P. Excised linear introns regulate growth in yeast. Nature. 2019 Jan;565(7741):606. doi: 10.1038/s41586-018-0828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parenteau J., Maignon L., Berthoumieux M., Catala M., Gagnon V., Elela S.A. Introns are mediators of cell response to starvation. Nature. 2019 Jan;565(7741):612. doi: 10.1038/s41586-018-0859-7. [DOI] [PubMed] [Google Scholar]
- 47.Rishishwar L., Wang L., Wang J., Soojin V.Y., Lachance J., Jordan I.K. Evidence for positive selection on recent human transposable element insertions. Gene. 2018 Oct 30;675:69–79. doi: 10.1016/j.gene.2018.06.077. [DOI] [PubMed] [Google Scholar]