Abstract
CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated) has been extensively exploited as a genetic tool for genome editing. The RNA guided Cas nucleases generate DNA double-strand break (DSB), triggering cellular repair systems mainly Non-homologous end-joining (NHEJ, imprecise repair) or Homology-directed repair (HDR, precise repair). However, DSB typically leads to unexpected DNA changes and lethality in some organisms. The establishment of bacteria and plants into major bio-production platforms require efficient and precise editing tools. Hence, in this review, we focus on the non-DSB and template-free genome editing, i.e., base editing (BE) and prime editing (PE) in bacteria and plants. We first highlight the development of base and prime editors and summarize their studies in bacteria and plants. We then discuss current and future applications of BE/PE in synthetic biology, crop improvement, evolutionary engineering, and metabolic engineering. Lastly, we critically consider the challenges and prospects of BE/PE in PAM specificity, editing efficiency, off-targeting, sequence specification, and editing window.
Keywords: CRISPR, Base editing, Prime editing, Cytosine base editor, Adenine base editor
1. Introduction
Scientists have for long worked to develop a facile, precise, and efficient DNA editing tool. Though the efforts led to the development of some significant editing tools like zinc finger nuclease (ZFN) and transcription activator-like effector nucleases (TALEN), they are not simple enough to have widespread use [1]. The discovery of CRISPR-Cas [[2], [3], [4]] and its development into a genome editing tool [5] have revolutionized the field of biotechnology, therapeutics, DNA sequencing, and biology as a whole. CRISPR-Cas systems that naturally evolved as a prokaryotic adaptive immune system against phage infection and invasion of mobile genetic elements (MGEs), are found in around 45% bacterial and 90% archaeal genomes [6]. CRISPR-Cas systems generally consist of CRISPR array and Cas proteins. Upon the infection, CRISPR array is transcribed, producing precursor-CRISPR RNA (pre-crRNA), which is further processed to form CRISPR RNA (crRNA). Later, for simple use and design, crRNA was engineered to form a single-guide RNA (sgRNA or gRNA). crRNA complementary to the foreign nucleotide sequence guides the Cas proteins for DNA cleavage via recognition of Cas-specific protospacer adjacent motif (PAM) [7,8]. This strategy has been repurposed to develop editing tools by programming sgRNA to direct Cas proteins to the target site for DNA break, subsequently, initiating cellular repair machinery mainly either imprecise repair system NHEJ or precise repair system HDR. NHEJ has been used for gene disruption by the introduction of indels or translocations, while HDR has been used in combination with donor DNA templates for precise replacements, deletions, insertions or point mutations [9].
However, DSB can be lethal for some cells, especially for bacterial cells. While, in the case of eukaryotic cells, the DSB repair mechanisms compete with each other like NHEJ vs. HDR, the former being a preferred choice of repair [10]. The competition mostly leads to low HDR efficiency for precise editing. Especially in plants, DSB-induced HDR editing suffers low efficiency because of weak HDR in somatic plant cells. The requirement of donor DNA template makes matters worse due to the challenging delivery process and insufficient availability of donor DNA template during DSB repairing [11]. On the other hand, most bacteria lack NHEJ [[12], [13], [14]], leaving DSBs to be repaired mainly by HDR. However, HDR in most bacteria (except some bacteria such as Streptomyces [15]) is not effective enough and shows low editing efficiencies. Recombinases like RecET [16] and Lambda-Red [17] systems have been combined with DSB-induced HDR to increase editing efficiency. Additionally, to increase editing efficiency, counter-selection techniques such as Cas induced DSB selection markers have been applied [18]. Still, the requirement of high dose of donor DNA template and low editing efficiencies between 10−6 to 10−1 in the absence of counter-selection, means that more efficient editing tools are required [19,20]. Development of base editor (BE) [[21], [22], [23]] and prime editors (PE) [24] answers the call for the need of precise and efficient non-DSB and template-free editing systems.
BE and PE use catalytically nuclease deficient Cas proteins (For example, dCas9(D10A and H840A) or nCas9(D10A or H840A), all generated from Streptococcus pyogenes Cas9 (SpCas9)), allowing DSB-free editing. In the case of BE, dCas/nCas(D10A) are fused to deaminase domains that allow C-to-T (by cytosine base editor, CBE) or A-to-G (by adenine base editor, ABE) substitution [[21], [22], [23]]. While PE consists of nCas9(H840A) fused to reverse transcriptase, which allows the insertions, deletions, and point mutations at specific loci [24]. BE and PE technologies have, to some extent, overcome the drawbacks of the DSB-dependent CRISPR editing systems. Plenty of groups have already adopted base editing systems for animal, plant and bacterial cells. CRISPR technologies and specifically base editing in animal cells have been extensively reviewed by Liu and Rees [25], Hees et al. [26], Molla and Yang [27], and Wu et al. [28]. Here, we comprehensively review the current developments in non-DSB and template-free editing systems in bacteria and plants.
Bacteria have for long been considered major platforms for bio-production and there has been an increasing interest in developing plants into key bio-production platforms. The development of a broad set of biological tools for bacteria and plants releases their potential to produce a wide array of valuable chemicals [29,30], secondary metabolites [31,32], recombinant proteins [33,34], biofuels [35,36] and food additives [37]. The regulation of interconnected metabolic pathways and synthetic multi-gene programmes require efficient, precise, and simple gene editing tools. Traditional CRISPR tools have been adopted in bacteria and plants for crop improvement [38], synthetic biology, and metabolic engineering to improve bio-production [[39], [40], [41], [42]]. However, efforts to develop bacteria and plants into major cell factories for bio-production have been halted because of low editing efficiencies and the requirement of the donor DNA template. The adoption of the BE and PE systems should overcome the shortcomings, and these systems hold promise to further enhance microbial and plant bio-production abilities.
2. Development of base and prime editors
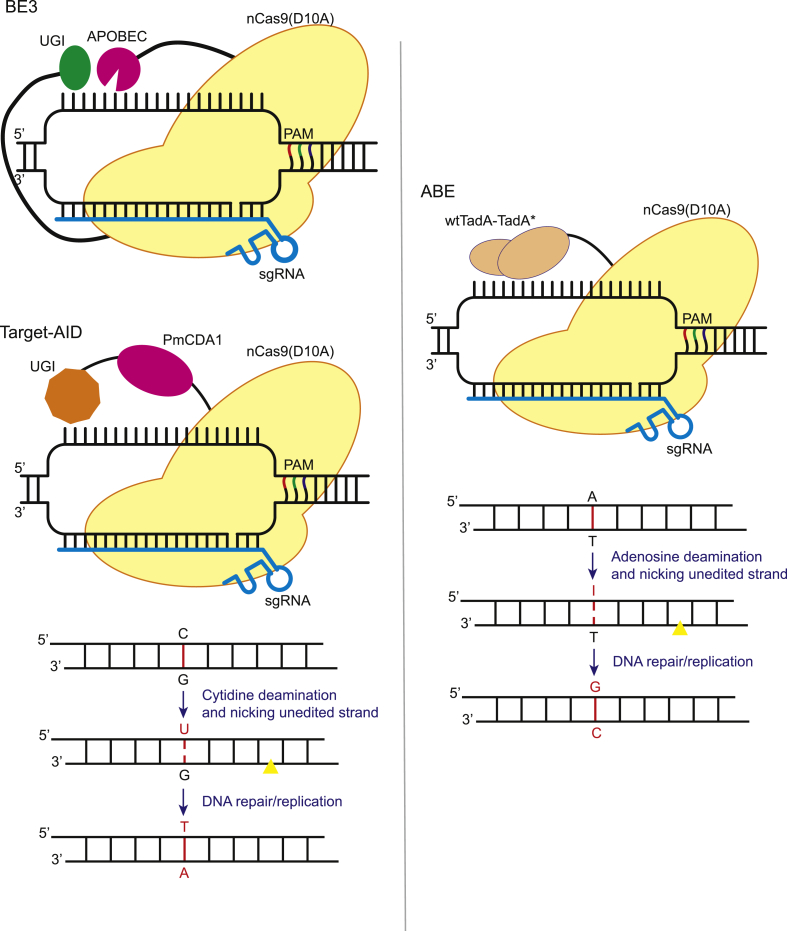
Base editors are chimeric complexes with the ability to deaminate cytosine and adenine bases. The catalytically inactive or nuclease deficient Cas proteins are utilized by BE to direct deaminase machinery to the target locus and carry out precise nucleotide substitution (Fig. 1) [[21], [22], [23]]. Apart from BE, the recently developed PE also allows non-DSB and template-free insertion, deletion or nucleotide substitution of arbitrary sequence (Fig. 2A) [24].
Fig. 1.
Deaminase based Cas9 base editing. A schematic model of deaminase based Cas9 base editing systems: cytosine base editors (CBE, for example, BE3 & Target-AID) and adenine base editor (ABE). CBE consists of rat apolipoprotein B mRNA editing enzyme (APOBEC), and uracil glycosylase inhibitor (UGI) fused to N and C terminus of nCas9(D10A), respectively. Target-AID consists of activation-induced cytosine deaminase ortholog PmCDA1 and UGI fused to N terminus of nCas9(D10A). CBE involves deamination of cytosine by the deaminase which converts it into uracil, making U•G wobble base. UGI prevents its conversion back to C. Mismatch repair machinery (MMR) recognizes it forming U•A which is then converted to T•A by replication machinery leading C•G-to-T•A substitution. ABE consists of heterodimeric wtTadA-TadA* fused to nCas9(D10A). ABE performs deamination converting T•A-to-T•I which is then recognized by DNA repair and replication machinery and converted to C•G base pair (Yellow triangle: nick site).
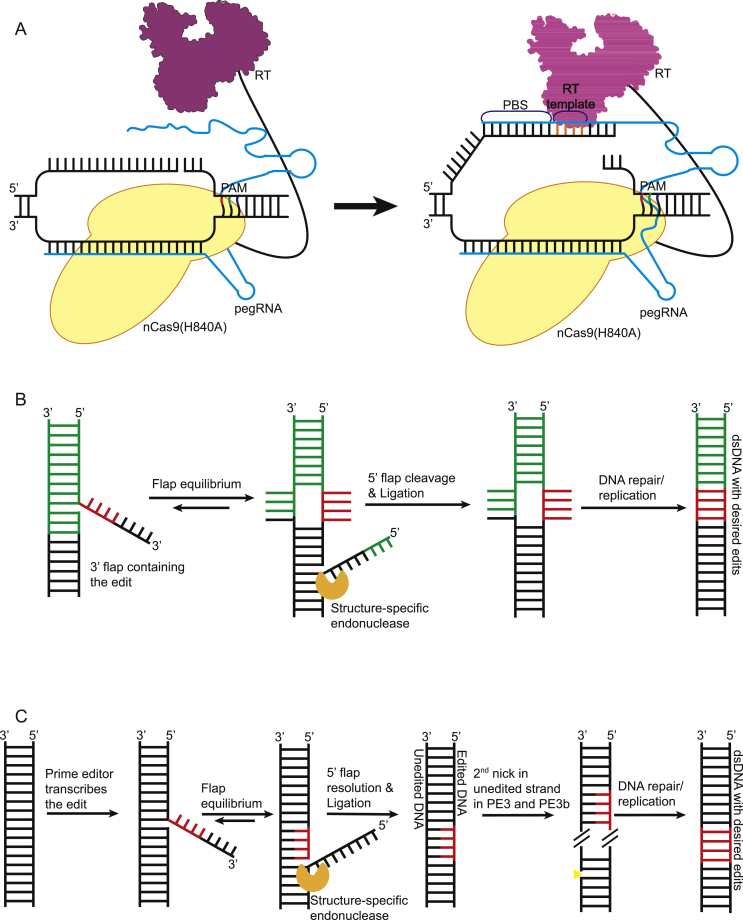
Fig. 2.
Schematic representation of prime editing system. (A) The basic construct of PE consists of nCas9(H840A) fused to reverse transcriptase (RT) via a linker. The prime guide RNA (pegRNA) in PE consists of gRNA, primer binding site (PBS) and RT template. Once nCas9(H840A) nicks the strand, RT uses PBS fused to 3′ flap as a primer to encode RT DNA template. (B) After reverse transcription, PE1 and PE2 systems involve flap equilibrium of edited 3′ and unedited 5′ flap. The unedited 5′ flap is then cleaved off by structure-specific endonucleases followed by ligation and DNA repair/replication leading to permanent editing (Green bases: pegRNA binding regions, Red bases: desired edits). (C) After 5′ flap cleavage, a heteroduplex of the edited and unedited strand is formed. PE3 and PE3b involve a 2nd nick in the unedited strand 14-116 nt away from the initial pegRNA nick. This increases the chances of generation of dsDNA with the desired edit by favouring the repair of unedited strand by repair/replication machinery (Black strand: original DNA, Red strand: edited strand, Yellow triangle: nick site).
The first base editors were individually developed by Liu [22] and Kondo [23] groups for C-to-T substitution. The editors convert cytosine to uracil by deamination of exocyclic amine, which leads to U•G, a wobble base pair. U being an illegitimate base would generally be recognized by uracil-DNA glycosylase (UDG), which initiates nucleotide excision repair (NER) converting U back to the original base C [43,44]. However, the CBE uses uracil glycosylase inhibitor (UGI) to prevent the conversion of U back to C and activate mismatch repair system (MMR) which converts U•G-to-U•A base pair which is then converted to T•A base pair leading to C•G-to-T•A substitution [45]. The CBEs from both the groups (Liu and Kondo) are functionally similar but differ in the deaminase types and the arrangement of the domains. The BE system from Liu group uses rat apolipoprotein B mRNA editing enzyme (rAPOBEC1) fused via 16-residue XTEN linker to N-terminus, and the UGI fused via 4-amino acid linker to the C-terminus of the Cas9 (Fig. 1). Whereas the Target-AID system from Kondo group uses activation induced cytosine deaminase (AID) ortholog PmCDA1 from sea lamprey. The PmCDA1 is fused to N-terminus of Cas9 via 100 amino acid linker, while UGI is fused to PmCDA1 (Fig. 1). Both the groups initially used dCas9 fused deaminase complex and reported humble editing efficiency of less than 5%. The low editing efficiency of the dCas9-dependent CBE systems is regarded to their ability to edit only a single DNA strand. The deaminated base can be corrected back to the original base by the cells; subsequently, only a handful of deamination results in the desired editing. Therefore, to yield higher editing efficiency, authors adopted nCas9(D10A) which nick's the unedited strand. Nick in the unedited strand favours the conversion of U•G-to-U•A base pair during the DNA repair process, ultimately increasing the editing efficiency (Fig. 1) [22,23].
Liu group was the first one to develop an adenine base editor [21]. There is no known naturally existing DNA adenosine deaminase. Therefore, the group evolved an E. coli TadA tRNA adenosine deaminase. TadA catalyzes the conversion of adenosine to inosine in the single-stranded anticodon loop of tRNA-Arg. The inosine is recognized by the polymerase as G and subsequently, G•C base pair is introduced [46]. The authors used E. coli to evolve TadA to accept ssDNA by initially making unbiased libraries with mutated wild-type ecTadA fused to dCas9. The complex was used along with target sgRNA to transfect E. coli containing chloramphenicol resistance gene H193Y mutant, which lost the resistance. The colonies that survived on chloramphenicol showed certain TadA mutations that were incorporated in future AB editors. The same strategy was repeated by applying stringent conditions and incorporation of observed mutations in the later series of ABEs. Finally, evolved TadA (TadA*) (W23R + H36L + P48A + R51L + L84F + A106V + D108 N + H123Y + S146C + D147Y + R152P + E155V + I156F + K157 N) was developed and ABE7.10, wtTadA‐TadA*‐nCas9 fused together via two 32-amino acid linkers, was established (Fig. 1) [21].
Apart from CBE and ABE, Liu group recently developed the search and replace editing, i.e., prime editing [24]. Contrary to BE that use deaminase, PE works on a completely different principle, which involves reverse transcription. PE consists of nCas9(H840A)-reverse transcriptase fusion complex which uses prime guide RNA (pegRNA, consisting of primer binding site (PBS) which is complementary to the sequence upstream of PAM site of the nicked strand and reverse transcriptase template (RT template)) to form a primer with the 3′ flap of the nicked strand for reverse transcription to encode desired edits (Fig. 2A). PE then uses the structure-specific flap endonucleases found in the cell that prefer 5′ flap as a substrate to its advantages by letting them digest the unedited 5′ flap while 3′ flap with edited DNA is ligated, forming a heteroduplex of DNA containing edited and unedited strand (Fig. 2B). This heteroduplex is then resolved by the cell repair machinery as it copies the edited strand to the complementary unedited strand leading to permanent incorporation of the edit. However, the mechanism of the 5′ flap digestion and the heteroduplex resolution are not thoroughly understood. PE initially used mouse-murine leukemia virus (M-MLV RT). The group increased the PE editing efficiency by several folds by developing an engineered M-MLV RT (D200 N + L603W + T330P + T306K + W313F), thus developing the PE2 system. The authors further improved the editing efficiency by 1.5–4.2 folds compared to PE2 by developing the PE3 system, which involves the induction of a second nick in the unedited strand 14 to 116 nt away from original pegRNA nick (Fig. 2C) [24]. The induction of the second nick increases the chances of the unedited strand, rather than the edited strand, to be repaired by MMR machinery, thus, increasing the possibility of getting the duplex DNA with the desired edits. To reduce the indel formation by PE3, PE3b strategy was developed in which the second nick is carried out after the flap resolution and the successful editing of the initial editing strand. This was achieved by using sgRNA containing mismatches between the spacer and the unedited allele, and making it complementary to the edited strand and not the original allele (Fig. 2C). PE3b results in 13-folds lower indels compared to PE3. PE's strength to insert or delete arbitrary DNA sequence and allow all possible point mutations at target locus with high precision makes it a highly attractive editing system [24]. Although BE and PE have developed and applied broadly in animal cells, their adoption for bacteria and plants has provided a new avenue to advance their genome editing.
3. DSB & template-free editing in bacteria
CBE and ABE have been applied in several bacteria such as Escherichia coli [21,[47], [48], [49], [50], [51], [52]], Brucella melitensis [48], Clostridium beijerinckii [53], Corynebacterium glutamicum [54], Klebsiella pneumoniae [55], Pseudomonas [56], Staphylococcus [52,57] and Streptomyces [[58], [59], [60]] (Table 1). However, PE has not been set up in bacteria yet. The development of PEs for bacterium would be a crucial milestone in the bacterial gene-editing toolbox. Here we review the base editing systems in bacteria.
Table 1.
Base editing systems in bacteria.
| Organism | Editor type | Base editing systema | Target genes | Editing efficiency | Editing window (upstream of PAM) | Ref. |
|---|---|---|---|---|---|---|
| Escherichia coli | ||||||
| E. coli | BE3 | pEcBE3 | tetA, lacZ, rppH | 100% | −12 to −16 | [48] |
| E. coli | Target-AID | dCas9-CDA-UL | galK, rpoB, xylB, manA, pta, tipaA | 61–95% | −16 to −20 (extended sgRNA = −18 to −24) | [49] |
| E. coli | Cpf1-based BE | dLB-Cpf1-BE | SupF in shuttle vector | 45–80% | −7 to −13 (downstream of PAM) | [47] |
| E. coli | ABE | pABE | lacZ, yagl, murR | 66–100% | −13 to −17 | [52] |
| E. coli | ABE | pnCas9-TadA | lacZ, poxb | 9.3–65.4% | −12 to −17 | [50] |
| pdCas9-TadA | 1.3–8.3% | |||||
| pnCas9-TadA + Cas9 | 12.7–99% | |||||
| Streptomyces | ||||||
|
S. coelicolor S. rapamycinicus |
Target-AID | dCas9-CDA-ULstr | redw, redL, redX, | 50–100%, 60%b 20%c |
−16 to −20 | [59] |
|
S. coelicolor S. griseofuscus |
BE3 | CRISPR-cBEST | actinorhodin gene cluster, CDA gene cluster, RED gene cluster | 30–100% | −11 to −17 | [58] |
| S. coelicolor | BE2 | BE2 | red, actl-ORF2 | 43–70% 43%b |
−13 to −17 | [60] |
| S. coelicolor | BE3 | BE3 | red, actl-ORF2 | 100% 100%b |
−13 to −17 | [60] |
| S. avermitilis | ave | 0–100% multiplex editing | ||||
| S. avermitilis | HF-BE3 | HF-BE3 | ave | 60–80% | −13 to −17 | [60] |
| S. coelicolor | ABE | CRISPR-aBEST | SCO5087 | 0–40% | −12 to −17 | [58] |
| S. coelicolor | ABE | ABEd | actVB | – | −14 to −17 | [60] |
| S. coelicolor | ABE | ABEn | actvB | 100% | −14 to −17 | [60] |
| S. hygroscopicus | BE3 | pWHU77-BE | hygD, hygL, hygJ, hygY, hygM | – | – | [151] |
| Staphylococcus | ||||||
| S. aureus | BE3 | pnCasSA-BEC | agrA, cntA, esaD | 100% | −13 to −17 | [57] |
| S. aureus | ABE | pABE | agrA, murR | 50–100% | −13 to −17 | [52] |
| Pseudomonas | ||||||
| P. aeruginosa | BE3 | pnCasPA-BEC | rhlR, rhlB | >90% | −13 to −18 | [56] |
| P. putida | cadR, ompR | >90% | ||||
| P. fluorescens | per, aspC | >84% | ||||
| P. syringae | gacA, hrpL | >90% | ||||
| Klebsiella pneumoniae | ||||||
| K. pneumonia | BE3 | pBECKP | fosA, dhak, blaKPC-2, blaCTX-M-65 | 25–100% | −13 to −18 | [55] |
| Clostridium beijerinckii | ||||||
| C. beijerinckii | BE3 | pCBEclos-opt | pyre, xylR, spo0A, araR | 40–100% | – | [53] |
| Brucella melitensis | ||||||
| B. melitensis | BE3 | BE3 | virB10 | 100% | – | [48] |
| Corynebacterium glutamicum | ||||||
| C. glutamicum | Target-AID | CRISPR/dCas9-AID | upp | 11.2% | – | [54] |
| C. glutamicum | CRISPR/nCas9(D10A)-AID |
Upp, rfp, ald, csp, ldhA, adhA, odhA, 94 transcription factor genes |
>48%, 41–100%b 23.3%c |
−16 to −20 | ||
Base editing system refers to the specific name of the editor used in the specific papers.
Dual loci targeting efficiency.
Triple loci targeting efficiency.
3.1. Escherichia coli
Escherichia coli is an extensively studied gram-negative bacterium with well-established genetic literature and an ever-expanding toolbox that includes expression vectors, fermentation techniques, biochemical and biofuels production strains, and CRISPR tools [41,[61], [62], [63], [64]]. It is the preferred bacteria for large scale production of chemicals such as fatty acids and 1,4-butanediol, and about 30% of approved therapeutics proteins are industrially produced using E. coli [[63], [64], [65], [66], [67]]. To enhance productivity, several gene-editing tools have been developed, however, none of the developed tools allow precise single base editing except BEs [68].
Zheng et al. used the BE3 system to develop pEcBE3 cytosine base editing system for E. coli [48]. The pEcBE3 consists of nCas9(D10A) fused to rAPOBEC1 and UGI. The editing system was used to induce a stop codon (TAA) by C-to-T conversion of Gln codon (CAA) with 100% efficiency in tetA in E. coli strain XL1-Blue. pEcBE3 system showed near 100% editing efficiency as it introduced base conversion at lacZ and rppH in E. coli [48]. Banno et al. [49] adopted another version of CBE, i.e., Target-AID which has a more distal activity window at −16 to −20 bases upstream of PAM compared to BE3, which shows activity at −12 to −16 bases upstream of PAM [22,23]. The authors initially used nCas9(D10A) but observed poor transformation efficiency and speculated growth defect or cell death similar to Cas9. Therefore, they adopted dCas9. The dCas9 was fused to PmCDA1, UGI, and protein degradation tag (LVA) developing dCas9-CDA-UL. The editing system successfully performed multiplex editing as six out of eight sequenced clones showed simultaneous editing of galK, xylB, manA, pta, adhE, and tpiA. dCas9-CDA-UL also successfully edited 41 loci simultaneously depicting the multiplex editing strength of the system [49]. However, increased Cas9 tolerance for nonseed mismatches increases the occurrence of unwanted C-to-T base substitutions [69,70], and it's G/C-rich PAM sequence requirement limits the BE applications [71]. Therefore, Chen and colleagues developed a Cas12a based CBE system (dLB-Cpf1-BE) by fusing catalytically inactive L. bacterium Cpf1 (dLb-Cpf1) with rAPOBEC1 [47]. They successfully achieved 44%–74% C-to-T substitution efficiency in E. coli [47]. This system widens the genome target area by CBE as Cas12a requires a T-rich PAM sequence (For example, TTTV) [72,73].
Apart from CBE, ABE has also been adopted for E. coli [52]. ABE7.10 system successfully edited yagl, murR, and lacZ in MG1655 and DH5α strains, achieving high editing efficiency between 66% and 100% [52]. Additionally, Zhang and colleagues developed a double-check base editing system, which uses the DSB capability of Cas9 to eliminate unedited cells [50]. They were able to increase editing efficiency by 3–6 folds at specific loci, compared to ABE [50]. This provides an efficient way to enhance ABE editing capabilities in E. coli.
3.2. Streptomyces
Streptomyces is a gram-positive GC-rich Actinobacteria consisting of over 500 species. It is known for its industrial applications, as it produces 39% of all microbial metabolites [74,75] and 70% of the current antibiotics [76]. Applying metabolic engineering to enhance biosynthesis requires the development of novel Streptomyces editing tools. However, like other Actinomycetes, very GC-rich (~70%) and instable genome make genome editing difficult [77,78]. Development of BE for Streptomyces expands its genome editing toolbox, helping the advancement of efforts to increase its utility.
Zhao et al. developed a Target-AID CBE system, i.e., dCas9-CDA-ULstr for Streptomyces [59]. The system consists of dCas9 controlled by tipAp leaky promoter fused with cytosine deaminase (CDA), UGI (U), and protein degradation tag (L). The editing system achieved 100%, 60%, and 20% editing efficiency with single, double, and triple target locus editing, respectively [59]. To expand the genetic toolbox of Streptomyces, Lee group developed two CRISPR-Base Editing SysTems, i.e., CRISPR-cBEST (CBE system) and CRISPR-aBEST (ABE system) consisting of nCas9(D10A) fused with either rAPOBEC1-UGI or TadA*, respectively [58]. The CRISPR-cBEST achieved 30%–100% editing efficiency in S. griseofuscus and S. coelicolor, while 0%–40% editing efficiency was reported by CRISPR-aBEST in S. coelicolor. Interestingly, the group reported varying editing efficiency of CRISPR-BEST depending on the adjoining base to the target base, showing 5′-TC > CC > AC > GC substrate priority [58]. In order to facilitate sgRNA designing for CRISPR-BEST, Weber group put forward an updated version of guide RNA design tool, i.e., CRISPy-web single guide RNA design tool [79].
The above described Streptomyces base editing systems are similar in the aspect that they efficiently perform C-to-T or A-to-G substitutions. However, they differ in the editing efficiencies which are described above, and the editing window. The Target-AID based, dCas9-CDA-ULstr performs C-to-T substitutions further upstream of the PAM site at −16 to −20, whereas CRISPR-cBEST performs CB editing at −11 to −17 upstream of PAM site [[58], [59], [60]] (Table 1). The variance in activity spectra of editing systems can be useful depending on specific experimental requirements.
3.3. Staphylococcus
Staphylococcus is a gram-positive facultative anaerobic, highly common human pathogen. It colonizes 30% of the human population and is a leading cause of bacteremia [80]. The major public health crisis is unfolding with the emergence of antibiotic-resistant Staphylococcus strains, such as methicillin-resistant strains (MRSA) [81]. Countering the crisis requires studying virulence factors, chronicity, and pathogenicity, but these studies have been hindered due to a lack of availability of genetic tools [82]. The existing genetic tools rely heavily on HDR, which makes them inefficient [83]. Enhancement in CRISPR tools and the development of BE have helped to engineer non-DSB and HDR-free genetic manipulation tools for Staphylococcus.
Ji group developed CBE [57] and ABE [52] systems for Staphylococcus. For CBE, pnCasSA-BEC was constructed by fusing nCas9(D10A) to rAPOBEC1 via XTEN linker [57]. pnCasSA-BEC was used in S. aureus to introduce premature stop codon by C-to-T substitution in agrA, cntA, and esaD with 100% editing efficiency between −13 and −17 bases upstream of PAM. The editing system was then tested in clinically isolated S. aureus strains such as MRSA, Newman, and USA300, achieving 100% editing efficiency. The group also performed bioinformatics analysis to calculate total editable sites in MRSA and Newman strains. The results showed that at least 95% of the genes possessed editable C(s), while ~70% of the genes contained editable stop codon in both the strains [57]. Moreover, an ABE system, pABE, was developed by the fusion of nCas9(D10A) with ABE7.10 [52]. pABE based A-to-G substitution resulted in 50%–100% editing efficiency between −13 and −17 bases upstream of PAM. The group used whole-genome sequencing of edited S. aureus RN4220 strains to study the off-target effect and reported no off-targeting at any of the potential off-target sites.
3.4. Pseudomonas
Pseudomonas belongs to the genus of gram-negative bacteria with 144 different species, largest among gram-negative bacteria [84]. Research in Pseudomonas has been expanding due to its biomedical and ecological importance, and biotechnological applications. For instance, P. aeruginosa, an opportunistic human pathogen, is the leading cause of morbidity in patients with cystic fibrosis, acquired immune deficiency syndrome, and cancer [85,86]. Whereas P. putida, a soil microbiome, has been engineered for the production of high-value chemicals and used for bioremediation of polluted sites [75,[87], [88], [89]]. While P. fluorescens, which inhabits the surface of plants, is engineered for the production of recombinant proteins [90]. CRISPR editing tools have been developed for Pseudomonas species to enhance their productivity [85,[91], [92], [93]]. However, the lack of effective DNA repair systems and high GC content (~62%) in some Pseudomonas species make it hard to efficiently use DSB-induced HDR-CRISPR tools [94].
Chan et al. developed pnCasPA-BEC, a CBE system for Pseudomonas species [56]. It was constructed by fusing nCas9(D10A) with rAPOBEC1 with XTEN linker. pnCasPA-BEC system was used to introduce stop codons in rhlR and rhlB with high efficiency (>90%) in P. aeruginosa strains. pnCasPA-BEC was then used to edit genes in P. putida, P. fluorescens, and P. syringae, demonstrating the system's ability to be applied in a wide range of Pseudomonas species. The authors reported the editing window to be −13 to −18 bases upstream of PAM and substrate preference to be TC ≥ CC > AC > GC similar to previously described CBE systems [56].
3.5. Corynebacterium glutamicium
Corynebacteria are gram-positive industrially important soil bacteria. They are widely used for the production of bioproducts. Especially, C. glutamicium whose ability to produce amino acids from sugar and ammonia has been utilized for industrial-scale production of several amino acids such as glutamate, lysine, isoleucine, tryptophan and threonine [95,96]. C. glutamicium has also been engineered to produce a wide variety of biochemicals, such as polymer subunits and biofuels [97,98]. The engineering of C. glutamicium is laborious and challenging as the editing tools show low recombination efficiency and lack of positive selection for mutations. Therefore, an efficient editing tool that allows robust and easy genetic engineering is needed.
Wang et al. adopted Target-AID to develop an automated base editing method, i.e., MACBETH for C. glutamicium [54]. The editing system consists of nCas9(D10A) fused to AID under tac promoter and a sgRNA cassette under a strong constitutive PcspB promoter. The editing process was automated by using an integrated robotics system for plasmid construction and PCR amplification, and automated high-throughput colony picker for plating and colony picking. The editing system successfully performed single, dual, and triple locus editing with editing efficiencies between 23% and 100%. Additionally, the authors carried out genome-wide bioinformatics analysis for C. glutamicium ATCC 13032. They reported that the editing system could introduce an early stop codon in 88% of the total 3099 genes.
3.6. Other species
Base editing has been developed for a few more bacterial species like Klebsiella pneumonia [55] and Clostridium beijerinckii [53].
K. pneumonia is a human pathogen and an industrially relevant bacterium. Research in K. pneumonia has become increasingly significant because of the increase in its pathogenicity due to the emergence of multi-drug resistant strains. K. pneumonia strains resistant to carbapenems and extended-spectrum β-lactams (ESBL) have emerged in various parts of the world [99,100]. Ji group developed a CBE system for K. pneumonia, pBECKP, a single-plasmid editing system consisting of nCas9(D10A) fused with rAPOBEC1 via XTEN linker, and sgRNA under the control of strong constitutive promoter (J23119) [55]. pBECKP system was used to introduce a premature stop codon in fosA in a clinically isolated strain of K. pneumonia with 100% editing efficiency. The group successfully tested the ability of the editing system to edit a GC-rich spacer in dhak. Moreover, the group enhanced the carbapenems susceptibility of carbapenem-resistant K. pneumoniae strain's KP-CRE23 by effectively using pBECKP system to inactivate carbapenemase gene, blaKPC-2 [55].
C. beijerinckii is an industrially important gram-positive solventogenic bacterium. It produces biochemicals such as acetone, butanol, ethanol (ABE chemicals) utilizing various carbon sources [[101], [102], [103]]. Interestingly, during the fermentation of pentose sugar, C. beijerinckii does not show glucose repression effect in the presence of glucose, which makes it more attractive than other Clostridium species [104]. Li et al. developed a CBE system, pCBEclos-opt for C. beijerinckii [53]. pCBEclos-opt is a single plasmid editing system consisting of nCas9(D10A) fused with rAPOBEC1 and UGI, and sgRNA expression cassette. pCBEclos-opt was used to disrupt pyrE in C. beijerinckii leading to uracil analog 5-FOA resistance, which was used as a selective marker. Half of the colonies were edited in pyrE and other gene sites araR, xylR, and spo0A [53].
4. DSB & template-free editing in plants
Apart from bacteria, BE has been adopted for several plant species and more recently prime editors have been developed for plants to carry out efficient and precise editing. The DSB-induced HDR editing techniques have been applied in plants to improve the crops. However, its applications have been limited as most acceleration instances in agriculture are conferred by single-nucleotide mutations. The low frequency of HDR in plants due to their gene-repair system [105] and the installation of undesired by-products by the DSB-induced Cas9 system [106] render the editing technique an unfeasible approach. The adoption of BEs and PEs in the primary DNA sequences has enabled the precise engineering of crops with novel characteristics (Table 2).
Table 2.
Base editing and prime editing systems in plants.
| Organism | Editor type | Base editing systema | Target genes | Editing frequencyb | Editing window (upstream of PAM) | Ref. |
|---|---|---|---|---|---|---|
| Rice | BE3 | pnCas9-PBE | OsCDC48, OsNRT1.1B, OsSPL14 | 43.48c 0.39–7.07%f |
−18 to −12 | [108] |
| Rice | BE2 | APOBEC1-XTEN-Cas9(D10A) | NRT1.1B, SLR1 | ≤13.3%c | −17 to −14 | [111] |
| Rice | BE3 | APOBEC1-XTEN-Cas9n-UGI-NLS | OsCERK1, OsSERK1, OsSERK2, ipa1 | 10–38.9%c | −19 to −13 | [113] |
| Rice | BE4 | APOBEC1-XTEN-Cas9n (VQR)-UGI-NLS | pi-ta | 18.2%c | −19 to −13 | |
| Rice | Target-AID | nCas9Os-PmCDA1At | ALS, FTIP1e | 6–89%c | −19 to −17 | [109] |
| Rice | ABE | PABE | OsALS, OsCDC48, OsAAT, OsDEP1, OsACC, OsNRT1.1B, OsEV, OsOD | 3.2–59.1%c | −17 to −13 | [114] |
| Rice | ABE | TadA-TadA7.10-Cas9n | OsMPK6, OsMPK13, OsSERK2, OsWRKY45 | 0–62.26%c | −17 to −14 | [115] |
| Rice | ABE | pRABEsp-OsU6 | OsSPL14, SLR1, OsSPL16, OsSPL18, LOC_Os02g24720 | >4.8%c | −17 to −14 | [161] |
| Rice | ABE | pRABESA-OsU6sa | OsSPL14, OsSPL17, OsSPL16, OsSPL18 | >17%c | – | |
| Rice | ABE | pHUN411-ABE | Wx, GL2/OsGRF4, OsGRF3 | <10%f | −16 to −12 | [116] |
| Rice | APOBEC3A | A3A-PBE | OsAAT, OsCDC48, OsDEP1, OsNRT1.1B, OsOD, OsEV | 44–83%c | −20 to −4 | [142] |
| Rice | hAID∗Δ | hAID∗Δ-XTEN-Cas9n-NLS | OsRLCK185, OsCERK1, Pi-d2, OsFLS2, OsAOS1, OsJAR1, OsJAR2, OsCOI2 | – | −18 to −14 | [112] |
| Rice | ABE | VQR-Cas9 (D10A)/VRER-Cas9 (D10A) | OsSPL14, OsSPL17, OsSPL16, OsSPL18, OsIDS1, OsTOE1 | ≤74.3%c | −19 to −11 | [162] |
| Rice | ABE | SaKKH-Cas9 (D10A) | SNB | 6.5%c | – | |
| Rice | BE | VQR-Cas9 (D10A) | PMS3 | ≤61.1%c | – | |
| Rice | BE3 | xCas9(D10A)-rAPOBEC1 | OsDEP1 | ≤30%f | −19 to −16 | [175] |
| Rice | Target-AID | xCas9(D10A)-PmCDA1 | OsDEP1 | <20%c | −19 to −16 | |
| Rice | Target-AID | Cas9-NG (D10A)-PmCDA-UGI | OsDEP1, OsCDC48, OsPDS | 0–56.3%c | −20 to −7 | |
| Rice | Target-AID | NGv1 (D10A) | EPSPS, ALS, DL | 5–95.5%c 4.2–86.3%f |
−20 to −9 | [176] |
| Rice | BE | NGv1 (D10A) | EPSPS, ALS, DL | 4.3–21.8%f | −19 to −14 | |
| Rice | BE3 | pCXUN-BE3 | OsPDS, OsSBEIIb | 0.1–20%c | −17 to −13 | [110] |
| Rice | BE3 Target-AID ABE |
eBE3, eCDA, eABE | OsACC | – | – | [149] |
| Rice | BE3 ABE |
Base-Editing-mediated Gene Evolution (BEMGE) | OsALS1, OsALS2, OsALS3 | – | – | [150] |
| Rice | BE3 Target-AID |
xCas9-epBE | OsMPK2, OsMPK5, OsMPK5, OsALS, OsNRT1.1B | 5–64.3%c | −20 to −10/-7 | [163] |
| Rice | BE4 ABE |
xCas9n-CBE, Cas9n-NG-CBE, eCas9n-NG-CBE xCas9n-ABE, Cas9n-NG-ABE, eCas9n-NG-ABE |
OsWaxy, OsEUI1, OsCKX2 OsWaxy, OsEUI1, OsCKX2 |
9.1–45.5%c 2–6.5%c |
−18 to −13 | [177] |
| Rice | ecTadA∗7.10-nCas9 | ABE-P1S | OsSPL14, SLR1, OsSERK2, Tms9-1, OsNRT1.1B, OsACC1, OsDEP1 | 11.4–96.3%c | −20 to −9 | [168] |
| Rice | ecTadA∗7.10-nSaCas9, | ABE-P2S | SPX-MSF2, OsSPL14, OsSPL17, OsSPL14, OsSPL17, OsSPL16, OsSPL18 | 15.9–61.1%c | −20 to −18 | |
| Rice | ecTadA∗7.10-nSaKKH-Cas9 | ABE-P5S | OsSPL13, SNB | 6.1–33.9%c | −17 to −10 | |
| Rice | BE3 | nCas9-PBE | GL1-1, NAL1 | 58–68%c | −18 to −12 | [125] |
| Rice | ABE | ABE7.10-nSpCas9-NGv1 | sgOs-siteG1, sgOs-site2, sgOs-site3, sgOs-site4 | 29.2–45.8%c | −16 to −13 | [178] |
| Rice | ABE | pPUN411-HABE, pPUN411-ABEH | Pid3, WX | >97.9%c | – | [117] |
| Rice | PE | PPE2, PPE3, PPE3b | OsCDC48, OsALS, OsDEP1, OsEPSPS, OsLDMAR, OsGAPDH, OsAAT, TaUbi10, TaGW2, TaGASR7, TaLOX2, TaMLO, TaDME1 | ≤21.8%c | – | [118] |
| Rice | PE | pPE2, pPE3, pPE3b | OsPDS, OsACC1, OsWx | 0–31.3%c | – | [121] |
| Rice | PE | PE-P1, PE-P2 | OsALS, OsACC, OsDEP1 | ≤26%c | – | [119] |
| Rice | PE | pCXUN-Ubi-NLS-nCas9(H840A)-Linker1 (33aa)-M-MLV-RT-Linker2 (14aa)-NLS-PolyA-E9-Actin-Nos | hptll, OsEPSPS | 2.22–9.38%c | – | [120] |
| Rice | PE | Sp-PE2, Sp-PE3, Sa-PE3 | ALS, APO1, SLR1, OsSPL14, APO2 | 0–17.1%c | – | [122] |
| Wheat | BE3 | pnCas9-PBE | TaLOX2 | 1.25%d | −18 to −12 | [108] |
| Wheat | ABE | PABE | TaDEP1, TaGW2 | 0.4–1.1%c | −17 to −13 | [114] |
| Wheat | APOBEC3A | A3A-PBE | TaALS, TaMTL, TaLOX2 | 16.7–22.5%d | −20 to −4 | [142] |
| Wheat | BE3 | PBE | TaALS-P174 | 33–75%d | −18 to −12 | [141] |
| Maize | BE3 | pnCas9-PBE | ZmCENH3 | 10%c | −18 to −12 | [108] |
| Tomato | Target-AID | nCas9At-PmCDA1Hs/nCas9At-PmCDA1At | DELLA, ETR1 | 41–92%c | −20 to −18 | [109] |
| Tomato | Target-AID | pDeSpnCas9-NG_PmCDA1_UGI | SIALS1 | 32%c | −20 to −13 | [179] |
| Potato | Target-AID | pDeSpnCas9-NG_PmCDA1_UGI | StDMR6-1, StGBSSI | 9–64%c | −18 to −16 | |
| Potato | APOBEC3A | pDeSpnCas9-NG_hAPOBEC3A_PmCDA1_UGI | StDMR6-1, StGBSSI | 8–42%c | −18 to −11 | |
| Potato | APOBEC3A | A3A-PBE | StALS, StGBSS | 6.5%e | −20 to −4 | [142] |
| Potato | Target-AID | – | SlALS1 | 100%c | – | [128] |
| Arabidopsis | ABE | pcABEs | AtALS, AtPDS, AtFT, AtLFY | 0–85%c | −16 to −12 | [127] |
| Arabidopsis | BE3 | BE3 | ALS | 2.7–40%c | −17 to −12 | [124] |
| Arabidopsis | BE3 | nCas9-PBE | MTA | 39.3%c | −18 to −12 | [125] |
| Arabidopsis | BE3 | BE3 | eIF4E1 | 50%c | – | [126] |
| Rapeseed | ABE | pcABEs | BnALS, BnPDS | 8.8% e | −16 to −12 | [127] |
| Watermelon | BE3 | CBE3 | ALS | 23%c | −14 to −13 | [130] |
| Cotton | BE3 | GhBE3 | GhCLA, GhPEBPc | 26–58%c ≤18.63%f |
−17 to −12 | [129] |
| Soybean | BE3 | pTF101.1-sgRNA-BE | GmFT2a, GmFT4 | ≤18.2%c | – | [180] |
| Oilseed rape | BE3 | CBE | BnALS1 | 1.8%c | – | [181] |
Base editing system refers to the specific name of the editor in the specific papers.
Editing frequency refers to the ratio of edited to unedited plants.
Agrobacterium mediated system.
Particle bombardment.
Protoplast transformation.
Efficiency in targeted sequence.
4.1. Rice, wheat and maize
Rice, wheat, and maize are major monocot cereal crops that collectively account for about 50% of the total world's calorie consumption. This makes it critical to carry out research in them. CRISPR tools have been widely developed for these crops for the enhancement of bio-production and trait improvement [107]. However, BE and PE are more reliable, facile, and efficient tools compared to DSB-dependent CRISPR tools, and they have been effectively applied in rice, wheat, and maize (Table 2).
Gao group developed a CBE system composed of rAPOBEC1 and nCas9(D10A) for rice, wheat, and maize, and reported prominent editing efficiency [108]. The editing system showed 0.39–7.07% C-to-T substitutions in which the highest frequency appeared at or near −13 upstream of PAM with very few indels (0.01–0.22%). They attempted to use dCas9 instead of nCas9 in CBE; however, it led to decreased editing efficiency (0.00–1.29%). In the base-edited plants, 40 out of 92 transformed plants (43.48%) revealed at least one substitution in the editing window. At the same time, Kondo group also published a study in which they successfully applied Target-AID for point mutagenesis in rice [109]. They used the ALS A96 target to develop herbicide imazamox (IMZ) resistant rice by the C-to-T substitution and two other target sites (G590 and W483) of FTIP1e (LOC_Os07g30020) of rice to test the CBE system. Finally, 8 of 66 clones were obtained, which carried resistance to IMZ and had both point mutations in FTIP1e. Similarly, BE3 was reported by Li et al. that could introduce precise point mutations in rice with minimal indels [110]. Lu et al. gained a frequency of 1.4%–11.5% in which the highest editing efficiency location was at or near −13 upstream of PAM, targeting NRT1.1B and SLR1 in rice [111]. Moreover, human AID (hAID) was used to enhance CBE editing efficiency at GC-rich regions in rice [112]. The authors chose the hAID variant (K10E + T82I + E156G) to construct their hAID*Δ-XTEN-Cas9n-NLS chimeric complex, which showed high editing efficiency in GC-rich regions. However, the hAID*Δ-XTEN-Cas9n-NLS system led to more undesired indels compared to rBE3 and rBE4 [113].
Apart from CBE, Gao and colleagues constructed the ABE system, i.e., PABE system consisting of the wtTadA-TadA* adenosine deaminase, nCas9, three NLSs and esgRNA (sgRNA with improved expression) to generate A•T-to-G•C conversions in rice and wheat [114]. They achieved up to 7.5% A-to-G substitution frequency in protoplasts and 15.8%–59.1% editing efficiency in regenerated plants. Yan et al. reported that ABE could gain 16.67% efficiency at OsMPK6 site in rice [115]. Furthermore, Wei and his group also reported an ABE system but observed the point mutation frequency of <10% [116]. They increased the ABE editing efficiency by double selection (selection of edited cells using hygromycin and herbicide) [117], in which >50% plants were edited.
Besides BE, the PE system has also been introduced in rice and wheat. Lin et al. used a maize Ubiquitin-1 (Ubi-1) promoter to develop the fusions of Cas9(H840) and M-MLV RT, CaMV RT, or RT-retron [118]. They reported that PPE3 and PPE3b had the same efficiency as PPE2, and obtained up to 21.8% regenerated prime-edited rice plants. Xu et al. [119] and Li et al. [120] adopted PE3 in rice T0 lines and rice calli, respectively, with the former achieving 26% frequency compared to 9% for the latter. Meanwhile, Wei group reported that PE2 could generate 0%–31.3% editing frequency in T0 plants HPT−ATG reporter in rice while the PE3 systems did not show preferential editing efficiency [121]. Apart from this, Zhu and his group replaced SpCas9(H840A) with SaCas9(N580A) to enhance PE3 efficiency, but Sa-PE3 exhibited a lower editing activity compared to Sp-PE3 [122].
4.2. Arabidopsis
Arabidopsis is the most thoroughly studied model flowering plant. Since the realization of the requirement of an efficient plant model organism, Arabidopsis has been seen as a go-to plant. Its fast life cycle, ease of handling, and limited minerals requirement make it a flawless model organism. Several decades of research have led to the development of a range of genetic tools. Since the first reported CRISPR-Cas based editing in Arabidopsis [123], several groups have developed CRISPR tools for it [107]. Plenty of groups have expanded Arabidopsis genetic toolbox using BE.
Jiang group developed the CBE system (BE3) in Arabidopsis [124]. They used Agrobacterium-mediated transformation to introduce the editing system in Arabidopsis. BE3 was used to confer herbicide resistance by C-to-T conversion in the ALS at Pro197. A low editing efficiency (1.7%) was achieved in the T1 generation, and the resistance was successfully passed onto the next generation. In another study, BE3 was used for gene inactivation by mRNA mis-splicing [125]. Maize Ubiquitin-1 (ZmUbi) promoter-driven nCas9-PBE system was used to introduce C-to-T mutation at the donor site of Arabidopsis MTA intron 1 which led to the development of dwarfism (heterozygous mutant) and embryo-lethality (homozygous mutant) phenotype. The study reported successful mutation in 39.3% of transgenic T1 plants [125]. CBE typically leads to C-to-T mutations; however, at times, it can also produce C-to-G or C-to-A mutations. Bastet et al. used this ability to develop Arabidopsis with resistance against the potyvirus family by editing elF4E [126].
Moreover, Kang et al. were first to report that ABE (ABE7.10) in Arabidopsis [127]. They initially directly introduced the editing system (pcABE7.10) into the Arabidopsis protoplast and achieved editing efficiency of up to 4.1% and reported insignificant off-targeting at the potential off-target sites. They later developed phenotypically variant Arabidopsis by targeting FT to develop late-flowering phenotype. They reported that as a promoter, RPS5A led to more efficient A-to-G conversions compared to the 35 S promoter-ABE system [127].
4.3. Other species
Base editing has been applied to some other agriculturally and economically significant plant species (Table 2). Shimatani et al. applied codon-optimized Target-AID in tomato and demonstrated the nucleotide substitutions in T1 plants, with rare off-target mutations (0.14–0.38%) [109]. They tested the on-target frequency by introducing C-to-T or C-to-G substitutions in DELLA and reported editing frequency to be 26.2%–53.8%. A CBE system (fusion of PmCAD1 and nSpCas9) was developed in tomato and potato and produced 12.9%, and 10% C-to-T mutated transgene-free plants in the first generation, respectively [128].
BE3 system has been adopted for the cotton crop by Qin et al. [129]. They developed a GhBE3 editing system consisting of nCas9(D10A) fused to cytosine deaminase and UGI domain. It was used to edit GhCLA and GhPEBP with mutation frequencies between 26.67% and 57.78% in T0 plants, and off-targeting as low as 0.1%. Xu and his team engineered the ability of herbicide resistance in watermelon (up to 23% in T0), using CBE3 by converting C-to-T in the codon of Pro190 (CCG) of ALS gene [130].
5. Applications: current & future
CRISPR-Cas editing has evolved exponentially since its discovery as an adaptive immune system of bacteria. Development of BE and more recently PE in plants and bacteria (to date, PE is not reported in bacteria yet) have enhanced their genetic toolbox and provided the ability to perform precise and efficient nucleotide substitutions at target locus (Table 1, Table 2). These tools can be exploited to develop efficient strategies to enhance bio-production and advance the establishment of bacteria and plants as major bio-production platforms.
5.1. Synthetic biology
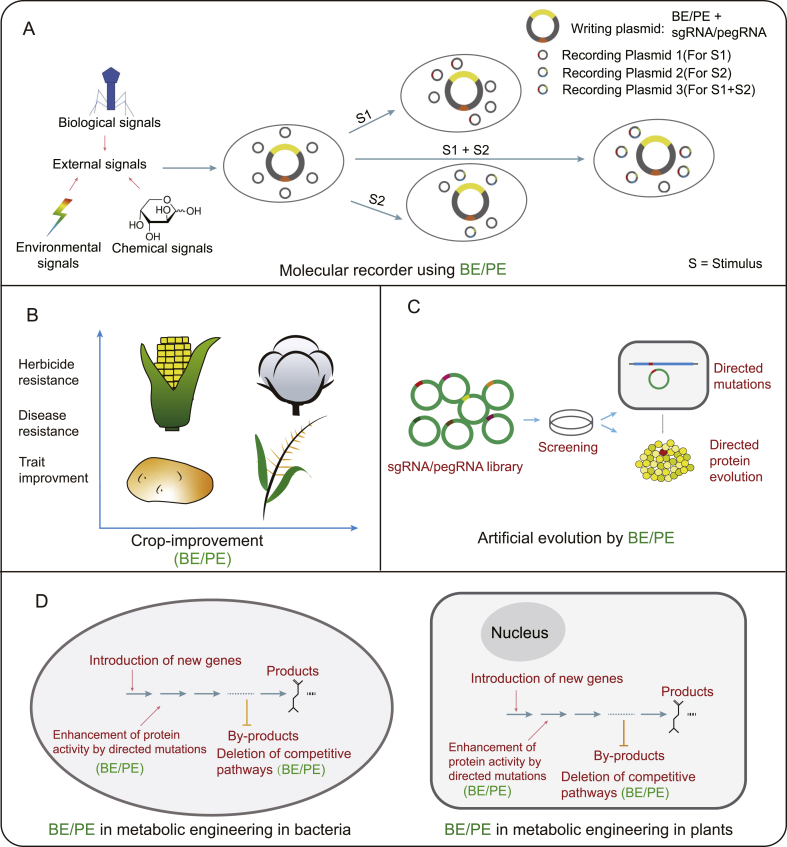
Technologies to incorporate and record biological information in living cells have started to emerge recently [[131], [132], [133]]. BEs can be exploited to develop synthetic devices for external stimulus, event, and memory recording (Fig. 3A). Liu and Tang demonstrated the use of BE in E. coli in an analog event recorder, CRISPR-mediated analog multi-event recording apparatus (CAMERA2) [134]. They incorporated the BE2 system in a writing plasmid in which BE or guide RNA (gRNA) was under stimulus-dependent promoter induced by an exogenous signal. Upon the introduction of the exogenous signal, the dCas9/gRNA are transcribed, leading to base editing in the recording plasmid. Detection of base editing at the target site in recording plasmid represents the presence of the specific stimulus. CAMERA2 showed the ability to respond and record multiple independent stimuli, as incorporating multiple small-molecule responsive guide RNAs into a single writing plasmid led to the detection of targeted editing corresponding to the relative stimulus. Apart from detecting the presence of small-molecule dependent stimuli, other external stimuli like light, antibiotics, nutrient, and phage infection can be detected via CAMERA2 [134].
Fig. 3.
Base editing and Prime editing applications (current & future). (A) Molecular recorders use BE to carry out nucleotide substitution relative to a specific stimulus. The base editing takes place in the presence of an external signal. The output signal either be phenotypic which can be recorded in the real-time or genotypic read via sequencing. DNA-writing technology can be expanded by the incorporation of PEs. (B) BE and PE can play a critical role in crop improvement. It has already been used for herbicide and disease resistance and trait improvement. (C) Artificial evolution can be achieved by BE/PE via the development of sgRNA/pegRNA libraries. UGI deprived CBE can lead to versatile mutations like C-to-G and C-to-A, this ability can be exploited for direct protein evolution. (D) Metabolic engineering in plants and bacteria can be improved by enhancing the enzymatic activity and deleting the competitive pathways by BE/PE tools.
Another molecular recorder was put forward by Farzadfard et al., a read and write DNA-state reporter genetic circuit, DNA-based Ordered Memory, and Iteration Network Operator (DOMINO) [135]. Similar to CAMERA2, DOMINO uses base editing, CDA-nCas9-UGI system, which results in C-to-T substitution at the target site for permanent DNA recording. In the circuit, CBE-nCas9-UGI expression is controlled by small-molecule induced promoter termed as operational signal while the small-molecule inducing gRNA is termed as the input signal. The introduction of an operation signal enables reading, while the introduction of the input signal enables writing. The device allows independent, sequential, and temporal operation of logic and memory operations. DOMINO system can also provide phenotypical readouts to the input signal as demonstrated by the use of GFP, where the sequential addition of operational and then input signal led to the exclusion of stop codon at target sites [135].
The readable, writeable, and recordable systems can play a vital role if developed for industrially significant bacterial strains where overtime accumulation of by-products can affect the yield. Such stimulus detecting systems can help enhance productivity by early detection of by-products. For example, E. coli strains are widely used for the industrial-scale production of several therapeutic proteins; however, protein production is affected by the accumulation of acetate [136,137]. The CAMERA2 and DOMINO systems can be used for dose and time-dependent recording of acetate accumulation by using acetate induced promoters like glnAP2 promoter [138]. Moreover, BE dependent recording systems could also work as a bacterial synthetic population quality control device to record and detect unwanted variations and production of undesired molecules in real-time [139]. Such techniques may have broad industrial utility.
Besides industrial applications, BE or PE-based DNA writers and molecular recorders could be adopted for medical purposes. Their non-DSB and template-free feature make them a perfect candidate for medical use. DNA recorders with the ability to detect disease-specific molecular stimuli may lead to the development of ingestible live biosensors [133]. These live biosensors could assist in the early diagnosis of major diseases like cancer and Parkinson's. The live biosensors could also be programmed to detect environmental signals like chemicals and toxins, thus offering a whole new direction to such technology. In the future, we expect to see more DNA-writing technology and especially information recording and storage systems, with the use of not just BE but also PE. PEs dependent analog and digital information storage systems may be a game-changer in the field for the fact that they can insert arbitrary sequences.
5.2. Crop improvement
BE systems have widely been used in agriculture, in which single-nucleotide mutations generally lead to the acceleration of the crops in most cases. With the functional SNPs, crops could have a significant improvement in their traits directly related to bioproduction, such as growth rates, yield, and sensitivity to environmental stress (Fig. 3B) [140]. BEs can be used to introduce such desired SNPs to introduce preferred traits without the concern of linkage drag. BEs can also be utilized to produce herbicide-resistant crops that have superiority in agriculture, as demonstrated in plenty of studies [108,109,114,124,130,141,142]. Besides herbicide-resistant, disease-resistant crops are another popular trend, for that the plants would have the ability to impart resistance if the allelic pseudogenes are corrected [27]. Specific disease resistance targets like elF4E (resistant alleles provide resistance to a wide range of single-stranded RNA viruses) [143] require several amino acid changes to develop resistance to a wide range of viruses [144]. Studies favor the use of resistant alleles that can still encode for functional translational initiation factors over loss-of-function alleles, which lead to a narrower resistance spectrum [[144], [145], [146]]. Achieving multiple amino acid changes using DSB-dependent genome editing tools and mutagenesis techniques can be challenging. Fortunately, BE provides a better technique to carry out such mutations. Therefore, Bastet et al. adopted CBE to develop enhanced resistance to potyviruses in Arabidopsis [126]. The study emphasizes the utilization of BE in developing disease-resistant crops. Despite some limitations, like targeting site, editing window, or off-target editing, BE/PE systems have powerful potential to facilitate crop bio-production or improvement through high precision and efficiency.
5.3. Evolutionary engineering
BE could play a vital role in the introduction of gene mutations and directed protein evolution. CBE lacking UGI has been shown to have the capacity to make diverse mutations other than C-to-T [22,147,148]. This ability has already been used in CRISPR-X and targeted AID-mediated mutagenesis (TAM) based studies to identify known and novel mutations in mammalian cells in cancer therapeutics targets PSMB5, and BCR-ABL, respectively [147,148]. The identified mutations produce PSMB5 and BCR-ABL variants that are resistant to widely used cancer treatments like bortezomid and imatinib, respectively. Similarly, BE systems have effectively been used in plants to enable protein evolution, e.g., screening of ACCase variants [149] and genomic diversification, e.g., accelerated evolution of OsALS1 [150] with gRNA library. Development of bacterial or plant protein and genetic libraries using UGI absent CBE or PE could provide functionally diverse protein variants with industrial importance (Fig. 3C). A bioinformatics study into targeted point mutations using BE/PE that leads to variation in protein activity and, ultimately, metabolic pathways, may widely assist in bacterial and plant metabolic engineering.
5.4. Metabolic engineering
The precise editing ability of BEs has been exploited for metabolic engineering studies to understand the biosynthesis pathways in bacteria (Fig. 3D). Li et al. studied the Hygromycin B biosynthesis pathway in the industrially significant S. hygroscopicus subsp. hygroscopicus bacterium [151]. The group used BE for in vivo gene inactivation to enhance the understanding of the pathway. They studied HygD, HygJ, HygL, HygY, and HygM, and found their critical roles in the pathway [151]. This work demonstrates the significance of base editing tools in gene characterization of biosynthesis pathway studies. Additionally, Ji group used pABE system for S. aureus to screen for key residues of CntBC, an essential transporter that plays a role as a transporter for staphylopine/transition metal complex acquisition [52]. Using the pABE-based screening method, authors were able to identify four key residues of CntBC. This depicts the efficacy of BEs in studies related to the identification of functional protein residues. Similar studies into industrially relevant bacterial biosynthesis pathways using BE will enhance our understanding of bio-production networks and assist in improving the yield. Metabolic pathways in bacteria are vastly interconnected as the central and branched pathways form a complex network for synthesizing and breaking of the molecules. Such interrelated networks make it tough to tweak metabolic flux to achieve the highest yield of target products. The BE/PE could assist in a better understanding of how the network affects yield with high precision at a broader scale, eventually facilitating enhanced biosynthesis.
Plant as workhorse has a more complex cell system compared to the bacteria, facilitating more complicated biosynthesis [152]. Besides introducing new metabolic pathways and enhancing related enzyme activities, knock-out/down of competitive pathway branched genes [153] is critical in plant metabolic engineering (Fig. 3D). By destroying the competitive polyunsaturated fatty acid FAD2 alleles using DSB-dependent CRISPR-Cas9, Jiang et al. increased the oleic acid content in Camelina seeds from 16% to >50% of the fatty acid composition [154]. In their research, less desirable polyunsaturated fatty acids still occupied a certain percentage, probably because of the rudimental N-terminal FAD2 domains. Such an issue can be resolved by BE system, as demonstrated by the introduction of premature stop codons (iSTOP or CRISPR-STOP) [155,156] to generate catalytically inactive proteins. Hence, BE and PE have the capacity to play a leading role in plant metabolic engineering.
5.5. RNA base editing
Contrary to DNA editing, RNA editing, including insertion, deletion, or substitution of nucleotides, allows accurate and efficient editing of RNA molecules. The development of RNA base editors exponentially expands the available RNA editing tools by allowing precise single base substitution.
In mammals, the most prevalent post-transcription RNA editing case is catalyzed by the adenosine deaminase enzymes (ADARs). ADAR binds to dsRNA and catalyzes adenosine to inosine (A-to-I), and ultimately, I is read as G by the cellular machinery [157]. Recently, RNA-guided RNA-targeting CRISPR nuclease C2C2 (later named as Cas13a) from Leptotrichia shahii was illustrated [158]. Cas13 generally encompasses two higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains, which contribute to the RNA-targeted nucleolytic activity. Mutations of HEPNs abolish RNA cleavage activity while maintaining RNA targeting activity, which was used by Zhang group to create an RNA base editing tool, namely, RNA Editing for Programmable A-to-I Replacement (REPAIR) [159]. They combined a catalytically inactive dCas13 variant with a RNA deaminase ADAR2 (E488Q) to form a fusion protein dCas13-ADAR2DD, which can execute RNA editing for programmable A-to-I (G) replacement. To expand the RNA base editors, a new tool, RNA Editing for Specific C-to-U Exchange (RESCUE) was later developed [160].
Till now, neither REPAIR nor RESCUE systems have been adopted for plants or bacteria. The systems can be safely used in cells for the fact that they do not lead to permanent genomic DNA mutations, which may be used for research into understanding the metabolic pathways and crop improvement.
6. Concluding remarks, challenges & prospects
Availability of the tools that allow precise, efficient, and DSB-free editing ability to resolution as low as a single base was unthought-of until recently. Development of BE and more recently PE have handed scientists with powerful gene-editing tools. The formidable tools have been deployed in diverse fields, from synthetic metabolic engineering to agriculture and medicine. Today with ever-increasing research in life-sciences, these tools have quickly been adopted for animal, plant, and bacterial models. Exploitation of these tools is required to further advance the bacterial and plant bio-production. Rapid advancements in the fields make it hard to keep up with the studies; therefore, this review summarizes the developments in BE and PE technology in bacteria and plants.
Though BE and PE editing tools are seeing an ever-increasing interest, there remain challenges that require further research. One major challenge surrounding BE and PE is their constrained target sites due to the PAM specificity of Cas proteins. BE fused to different Cas proteins like SaCas9(D10A) (PAM recognized, NNGRRT) [161], VQR-Cas9(D10A) (PAM recognized, NGA, AGTG, AGCG) [162], VRER-Cas9(D10A) (PAM recognized, NGCG) [162], SaKKH-Cas9(D10A) (PAM recognized, NNNRRT) [162], xCas9 (PAM recognized, NG and more) [163] and Cas12a (PAM recognized, TTTV and more) [47] have been developed, however, they are still limited to narrow PAM accessibility. The recently developed Cas proteins, SpG (PAM recognized, NGN) [164], SpRY (PAM recognized, NRN (R being A or G) > NYN (Y being C or T)) [164], Sc++ (PAM recognized, NNG) [165] and HiFi-Sc++ (PAM recognized, NNG) [165] are reported to have significantly relaxed PAM dependence, providing the ability to target previously inaccessible locus. The fusion of SpG/SpRY/Sc++/HiFi-Sc++ Cas proteins with either BE or PE will help advance the available editing tools.
The next limitation surrounding BE/PE is editing efficiency. Although BE has been fused to several Cas proteins, their efficiency remains lower than the original BE. Liu group has worked on developing advanced base editors with higher Cas compatibility and increased editing efficiencies like ABE8e, which is reported to have higher editing efficiency compared to the original ABE7.10 [166], and fourth-generation CBE BE4 [167]. Hua et al. enhanced the efficiency by simplifying the adenine deaminase in rice [168]. Contrarily, PEs are still very young and therefore have limited studies. Lin et al. adopted PEs for plants and used different RTs in the hope of increasing efficiency [118]. They used RT-CaMV and RT-retron but reported either comparable or lower editing efficiency. Further studies into engineered and evolved RTs will help in the development of advanced PEs with higher editing efficiencies.
Another challenge facing BE/PE is off-target editing. BE and PE allow precise editing with lower indels and off-target effects compared to DSB-dependent CRISPR editing. CBE and ABE lead to 1.1% and ≤0.1% indels, respectively, compared to much higher 4.3% indels from Cas9-HDR editing [21,22]. Research into the characterization of the observed undesired indels, bystander, and off-target editing as either because of BEs or the Cas protein is required. CBE shows more off-target mutations than ABE [161,[169], [170], [171]]. Therefore, Doman et al. studied the Cas9-independent off-targeting by CBE and engineered CBE variants, which show a considerably lower level of Cas9-independent off-targeting mutations (~10–100 folds lower) and comparable or lower indels [51]. To limit the Cas9 dependent off-targeting, high-fidelity Cas9 was fused to BE3 and BE2 to develop HF-BE3 and HF-BE2, respectively [172]. Both of the developed HF-BEs showed several folds lower off-targeting, with HF-BE3 showing 37-fold lower off-targeting relative to BE3. Other efforts to reduce BE off-targeting include co-expression of free UGI with BE3 containing triple UGI [173] and fusion of bacteriophage Gam protein with BE3 and BE4 [167]. These studies depict that the uracil base excision repair system (BER) has an important role in BE induced off-target effect as multi-copy UGI based systems generally lead to lower off-targeting. Further studies into off-target characterization and understanding the mechanisms that lead to it are required. In the case of PE, the initial work shows significantly lower off-targeting compared to DSB-dependent Cas9 editing [24]. PE, on average showed ≤0.6% off-target changes at four major Cas9 off-target loci compared to an average 32% off-targeting by Cas9+sgRNA at the same four loci [24]. However, further research in the genome-wide off-target analysis is required to better understand the PE off-targeting as it may show undesired mutations at previously unknown off-target sites.
Other challenges facing base editing include sequence specification and editing window. Studies have reported 5′-TC ≥ CC > AC > GC sequence preference by BE3 [22,47,58]. Whereas, the editing window of BEs typically limits the editing within −12 to −16 and −16 to −20 distal to the PAM. Efforts to increase the editing window include the use of extended guide RNA [49], the development of advanced BE systems like BE-PLUS [174], and using human APOBEC3A (hA3A) in CBE [142]. We believe that studies into developing engineered or evolved BEs with lower sequence specificity and the wider or narrower editing windows will be important for future BE applications. Whereas PE requires further work into understanding its working mechanism and in-depth study of edit range.
Base editing and prime editing are young editing tools poised to play a critical role in basic research, crop improvement, synthetic biology, metabolic engineering, and evolutionary engineering. Both the editing tools require extensive research to reach their full potential, especially PE, whose adoption in bacteria/plants will be crucial for advancement of their bio-production capabilities.
Declaration of competing interest
None.
Acknowledgments
This work was sponsored by National Key R&D Program of China (2018YFA0901200), Science and Technology Commission of Shanghai Municipality (18JC1413600) and National Natural Science Foundation of China (31870071).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 3.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., van der Oost J., Koonin E.V. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinf. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E. The biology of CRISPR-cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Yao R., Liu D., Jia X., Zheng Y., Liu W., Xiao Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth Syst Biotechnol. 2018;3:135–149. doi: 10.1016/j.synbio.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 10.Miyaoka Y., Berman J.R., Cooper S.B., Mayerl S.J., Chan A.H., Zhang B., Karlin-Neumann G.A., Conklin B.R. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci Rep. 2016;6:23549. doi: 10.1038/srep23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schindele P., Wolter F., Puchta H. Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett. 2018;592:1954–1967. doi: 10.1002/1873-3468.13073. [DOI] [PubMed] [Google Scholar]
- 12.Shuman S., Glickman M.S. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 13.Della M., Palmbos P.L., Tseng H.M., Tonkin L.M., Daley J.M., Topper L.M., Pitcher R.S., Tomkinson A.E., Wilson T.E., Doherty A.J. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 14.Bertrand C., Thibessard A., Bruand C., Lecointe F., Leblond P. Bacterial NHEJ: a never ending story. Mol Microbiol. 2019;111:1139–1151. doi: 10.1111/mmi.14218. [DOI] [PubMed] [Google Scholar]
- 15.Tao W., Yang A., Deng Z., Sun Y. CRISPR/Cas9-Based editing of Streptomyces for discovery, characterization, and production of natural products. Front Microbiol. 2018;9:1660. doi: 10.3389/fmicb.2018.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh J.H., van Pijkeren J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran G., Bikard D. Editing the microbiome the CRISPR way. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180103. doi: 10.1098/rstb.2018.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Coupling the CRISPR/Cas9 system with Lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol. 2015;81:5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y., Shimatani Z., Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 24.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molla K.A., Yang Y. CRISPR/Cas-Mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Wu W., Yang Y., Lei H. Progress in the application of CRISPR: from gene to base editing. Med Res Rev. 2019;39:665–683. doi: 10.1002/med.21537. [DOI] [PubMed] [Google Scholar]
- 29.Donnez D., Jeandet P., Clement C., Courot E. Bioproduction of resveratrol and stilbene derivatives by plant cells and microorganisms. Trends Biotechnol. 2009;27:706–713. doi: 10.1016/j.tibtech.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Zhou M.L., Zhu X.M., Shao J.R., Tang Y.X., Wu Y.M. Production and metabolic engineering of bioactive substances in plant hairy root culture. Appl Microbiol Biotechnol. 2011;90:1229–1239. doi: 10.1007/s00253-011-3228-0. [DOI] [PubMed] [Google Scholar]
- 31.Pham J.V., Yilma M.A., Feliz A., Majid M.T., Maffetone N., Walker J.R., Kim E., Cho H.J., Reynolds J.M., Song M.C., Park S.R., Yoon Y.J. A review of the microbial production of bioactive natural products and biologics. Front Microbiol. 2019;10:1404. doi: 10.3389/fmicb.2019.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz B., Chavez A., Forero A., Garcia-Huante Y., Romero A., Sanchez M., Rocha D., Sanchez B., Rodriguez-Sanoja R., Sanchez S., Langley E. Production of microbial secondary metabolites: regulation by the carbon source. Crit Rev Microbiol. 2010;36:146–167. doi: 10.3109/10408410903489576. [DOI] [PubMed] [Google Scholar]
- 33.Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv. 2012;30:1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Schillberg S., Raven N., Spiegel H., Rasche S., Buntru M. Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front Plant Sci. 2019;10:720. doi: 10.3389/fpls.2019.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao J.C., Mi L., Pontrelli S., Luo S. Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat Rev Microbiol. 2016;14:288–304. doi: 10.1038/nrmicro.2016.32. [DOI] [PubMed] [Google Scholar]
- 36.Furtado A., Lupoi J.S., Hoang N.V., Healey A., Singh S., Simmons B.A., Henry R.J. Modifying plants for biofuel and biomaterial production. Plant Biotechnol J. 2014;12:1246–1258. doi: 10.1111/pbi.12300. [DOI] [PubMed] [Google Scholar]
- 37.Lee P.C., Schmidt-Dannert C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol. 2002;60:1–11. doi: 10.1007/s00253-002-1101-x. [DOI] [PubMed] [Google Scholar]
- 38.Wang T., Zhang H., Zhu H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic Res. 2019;6:77. doi: 10.1038/s41438-019-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee A., Banerjee C., Negi S., Chang J.S., Shukla P. Improvements in algal lipid production: a systems biology and gene editing approach. Crit Rev Biotechnol. 2018;38:369–385. doi: 10.1080/07388551.2017.1356803. [DOI] [PubMed] [Google Scholar]
- 40.Liang Y., Jiao S., Wang M., Yu H., Shen Z. A CRISPR/Cas9-based genome editing system for Rhodococcus ruber. TH. Metab Eng. 2020;57:13–22. doi: 10.1016/j.ymben.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Cho S., Shin J., Cho B.K. Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarnaik A., Liu A., Nielsen D., Varman A.M. High-throughput screening for efficient microbial biotechnology. Curr Opin Biotechnol. 2020;64:141–150. doi: 10.1016/j.copbio.2020.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Schormann N., Ricciardi R., Chattopadhyay D. Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014;23:1667–1685. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Houten B., Croteau D.L., DellaVecchia M.J., Wang H., Kisker C. 'Close-fitting sleeves': DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Eisen J.A., Hanawalt P.C. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasui M., Suenaga E., Koyama N., Masutani C., Hanaoka F., Gruz P., Shibutani S., Nohmi T., Hayashi M., Honma M. Miscoding properties of 2'-deoxyinosine, a nitric oxide-derived DNA Adduct, during translesion synthesis catalyzed by human DNA polymerases. J Mol Biol. 2008;377:1015–1023. doi: 10.1016/j.jmb.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 47.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X., Yang L., Chen J. Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 48.Zheng K., Wang Y., Li N., Jiang F.F., Wu C.X., Liu F., Chen H.C., Liu Z.F. Highly efficient base editing in bacteria using a Cas9-cytidine deaminase fusion. Commun Biol. 2018;1:32. doi: 10.1038/s42003-018-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banno S., Nishida K., Arazoe T., Mitsunobu H., Kondo A. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat Microbiol. 2018;3:423–429. doi: 10.1038/s41564-017-0102-6. [DOI] [PubMed] [Google Scholar]
- 50.Xin X., Li J., Zhao D., Li S., Xie Q., Li Z., Fan F., Bi C., Zhang X. Double-check base editing for efficient A to G conversions. ACS Synth Biol. 2019;8:2629–2634. doi: 10.1021/acssynbio.9b00284. [DOI] [PubMed] [Google Scholar]
- 51.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Zhang H., Wang Z., Wu Z., Wang Y., Tang N., Xu X., Zhao S., Chen W., Ji Q. Programmable adenine deamination in bacteria using a Cas9-adenine-deaminase fusion. Chem Sci. 2020;11:1657–1664. doi: 10.1039/c9sc03784e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q., Seys F.M., Minton N.P., Yang J., Jiang Y., Jiang W., Yang S. CRISPR-Cas9(D10A) nickase-assisted base editing in the solvent producer Clostridium beijerinckii. Biotechnol Bioeng. 2019;116:1475–1483. doi: 10.1002/bit.26949. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Liu Y., Liu J., Guo Y., Fan L., Ni X., Zheng X., Wang M., Zheng P., Sun J., Ma Y. MACBETH: multiplex automated Corynebacterium glutamicum base editing method. Metab Eng. 2018;47:200–210. doi: 10.1016/j.ymben.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Wang S., Chen W., Song L., Zhang Y., Shen Z., Yu F., Li M., Ji Q. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W., Zhang Y., Zhang Y., Pi Y., Gu T., Song L., Wang Y., Ji Q. CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience. 2018;6:222–231. doi: 10.1016/j.isci.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu T., Zhao S., Pi Y., Chen W., Chen C., Liu Q., Li M., Han D., Ji Q. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci. 2018;9:3248–3253. doi: 10.1039/c8sc00637g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tong Y., Whitford C.M., Robertsen H.L., Blin K., Jorgensen T.S., Klitgaard A.K., Gren T., Jiang X., Weber T., Lee S.Y. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc Natl Acad Sci U S A. 2019;116:20366–20375. doi: 10.1073/pnas.1913493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y., Tian J., Zheng G., Chen J., Sun C., Yang Z., Zimin A.A., Jiang W., Deng Z., Wang Z., Lu Y. Multiplex genome editing using a dCas9-cytidine deaminase fusion in Streptomyces. Sci China Life Sci. 2020;63:1053–1062. doi: 10.1007/s11427-019-1559-y. [DOI] [PubMed] [Google Scholar]
- 60.Zhong Z., Guo J., Deng L., Chen L., Wang J., Li S., Xu W., Deng Z., Sun Y. BioRxiv; 2019. Base editing in Streptomyces with cas9-deaminase fusions. [DOI] [Google Scholar]
- 61.Cabrita L.D., Dai W., Bottomley S.P. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 2006;6:12. doi: 10.1186/1472-6750-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C.J., Lin H., Yang X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- 63.Yang J.E., Park S.J., Kim W.J., Kim H.J., Kim B.J., Lee H., Shin J., Lee S.Y. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat Commun. 2018;9:79. doi: 10.1038/s41467-017-02498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J.D., Osterhout R.E., Stephen R., Estadilla J., Teisan S. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 65.Rude M.A., Schirmer A. New microbial fuels: a biotech perspective. Curr Opin Microbiol. 2009;12:274–281. doi: 10.1016/j.mib.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Xu P., Gu Q., Wang W., Wong L., Bower A.G., Collins C.H., Koffas M.A. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 67.Baeshen M.N., Al-Hejin A.M., Bora R.S., Ahmed M.M., Ramadan H.A., Saini K.S., Baeshen N.A., Redwan E.M. Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J Microbiol Biotechnol. 2015;25:953–962. doi: 10.4014/jmb.1412.12079. [DOI] [PubMed] [Google Scholar]
- 68.Peters J.M., Silvis M.R., Zhao D., Hawkins J.S., Gross C.A., Qi L.S. Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol. 2015;27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swarts D.C., Jinek M. Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. Wiley Interdiscip Rev RNA. 2018;9:e1481. doi: 10.1002/wrna.1481. [DOI] [PubMed] [Google Scholar]
- 70.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R., Aryee M.J., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V., Ishitani R., Zhang F. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 2012;65:385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 75.Tong Y., Weber T., Lee S.Y. CRISPR/Cas-based genome engineering in natural product discovery. Nat Prod Rep. 2019;36:1262–1280. doi: 10.1039/c8np00089a. [DOI] [PubMed] [Google Scholar]
- 76.Nett M., Ikeda H., Moore B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M., Takahashi Y., Horikawa H., Nakazawa H., Osonoe T., Kikuchi H., Shiba T. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohnishi Y., Ishikawa J., Hara H., Suzuki H., Ikenoya M., Ikeda H., Yamashita A., Hattori M., Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blin K., Shaw S., Tong Y., Weber T. Designing sgRNAs for CRISPR-BEST base editing applications with CRISPy-web 2.0. Synth Syst Biotechnol. 2020;5:99–102. doi: 10.1016/j.synbio.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wertheim H.F., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A., Nouwen J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 81.Espedido B.A., Gosbell I.B. Chromosomal mutations involved in antibiotic resistance in Staphylococcus aureus. Front Biosci. 2012;4:900–915. doi: 10.2741/s307. [DOI] [PubMed] [Google Scholar]
- 82.Cullen L., McClean S. Bacterial adaptation during chronic respiratory infections. Pathogens. 2015;4:66–89. doi: 10.3390/pathogens4010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Q., Jiang Y., Shao L., Yang P., Sun B., Yang S., Chen D. CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Acta Biochim Biophys Sin. 2017;49:764–770. doi: 10.1093/abbs/gmx074. [DOI] [PubMed] [Google Scholar]
- 84.Parte A.C. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hauser A.R., Jain M., Bar-Meir M., McColley S.A. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev. 2011;24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sadikot R.T., Blackwell T.S., Christman J.W., Prince A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samuel M.S., Sivaramakrishna A., Mehta A. Bioremediation of p-Nitrophenol by Pseudomonas putida 1274 strain. J Environ Health Sci Eng. 2014;12:53. doi: 10.1186/2052-336X-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nikel P.I., Chavarria M., Danchin A., de Lorenzo V. From dirt to industrial applications: Pseudomonas putida as a Synthetic Biology chassis for hosting harsh biochemical reactions. Curr Opin Chem Biol. 2016;34:20–29. doi: 10.1016/j.cbpa.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 89.Samin G., Pavlova M., Arif M.I., Postema C.P., Damborsky J., Janssen D.B. A Pseudomonas putida strain genetically engineered for 1,2,3-trichloropropane bioremediation. Appl Environ Microbiol. 2014;80:5467–5476. doi: 10.1128/AEM.01620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Retallack D.M., Jin H., Chew L. Reliable protein production in a Pseudomonas fluorescens expression system. Protein Expr Purif. 2012;81:157–165. doi: 10.1016/j.pep.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 91.Tan S.Z., Reisch C.R., Prather K.L.J. A robust CRISPR interference gene repression system in Pseudomonas. J Bacteriol. 2018;200 doi: 10.1128/JB.00575-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun J., Wang Q., Jiang Y., Wen Z., Yang L., Wu J., Yang S. Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb Cell Factories. 2018;17:41. doi: 10.1186/s12934-018-0887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jia X., Zhang S., Li J., Xia J., Yao R., Zhao X., Wu B., Bai F., Xiao Y. Engineered bacterial biofloc formation enhancing phenol removal and cell tolerance. Appl Microbiol Biotechnol. 2020;104:1187–1199. doi: 10.1007/s00253-019-10289-0. [DOI] [PubMed] [Google Scholar]
- 94.Tang H., Yao Y., Wang L., Yu H., Ren Y., Wu G., Xu P. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep. 2012;2:377. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wendisch V.F., Jorge J.M.P., Perez-Garcia F., Sgobba E. Updates on industrial production of amino acids using Corynebacterium glutamicum. World J Microbiol Biotechnol. 2016;32:105. doi: 10.1007/s11274-016-2060-1. [DOI] [PubMed] [Google Scholar]
- 96.Eggeling L., Bott M. A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2015;99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 97.Lee J.Y., Na Y.A., Kim E., Lee H.S., Kim P. The actinobacterium Corynebacterium glutamicum, an industrial workhorse. J Microbiol Biotechnol. 2016;26:807–822. doi: 10.4014/jmb.1601.01053. [DOI] [PubMed] [Google Scholar]
- 98.Heider S.A., Wendisch V.F. Engineering microbial cell factories: metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products. Biotechnol J. 2015;10:1170–1184. doi: 10.1002/biot.201400590. [DOI] [PubMed] [Google Scholar]
- 99.Souli M., Galani I., Antoniadou A., Papadomichelakis E., Poulakou G., Panagea T., Vourli S., Zerva L., Armaganidis A., Kanellakopoulou K., Giamarellou H. An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010;50:364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 100.Quan J., Li X., Chen Y., Jiang Y., Zhou Z., Zhang H., Sun L., Ruan Z., Feng Y., Akova M., Yu Y. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17:400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 101.Thakker C., Martinez I., Li W., San K.Y., Bennett G.N. Metabolic engineering of carbon and redox flow in the production of small organic acids. J Ind Microbiol Biotechnol. 2015;42:403–422. doi: 10.1007/s10295-014-1560-y. [DOI] [PubMed] [Google Scholar]
- 102.Lee S.Y., Park J.H., Jang S.H., Nielsen L.K., Kim J., Jung K.S. Fermentative butanol production by Clostridia. Biotechnol Bioeng. 2008;101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 103.Chen C.K., Blaschek H.P. Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl Microbiol Biotechnol. 1999;52:170–173. doi: 10.1007/s002530051504. [DOI] [PubMed] [Google Scholar]
- 104.Ezeji T., Qureshi N., Blaschek H.P. Butanol production from agricultural residues: impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol Bioeng. 2007;97:1460–1469. doi: 10.1002/bit.21373. [DOI] [PubMed] [Google Scholar]
- 105.Que Q., Chen Z., Kelliher T., Skibbe D., Dong S., Chilton M.D. Plant DNA repair pathways and their applications in genome engineering. Methods Mol Biol. 2019;1917:3–24. doi: 10.1007/978-1-4939-8991-1_1. [DOI] [PubMed] [Google Scholar]
- 106.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma X., Zhu Q., Chen Y., Liu Y.G. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant. 2016;9:961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 108.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 109.Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K., Ezura H., Nishida K. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 110.Li J., Sun Y., Du J., Zhao Y., Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 111.Lu Y., Zhu J.K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 112.Ren B., Yan F., Kuang Y., Li N., Zhang D., Zhou X., Lin H., Zhou H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-Guided hyperactive hAID mutant. Mol Plant. 2018;11:623–626. doi: 10.1016/j.molp.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 113.Ren B., Yan F., Kuang Y., Li N., Zhang D., Lin H., Zhou H. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci China Life Sci. 2017;60:516–519. doi: 10.1007/s11427-016-0406-x. [DOI] [PubMed] [Google Scholar]
- 114.Li C., Zong Y., Wang Y., Jin S., Zhang D., Song Q., Zhang R., Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan F., Kuang Y., Ren B., Wang J., Zhang D., Lin H., Yang B., Zhou X., Zhou H. Highly efficient A.T to G.C base editing by cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 116.Hao L., Ruiying Q., Xiaoshuang L., Shengxiang L., Rongfang X., Jianbo Y., Pengcheng W. CRISPR/Cas9-Mediated adenine base editing in rice genome. Rice Sci. 2019;26:125–128. doi: 10.1016/j.rsci.2018.07.002. [DOI] [Google Scholar]
- 117.Li J., Qin R., Zhang Y., Xu S., Liu X., Yang J., Zhang X., Wei P. Optimizing plant adenine base editor systems by modifying the transgene selection system. Plant Biotechnol J. 2020;18:1495–1497. doi: 10.1111/pbi.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A.V., Raguram A., Doman J.L., Liu D.R., Gao C. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- 119.Xu W., Zhang C., Yang Y., Zhao S., Kang G., He X., Song J., Yang J. Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant. 2020;13:675–678. doi: 10.1016/j.molp.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 120.Li H., Li J., Chen J., Yan L., Xia L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol Plant. 2020;13:671–674. doi: 10.1016/j.molp.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 121.Xu R., Li J., Liu X., Shan T., Qin R., Wei P. Development of plant prime-editing systems for precise genome editing. Plant Communications. 2020;1 doi: 10.1016/j.xplc.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hua K., Jiang Y., Tao X., Zhu J.K. Precision genome engineering in rice using prime editing system. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen Y., Wang Z., Ni H., Xu Y., Chen Q., Jiang L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci China Life Sci. 2017;60:520–523. doi: 10.1007/s11427-017-9021-5. [DOI] [PubMed] [Google Scholar]
- 125.Li Z., Xiong X., Wang F., Liang J., Li J.F. Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol. 2019;222:1139–1148. doi: 10.1111/nph.15647. [DOI] [PubMed] [Google Scholar]
- 126.Bastet A., Zafirov D., Giovinazzo N., Guyon-Debast A., Nogue F., Robaglia C., Gallois J.L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol J. 2019;17:1736–1750. doi: 10.1111/pbi.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kang B.C., Yun J.Y., Kim S.T., Shin Y., Ryu J., Choi M., Woo J.W., Kim J.S. Precision genome engineering through adenine base editing in plants. Native Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- 128.Veillet F., Perrot L., Chauvin L., Kermarrec M.P., Guyon-Debast A., Chauvin J.E., Nogue F., Mazier M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qin L., Li J., Wang Q., Xu Z., Sun L., Alariqi M., Manghwar H., Wang G., Li B., Ding X., Rui H., Huang H. High-efficient and precise base editing of C*G to T*A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:45–56. doi: 10.1111/pbi.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tian S., Jiang L., Cui X., Zhang J., Guo S., Li M., Zhang H., Ren Y., Gong G., Zong M., Liu F., Chen Q. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018;37:1353–1356. doi: 10.1007/s00299-018-2299-0. [DOI] [PubMed] [Google Scholar]
- 131.Farzadfard F., Lu T.K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346:1256272. doi: 10.1126/science.1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McKenna A., Findlay G.M., Gagnon J.A., Horwitz M.S., Schier A.F., Shendure J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf7907. doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Farzadfard F., Lu T.K. Emerging applications for DNA writers and molecular recorders. Science. 2018;361:870–875. doi: 10.1126/science.aat9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tang W., Liu D.R. Rewritable multi-event analog recording in bacterial and mammalian cells. Science. 2018;360 doi: 10.1126/science.aap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Farzadfard F., Gharaei N., Higashikuni Y., Jung G., Cao J., Lu T.K. Single-nucleotide-resolution computing and memory in living cells. Mol Cell. 2019;75:769–780. doi: 10.1016/j.molcel.2019.07.011. e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee S.Y. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 137.Luli G.W., Strohl W.R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol. 1990;56:1004–1011. doi: 10.1128/AEM.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Feng J., Atkinson M.R., McCleary W., Stock J.B., Wanner B.L., Ninfa A.J. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xiao Y., Bowen C.H., Liu D., Zhang F. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol. 2016;12:339–344. doi: 10.1038/nchembio.2046. [DOI] [PubMed] [Google Scholar]
- 140.Thomson M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breeding Biotechnol. 2014;2:195–212. doi: 10.9787/pbb.2014.2.3.195. [DOI] [Google Scholar]
- 141.Zhang R., Liu J., Chai Z., Chen S., Bai Y., Zong Y., Chen K., Li J., Jiang L., Gao C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Native Plants. 2019;5:480–485. doi: 10.1038/s41477-019-0405-0. [DOI] [PubMed] [Google Scholar]
- 142.Zong Y., Song Q., Li C., Jin S., Zhang D., Wang Y., Qiu J.L., Gao C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 2018 doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]
- 143.Robaglia C., Caranta C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 144.Bastet A., Robaglia C., Gallois J.L. eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 2017;22:411–419. doi: 10.1016/j.tplants.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 145.Bastet A., Lederer B., Giovinazzo N., Arnoux X., German-Retana S., Reinbold C., Brault V., Garcia D., Djennane S., Gersch S., Lemaire O., Robaglia C. Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants. Plant Biotechnol J. 2018 doi: 10.1111/pbi.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ruffel S., Gallois J.L., Moury B., Robaglia C., Palloix A., Caranta C. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J Gen Virol. 2006;87:2089–2098. doi: 10.1099/vir.0.81817-0. [DOI] [PubMed] [Google Scholar]
- 147.Hess G.T., Fresard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 149.Liu X., Qin R., Li J., Liao S., Shan T., Xu R., Wu D., Wei P. A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol J. 2020;18:1845–1847. doi: 10.1111/pbi.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuang Y., Li S., Ren B., Yan F., Spetz C., Li X., Zhou X., Zhou H. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol Plant. 2020;13:565–572. doi: 10.1016/j.molp.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 151.Li S., Liu Q., Zhong Z., Deng Z., Sun Y. Exploration of hygromycin B biosynthesis utilizing CRISPR-cas9-associated base editing. ACS Chem Biol. 2020;15:1417–1423. doi: 10.1021/acschembio.0c00071. [DOI] [PubMed] [Google Scholar]
- 152.DellaPenna D. Plant metabolic engineering. Plant Physiol. 2001;125:160–163. doi: 10.1104/pp.125.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fu R., Martin C., Zhang Y. Next-generation plant metabolic engineering, inspired by an ancient Chinese irrigation system. Mol Plant. 2018;11:47–57. doi: 10.1016/j.molp.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 154.Jiang W.Z., Henry I.M., Lynagh P.G., Comai L., Cahoon E.B., Weeks D.P. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J. 2017;15:648–657. doi: 10.1111/pbi.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., Ciccia A. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell. 2017;67:1068–1079. doi: 10.1016/j.molcel.2017.08.008. e1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kuscu C., Parlak M., Tufan T., Yang J., Szlachta K., Wei X., Mammadov R., Adli M. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 157.Solomon O., Oren S., Safran M., Deshet-Unger N., Akiva P., Jacob-Hirsch J., Cesarkas K., Kabesa R., Amariglio N., Unger R., Rechavi G., Eyal E. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abudayyeh O.O., Gootenberg J.S., Franklin B., Koob J., Kellner M.J., Ladha A., Joung J., Kirchgatterer P., Cox D.B.T., Zhang F. A cytosine deaminase for programmable single-base RNA editing. Science. 2019;365:382–386. doi: 10.1126/science.aax7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hua K., Tao X., Yuan F., Wang D., Zhu J.K. Precise A.T to G.C base editing in the rice genome. Mol Plant. 2018;11:627–630. doi: 10.1016/j.molp.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 162.Hua K., Tao X., Zhu J.K. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J. 2019;17:499–504. doi: 10.1111/pbi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang C., Xu W., Wang F., Kang G., Yuan S., Lv X., Li L., Liu Y., Yang J. Expanding the base editing scope to GA and relaxed NG PAM sites by improved xCas9 system. Plant Biotechnol J. 2020;18:884–886. doi: 10.1111/pbi.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chatterjee P., Jakimo N., Lee J., Amrani N., Rodriguez T., Koseki S.R.T., Tysinger E., Qing R., Hao S., Sontheimer E.J., Jacobson J. An engineered ScCas9 with broad PAM range and high specificity and activity. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0517-0. [DOI] [PubMed] [Google Scholar]
- 166.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., Doudna J.A., Liu D.R. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017;3 doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hua K., Tao X., Liang W., Zhang Z., Gou R., Zhu J.K. Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol J. 2020;18:770–778. doi: 10.1111/pbi.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee H.K., Willi M., Miller S.M., Kim S., Liu C., Liu D.R., Hennighausen L. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat Commun. 2018;9:4804. doi: 10.1038/s41467-018-07322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 171.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang L., Xue W., Yan L., Li X., Wei J., Chen M., Wu J., Yang B., Yang L., Chen J. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 2017;27:1289–1292. doi: 10.1038/cr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang W., Feng S., Huang S., Yu W., Li G., Yang G., Liu Y., Zhang Y., Zhang L., Hou Y., Chen J., Chen J. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res. 2018;28:855–861. doi: 10.1038/s41422-018-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhong Z., Sretenovic S., Ren Q., Yang L., Bao Y., Qi C., Yuan M., He Y., Liu S., Liu X., Wang J., Huang L. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting cas9-NG. Mol Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 176.Endo M., Mikami M., Endo A., Kaya H., Itoh T., Nishimasu H., Nureki O., Toki S. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Native Plants. 2019;5:14–17. doi: 10.1038/s41477-018-0321-8. [DOI] [PubMed] [Google Scholar]
- 177.Zeng D., Li X., Huang J., Li Y., Cai S., Yu W., Li Y., Huang Y., Xie X., Gong Q., Tan J., Zheng Z. Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol J. 2020;18:1348–1350. doi: 10.1111/pbi.13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Negishi K., Kaya H., Abe K., Hara N., Saika H., Toki S. An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol J. 2019;17:1476–1478. doi: 10.1111/pbi.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Veillet F., Perrot L., Guyon-Debast A., Kermarrec M.P., Chauvin L., Chauvin J.E., Gallois J.L., Mazier M., Nogue F. Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in solanaceae crops. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21031024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cai Y., Chen L., Zhang Y., Yuan S., Su Q., Sun S., Wu C., Yao W., Han T., Hou W. Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu J., Chen C., Xian G., Liu D., Lin L., Yin S., Sun Q., Fang Y., Zhang H., Wang Y. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]