Abstract
Background
Malignant pleural effusion (MPE) is a common medical problem caused by multiple malignancies, especially lung cancers, and always comes along with a poor outcome. Early detection and diagnosis are important for improving the prognosis in patients with MPE. Salivary microRNAs (miRNAs) may represent a relatively convenient way for diagnosing MPE. We investigated the characteristics of salivary miRNAs of MPE patients, benign pleural effusion (BPE) patients, patients with a malignant tumor but without pleural effusion (MT), and healthy controls (HCs). We believe that they may show potential as a non-invasive and convenient biomarker for diagnosing MPE.
Methods
From January 1, 2019, to July 1, 2019, 57 MPE patients, 33 BPE patients, 50 MT patients, and 49 HCs were enrolled. To select candidate biomarkers, in the discovery phase, the salivary miRNA profiles were detected in three MPE patients and three HCs. Then, qPCR was used in the validation phase with 54 MPE patients, 33 BPE patients, 50 MT patients, and 46 HCs to assay the selected miRNAs.
Results
hsa-miR-4484 and hsa-miR-3663-3p were identified as potential biomarkers to diagnose MPE patients, with areas under the curve (AUC) of 0.768 and 0.666, respectively. The diagnostic efficacy was higher when the combination of both miRNAs was used, with an AUC of 0.802. No correlation was found between the volume of MPE and the expression of salivary miRNAs.
Conclusions
This study reports the characterization of salivary miRNAs collected from MPE patients. A combination of hsa-miR-4484 and hsa-miR-3663-3p showed potential discriminatory power for MPE detection, and it may be helpful for the early diagnosis of MPE, i.e., before the pleural effusion volume is too large.
Keywords: Human saliva, microRNA microarray, malignant pleural effusion, diagnostic biomarker
Introduction
Pleural effusions are common medical problems resulting from increased fluid in the pleural space (1). More than 50 diseases can cause pleural effusion, such as pleura disease, lung disease, organ dysfunction, systemic conditions, and also several drugs (2). Malignant pleural effusion (MPE) is a common type of pleural effusion. It can be observed in many malignant diseases, especially lung cancer (3-5). The existence of MPE always indicates systemic cancer dissemination and a decline in the life expectancy and quality of patients (6,7). Thus, having an effective and minimally invasive method to diagnose MPE as early as possible and accurately is urgently needed.
Many methods can be used to diagnose MPE. Pleural effusion cytological analysis is always the clinician’s first choice. However, cytological analysis’s diagnostic efficiency is far from ideal, with a sensitivity range between 40% and 70% (8-10). Closed pleural biopsy is another way to diagnose MPE, but this procedure cannot be performed when the depth of pleural effusion is below 10 mm (11). A less invasive way to diagnose MPE is to detect tumor markers in pleural effusions, but this method’s sensitivity is only 25–88% (12,13). All of these methods may lead to MPEs incorrectly identified or overlooked. Instead, medical thoracoscopy is considered as a highly sensitive (92.7%) method for diagnosing pleural effusions (3,14). This year, our group found that the PET-CT score can be an efficient method to diagnose MPE, with an area under the curve of 0.949 (15). However, both medical thoracoscopy and PET-CT require high technology support and have a high cost. Besides, most of the methods described so far need enough pleural effusion volume to allow drainage, which is impossible when pleural effusion volume is low.
Saliva is now considered a readily accessible body fluid containing diverse components, such as protein, DNA, RNA, and metabolites, that may potentially be effective biomarkers (16-20). Many studies have recently indicated the diagnostic value of salivary biomarkers, ranging from oral diseases to systemic diseases (21-31).
In this study, we investigate the characteristics of salivary microRNAs (miRNAs) of MPE patients and highlight their potential as non-invasive and convenient biomarkers for diagnosing MPE. We present this article following the STARD guideline checklist (available at http://dx.doi.org/10.21037/tlcr-19-530).
Methods
Study design
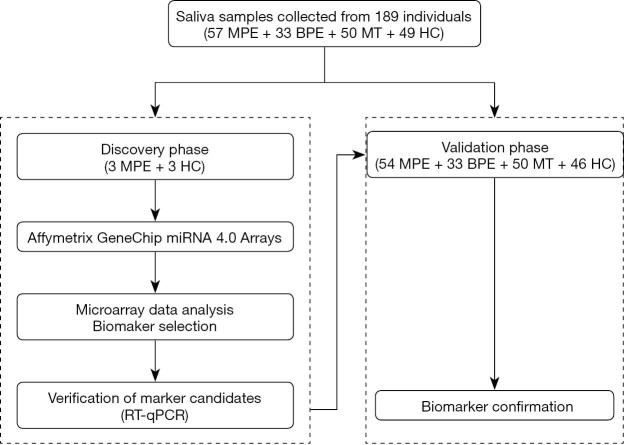
This work is a retrospective case-control study. Two phases were designed in this study, a discovery phase and a validation phase (Figure 1). In the discovery phase, six salivary samples from three MPE patients, and three healthy controls (HCs) were randomly enrolled. The miRNA profiles of these saliva samples were detected using the miRNA arrays for miRNA biomarker signature discovery. Then, in the validation phase, 54 MPE patients, 33 benign pleural effusion (BPE) patients, 50 malignant tumors without pleural effusion (MT) patients, and 46 HCs were included to assay the candidate miRNAs using qPCR.
Figure 1.
The study design for the two phases of salivary miRNA biomarker development for MPE. MPE, malignant pleural effusion; BPE, benign pleural effusion; MT, malignant tumor without pleural effusion; HC, healthy control.
Patients
The study protocol was conducted following the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Beijing, China (# 2018-ke-321). All of the patients enrolled completed the informed consent form.
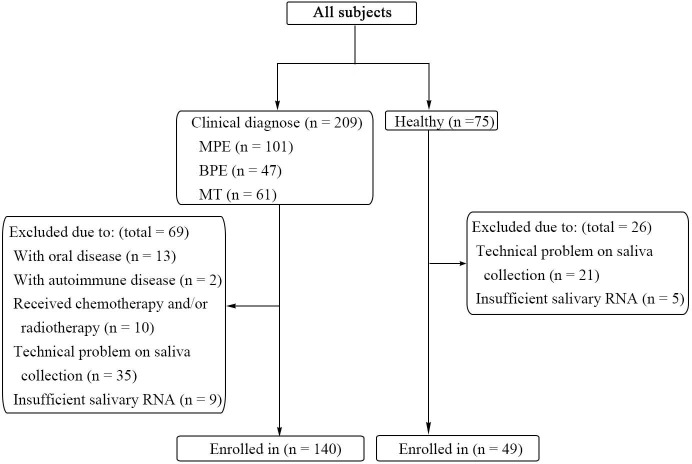
From January 1, 2019, to July 1, 2019, 209 patients with or without pleural effusion were treated in the Department of Respiratory and Critical Care Medicine of Beijing Chao-Yang Hospital, China. Initial diagnostic workups were performed for all patients, including a detailed medical evaluation, pleural radiological assessments, thoracic ultrasound, pleural fluid analyses, and/or pleural biopsy examination. All patients were consecutively enrolled in the study. Eventually, 57 patients with a definite diagnosis of MPE, 33 patients with a diagnosis of BPE, 50 patients with a diagnosis of malignancy without pleural effusion and 49 HCs with similar age, ethnicity, gender, and smoking history were enrolled in our study, and their saliva samples were collected. In the MPE and HC groups, a researcher, without knowing the groups’ information, participants’ information, and miRNA expression information, randomly selected three participants from each group for the discovery cohort. The other participants were enrolled in the validation cohort. MPE was diagnosed when cytological examination of pleural effusion showed malignant cells or when pleural histopathology showed malignant lesions. BPE was diagnosed when no malignant cells were observed in the pleural effusion, and pleural effusion was reduced after the treatment of primary disease. MT patients were diagnosed when the tumor histopathology showed malignant lesions but without pleural effusion. All healthy subjects were approached at public places such as parks and residential areas near Beijing Chao-Yang Hospital. Their demographic information and medical histories were asked, and saliva was collected voluntarily. Subjects with oral, infectious, or autoimmune diseases were excluded from this study. No patients received chemotherapy and radiotherapy before saliva collection. Inclusion and exclusion criteria are shown in Figure S1.
Figure S1.
Inclusion and exclusion criteria of all subjects enrolled in this study. MPE, malignant pleural effusion; BPE, benign pleural effusion; MT, malignant tumor without pleural effusion.
Saliva collection
Before saliva collection, all patients and HCs were required to avoid drinking, eating, or smoking for at least 2 h. At least 3 mL of saliva was collected from each subject. All samples were centrifuged at 3,000 ×g at 4 °C for 15 min to remove the cells. Only the supernatant was transferred to a 2 mL centrifuge tube. Then, to remove cellular components completely, centrifugation of 12,000 ×g at 4 °C for 10 min was followed. The supernatant was transferred into a 2 mL centrifuge tube and stored at −80 °C until use.
Microarray analysis
In the discovery phase, six salivary samples from three MPE patients and three HCs were randomly enrolled. The miRNA profiles of these saliva samples were detected using the Affymetrix® GeneChip® miRNA 4.0 Arrays (Affymetrix, Santa Clara, CA, USA) for miRNA biomarker signature’s discovery. In total, 2,578 miRNAs were assembled into our miRNA microarray, and two of them were selected as our candidate biomarkers. The selection criteria were as follows. First, the average miRNA expression level between the MPE group and control group were compared. A statistically significant difference was set when P values <0.05. Then, differentially expressed miRNAs that showed at least a 2-fold change were selected for the validation phase. We have uploaded the microarray data to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) (GSE135947).
Quantification of salivary miRNAs
Researchers were not aware of the participants’ and group information of all samples in advance. According to the manufacturer’s protocol, total RNA was extracted from 2 mL of saliva supernatant using TRIzol (Thermo Scientific, Waltham, MA, USA) extraction (32). After extraction, the RNA was resuspended in 20 µL of nuclease-free water, and cDNA was transcribed right away using the stem-loop reverse transcription reaction (PrimeScript™ RT reagent Kit with gDNA Eraser, Takara, Dalian, China). The stem-loop reverse transcription primer (supplementary materials) contains a highly stable stem-loop structure that lengthens the target cDNA to enhance the assay’s specificity (33). The cDNA solution was amplified using LightCycler® 480 Probes Master (Roche, Mannheim, DE), and quantitative PCR was run on a LightCycler® 480 System (Roche, Mannheim, DE). The reaction mixtures were incubated at 95 °C for 5 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 10 s. Each sample was analyzed in triplicate. Expression levels were calculated using the 2-ΔΔCt method (34), and the expression levels of miRNAs were normalized by U6 snRNA.
Statistical analysis
All analyses were performed using SPSS version 25.0 statistical software (SPSS Inc., Chicago, IL, USA) and MedCalc statistical software 19 (MedCalc Software, Ostend, BE). Descriptive statistical methods were used for summary statistics reporting. Mean ± standard deviation (SD) or number was presented as appropriate. The significance of differences between the two groups was analyzed using Student’s t-test. The correlation coefficient between variables was performed using Pearson’s correlation test. Receiver operating characteristic (ROC) curves and the areas under the curves (AUCs) were used to evaluate the miRNA’s diagnostic value. Sensitivity, specificity, positive predictive value, negative predictive value, positive likelihood ratio, and negative likelihood ratio were also analyzed. The 95% confidence intervals (CIs) of these parameters were also evaluated in this study. The optimal cutoff points were established based on their maximum Youden’s index. Sample size analysis was conducted by PASS version 11.0 software (NCSS Inc., Kaysville, UT). The power was calculated based on the ROC test. At the 5% level of significance, with a sample size of 54 in the MPE group, 33 in the BPE group, 50 in the MT group, and 46 in the healthy control group, the trial will have above 95% power. All P values were two-sided. A statistically significant difference was set when P values <0.05.
Results
Characteristics of MPE patients and HCs
In our study, 6 patients (3 MPE patients and 3 HCs) were included in the discovery phase and 183 patients (54 MPE patients, 33 BPE patients, 50 MT patients, and 46 HCs) in the validation phase. The table (Table 1) shows the baseline characteristics of these subjects. In the discovery phase, there were two males and one female in the MPE group and three females in the HC group. The average ages of the patients in the MPE and HC groups were 78±3 and 63±8.89 years, respectively. One of the three MPE patients was a smoker, and none of the HCs was a smoker. The histology type of all three MPE patients was adenocarcinoma. Then, in the validation phase, the percentages of males in the MPE group, the BPE group, the MT group, and the HC groups were 51.9%, 66.7%, 60.0%, and 41.3%, respectively; the average ages of the patients in these four groups were 61.87±13.13, 59.82±13.61, 62.02±13.24, and 55.80±14.38 years, respectively; the percentages of smokers in these four groups were 50%, 39.4%, 46.0%, and 58.7%, respectively. Among the 54 MPE patients in the validation phase, there were 26 adenocarcinomas, 12 small cell lung cancers, 9 squamous cell carcinomas, and 7 malignant mesotheliomas. Among the 33 members of the BPE group, 12 were parapneumonic effusion patients, 9 were tuberculous pleural effusion patients, and 5 were transudative pleural effusion patients. Six pleural effusions were caused by connective tissue disease, and one was caused by pulmonary embolism. Among the 50 MT patients, there were 20 adenocarcinomas, 19 small cell lung cancers, and 11 squamous cell carcinomas. There were no statistical differences in age, gender, and smoking history between these groups.
Table 1. Patient demographics and clinical profiles.
| Variables | Discovery phase | Validation phase | ||||||
|---|---|---|---|---|---|---|---|---|
| MPE (n=3) | HC (n=3) | P value | MPE (n=54) | BPE (n=33) | MT (n=50) | HC (n=46) | ||
| Age, yr, mean ± SD | 78±3 | 63±8.89 | 0.051 | 61.87±13.13 | 59.82±13.61 | 62.02±13.24 | 55.80±14.38 | |
| Gender | 0.083 | |||||||
| Male | 2 | 0 | 28 | 22 | 30 | 19 | ||
| Female | 1 | 3 | 26 | 11 | 20 | 27 | ||
| Smoking | 0.273 | |||||||
| Smoker | 1 | 0 | 27 | 13 | 23 | 27 | ||
| Non-smoker | 2 | 3 | 27 | 20 | 27 | 19 | ||
| Histology type | ||||||||
| Adenocarcinoma | 3 | 26 | 20 | |||||
| Squamous cell carcinoma | 0 | 9 | 11 | |||||
| Small cell lung cancer | 0 | 12 | 19 | |||||
| Malignant mesothelioma | 0 | 7 | 0 | |||||
| Parapneumonic effusion | 12 | |||||||
| Tuberculous pleural effusion | 9 | |||||||
| Connective tissue disease | 6 | |||||||
| Transudative pleural effusion | 5 | |||||||
| Pulmonary embolism | 1 | |||||||
MPE, malignant pleural effusion; BPE, benign pleural effusion; MT, malignant tumor without pleural effusion; HC, healthy control.
Salivary miRNA profiles of MPE patients and HCs
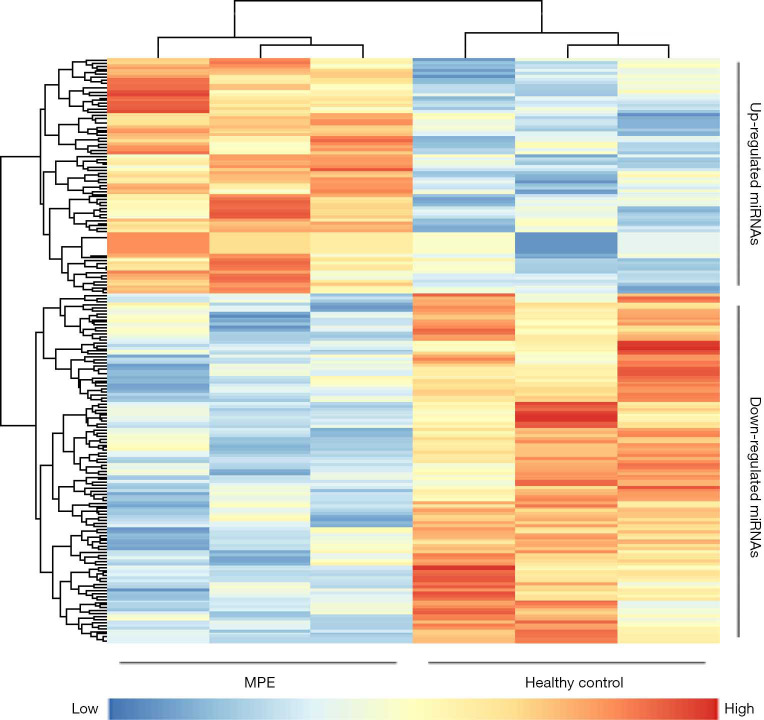
In the discovery phase, a miRNA microarray was used to examine miRNA expression in saliva samples from three MPE patients and three HCs. The microarray results showed that 29 miRNAs were upregulated, and 48 miRNAs were downregulated in the saliva of MPE patients. The heat map of these differentially expressed miRNAs is shown in Figure 2. The results showed that the MPE patients and HCs were divided into two groups, and these salivary miRNAs may have potential diagnostic power. Eventually, two miRNAs, hsa-miR-4484 (P=0.027, fold change =5.337) and hsa-miR-3663-3p (P=0.030, fold change =2.403), were selected for the followed validation phase. Hsa-miR-4484 showed significant upregulation in MPE patients, while hsa-miR-3663-3p showed downregulation.
Figure 2.
Heat map of the verified miRNAs (29 upregulated and 48 downregulated) based on the microarray data. MPE patients (n=3) and HCs (n=3) could be classified into distinct groups, preliminarily indicating the discriminatory power of these salivary miRNAs.
Validation of salivary biomarkers and biomarker combination analysis
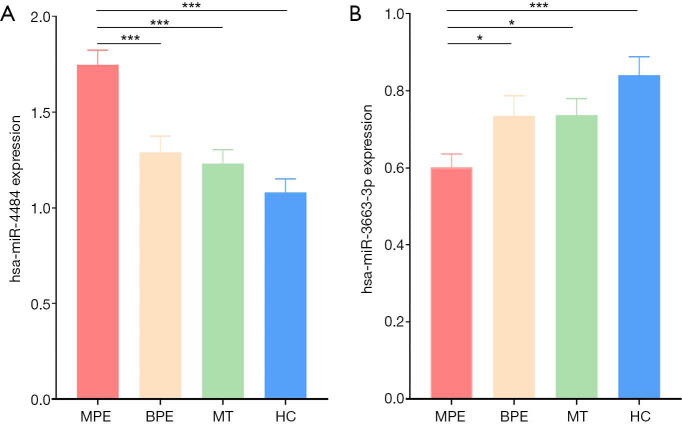
In the validation phase, quantitative PCR was used to validate the two candidate miRNAs in 54 MPE patients, 33 BPE patients, 50 MT patients, and 46 HCs. The results showed that hsa-miR-4484 was significantly upregulated, and hsa-miR-3663-3p was significantly downregulated in MPE patients (Figure 3). These results were consistent with the microarray analysis results. ROC curves were used to test if these two miRNAs can be used for MPE diagnosis. AUC for hsa-miR-4484 and hsa-miR-3663-3p were 0.768 and 0.666, respectively. The sensitivities were 74.1% and 50.4%, and the specificities were 74.4% and 79.6%, respectively (Figure 4A). A binary logistic regression model was built to evaluate the diagnostic value of the combination of hsa-miR-4484 and hsa-miR-3663-3p, and the ROC curve was computed. The results show a sensitivity of 82.2%, a specificity of 74.1%, and an AUC of 0.802 for the combination of the two validated salivary biomarkers (Figure 4B). The other diagnostic parameters of salivary hsa-miR-4484 and hsa-miR-3663-3p, including sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, positive predictive value, and negative predictive value, are presented in Table 2. Based on these parameters, it was clear that the combination of hsa-miR-4484 and hsa-miR-3663-3p had a high diagnostic performance on diagnosing MPE.
Figure 3.
Column diagram of salivary hsa-miR-4484 (A) and hsa-miR-3663-3p (B) based on qPCR data of the MPE group (n=54), BPE group (n=33), MT group (n=50) and HC group (n=46). *, P<0.05; ***, P<0.001.
Figure 4.
ROC curve analysis for the discriminatory power of salivary miRNA biomarkers. In MPE patients versus HCs, hsa-miR-4484 showed an AUC of 0.768, with a sensitivity of 74.1% and a specificity of 74.4%. The AUC for hsa-miR-3663-3p was 0.666, with a sensitivity of 50.4% and a specificity of 79.6% (A). The predictive power of combined salivary hsa-miR-4484 and hsa-miR-3663-3p also determined by ROC curve analysis (B). We obtained a sensitivity of 82.2% and a specificity of 74.1%. The calculated area under the ROC curve was 0.802.
Table 2. Diagnostic values of salivary hsa-miR-4484, hsa-miR-3663-3p and the combination of salivary hsa-miR-4484 and hsa-miR-3663-3p in diagnosing MPE.
| Variable | Cutoff | AUC (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PLR (95% CI) | NLR (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-4484 | 1.384 | 0.768 (0.689 to 0.874) | 74.1 (60.1 to 84.6) | 74.4 (65.8 to 81.5) | 2.9 (2.1 to 4.0) | 0.3 (0.2 to 0.5) | 54.8 (42.8 to 66.3) | 87.3 (79.2 to 92.6) |
| hsa-miR-3663-3p | 0.748 | 0.666 (0.581 to 0.750) | 50.4 (41.5 to 59.2) | 79.6 (64.1 to 87.5) | 2.3 (1.3 to 3.8) | 0.6 (0.5 to 0.8) | 84.4 (74.0 to 91.3) | 39.6 (30.4 to 49.6) |
| Combination of two miRNAs | 0.672 | 0.802 (0.729 to 0.875) | 82.2 (74.2 to 88.1) | 74.1 (58.1 to 83.1) | 3.0 (1.9 to 4.6) | 0.2 (0.1 to 0.3) | 87.6 (80.1 to 92.7) | 62.9 (49.7 to 74.6) |
AUC, area under the curve; PLR, positive likelihood ratio; NLR, negative likelihood ratio; PPV, positive predictive value; NPV, negative predictive value.
Salivary miRNAs may become useful biomarkers for MPE early detection
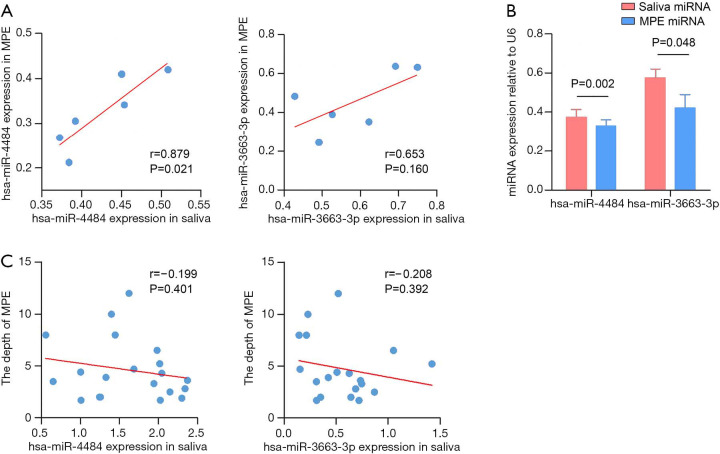
To explore the characterization of salivary miRNAs, we extracted the paired MPE and saliva from six patients to determine if the expression of miRNAs was correlated with MPE and saliva. We found that the hsa-miR-4484 level in saliva was correlated with MPE, but the same result was not found for hsa-miR-3663-3p (Figure 5A). Generally speaking, the miRNA level is higher in saliva than in MPE (Figure 5B). Then, we investigated the correlation between salivary miRNA levels and pleural effusion volume. The results shown in Figure 5C indicate that the miRNA levels did not correlate with the MPE volume, suggesting that salivary miRNAs may be detected in a low MPE volume and buy more time for early diagnosis.
Figure 5.
P=0.021 indicates that the hsa-miR-4484 level in saliva correlated with it in MPE, but the same result was not found in the level of hsa-miR-3663-3p with P=0.160 (A). The miRNA level is higher in saliva than in MPE for both hsa-miR-4484 and hsa-miR-3663-3p (B). The miRNA levels did not correlate with the pleural effusion volume with both P>0.05 (C). Pleural effusion volume was evaluated by the depth that the thoracic ultrasound detected.
Discussion
MPE is a common medical problem caused by multiple malignancies, especially lung cancers. The appearance of pleural effusion often indicates the late stage of the disease. Early detection and diagnosis are important for improving the prognosis in patients with MPE.
Many studies have proved that salivary components, such as proteins, transcripts, and metabolites, can be used for detecting oral, infectious, systemic, and cardiovascular diseases and cancers (21-31). In 2009, Park et al. found that miR-125a and miR-200a were present in significantly lower levels in the saliva of oral squamous cell carcinoma patients than in control subjects with a combined AUC of 0.66 (24). Then, in 2010, Zhang et al. found that the combination of four salivary mRNA biomarkers could differentiate pancreatic cancer patients from non-cancer subjects, yielding a ROC-plot AUC value of 0.971 (17). Thus, with all of these discoveries, the saliva is believed to mirror systemic health and reflect conditions of diseases (35). Can salivary miRNAs present differently between MPE patients and HCs, and provide a non-invasive way to diagnose MPE?
A less invasive way to diagnose MPE is to detect tumor markers in pleural effusions. In 2015, Nguyen et al. performed a meta-analysis showing the summary estimates of the sensitivity and specificity of pleural CEA, CA 15-3, CA 19-9, CA 125, and CYFRA 21-1 (36). The most efficient marker found in these five tumor markers was CEA, with a sensitivity of 0.549 and specificity of 0.962, which is not more effective than salivary hsa-miR-4484. In 2017, our laboratory detected the concentration of these tumor markers using a chemiluminescent microparticle immunoassay technology, resulting in a much better CEA sensitivity (84.7%) and specificity (90.9%) (37). These results may differ due to the heterogeneity of the population. So, which is better, traditional tumor markers in pleural effusion or salivary miRNAs?
With these questions, we profiled salivary miRNAs to explore if the differentially expressed genes could be used to diagnose MPE. The results are encouraging, given the high sensitivity and specificity. According to qPCR, hsa-miR-4484 and hsa-miR-3663-3p are potential biomarkers to distinguish MPE patients from BPE patients, MT patients, and HCs. The logistic regression model showed a better discriminatory power of the combination of hsa-miR-4484 and hsa-miR-3663-3p. These encouraging results suggest that salivary miRNAs might be used to diagnose molecular oncology events.
Moreover, in further research, we found that the miRNA levels did not correlate with the pleural effusion volume. This result indicates that salivary miRNAs detect MPE in a low MPE volume and overcome the difficult problem that low pleural volume cannot be extracted for traditional diagnostic methods such as pleural effusion cytological analysis or tumor genesis analysis. We know that early detection and diagnosis are important for improving the prognosis in patients with MPE, and salivary miRNA detection shows a new method for MPE early detection.
Comparisons of saliva miRNAs between MPE patients and other groups showed that hsa-miR-4484 was significantly upregulated, and hsa-miR-3663-3p was significantly downregulated in MPE patients. miR-4484 was previously reported as a deleted and downregulated miRNA in glioblastoma, and overexpression of miR-4484 reduced colony-forming ability and suppressed the migratory capacity of glioma cells (38). This result was contrary to our finding that hsa-miR-4484 was upregulated in MPE. The first possible explanation for these contradictory conclusions might be that hsa-miR-4484 showed a different effect on these two diseases, which may have led to opposite consequences at the expression level. A second reason could be related to the various effects of extracellular miRNA and cellular miRNA. Extracellular miRNAs from the saliva supernatant may be intercellular signaling molecules and transduce intercellular signals when flowing (39). Furthermore, according to our results, the salivary hsa-miR-4484 level significantly correlates with the hsa-miR-4484 level in MPE. Thus, a high miR-4484 level in MPE may lead to a high miR-4484 level in saliva. The biological function of hsa-miR-3663-3p has not been reported and remains to be investigated.
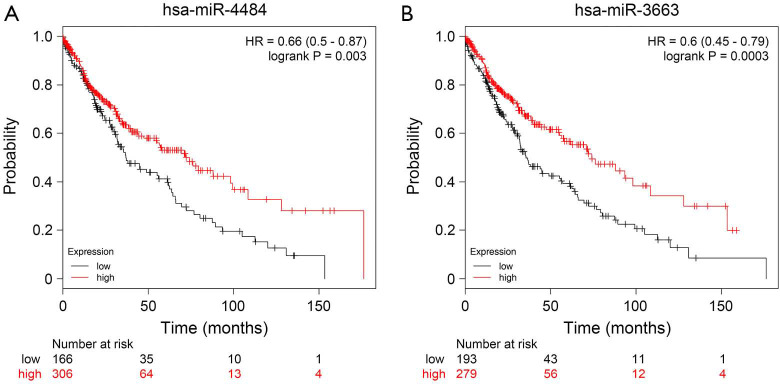
Although we did not obtain the survival information of the enrolled patients, to learn more about the functions of hsa-miR-4484 and hsa-miR-3663-3p, we checked the correlation between these two miRNAs and overall survival in Kaplan-Meier Plotter (http://www.KMplot.com). In lung squamous cell carcinoma patients, these two miRNAs overexpression was related to better overall survival (Figure S2). This result may indicate a protection function of hsa-miR-4484 and hsa-miR-3663 in patients with lung squamous cell carcinoma. It suggests that these two miRNAs have more roles in lung cancer and even in MPE that still need to be discovered. The mechanistic data that explain why MPE can lead to saliva biomarker profile changes and whether these miRNAs are correlated with the patients’ survival of MPE are emerging. Future studies should soon elucidate these mechanisms.
Figure S2.
Kaplan-Meier plots of hsa-miR-4484 (A) and hsa-miR-3663 (B) expression of lung squamous cell carcinoma patients using the program Kaplan-Meier plotter.
To our knowledge, this is the first study to profile salivary miRNAs from MPE patients using microarray analysis. Our analysis suggests a simple and non-invasive method to diagnose MPE early on. It also raises the possibility that salivary components can be used as potential biomarkers to detect systemic diseases. Furthermore, how MPE can lead to the salivary miRNAs profile change is still unclear and waiting to be explained.
Our work has some limitations. First, all MPE patients enrolled in our study were caused by lung cancer. MPE induced by other kinds of malignant diseases will be analyzed in future studies. Second, the sample size was relatively small in this study, with 57 MPE patients, 33 BPE patients, 50 MT patients, and 49 HCs. Finally, all the subjects came from the same country and belonged to the same ethnic group. Thus, multicenter prospective studies with larger sample sizes are needed.
Conclusions
In conclusion, this study showed the characterization of salivary miRNAs collected from MPE patients. Through the discovery and validation phases, we have demonstrated that hsa-miR-4484 and hsa-miR-3663-3p in the saliva are expressed differently between MPE patients and other groups. The combination of these two miRNAs showed a potential discriminatory power for MPE detection, and salivary miRNAs may be used to detect MPE in a low MPE volume. All of these findings suggest that salivary miRNAs can be biomarkers for early and non-invasive detection of MPE.
Supplementary
The article’s supplementary files as
Acknowledgments
We gratefully acknowledge Department of Respiratory and Critical Care Medicine, Beijing Institute of Respiratory Medicine and Beijing Chao-Yang Hospital, Capital Medical University for providing samples for our study and thank all members in this department for their invaluable contributions during this collaboration.
Funding: This work was supported by grants from National Natural Science Foundation of China (No. 81730046 and No. 31470883), Beijing Municipal Administration of Hospitals’ Mission Plan (No. SML20150301), Beijing Nova program (Z171100001117015), Beijing Talents Foundation (2017000021223ZK38) and “1351 Talents Program” of Beijing Chao-Yang Hospital (WXZXZ-2017-01 and CYXX-2017-35).
Supplementary
All primers used in the study
Stem-loop primer for reverse transcription:
❖ h-loop-4484: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGGGC.
❖ h-loop-3663: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCGCCC.
❖ h-loop-U6: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGTGCT.
Universal forward primer and probe for q-PCR:
❖ Universal forward primer: CAGTGCAGGGTCCGAGGTA.
❖ Universal probe: FAM-TCGCACTGGATACGAC-MGB.
Reverse primers for q-PCR:
❖ h-loop-4484-R: TGGCAAAAGGCGGGAGA.
❖ h-loop-3663-R: AGCTGAGCACCACACAGGC.
❖ h-loop-U6-R: AGCGTGCTCGCTTCGG.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of Beijing Chao-Yang Hospital, Beijing, China (# 2018-ke-321). All patients enrolled completed the informed consent form.
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-19-530
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-19-530
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-530). The authors have no conflicts of interest to declare.
References
- 1.Tarn AC, Lapworth R. BTS guidelines for investigation of unilateral pleural effusion in adults. Thorax 2004;59:358-9. [PMC free article] [PubMed] [Google Scholar]
- 2.Light RW. Pleural effusions. Med Clin North Am 2011;95:1055-70. 10.1016/j.mcna.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Wang XJ, Yang Y, Wang Z, et al. Efficacy and safety of diagnostic thoracoscopy in undiagnosed pleural effusions. Respiration 2015;90:251-5. 10.1159/000435962 [DOI] [PubMed] [Google Scholar]
- 4.Light RW. Clinical practice. Pleural effusion. N Engl J Med 2002;346:1971-7. 10.1056/NEJMcp010731 [DOI] [PubMed] [Google Scholar]
- 5.Hooper C, Lee YC, Maskell N. Investigation of a unilateral pleural effusion in adults: British Thoracic Society Pleural Disease Guideline 2010. Thorax 2010;65 Suppl 2:ii4-17. 10.1136/thx.2010.136978 [DOI] [PubMed] [Google Scholar]
- 6.Sugiura S, Ando Y, Minami H, et al. Prognostic value of pleural effusion in patients with non-small cell lung cancer. Clin Cancer Res 1997;3:47-50. [PubMed] [Google Scholar]
- 7.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007;2:706-14. 10.1097/JTO.0b013e31812f3c1a [DOI] [PubMed] [Google Scholar]
- 8.Loddenkemper R. Thoracoscopy--state of the art. Eur Respir J 1998;11:213-21. 10.1183/09031936.98.11010213 [DOI] [PubMed] [Google Scholar]
- 9.Alemán C, Sanchez L, Alegre J, et al. Differentiating between malignant and idiopathic pleural effusions: the value of diagnostic procedures. QJM 2007;100:351-9. 10.1093/qjmed/hcm032 [DOI] [PubMed] [Google Scholar]
- 10.Desai NR, Lee HJ. Diagnosis and management of malignant pleural effusions: state of the art in 2017. J Thorac Dis 2017;9:S1111-22. 10.21037/jtd.2017.07.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts ME, Neville E, Berrisford RG, et al. Management of a malignant pleural effusion: British Thoracic Society Pleural Disease Guideline 2010. Thorax 2010;65 Suppl 2:ii32-40. 10.1136/thx.2010.136994 [DOI] [PubMed] [Google Scholar]
- 12.Liang QL, Shi HZ, Qin XJ, et al. Diagnostic accuracy of tumour markers for malignant pleural effusion: a meta-analysis. Thorax 2008;63:35-41. 10.1136/thx.2007.077958 [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Liu YL, Shi HZ. Diagnostic Accuracy of Combinations of Tumor Markers for Malignant Pleural Effusion: An Updated Meta-Analysis. Respiration 2017;94:62-9. 10.1159/000468545 [DOI] [PubMed] [Google Scholar]
- 14.Wu YB, Xu LL, Wang XJ, et al. Diagnostic value of medical thoracoscopy in malignant pleural effusion. Bmc Pulm Med 2017;17:109. 10.1186/s12890-017-0451-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang MF, Tong ZH, Wang Z, et al. Development and validation of the PET-CT score for diagnosis of malignant pleural effusion. Eur J Nucl Med Mol Imaging 2019;46:1457-67. 10.1007/s00259-019-04287-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao H, Wong DT. Proteomics and its applications for biomarker discovery in human saliva. Bioinformation 2011;5:294-6. 10.6026/97320630005294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Farrell JJ, Zhou H, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010;138:949-57.e1. 10.1053/j.gastro.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Begne M, Lu B, Han X, et al. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res 2009;8:1304-14. 10.1021/pr800658c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc 2006;137:313-21. 10.14219/jada.archive.2006.0180 [DOI] [PubMed] [Google Scholar]
- 20.Pfaffe T, Cooper-White J, Beyerlein P, et al. Diagnostic potential of saliva: current state and future applications. Clin Chem 2011;57:675-87. 10.1373/clinchem.2010.153767 [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Arellano M, Boontheung P, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res 2008;14:6246-52. 10.1158/1078-0432.CCR-07-5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Wang J, Meijer J, et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum 2007;56:3588-600. 10.1002/art.22954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, St JM, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 2004;10:8442-50. 10.1158/1078-0432.CCR-04-1167 [DOI] [PubMed] [Google Scholar]
- 24.Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 2009;15:5473-7. 10.1158/1078-0432.CCR-09-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: a feasibility study. J Oral Pathol Med 2006;35:292-300. 10.1111/j.1600-0714.2006.00427.x [DOI] [PubMed] [Google Scholar]
- 26.Kaufman E, Lamster IB. The diagnostic applications of saliva--a review. Crit Rev Oral Biol Med 2002;13:197-212. 10.1177/154411130201300209 [DOI] [PubMed] [Google Scholar]
- 27.Bigler LR, Streckfus CF, Dubinsky WP. Salivary biomarkers for the detection of malignant tumors that are remote from the oral cavity. Clin Lab Med 2009;29:71-85. 10.1016/j.cll.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 28.Gao K, Zhou H, Zhang L, et al. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One 2009;4:e5875. 10.1371/journal.pone.0005875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Xiao H, Zhou H, et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 2012;69:3341-50. 10.1007/s00018-012-1027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z, Yin X, Gong B, et al. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res (Phila) 2015;8:165-73. 10.1158/1940-6207.CAPR-14-0192 [DOI] [PubMed] [Google Scholar]
- 31.Xie Z, Zhou F, Yang Y, et al. Lnc-PCDH9-13:1 Is a Hypersensitive and Specific Biomarker for Early Hepatocellular Carcinoma. Ebiomedicine 2018;33:57-67. 10.1016/j.ebiom.2018.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rio DC, Ares MJ, Hannon GJ, et al. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010;2010:t5439. [DOI] [PubMed]
- 33.Kramer MF. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol 2011;Chapter 15:10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Wong DT. Towards a simple, saliva-based test for the detection of oral cancer 'oral fluid (saliva), which is the mirror of the body, is a perfect medium to be explored for health and disease surveillance'. Expert Rev Mol Diagn 2006;6:267-72. 10.1586/14737159.6.3.267 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen AH, Miller EJ, Wichman CS, et al. Diagnostic value of tumor antigens in malignant pleural effusion: a meta-analysis. Transl Res 2015;166:432-9. 10.1016/j.trsl.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai K, Wang W, Wang Y, et al. Diagnostic accuracy of tumor markers for malignant pleural effusion: a derivation and validation study. J Thorac Dis 2017;9:5220-9. 10.21037/jtd.2017.11.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawaz Z, Patil V, Thinagararjan S, et al. Impact of somatic copy number alterations on the glioblastoma miRNome: miR-4484 is a genomically deleted tumour suppressor. Mol Oncol 2017;11:927-44. 10.1002/1878-0261.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol 2010;3:478-81. 10.4161/cib.3.5.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as