Abstract
Background
Robust imaging biomarkers are needed for risk stratification in stage I lung adenocarcinoma patients in order to select optimal treatment regimen. We aimed to construct and validate a radiomics nomogram for predicting the disease-free survival (DFS) of patients with resected stage I lung adenocarcinoma, and further identifying candidates benefit from adjuvant chemotherapy (ACT).
Methods
Using radiomics approach, we analyzed 554 patients’ computed tomography (CT) images from three multicenter cohorts. Prognostic radiomics features were extracted from computed tomography (CT) images and selected using least absolute shrinkage and selection operator (LASSO) Cox regression model to build a radiomics signature for DFS stratification. The biological basis of radiomics was explored in the Radiogenomics dataset (n=79) by gene set enrichment analysis (GSEA). Then a nomogram that integrated the signature with these significant clinicopathologic factors in the multivariate analysis were constructed in the training cohort (n=238), and its prognostic accuracy was evaluated in the validation cohort (n=237). Finally, the predictive value of nomogram for ACT benefits was assessed.
Results
The radiomics signature with higher score was significantly associated with worse DFS in both the training and validation cohorts (P<0.001). The GSEA presented that the signature was highly correlated to characteristic metabolic process and immune system during cancer progression. Multivariable analysis revealed that age (P=0.031), pathologic TNM stage (P=0.043), histologic subtype (P=0.010) and the signature (P<0.001) were independently associated with patients’ DFS. The integrated radiomics nomogram showed good discrimination performance, as well as good calibration and clinical utility, for DFS prediction in the validation cohort. We further found that the patients with high points (point ≥8.788) defined by the radiomics nomogram obtained a significant favorable response to ACT (P=0.04) while patients with low points (point <8.788) showed no survival difference (P=0.7).
Conclusions
The radiomics nomogram could be used for prognostic prediction and ACT benefits identification for patient with resected stage I lung adenocarcinoma.
Keywords: Radiomics, adjuvant chemotherapy (ACT), lung adenocarcinoma, disease-free survival (DFS), gene set enrichment analysis (GSEA)
Introduction
Lung adenocarcinoma remains a critical challenge for global public health as it gradually evolves into the largest subtype of non-small-cell lung cancer, with its proportion increasing to over 60% (1). Complete surgical excision is conducted as the primary intervention for patients with stage I lung adenocarcinoma, but the 5-year recurrence rate of these patients reaches 30% (2). Because postoperative recurrence results in an extremely poor survival outcome, adjuvant chemotherapy (ACT) has been used in an attempt to reduce its occurrence. The absolute survival improvement of ACT for patients with resected stage II-III lung cancer has been endorsed (3,4). But its effect in stage I disease remains undetermined, especially in the stage IB (5,6). Previous studies revealed that the adoption of ACT was beneficial for some patients with stage I disease but harmful for those who could be cured by surgery alone (7,8). Therefore, a more accurate stratification within the same stage may allow for identifying low and high-risk patients for recurrence and selecting those with more likelihoods to benefit from ACT.
Medical imaging is a vital technology in clinical management to aid decision making and direct personalized treatment (9). Radiomics, as an advanced imaging analytic process for extracting high dimensional features from medical images, has been demonstrated with promising performance in prognosis evaluation for various types of cancers (10-12). A nomogram integrating the radiomic biomarkers with clinicopathological features appears to improve the prognostic accuracy (13). To date, there are five radiomics nomograms have been developed for prognosis prediction in lung adenocarcinoma (Table S1). But a radiomics nomogram research which investigates its association with survival of stage I lung adenocarcinoma and further guides candidates’ selection for ACT, has not yet been fully reported.
Table S1. Radiomic nomograms for prognosis prediction of lung adenocarcinoma in the literatures.
| Literature | Year | Patient | Sample size | Variates included in nomogram |
|---|---|---|---|---|
| Huang et al. (13) | 2016 | Early stage (I and II) NSCLC | 282 | Radiomic signature, age, clinical stage, histologic grade, gender |
| Desseroit et al. (44) | 2016 | Stage I–IIIb NSCLC | 116 | Overall stage, selected radiomics features (metabolic volume, PET entropy, CT zone percentage) |
| Wang et al. (45) | 2019 | Advanced NSCLC | 118 | Radiomics signature, age, lymph node, lymphocyte2, NLR1 |
| Yang et al. (46) | 2019 | Stage I–IIIb NSCLC | 371 | Radiomics signature, age, sex, T stage, N stage |
| Akinci D’Antonoli et al. (47) | 2020 | stage Ia–IIb NSCLC | 124 | Radiomics signature, overall stage |
Therefore, the aim of present study was to construct and validate a nomogram based on radiomics and clinical features to estimate the DFS in resected stage I lung adenocarcinoma patients and to further explore its potential value for predicting survival benefit from ACT. Additionally, we intended to explore the potential biological basis of radiomics with imaging and gene expression data.
We present the following article in accordance with the STROBE Reporting Checklist (available at http://dx.doi.org/10.21037/tlcr-19-577).
Methods
Data collection
The study was approved by The Institutional Review Boards of Shanghai Pulmonary Hospital and the informed consent was waived for this retrospective study (No. k19-134Y). Consecutive patients who received complete resection for lung cancer in Shanghai Pulmonary Hospital from January 2011 to December 2012 were retrieved. The patients diagnosed with stage I adenocarcinoma according to the eighth edition TNM staging system were identified (14). Patients were excluded based on the following criteria: (I) patients without thin-slice CT images (1 mm) within one month prior to surgery; (II) patients with incomplete clinicopathologic data and follow-up records; (III) patients whose lesions cannot be accurately segmented. The patients undergone surgical resection before December 2011 were set as the training cohort (n=238), and after December 2011 as an independent validation cohort (n=237). In addition, a dataset comprised CT imaging data and matched RNA sequencing data of 79 resected stage I adenocarcinoma patients was obtained from the NSCLC Radiogenomics Dataset (15) of the Cancer Imaging Archive (TCIA) to evaluate the biological process of radiomics signature.
The follow-up data was obtained through electronic medical records and telephone interviews as a complete. We set the endpoint of this study as disease-free survival (DFS), which was defined as the duration from the date of surgery to that of recurrence, death, or the last follow-up. The histologic subtype was re-evaluated according to the current lung adenocarcinoma classification system (16).
Imaging data acquisition and radiomics processing
All details of CT image acquisition protocols in our study were described in Supplementary material I. After downloading CT images from the institutional Picture Archiving and Communication Systems with DICOM format, tumor segmentation was performed in an open-source software (3D-slicer, v4.10.1, www.slicer.org) (17). The regions of interest (ROI) on CT images were manually delineated layer-by-layer by two junior radiologists (T.W. and Y.Y., with 3 and 5 years of experience, respectively) and verified by a senior radiologist (X.S., with 30 years of experience in chest imaging diagnosis). An open-source platform, PyRadiomics in Python, was utilized to extract 107 radiomics features from the non-filtered segmented ROI (18). The details of radiomics features were descripted in Supplementary material II and Table S2.
Table S2. Extracted radiomics features.
| Intensity | Shape | Texture | ||||
|---|---|---|---|---|---|---|
| GLCM | GLRLM | GLSZM | NGTDM | GLDM | ||
| 10 Percentile | Elongation | Autocorrelation | Gray level nonuniformity | Gray level nonuniformity | Busyness | Busyness |
| 90Percentile | Flatness | Cluster Prominence | Gray level nonuniformity normalized | Gray level nonuniformity normalized | Coarseness | Coarseness |
| Energy | Least axis length | Cluster Shade | Gray level variance | Gray level variance | Complexity | Complexity |
| Entropy | Major axis length | Cluster Tendency | High gray level run emphasis | High gray level zone emphasis | Contrast | Contrast |
| Interquartile range | Maximum 2D diameter column | Contrast | Long run emphasis | Large area emphasis | Strength | Strength |
| Kurtosis | Maximum 2D diameter row | Correlation | Long run high gray level emphasis | Large area high gray level emphasis | Dependence entropy | |
| Maximum | Maximum 2D diameter slice | Difference Average | Long run low gray level emphasis | Large area low gray level emphasis | Dependence nonuniformity | |
| Mean absolute deviation | Maximum 3D diameter | Difference Entropy | Low gray level run emphasis | Low gray level zone emphasis | Dependence nonuniformity normalized | |
| Mean | Mesh volume | Difference Variance | Run entropy | Size zone nonuniformity | Dependence variance | |
| Median | Minor axis length | Id | Run length nonuniformity | Size zone nonuniformity normalized | Gray level nonuniformity | |
| Minimum | Sphericity | Idm | Run length nonuniformity normalized | Small area emphasis | Gray level variance | |
| Range | Surface area | Idmn | Run percentage | Small area high gray level emphasis | High gray level emphasis | |
| Robust mean absolute deviation | Surface volume ratio | Idn | Run variance | Small area low gray level emphasis | Large dependence emphasis | |
| Root mean squared | Voxel volume | Imc1 | Short run emphasis | Zone entropy | Large dependence high gray level emphasis | |
| Skewness | Imc2 | Short run high gray level emphasis | Zone percentage | Large dependence low gray level emphasis | ||
| Total energy | Inverse variance | Short run low gray level emphasis | Zone variance | Low gray level emphasis | ||
| Uniformity | Joint average | Small dependence emphasis | ||||
| Variance | Joint energy | Small dependence high gray level emphasis | ||||
| Joint entropy | Small dependence low gray level emphasis | |||||
| MCC | ||||||
| Maximum probability | ||||||
| Sum average | ||||||
| Sum entropy | ||||||
| Sum squares | ||||||
Radiomics feature selection and signature construction
We determined the optimum cutoff value for every radiomics feature based on its association with the patients’ DFS using X-tile program (version 3.6.1, Yale University School of Medicine, New Haven, CT, USA) (19). Accordingly, the significant features (P<0.05) which could divide patients into different risk groups were selected. To reduce the redundancy, the least absolute shrinkage and selection operator (LASSO) Cox regression model was adopted to select the optimal radiomics feature subset. Then a radiomics signature was obtained through the selected features and their respective weighted coefficients.
Survival analysis based on the radiomics signature
The radiomics signature stratified the patients of training cohort into low- and high-risk groups based on its optimal cutoff value which was identified by X-tile program. The Kaplan-Meier survival curves and log-rank test were delineated to seek the survival difference between the risk groups. These calculation and cutoff value were applied to the validation cohort. Meanwhile, subgroups analyses based on the clinicopathologic risk factors were performed for its robustness assessment.
Gene set enrichment analysis (GSEA)
We performed a GSEA to explore the biological basis of the developed radiomics signature for prognosis prediction. The RNA-seq expression data in Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values for all samples were obtained from GSE103584 and transformed to log2 (FPKM + 0.1). Genes with missing expression values in more than 50% of all samples were removed, resulting in 12,901 genes. Then we created a rank of these genes using a fold change which was defined as the difference value the mean gene expression between the high- and low-risk group. The pre-ranked GSEA was performed using the PIANO R package (20) with 10,000 permutations. As gene sets, we tested expert-curated pathways from the C2 Reactome collection version 7.0 available at MSigDB (21). Gene sets were restricted to sizes between 15 and 500, resulting in 805 tested gene sets. The Enrichment Score (ES) was utilized to quantify the association of the rank of genes with pathways and validated with false discovery rate (FDR) to corrected for multiple comparison.
Development and validation of the radiomics nomogram
The univariate and multivariate Cox regression analyses with stepwise selection were carried out in the training cohort to identify the independent prognostic factors. Then a nomogram was applied for DFS prediction visualization. Corresponding calibration curves was developed in the validation cohort; the Harrell concordance index (C-index) quantified its discrimination ability; a decision curve analysis exhibited its clinical utility by measuring the net benefits at different threshold probabilities.
ACT benefit analysis based on the nomogram
A polynomial equation extracted from the prognostic nomogram were used for calculating the total risk point, which might reflect the weight of each predictor and correspond to the survival probability. The optimal cutoff risk point of stage IB patients was identified to divide them into low- and high-risk groups and the survival analyses were performed to evaluate the benefits of ACT.
Statistical analysis
Patients’ baseline characteristics in two cohorts were compared using analyses of independent t test for continuous variates and a chi-squared test for categorical variates. Statistical analyses were accomplished via R software, version 3.5.3 (http://www.R-project.org) and SPSS for Windows, version 20.0 (IBM, Armonk, NY, USA). The packages used in R programming were showed in Supplementary material III. A two-sided P value below 0.05 was considered statistically significant.
Results
Clinicopathologic characteristics
The baseline characteristics of all included patients were listed in Table 1. The median follow-up time were 66.3 and 64.7 months, the 5-year DFS with 80.3% and 79.5% in the training and validation cohorts, respectively. There was no significant difference between two cohorts in terms of clinicopathologic factors or follow-up time.
Table 1. The baseline characteristics of 475 patients with resected stage I lung adenocarcinoma in the training and validation cohorts.
| Characteristic | Training cohort, N=238, n (%) | Validation cohort, N=237, n (%) | P value |
|---|---|---|---|
| Age, years | 0.436 | ||
| <65 | 165 (69.3) | 172 (72.6) | |
| ≥65 | 73 (30.7) | 65 (27.4) | |
| Gender | 0.966 | ||
| Female | 129 (54.2) | 128 (54.0) | |
| Male | 109 (45.8) | 109 (46.0) | |
| Smoking | 0.107 | ||
| No | 164 (68.9) | 179 (75.5) | |
| Yes | 74 (31.1) | 58 (24.5) | |
| p-TNM stage | 0.383 | ||
| IA | 121 (50.8) | 111 (46.8) | |
| IB | 117 (49.2) | 126 (53.2) | |
| Surgery types | 0.396 | ||
| Lobectomy | 221 (92.9) | 215 (90.7) | |
| Sub-lobar resection | 17 (7.1) | 22 (9.3) | |
| Histologic subtype | 0.589 | ||
| LPA | 83 (34.9) | 83 (35.0) | |
| APA | 91 (38.2) | 88 (37.1) | |
| PPA | 37 (15.5) | 46 (19.4) | |
| SPA | 17 (7.1) | 15 (6.3) | |
| MPA | 10 (4.2) | 5 (2.1) | |
| Pathologic tumor size, mm, mean ± SD | 20.2±8.2 | 20.1±8.4 | 0.948 |
| Follow-up time, median | 66.3 | 64.7 | 0.054 |
SD, standard deviation; LPA, lepidic predominant adenocarcinoma; APA, acinar predominant adenocarcinoma; PPA, papillary predominant adenocarcinoma; MPA, micropapillary pattern-predominant adenocarcinoma; SPA, solid predominant adenocarcinoma.
Radiomics feature-based signature construction
A total of 58 radiomics features were significant (P<0.05) for risk stratification in training cohort. Then an optimal feature subset consisting of eight radiomics features, including maximum, minimum, IDN, joint energy, long run low gray level emphasis, gray level variance, coarseness, and large dependence emphasis, were identified mostly related to the DFS by the LASSO Cox regression analysis (Figure S1). The radiomics score (rad-score), was constructed with the calculation formula generating in the LASSO model (Supplementary material IV).
Figure S1.
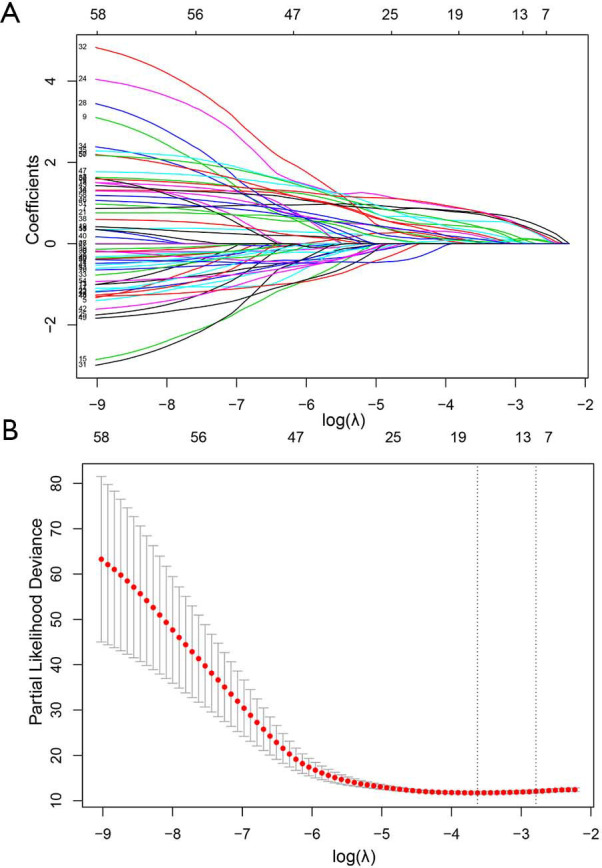
Radiomics feature selection using the least absolute shrinkage and selection operator (LASSO) regression model. (A) LASSO coefficient profiles of the radiomics features. As the tuning parameter (λ) increased using 5-fold cross-validation, more coefficients tended to approach 0 and the optimal non-zero coefficients generated, which yielded a set of the optimal radiomics features; (B) the partial likelihood deviance from the LASSO regression cross-validation procedure was plotted against log(λ).
Survival analysis based on the radiomics signature
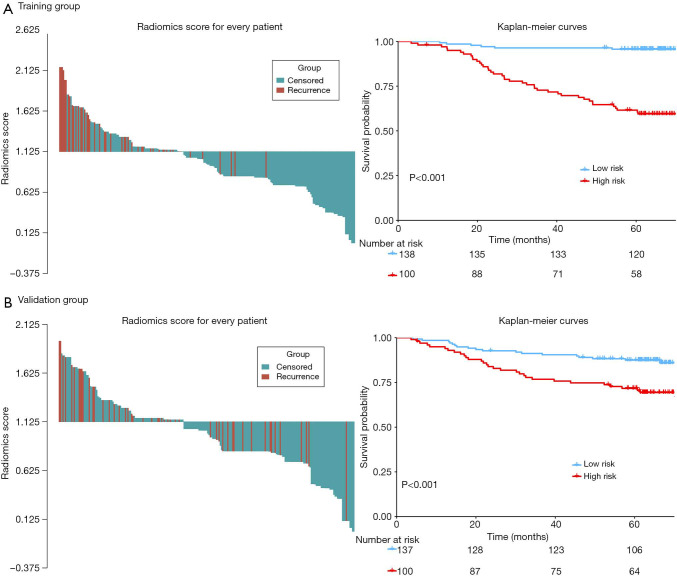
The distribution of rad-score and survival status indicated that patients with higher score generally had worse survival than did those with lower score (Figure 1, left panel). The optimal cutoff value of rad-score was 1.125, which divided all patients into low- (rad-score <1.125) and high-risk group (rad-score ≥1.125). The survival analyses showed that a significant difference existed between these groups (P<0.001) both in the training and validation cohorts (Figure 1, right panel). Furthermore, the survival analyses applied in the clinicopathologic subgroups of all patients, consisting of age, sex, pathological TNM stage and histologic subtype, further proved that this signature remained significant for survival stratification (P<0.01, Figure S2).
Figure 1.
The distribution of radiomics score (left panel) and KM survival (right panel) in the training (A) and validation cohorts (B). The P values of survival curves were calculated using the log-rank test.
Figure S2.
Kaplan-Meier survival analysis for all 475 patients with stage I lung adenocarcinoma according to the eight-feature-based radiomics signature stratified by clinicopathological risk factors.
Biological basis of the radiomics signature
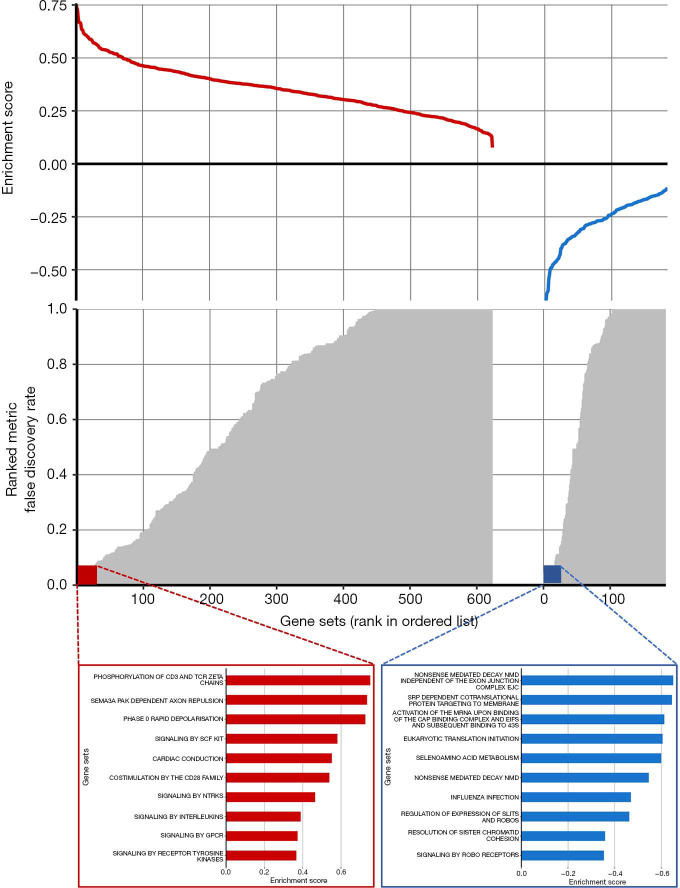
The biological basis of the radiomic signature was evaluated in the independent Radiogenomics dataset with CT images and RNA-sequence-based gene expression data. The pre-ranked GSEA showed that the significant enriched pathways (FDR <0.1) among the top associations with the radiomics signature were mostly correlated to various metabolic processes and immune system (Figure 2). This revealed that the developed imaging biomarker might reflect the different characteristic metabolic changes and immune system during the cancer progression, which could provide more potential for early prognosis prediction of lung adenocarcinoma.
Figure 2.
Gene set expression patterns—Radiogenomics dataset. The radiomics signature was linked to gene expression patterns using a pre-ranked gene set enrichment analysis (GSEA). Positive and negative enrichments are shown in red and blue, respectively. The top 10 enrichments in each category are highlighted.
Performance analysis of the radiomics nomogram
In the multivariate Cox regression analysis (Table 2), age (HR: 1.893; 95% CI: 1.061–3.379; P=0.031), pathologic TNM stage (HR: 2.109; 95% CI: 1.039–4.338; P=0.043), histologic subtype (LPA: reference; APA/PPA: HR: 2.703; 95% CI: 1.039–7.035, P=0.042; MPA/SPA: HR: 5.300; 95% CI: 1.782–15.767, P=0.003) and the radiomics signature (HR: 7.794; 95% CI: 3.185–19.078; P<0.001) were identified as independent predictors for DFS of patients with stage I adenocarcinoma.
Table 2. Univariate and multivariate analysis of disease-free survival for patients in training cohort.
| Variables | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age (≥65 vs. <65) | 2.506 (1.414–4.442) | 0.002 | 1.893 (1.061–3.379) | 0.031 | |
| Gender (Male vs. Female) | 2.120 (1.176–3.819) | 0.012 | 1.568 (0.850–2.890) | 0.150 | |
| Smoking status (Yes vs. No) | 1.602 (0.894–2.871) | 0.113 | NA | NA | |
| p-TNM stage (IB vs. IA) | 4.243 (2.109–8.535) | <0.001 | 2.109 (1.039–4.338) | 0.043 | |
| Operation type (Lobe vs. Sub-lobe) | 0.966 (0.300–3.115) | 0.954 | NA | NA | |
| Histologic subtype | 0.001 | 0.010 | |||
| LPA | 1.000 (Reference) | 1.000 (Reference) | |||
| APA/PPA | 4.720 (1.839–12.118) | 0.001 | 2.703 (1.039–7.035) | 0.042 | |
| MPA/SPA | 7.854 (2.682–22.998) | 0.002 | 5.300 (1.782–15.767) | 0.003 | |
| Pathologic tumor size | 1.763 (1.266–2.455) | <0.001 | 1.097 (0.779–1.545) | 0.596 | |
| Radiomics signature | 11.528 (4.890–27.178) | <0.001 | 7.794 (3.185–19.078) | <0.001 | |
HR, hazard ratio; CI, confidence interval; LPA, lepidic predominant adenocarcinoma; APA, acinar predominant adenocarcinoma; PPA, papillary predominant adenocarcinoma; MPA, micropapillary pattern-predominant adenocarcinoma; SPA, solid predominant adenocarcinoma.
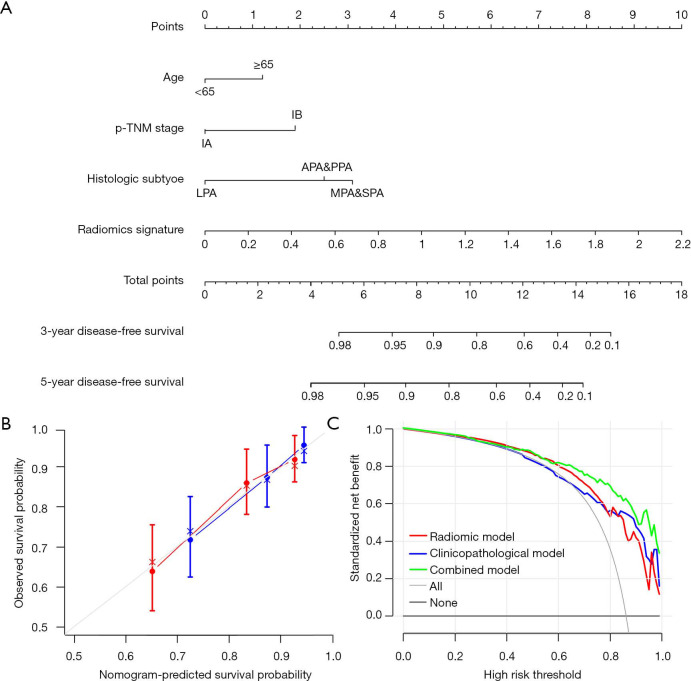
Then a nomogram that incorporated these independent clinical factors and radiomics signature was generated (Figure 3A). We found that the radiomics nomogram showed good discrimination performance (C-index: 0.713; 95% CI: 0.646–0.780) in the validation cohort, as well as satisfactory agreements among prediction outcomes and actual observations (Figure 3B). The decision curves analysis exhibited that the integrating radiomics nomogram achieved higher clinical usefulness relative to the clinicopathologic factors and radiomics signature alone (Figure 3C).
Figure 3.
A prediction performance analysis of patients with stage I lung adenocarcinoma. (A) The nomogram for predicting 3- and 5-year DFS after surgery; (B) plots depict the calibration of the nomogram in terms of agreement between predicted and observed 3- and 5-year DFS. Performance of the validation cohort was shown on the plot relative to the 45-degree line, which represented perfect prediction; (C) decision curve analysis for the comparison of prognostic factors. The y-axis measures the net benefit.
ACT benefit analysis based on the radiomics nomogram
The adoption of ACT (n=129/243) did not show survival benefit in all patients with stage IB lung adenocarcinoma (P=0.11, Figure S3). The polynomial equation (Supplementary material V) extracted from the radiomics nomogram was used to calculate the total risk point for all stage IB patients and divided them into low- and high-point groups with its optimal cutoff value. Compared with the patients in the low-group, the high-point group accounted for more patients with older age, male, micropapillary and solid predominant adenocarcinoma, larger tumor size and higher radiomics signature (P<0.05, Table 3). Interestingly, while patients with a low risk point (risk point <8.788) defined by the radiomics nomogram showed no survival difference with or without ACT (P=0.7; Figure 4A), patients with high point (total point ≥8.788) obtained a favorable response to ACT (P=0.04; Figure 4B). These results indicated that the integrated radiomics nomogram had great potential to successfully select these high-risk patients with stage IB lung adenocarcinoma who were suitable candidates for ACT.
Figure S3.
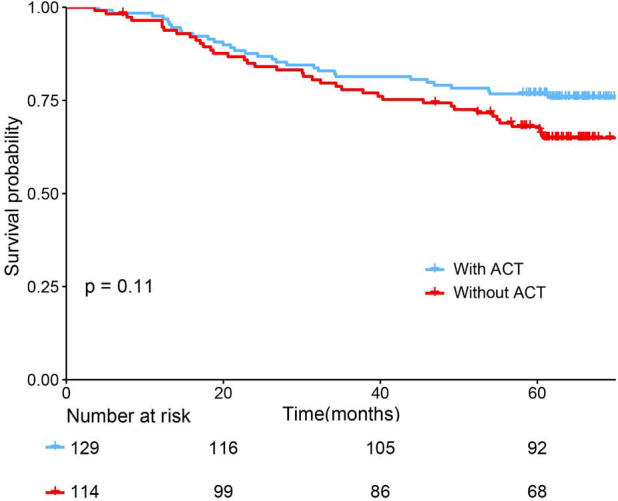
The Benefit Analysis of adjuvant chemotherapy (ACT) in All Patients with Stage IB Adenocarcinoma. These patients showed no survival difference between with and without ACT.
Table 3. The Clinicopathological features of 243 patients with resected stage IB lung adenocarcinoma in the low- and high-risk groups defined by the radiomics nomogram.
| Characteristic | Low-group, N=129, n (%) | High-group, N=114, n (%) | P value |
|---|---|---|---|
| Age, years | <0.001 | ||
| <65 | 102 (79.1) | 62 (54.4) | |
| ≥65 | 27 (20.9) | 52 (45.6) | |
| Gender | 0.004 | ||
| Female | 86 (66.7) | 55 (48.2) | |
| Male | 43 (33.3) | 59 (51.8) | |
| Smoking | 0.467 | ||
| No | 97 (75.2) | 81 (71.1) | |
| Yes | 32 (24.8) | 33 (28.9) | |
| Surgery types | 0.225 | ||
| Lobectomy | 125 (96.9) | 107 (93.9) | |
| Sub-lobar resection | 4 (3.1) | 7 (6.1) | |
| Histologic subtype | <0.001 | ||
| LPA | 55 (42.6) | 17 (14.9) | |
| APA | 48 (37.2) | 51 (44.7) | |
| PPA | 25 (19.4) | 20 (17.5) | |
| SPA | 0 (0.0) | 18 (15.8) | |
| MPA | 1 (0.8) | 8 (7.0) | |
| Pathologic tumor size, mm, mean ± SD | 21.9±7.8 | 24.8±8.9 | 0.007 |
| Radiomics signature, mean± SD | 0.789±0.336 | 1.373±0.297 | <0.001 |
SD, standard deviation; LPA, lepidic predominant adenocarcinoma; APA, acinar predominant adenocarcinoma; PPA, papillary predominant adenocarcinoma; MPA, micropapillary pattern-predominant adenocarcinoma; SPA, solid predominant adenocarcinoma.
Figure 4.
The Benefit analysis of adjuvant chemotherapy (ACT) in different subgroups. (A) Patients in the low-risk groups defined by the nomogram showed no survival difference between with and without ACT; (B) patients in the high-risk groups defined by the nomogram could obtain survival benefits from ACT.
Discussion
Patients with stage I lung adenocarcinoma have substantial risks for recurrence even after complete resection, and whether ACT could provide survival benefit remains controversial. Postoperative DFS evaluation is vital for guiding a patient’s individualized follow-up strategies and subsequent treatment option. In this research, the constructed radiomics signature could successfully categorize stage I adenocarcinoma patients into high- and low-risk groups with significant different in DFS and the integrated radiomics nomogram could identify the suitable patients benefit from ACT. Furthermore, the genomic studies revealed the correlations between radiomics signature and tumor metabolic changes and immune system.
The analysis of medical images has gradually evolved from subjective operator-dependent criteria to a more objective and quantitative evaluation (22,23). Radiomics could provide additional information in oncologic practice related to benign and malignant nodules differentiation (24), mutation types identification (10), subtype classification (25) and treatment response assessment (26). As for prognostic prediction based on lung CT, Lee et al. (27) found that two radiomics features of 339 patients were independent prognostic factor of lung adenocarcinoma survival. However, this method of describing gross lesions with few single features required further improvement as it might cause an underestimation of the application of radiomics (28). Hence, in radiomics analysis, a multi-feature-based radiomics signature has significantly greater prognostic accuracy than that of each selected feature alone, considering the interactions between different features (12,13,29). In our study, eight-feature-consisted radiomics signature based on multicenter cohorts of 475 patients reached a favorable performance for survival assessment of stage I lung adenocarcinoma.
As for the administration of ACT in lung adenocarcinoma, current guidelines do not recommend its adoption in stage IA disease but remain more ambiguous in stage IB disease (30,31). Our data supported that the adoption of ACT in all stage IB patients without selection could not bring survival benefit (P=0.11). Numerous studies have demonstrated that stage IB patients with older age are associated with greater recurrence risk (8,32,33). The histologic features of lung adenocarcinoma, especially the micropapillary and solid predominant patterns, remained independent predictor for evaluating tumor recurrence and ACT survival benefits (3,34-36). Our study is consistent with these studies. Moreover, we add to these findings by demonstrating that the radiomics nomogram integrating the proposed imaging signature with these clinicopathologic predictors is more clinically relevant because accurate survival prediction allows for good identification of patients who derive therapeutic benefit from ACT. The radiomics signature provides the incremental value for guiding the adoption of ACT in patients with stage IB disease that complemented clinicopathologic factors (37).
With respect to elaborating the underlying mechanism that drives biological process ultimately reflected in imaging phenotypes, series studies published by Aert et al. (38-40) have found that some biological pathways related to lung cancer prognosis, including mitosis, cell cycle and transcription, can be captured by imaging biomarkers. Our study used a GSEA in an external cohort to reveal how imaging biomarkers can be connected to postoperative prognosis, suggesting that different tumor metabolism and immune system change may result in different survival outcomes. Cancers have much more complex metabolic pathways to support tumor cell growth and proliferation (41). The radiomics features for constituting the signature can correlate with these metabolic pathways and immune system editing, which allows for tumor development and progression, and thereby worsening the prognosis (40,42).
Some limitations existed in our study besides for those commonly associated with retrospective studies. First, despite the constructed signature has been verified a favorable performance in identifying high-risk patients for recurrence and suitable candidates for ACT benefit on an independent validation cohort, multicenter datasets were still necessary for validating its robustness and generalization. Second, the task of ROIs delineation was completed in a manual method which was commonly considered as a gold criterion for images processing at present (43). However, the exploitation of a more efficient and perfectly accurate method for lesion segmentation will be an important direction in the field of future radiomics research. Third, only adenocarcinoma patients were enrolled in this study. Due to its enormous proportion in lung cancer and the heterogeneity in tumor behavior, a homogeneous study cohort only including lung adenocarcinoma patients may be better for the model analysis of radiomics.
In conclusion, our results suggest that the developed radiomics signature may successfully categorize stage I adenocarcinoma patients into high- and low-risk groups of disease recurrence, and the integrated radiomics nomogram may further identify the suitable patients benefit from ACT. Moreover, we evaluated the biological basis of the proposed signature and found to be correlated with different characteristic metabolic changes and immune system. These findings may potentially facilitate the clinical impact in guiding pertinent therapeutic administration and improved survival outcomes of patients with stage I lung adenocarcinoma.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors wish to thank Prof. Zhang Aihong for the design and guidance of statistical analysis in this study.
Funding: This study was supported by Shanghai Health Commission (2019SY072 & 2018ZHYL0102), Shanghai Pulmonary Hospital Research Fund (FK18001 & FKGG1805), Natural Science Foundation of Shanghai (19ZR1443100), and Shanghai municipal hospital (SHDC22015037).
Supplementary
Supplementary material I. CT Image acquisition protocol
All patients in our study underwent by a non-enhanced contrast CT scans by a Somatom Definition AS (Siemens Medical Systems, Germany) or Brilliance 40 (Philips Medical Systems, Netherlands). The parameters of above two scanners were listed as follows respectively: a. tube energy as 120 kVp, tube current as 130mA, rotation time as 0.5 s, detector collimation as 64 × 0.625 mm2; b. tube energy as 120 kV, tube current as 200 mA, rotation time as 0.75 s, detector collimation as 32 × 1.25 mm2. The images were reconstructed at 1.0 mm slice thickness and 0.7 mm increment by the standard soft kernel (Siemens B31 filter, Siemens Medical Solutions, Forchheim, Germany) and sharp reconstruction kernel (C filter, Philips, Cleveland, OH).
Supplementary material II. Extracted radiomics feature
The quantified image features were classified into three categories: (I) intensity(n=18): these features quantify the density characteristics of tumor region using first order histogram statistics of all voxel intensities, (II) shape (n=14): shape features describe the 3D geometric properties of the tumor, and (III) texture (n=75): these features were computed to analyze the spatial distribution of voxel intensities and then describe the intratumor heterogeneity. Texture features are derived from gray level co-occurrence matrix (GLCM) (n=24), run length matrices (GLRLM) (n=16), gray level size zone matrix (GLSZM) (n=16), neighboring gray tone difference matrix (NGTDM) (n=5) and gray level dependence matrix (GLDM) (n=14) (18).
Supplementary material III. Packages used in R programming
LASSO Cox regression was performed using the “glmnet” package. The waterfall plots were obtained by the “RColorBrewer” package. Survival analysis was performed with the “survival” package. Time-dependent receiver operating characteristic (ROC) analysis was achieved by the “pROC” package. Nomograms and calibration plots were generated with the “rms” package. Comparisons between C-indexes were performed with the “survcomp” package. Decision curve analysis was performed with the “RMDA” package. The equations extraction from a nomogram was performed with the “nomogramEX” package.
Supplementary material IV. Calculation formula for radiomics signature
Radiomics score = 0.038309518 × First Order_ Maximum + 0.521437365 × First Order_ Minimum + 0.119876232 × GLCM_Idn + 0.336529265 × GLCM_Joint energy + 0.321377301 × GLRLM_Long run low gray level emphasis + 0.378193705 × GLSZM_Gray level variance + 0.108937141 × NGTDM_Coarseness + 0.337786524 × GLDM_Large dependence emphasis.
Supplementary material V. The polynomial equation extracted from the radiomics nomogram
Total point = 0.0 × 0 (Age <65) + 1.210228 × 1 (Age >=65) + 0.0 × 0 (Pathological stage: IA) + 1.896196 × 1 (Pathological stage IB) + 0.0 × 0 (Histologic subtype: LPA) + 2.496489 × 1 (Histologic subtype: APA & PPA) + 3.092025 × 1 (Histologic subtype: MPA & SPA + 4.545455 * Radiomics signature.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by The Institutional Review Boards of Shanghai Pulmonary Hospital and the informed consent was waived for this retrospective study (No. k19-134Y).
Footnotes
Reporting Checklist: The authors have completed the STROBE Reporting Checklist. Available at http://dx.doi.org/10.21037/tlcr-19-577
Data Sharing Statement: available at http://dx.doi.org/10.21037/tlcr-19-577
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-577). The authors have no conflicts of interest to declare.
References
- 1.Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438-46. 10.1200/JCO.2011.37.2185 [DOI] [PubMed] [Google Scholar]
- 2.Hung JJ, Jeng WJ, Hsu WH, et al. Predictors of Death, Local Recurrence, and Distant Metastasis in Completely Resected Pathological Stage-I Non–Small-Cell Lung Cancer. J Thorac Oncol 2012;7:1115-23. 10.1097/JTO.0b013e31824cbad8 [DOI] [PubMed] [Google Scholar]
- 3.Kris MG, Gaspar LE, Chaft JE, et al. Adjuvant Systemic Therapy and Adjuvant Radiation Therapy for Stage I to IIIA Completely Resected Non-Small-Cell Lung Cancers: American Society of Clinical Oncology/Cancer Care Ontario Clinical Practice Guideline Update. J Clin Oncol 2017;35:2960-74. 10.1200/JCO.2017.72.4401 [DOI] [PubMed] [Google Scholar]
- 4.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351-60. 10.1056/NEJMoa031644 [DOI] [PubMed] [Google Scholar]
- 5.Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: updated survival analysis of JBR-10. J Clin Oncol 2010;28:29-34. 10.1200/JCO.2009.24.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss GM, Herndon JE, 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. 10.1200/JCO.2008.16.4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alam N, Shepherd FA, Winton T, et al. Compliance with post-operative adjuvant chemotherapy in non-small cell lung cancer. An analysis of National Cancer Institute of Canada and intergroup trial JBR.10 and a review of the literature. Lung Cancer 2005;47:385-94. 10.1016/j.lungcan.2004.08.016 [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Wu N, Lv C, et al. Should patients with stage IB non-small cell lung cancer receive adjuvant chemotherapy? A comparison of survival between the 8th and 7th editions of the AJCC TNM staging system for stage IB patients. J Cancer Res Clin Oncol 2019;145:463-9. [DOI] [PubMed] [Google Scholar]
- 9.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 10.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. 10.1158/0008-5472.CAN-17-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park H, Lim Y, Ko ES, et al. Radiomics Signature on Magnetic Resonance Imaging: Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clin Cancer Res 2018;24:4705-14. 10.1158/1078-0432.CCR-17-3783 [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Chen C, Xie J, et al. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine 2018;36:171-82. 10.1016/j.ebiom.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Liu Z, He L, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. 10.1148/radiol.2016152234 [DOI] [PubMed] [Google Scholar]
- 14.Detterbeck FC, Chansky K, Groome P, et al. The IASLC Lung Cancer Staging Project: Methodology and Validation Used in the Development of Proposals for Revision of the Stage Classification of NSCLC in the Forthcoming (Eighth) Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2016;11:1433-46. 10.1016/j.jtho.2016.06.028 [DOI] [PubMed] [Google Scholar]
- 15.Bakr S, Gevaert O, Echegaray S, et al. Data for NSCLC Radiogenomics Collection. The Cancer Imaging Archive. Available online: http://doi.org/ 10.7937/K9/TCIA.2017.7hs46erv [DOI]
- 16.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 17.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. 10.1158/1078-0432.CCR-04-0713 [DOI] [PubMed] [Google Scholar]
- 20.Väremo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 2013;41:4378-91. 10.1093/nar/gkt111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739-40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. 10.1016/j.jtho.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She Y, Zhang L, Zhu H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol 2018;28:5121-8. 10.1007/s00330-018-5509-9 [DOI] [PubMed] [Google Scholar]
- 26.Coroller TP, Agrawal V, Huynh E, et al. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J Thorac Oncol 2017;12:467-76. 10.1016/j.jtho.2016.11.2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee G, Park H, Sohn I, et al. Comprehensive Computed Tomography Radiomics Analysis of Lung Adenocarcinoma for Prognostication. Oncologist 2018;23:806-13. 10.1634/theoncologist.2017-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirienko M, Cozzi L, Rossi A, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur J Nucl Med Mol Imaging 2018;45:1649-60. 10.1007/s00259-018-3987-2 [DOI] [PubMed] [Google Scholar]
- 29.Kirienko M, Cozzi L, Antunovic L, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging 2018;45:207-17. 10.1007/s00259-017-3837-7 [DOI] [PubMed] [Google Scholar]
- 30.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. 10.1200/JCO.2007.13.9030 [DOI] [PubMed] [Google Scholar]
- 31.Carlson RW, Larsen JK, McClure J, et al. International adaptations of NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2014;12:643-8. 10.6004/jnccn.2014.0068 [DOI] [PubMed] [Google Scholar]
- 32.Ost D, Goldberg J, Rolnitzky L, et al. Survival after surgery in stage IA and IB non-small cell lung cancer. Am J Respir Crit Care Med 2008;177:516-23. 10.1164/rccm.200706-815OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgensztern D, Du L, Waqar SN, et al. Adjuvant Chemotherapy for Patients with T2N0M0 NSCLC. J Thorac Oncol 2016;11:1729-35. 10.1016/j.jtho.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung JJ, Wu YC, Chou TY, et al. Adjuvant Chemotherapy Improves the Probability of Freedom From Recurrence in Patients With Resected Stage IB Lung Adenocarcinoma. Ann Thorac Surg 2016;101:1346-53. 10.1016/j.athoracsur.2015.10.075 [DOI] [PubMed] [Google Scholar]
- 35.Qian F, Yang W, Wang R, et al. Prognostic significance and adjuvant chemotherapy survival benefits of a solid or micropapillary pattern in patients with resected stage IB lung adenocarcinoma. J Thorac Cardiovasc Surg 2018;155:1227-35.e2. 10.1016/j.jtcvs.2017.09.143 [DOI] [PubMed] [Google Scholar]
- 36.Lee HY, Lee SW, Lee KS, et al. Role of CT and PET Imaging in Predicting Tumor Recurrence and Survival in Patients with Lung Adenocarcinoma: A Comparison with the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society Classification of Lung Adenocarcinoma. J Thorac Oncol 2015;10:1785-94. 10.1097/JTO.0000000000000689 [DOI] [PubMed] [Google Scholar]
- 37.Verma V, Simone CB, 2nd, Krishnan S, et al. The Rise of Radiomics and Implications for Oncologic Management. J Natl Cancer Inst 2017. doi: . 10.1093/jnci/djx055 [DOI] [PubMed] [Google Scholar]
- 38.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting Response to Cancer Immunotherapy using Non-invasive Radiomic Biomarkers 2019;30:998-1004. [DOI] [PMC free article] [PubMed]
- 39.Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018;15:e1002711. 10.1371/journal.pmed.1002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017;6:e23421. 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17:351-9. 10.1038/ncb3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev 2018;32:1267-84. 10.1101/gad.314617.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Chen B, Liu X, et al. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl Oncol 2018;11:94-101. 10.1016/j.tranon.2017.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desseroit MC, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with (18)F-FDG PET/CT image features in non-small-cell lung cancer stage I-III. Eur J Nucl Med Mol Imaging 2016;43:1477-85. 10.1007/s00259-016-3325-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Dong T, Xin B, et al. Integrative nomogram of CT imaging, clinical, and hematological features for survival prediction of patients with locally advanced non-small cell lung cancer. Eur Radiol 2019;29:2958-67. 10.1007/s00330-018-5949-2 [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Yang J, Zhou X, et al. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur Radiol 2019;29:2196-206. 10.1007/s00330-018-5770-y [DOI] [PubMed] [Google Scholar]
- 47.Akinci D'Antonoli T, Farchione A, Lenkowicz J, et al. CT Radiomics Signature of Tumor and Peritumoral Lung Parenchyma to Predict Nonsmall Cell Lung Cancer Postsurgical Recurrence Risk. Acad Radiol 2020;27:497-507. 10.1016/j.acra.2019.05.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as