Abstract
Background
Recently, accumulating data have supported that long non-coding RNAs (lncRNAs) may contribute to tumorigenesis. LncRNA LINC00511 (LINC00511) has been proved to serve as an oncogene in several tumors. However, as a novel lncRNA, the crucial role and potential mechanism of LINC00511 in LUSC is largely unknown.
Methods
Here, we performed a differential gene expression analysis of the LINC00511 in LUSC using data from TCGA database. Loss-of-functional assays were used to gain further insights into the latter function of LINC00511 on the malignant phenotypes in vitro. Meanwhile, qRT-PCR, western blot, dual-luciferase reporter, and RIP assays were utilized to highlight the molecular basis of LINC00511 in LUSC.
Results
LINC00511 was upregulated in LUSC tissues in TCGA database compared to adjacent non-tumor counterparts, and its expression level was strongly associated with tumor stage. LINC00511 deficiency significantly suppressed LUSC cell proliferation and migration. Furthermore, mechanistic investigation demonstrated that LINC00511 accelerated LUSC progression partially through its up-regulation of TADA1 via targeting miR-150-5p.
Conclusions
our study highlights that LINC00511 facilitates LUSC progression via sequestering miR-150-5p and targeting TADA1, suggesting a need for development of a strategy for therapeutic targeting of LINC00511 in LUSC.
Keywords: LncRNA LINC00511 (LINC00511), miR-150-5p, TADA1, lung squamous cell carcinoma
Introduction
Lung cancer is by far the largest contributor to cancer death with a 5-year relative survival rate of only 19%, and lung squamous cell carcinoma (LUSC) is the second common form of lung cancer (1,2). The current treatment for LUSC patients diagnosed at an early stage is surgical resection followed by the administration of adjuvant chemotherapy. However, majority of the patients will ultimately develop disease progression and tumor metastasis, which are the underlying leading cause of death in LUSC. Therefore, elucidating the molecular basis underlying LUSC progression may provide crucial information to achieve a better outcome for LUSC.
Recent Advances in next-generation sequencing and bioinformatics analysis have resulted in the discovery of the significant roles of long non-coding RNAs (lncRNAs) (3). LncRNAs refer to the non-coding RNAs >200 nt with little or no protein-coding potential (4). LncRNAs often exhibit spatio-temporal, and tissue- and cell-type expression patterns (5). To date, lncRNAs have been identified as a crucial modulator, involving in numerous cellular processes that include chromatin modification, signaling pathways, cancer stem cells and regulation of target gene transcription (6-9). More importantly, subsequent studies suggest that lncRNAs could contribute to cancer development and progression acting as potential oncogenes or tumor suppressors (10,11). For instance, MALAT1 regulates gene expression at the level of transcription, post-transcription, as well as epigenetics and serves as an oncogene in several tumors (12-14). We observed that overexpression of GAS5-AS1 inhibited expression of a serious of mesenchymal gene markers in NSCLC cells (15). Nevertheless, the physiological functions and potential molecular basis of dysregulated lncRNAs in LUSC have not been elucidated in detail. In this regard, identifying LUSC-associated lncRNAs may gain a better understanding of LUSC development and progression at molecular level.
Also known as onco-LncRNA-12, LINC00511 is located on chromosome17 and has a transcript length of 2,265 nt (https://www.ncbi.nlm.nih.gov/gene/?term=LINC00511). LINC00511 has been previously implicated in breast cancer oncogenesis (16,17). LINC00511 also mediated oncogenic effects through modulating VEGFA expression by sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma (PDAC) (18). In agreement with these findings, Sun et al. have recently shown that high levels of LINC00511 expression promote cell growth, cell migration and invasion in NSCLC (19). They also proved that LINC00511 functioned as an oncogene via directly binding with EZH2 to epigenetically silence of the target genes. However, the functional experiments and mechanistical study were performed in the lung adenocarcinoma SPC-A1 and A549 cells. Thus, the oncogenic role and mechanism of governing the role of LINC00511 in LUSC need further exploration.
Here, we showed that LINC00511 was upregulated in LUSC tissues in TCGA compared with that in non-LUSC samples, and the level of LINC00511 expression was conspicuously associated with TNM stage. Thus, we chose LINC00511 to be the research focus. Functional assays were carried out to determine the role of LINC00511 in mediating LUSC cell proliferation and migration. Bioinformatics analysis followed by mechanism assays revealed the downstream miRNA/mRNA axis of LINC00511 in LUSC. Thus, our study focused on a novel molecular pathway in LUSC, thus finding functional molecules in LUSC initiation and progression.
Methods
Public data and tools
The Gene Expression Profiling Interactive Analysis (GEPIA) is a web tool based on the TCGA data, which are computed by a standard pipeline (20). The expression profile of LINC00511 in LUSC tissue samples was obtained from TCGA database (http://gepia.cancer-pku.cn/index.html). The potential miRNAs of LINC00511 and gene expression correlation analysis were obtained from Starbase (http://starbase.sysu.edu.cn/), along with the possible binding sites of miR-150-5p to LINC00511 sequence. The putative mRNAs of miR-150-5p were screened out using microT, miRmap, TargetScan, PicTar and miRanda. The interacting sequence of miR-150-5p to TADA1 was acquired from Starbase.
Cell lines and culture conditions
Human LUSC cell lines (SK-MES-1, H226) and the 16-HBE (Human bronchial epidermal cells) were provided by Shanghai Institute of Biochemistry and Cell Biology. Those cells were adapted to RPMI1640 medium contained 10% fetal bovine serum (FBS). The purity of all cell lines was routinely tested for mycoplasma contamination (MycoAlert™ Mycoplasma Detection Kit, Lonza Group Ltd.). All cell culture incubations were performed at 37 °C with 5% CO2 in a humidified chamber.
Extraction of RNA and qualitative analyses of RNA
The extracted total RNA derived from cell lines was extracted by TRIZOL reagent (Invitrogen, Shanghai, China) by TRIzol method principle. The cDNA synthesis was conducted using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China). Quantitative reverse transcriptase polymerase chain reaction analyses (qRT-PCR) was carried out using the Roche FastStart Universal SYBR Green Master (Rox) in an Applied Biosystems 7900 Real Time PCR machine (Applied Biosystems, Foster City, CA). We used GAPDH and U6 to normalize the levels of expression of target genes. The primers used in qRT-PCR experiments were list as follows: GAPDH forward 5'-CCACCTGGTGCTCAGTGTAG-3', reverse 5'-CCACCTGGTGCTCAGTGTAG-3'; LINC00511 forward 5'-GCCCTCGGAACACTAAAGCT-3', LINC00511 reverse 5'-TGGTGGAGGAGGAAGTGGA-3'.
Cell transfection
Short hairpin RNAs (shRNAs) directed against LINC00511 (sh/LINC00511#1, sh/LINC00511#2) were designed to silence linc00511 into the vector. Non-targeted shRNA (sh-Ctrl) was used as the negative control. According to the manufacturer’s instructions, cells were transfected with siRNAs using Invitrogen Lipofectamine 2000 Reagent on six-well plates. MiR-150-5p mimics and negative control (miRNA NC) were synthesized to upregulate miR-150-5p. The whole sequence of TADA1 was cloned into pcDNA3.1 vector to overexpress TADA1 in LUSC cells. Empty vector was transfected as the negative control. All the plasmids and synthetic vectors were purchased from Genepharma (Genepharma, Shanghai, China).
Cell proliferation assay
The quantification of cell growth and viability was measured by Cell Proliferation Reagent Kit I (MTT; Roche Applied Science) and then confirmed by an EdU assay kit (Life Technologies). All the procedure followed the manufacturer’s protocol.
Wound healing migration assay
Cells were seeded at 1.0×106 cells/well in 6-well culture plates. After they had grown to at least 95% of confluence, monolayer cells were washed with PBS and scraped with a pipette tip to create a scratch, and then incubated with the FBS-free medium. The scratch areas were photographed by microscopy at 0 or 48 hours’ time points.
Western blot analysis
After transfection, H226 and SK-MES-1 were harvested and lysed in a lysis RIPA buffer containing 1 mM PMSF and a protease inhibitor cocktail (Roche, IN, USA). The concentrations of protein were measured with the BCA method. Proteins were separated by 10% SDS-PAGE, transferred to 0.22 µm NC membranes (Sigma-Aldrich) and then incubated with specific antibodies including TADA1 (1:1,000, Cell Signaling Technology, Inc), GAPDH (1:1,000). Last, the immunoreactive protein bands were visualized by an enhanced chemiluminescence (ECL) Kit (Pierce, Thermo Fisher Scientific, IL, USA).
Subcellular fractionation assay
Nucleus-cytoplasm separation in H226 cells was performed by use of Cytoplasmic and Nuclear RNA Purification Kit (Norgenbiotek Corporation, Thorold, ON, Canada). qRT-PCR was applied to explore the levels of LINC00511, U6 (nuclear control) and GAPDH (cytoplasmic control).
Fluorescence in situ hybridization (FISH) assay
LINC00511 subcellular localization was examined using a FISH Kit (Roche, Basel, Switzerland). Cells were added with 4% paraformaldehyde (PFA; Solarbio, Beijing, China) for fixation, incubated with hybridization solution containing LINC00511 probe (Sigma Aldrich, Saint-Louis, Missouri, USA). DAPI (Sigma‐Aldrich) was applied for staining nucleus for 10 min. Laser confocal scanning microscopy (Olympus, Tokyo, Japan) was used for obtaining the fluorescence images.
RNA protein immunoprecipitation (RIP) assay
RIP assay was carried out by the utilization of EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). LUSC cells were lysed using RNA lysis buffer containing magnetic beads conjugated with human Ago2 antibody or negative control IgG (Millipore). qRT-PCR was used to analyze the coprecipitated RNAs of LINC00511, miR-150-5p and TADA1 after overnight incubation.
Luciferase reporter assay
The pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) containing LINC00511 sequence or TADA1 3'-UTR was co-transfected with miRNA mimic into 293T or H226 cells. Then the constructed LINC00511-Wt/Mut or TADA1-Wt/Mut vectors were co-transfected with miR-150-5p mimics or miRNA NC. The activities of the luciferases were quantified via the Dual Luciferase Reporter Assay System (Promega, Mannheim, Germany).
RNA pull-down assay
The 3'end biotinylated miR-150-5p-Wt, miR-150-5p-mut or negative control (NC) was transfected into H226 cells. The biotinylated RNA was co-incubated with cell lysis buffer overnight and then magnetic beads were added. After purification, RNA complex was evaluated by qRT-PCR.
Statistical analysis
Statistical analysis was performed using the SPSS package, version 22 for Windows (Chicago, IL, USA). Statistical significance between experimental groups was assessed using ANOVA or Student’s t-test (two-tailed). A P value of less than 0.05 shall be regarded as indicative of significance.
Results
LINC00511 is upregulated in human LUSC tissues and cell lines and enhances LUSC development
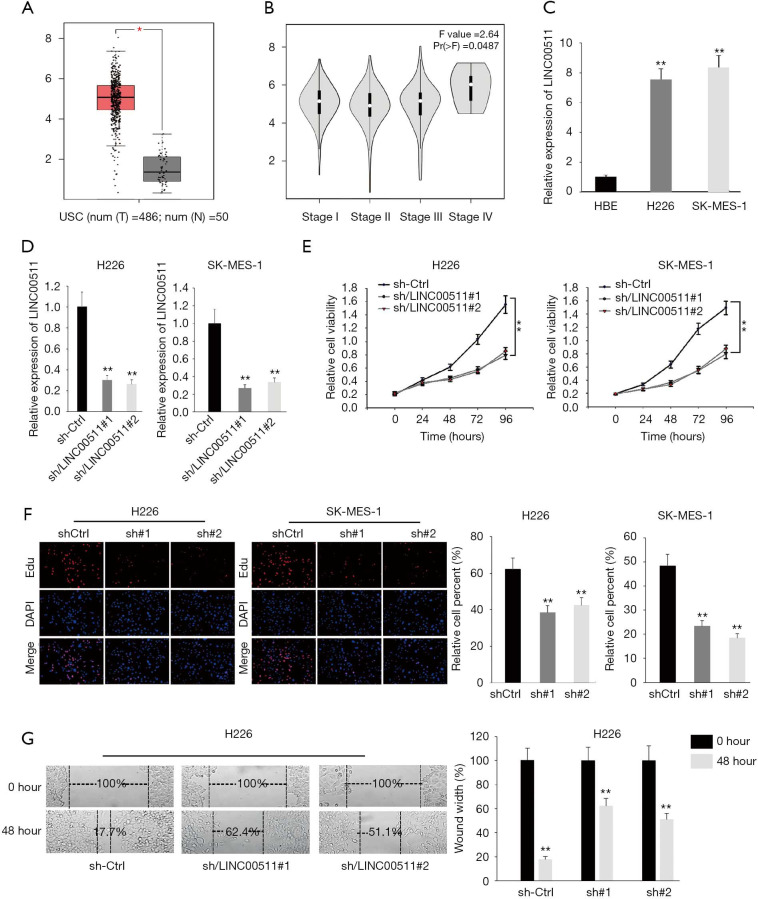
To uncover the functions of LINC00511 in LUSC, we first determined the expression of LINC00511 through TCGA database. As shown in Figure 1A, LINC00511 manifested remarkable higher expression in LUSC tissues. Next, we accessed the association of LINC00511 expression with tumor stages in LUSC from TCGA database. It was discovered that the expression of LINC00511 was intimately correlated with tumor stages in LUSC patients (Figure 1B). Moreover, qRT-PCR results demonstrated that the expression of LINC00511 was evidently augmented in H226 and SK-MES-1 than in HBE cells (Figure 1C). To investigate the biological functions of LINC00511 in LUSC, H226 and SK-MES-1 cells were stably transfected with LINC00511-specific shRNAs (sh-LINC00511#1/2) and control shRNA (sh-NC) (Figure 1D). Satisfactory transfection efficiency was obtained at 48 h post-transfection. Compared to negative control, LINC00511 depletion resulted in an evident decrease in cell proliferative abilities of H226 and SK-MES-1 cells, as monitored by MTT assay (Figure 1E). These results were further confirmed by EdU assay, which indicated that silence of LINC00511 impaired cell proliferation in LUSC (Figure 1F). Moreover, wound healing assay reflected a considerable inhibition in cell migration in sh-LINC00511 groups. Clearly, H226 cell migration was decreased by 44.7% and 33.4% in sh-LINC00511#1-transfected cells and sh-LINC00511#2-transfected cells, respectively (Figure 1G). Consequently, these findings support that LINC00511 knockdown exerts suppressive effects on cell proliferative and migratory abilities of LUSC cells.
Figure 1.
LINC00511 is upregulated in human LUSC tissues and cell lines and enhances LUSC development. (A) The expression of LINC00511 in LUSC tissues was figured out in TCGA database. (B) The correlation between LINC00511 expression and TNM stages was analyzed by Pearson’s correlation analysis. (C) qRT-PCR measured the expression of LINC00511 in LUSC cells. (D) The knockdown efficiency of LINC00511 was quantified by qRT-PCR analysis. (E,F) MTT and EdU assays were performed to reflect cell proliferation in cells transfected with sh-LINC00511#1/2. (G) The influences of LINC00511 deficiency on cell migration were investigated by wound healing assay. *P<0.05, **P<0.01.
LINC00511 suppresses expression of miR-150-5p
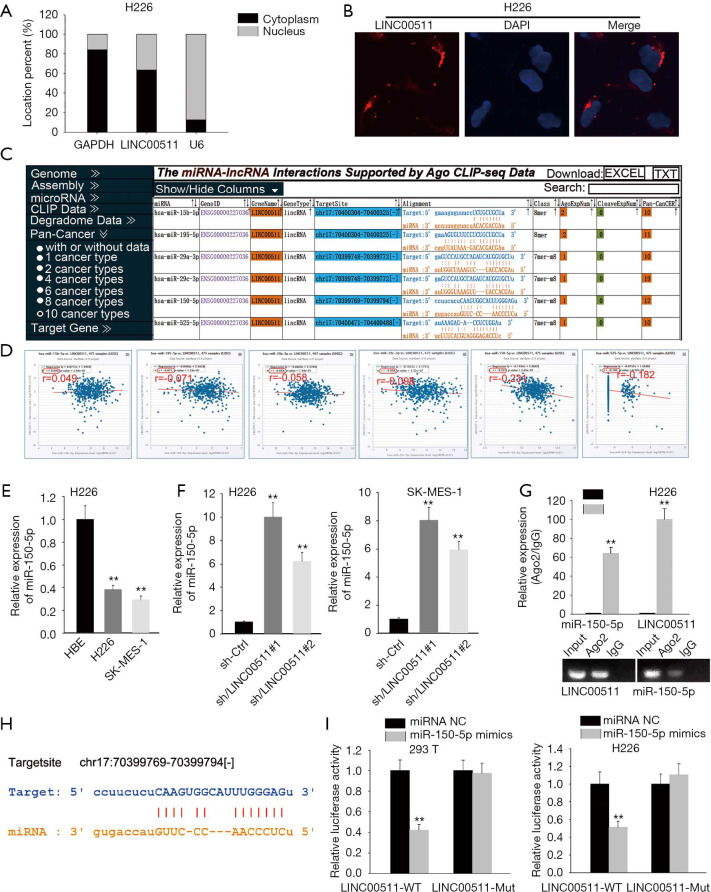
To clarify the molecular mechanism of LINC00511 in LUSC, subcellular fractionation and FISH assays were carried out to determine the localization of LINC00511. And the results indicated that LINC00511 was chiefly occupied in cytoplasm (Figure 2A,B). LINC00511-mediated ceRNA mechanism has been demonstrated in cancer. The candidate miRNAs targeted by LINC00511 consist of miR-765, miR-185-3p and miR-29b-3p (11,13,15-19). To search for specific target miRNA of LINC00511 in LUSC cells, starBase website was utilized. A cohort of 6 potential miRNAs was spot may be able to bind with LINC00511, among which miR-150-5p expression was most inversely correlated with LINC00511 expression (Figure 2C,D). To move on, the level of miR-150-5p in LUSC cells was quantified by qRT-PCR. It was detected that miR-150-5p expression was considerably low in H226 and SK-MES-1 cells (Figure 2E). As expected, the expression level of miR-150-5p was significantly elevated in the LINC00511 knockdown group, suggesting that miR-150-5p can be modulated by LINC00511 (Figure 2F). Furthermore, RIP assay revealed that LINC00511 and miR-150-5p coexisted in RISC (Figure 2G). To verify the binding between LINC00511 and miR-150-5p, we next analyzed the potential binding sites between LINC00511 and miR-150-5p by starBase website (Figure 2H). Subsequently, luciferase reporter assay pointed out that enforced expression of miR-150-5p could attenuate the luciferase activity of LINC00511-Wt reporter rather than LINC00511-Mut reporter, implying that LINC00511 could bind with miR-150-5p (Figure 2I). Taken together, miR-150-5p serves as a direct downstream target of LINC00511.
Figure 2.
LINC00511 suppresses expression of miR-150-5p. (A,B) Subcellular fractionation and FISH assays determined the localization of LINC00511 in LUSC cells. (C) Six miRNAs that might bind with LINC00511 was screened out from starBase website. (D) Pearson’s correlation analysis examined the correlation of the six candidate miRNAs expression and LINC00511 expression in LUSC tumors one by one. (E) The expression of miR-150-5p in LUSC cells was measured by qRT-PCR. (F) qRT-PCR revealed miR-150-5p level after LINC00511 being silenced in H226 and SK-MES-1 cells. (G) The relationship between LINC00511 and miR-150-5p was disclosed by RIP assay. (H) The underlying binding sites between LINC00511 and miR-150-5p were probed by starBase website. (I) The confirmation between LINC00511 and miR-150-5p was testified by luciferase reporter assay. **P<0.01.
TADA1 functions as a direct target of miR-150-5p
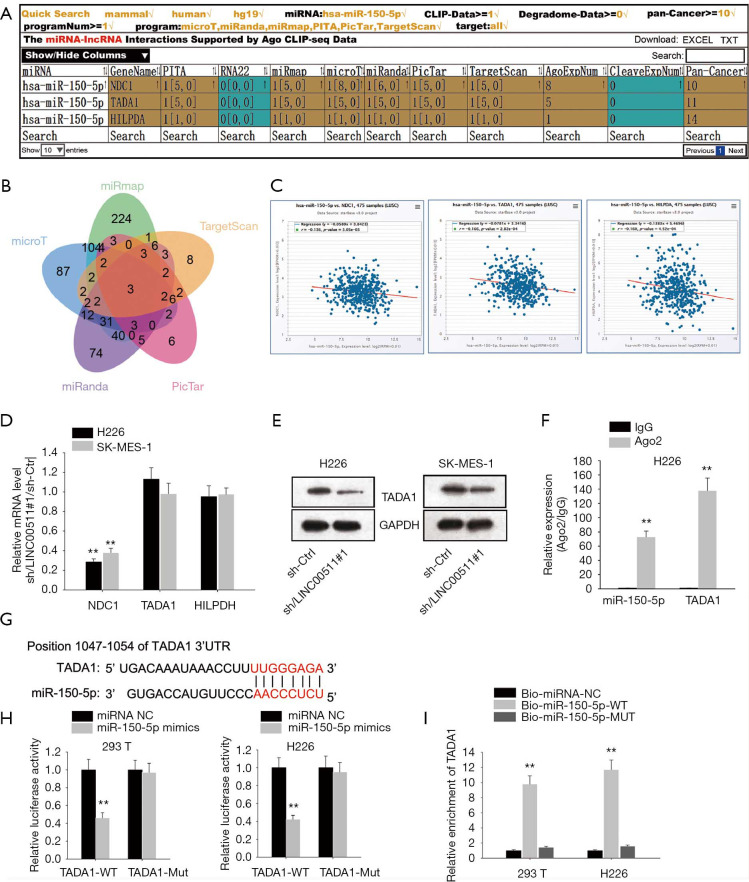
To determine potential target genes of miR-150-5p, we interrogated and analyzed via starBase websites (Figure 3A,B), of which TADA1 level was most inversely correlated with miR-150-5p level (Figure 3C). Then, qRT-PCR analyses demonstrated that only the mRNA level of TADA1 was markedly suppressed by LINC00511 silencing (Figure 3D). Besides, it was observed in western blot assay that absence of LINC00511 significantly lowered the protein level of TADA1 (Figure 3E). Furthermore, it was detected by RIP assay that TADA1 and miR-150-5p coexisted in RISC (Figure 3F). To move on, the binding sites between TADA1 and miR-150-5p were obtained and luciferase reporter assay confirmed the combination between TADA1 and miR-150-5p (Figure 3G,H). In the end, RNA pulls down assay suggested that TADA1 could only be pulled down by biotinylated miR-150-5p-Wt (Figure 3I). In sum, TADA1 acts as a functional target of miR-150-5p.
Figure 3.
TADA1 is a functional target of miR-150-5p. (A,B) Three candidate targets of miR-150-5p were chosen via starBase websites. (C) Pearson’s correlation analysis examined the correlation of the three candidate mRNAs expression and LINC00511 expression in LUSC tissues one by one. (D) The level of those three candidate mRNAs in sh-LINC00511#1-transfected cells were revealed by qRT-PCR. (E) Western blot assay illustrated the protein level of TADA1 after LINC00511 being knocked down in H226 and SK-MES-1 cells. (F) RIP assay researched the relationship between TADA1 and miR-150-5p. (G) StarBase website was applied to predict the underlying binding sites between TADA1 and miR-150-5p. (H,I) The binding ability between TADA1 and miR-150-5p was certified by luciferase reporter and RNA pull down assays. **P<0.01.
TADA1 is involved in LINC00511-mediated LUSC progression
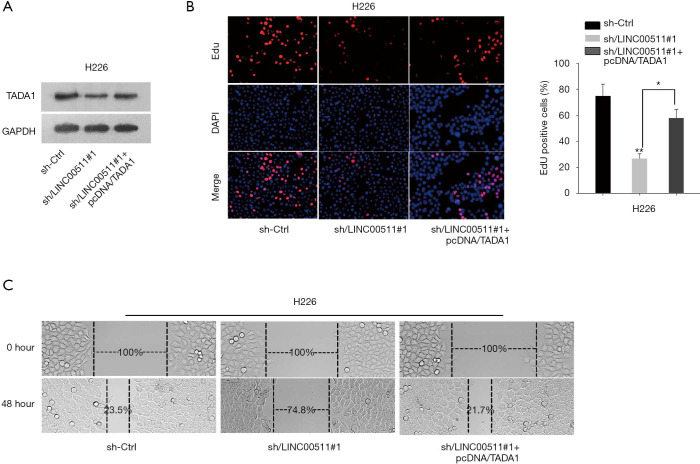
To explore the role of TADA1 in LINC00511-mediated LUSC development, rescue assays were performed. As showed in Figure 4A, the protein level of TADA1 decreased by LINC00511 knockdown was rescued after TADA1 overexpression. EdU assay validated that the impaired cell proliferation induced by LINC00511 depletion could be compensated by TADA1 overexpression (Figure 4B). Furthermore, enforced expression of TADA1 could restore the inhibited cell migration in sh-LINC00511#1-transfected cells (Figure 4C). These evidences illustrated that LINC00511 promotes LUSC progression via upregulating TADA1.
Figure 4.
TADA1 is involved in LINC00511-mediated LUSC progression. (A) Protein level of TADA1 decreased by LINC00511 knockdown was rescued after TADA1 overexpression. (B) Cell proliferation in cells co-transfected with sh-LINC00511#1 and pcDNA/TADA1 was reflected by MTT assay. (C) Wound healing assay evaluated cell migration in indicated cells. **P<0.01.
Discussion
Some lncRNA such as LncRNAs RP5-821D11.7, HULC have been implicated to contribute to the unfavourable prognosis of LUSC (21,22). Recently, LINC00511 has been identified to be the critical factors for carcinogenesis and metastasis in tumors, including glioma, bladder cancer, breast cancer, non-small cell lung cancer (19,23-25). A recent study revealed that LINC00511 could function as oncogene through suppressing p57 expression via binding with EZH2 in lung adenocarcinoma cells (A549 and SPC-A-1) in vitro and in vivo (19). In the present study, we discovered that LINC00511 was evidently overexpressed in LUSC tissues and cells, and the expression of LINC00511 was closely correlated with TNM stages in LUSC patients. More importantly, LINC00511 posed pro-proliferation and pro-migration influences on LUSC progression. Thus, these results validated that LINC00511 may serve as a potential oncogene contributed to LUSC progression.
LncRNAs work through multiple mechanisms to regulate cancer states. Studies so far suggest that lncRNA may act as ceRNA to participate in post-transcriptional regulation by suppressing the biological functions of miRNAs (26). Some studies have demonstrated that LINC00511 can reverse the degrading between miRNAs and their mRNA target via the ceRNA mechanism in tumors. In osteosarcoma, LINC00511 is reported to promoted cells growth and migration by suppressing miR-765 (27). Furthermore, Ding et al. found that LINC00511 promoted LAMC2 expression by sponging miR-765 in tongue squamous cell carcinoma (28). LINC00511 also acts as a ceRNA to recover E2F1 expression through interacting with miR-185-3p in breast cancer (25). The underlying ceRNA mechanisms for LINC00511 deregulation in LUSC remain to be elucidated. To gain mechanistic details, we sought to determine that LINC00511 mainly localized in cytoplasm and bioinformatics analysis identified miR-150-5p as a candidate miRNA. We observed that the decreased LINC00511 expression induced miR-150-5p both in H226 and SK-MES-1 cells. We thus hypothesized that LINC00511 might function in a miR-150-5p-dependent manner in LUSC. Then, RIP and luciferase report assays were taken out and confirmed that LINC00511 directly targets miR-150-5p. Thus, we conclude that LINC00511 might function as a ceRNA for miR-150-5p in LUSC.
There are evidence that miR-150-5p could inhibit metastasis and aggressiveness of lung cancer (29). For instance, miR-150-5p has been known to promote NSCLC metastasis by targeting gene expression of high mobility group AT-hook 2 (HMGA2) and β-Catenin (30). In accordance with this finding is an earlier report that miR-150-5p could contribute to cancer aggressiveness by matrix metalloproteinase 14 (MMP14) (31). Moreover, linc00673 could regulate NSCLC proliferation, migration and invasion through sponging miR-150-5p and modulating the expression of ZEB1 (32). Here, expected targets of miR-150-5p were identified by online miRNA target databases. TADA1, inversely related to miR-150-5p, was highlighted as a potential downstream target of miR-150-5p. As shown in previously study, TADA1 affects the invasion and migration in CRC (33). In the present study, TADA1 is shown to function as a miR-150 target and promote migration in LUSC cells. Furthermore, our rescue assays disclosed that the inhibited LUSC cell proliferation and migration caused by LINC00511 knockdown was partially recovered by TADA1 overexpression. Our findings indicate that LINC00511 contributes to the LUSC cell proliferation and migration via targeting miR-150-5p and regulating TADA1 expression.
Conclusions
Taken all together, these data have advanced our understanding of LINC00511 functions as an oncogene in LUSC. Intriguingly, we first identified that dysregulation of LINC00511/miR-150-5p/TADA1 signaling pathway was associated with abnormal proliferation and migration in LUSC cells. A detailed understanding of the accurate role of this molecular axis will suggests a novel strategy for therapeutic targeting of lncRNAs in LUSC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (No. 81702279).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-701). HL serves as an unpaid editorial board member of Translational Lung Cancer Research. The other authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Chi XJ, Wei LL, Bu Q, et al. Identification of high expression profiles of miR-31-5p and its vital role in lung squamous cell carcinoma: a survey based on qRT-PCR and bioinformatics analysis. Transl Cancer Res 2019;8:788-801. 10.21037/tcr.2019.04.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Wu Q, Li T, Zhu D, et al. Altered expression of long noncoding RNAs in peripheral blood mononuclear cells in patients with impaired leptomeningeal collaterals after acute anterior large vessel occlusions. Ann Transl Med 2019;7:523. 10.21037/atm.2019.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 9.Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu W, Ding J, He M, et al. Estrogen receptor beta promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 2019;38:1225-38. 10.1038/s41388-018-0463-1 [DOI] [PubMed] [Google Scholar]
- 13.Malakar P, Stein I, Saragovi A, et al. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res 2019;79:2480-93. 10.1158/0008-5472.CAN-18-1432 [DOI] [PubMed] [Google Scholar]
- 14.Pruszko M, Milano E. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep 2017;18:1331-51. 10.15252/embr.201643370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Lyu H, Liu H, et al. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep 2016;6:31093. 10.1038/srep31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, Lyu S, Dong S, et al. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther 2016;9:761-72. 10.2147/OTT.S97664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu S, Kong D, Chen Q, et al. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer 2017;16:129. 10.1186/s12943-017-0696-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Liu Y, Li Z, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med 2018;22:655-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun CC, Li SJ, Li G, et al. Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non-small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol Ther Nucleic Acids 2016;5:e385. 10.1038/mtna.2016.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Li J, Wang P, et al. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-kappaB. J Cell Biochem 2019;120:19415-21. 10.1002/jcb.29119 [DOI] [PubMed] [Google Scholar]
- 22.Luo Y, Xuan Z, Zhu X, et al. Long non-coding RNAs RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 were associated with the prognosis of lung squamous cell carcinoma. Mol Med Rep 2018;17:7238-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Liu H, Yang J, et al. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J Cell Mol Med 2019;23:4386-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Li Y, Meng F, et al. Knockdown of long non-coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/beta-catenin signaling pathway. Biosci Rep 2018;38:BSR20171701. 10.1042/BSR20171701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res 2018;37:289. 10.1186/s13046-018-0945-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. 10.1016/j.gpb.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Wu X, Liu Y, et al. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem 2018. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 28.Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med 2018;47:468-76. 10.1111/jop.12677 [DOI] [PubMed] [Google Scholar]
- 29.Lan T, Yuan K. LncRNA SNHG10 Facilitates Hepatocarcinogenesis and Metastasis by Modulating Its Homolog SCARNA13 via a Positive Feedback Loop. Cancer Res 2019;79:3220-34. [DOI] [PubMed] [Google Scholar]
- 30.Dai FQ, Li CR, Fan XQ, et al. miR-150-5p Inhibits Non-Small-Cell Lung Cancer Metastasis and Recurrence by Targeting HMGA2 and beta-Catenin Signaling. Mol Ther Nucleic Acids 2019;16:675-85. 10.1016/j.omtn.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suetsugu T, Koshizuka K, Seki N, et al. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int J Oncol 2018;52:913-24. [DOI] [PubMed] [Google Scholar]
- 32.Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer 2017;16:118. 10.1186/s12943-017-0685-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Li D, Fang H, et al. Species-specific function of microRNA-7702 in human colorectal cancer cells via targeting TADA1. Am J Transl Res 2018;10:2579-89. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as