Abstract
The Staphylococcus aureus type VII secretion system (T7SS) exports several proteins that are pivotal for bacterial virulence. The mechanisms underlying T7SS-mediated staphylococcal survival during infection nevertheless remain unclear. Here we report that S. aureus lacking T7SS components are more susceptible to host-derived antimicrobial fatty acids. Unsaturated fatty acids such as linoleic acid (LA) elicited an increased inhibition of S. aureus mutants lacking T7SS effectors EsxC, EsxA and EsxB, or the membrane-bound ATPase EssC, compared to the wild-type (WT). T7SS mutants generated in different S. aureus strain backgrounds also displayed an increased sensitivity to LA. Analysis of bacterial membrane lipid profiles revealed that the esxC mutant was less able to incorporate LA into its membrane phospholipids. Although the ability to bind labelled LA did not differ between the WT and mutant strains, LA induced more cell membrane damage in the T7SS mutants compared to the WT. Furthermore, proteomic analyses of WT and mutant cell fractions revealed that, in addition to compromising membranes, T7SS defects induce oxidative stress and hamper their response to LA challenge. Thus, our findings indicate that T7SS contribute to maintaining S. aureus membrane integrity and homeostasis when bacteria encounter antimicrobial fatty acids.
Subject terms: Microbiology, Molecular biology
Introduction
Staphylococcus aureus is a facultative pathogen that can colonize the skin and nares of healthy individuals. The asymptomatic carriage of S. aureus is a major risk for subsequent infections1. S. aureus infections, which can be healthcare or community-associated, range from benign impetigo to life-threatening bacteraemia2. Clinical management of staphylococcal infections is complicated by the increasing prevalence of multidrug resistant strains3.
The success of S. aureus as a deadly pathogen is attributed to an array of virulence factors that facilitate host tissue adhesion and immune response evasion4. One of these virulence factors is the type VII secretion system (T7SS), also known as the ESAT-6 secretion system (ESS). The orthologous ESX-1 system was initially discovered in Mycobacterium tuberculosis, where it is essential for bacterial virulence5. T7SSs (T7SSb) are found in both Gram-positive and Gram-negative bacteria, although these systems and their secretion machineries appear to be distinct to their mycobacterial counterparts6. In S. aureus, the T7SS displays modularity and heterogeneity in expression between different strains7,8. In extensively studied strains (COL, RN6390, USA300 and Newman), the T7SS consists of four integral membrane proteins (EsaA, EssA, EssB and EssC), two cytosolic proteins (EsaB and EsaG), five secreted substrates (EsxA, EsxB, EsxC, EsxD and EsaD), and EsaE, which interacts with the T7SS substrates to target them to the secretion apparatus9. A peptidoglycan hydrolase, EssH, was reported to mediate T7SS transport across the bacterial cell wall envelope10.
The molecular architecture of the staphylococcal T7SS has not yet been fully characterized. T7SS integral membrane proteins EsaA, EssA, EssB, and EssC are thought to be the core of the T7 secretion machinery, with EssC being the central membrane transporter11–13. Interactions between secreted substrates and co-dependent secretion of substrates have been demonstrated7,9,14,15. A recent study showed that the functional assembly of the T7SS machinery in S. aureus is supported by the flotillin homolog FloA, within functional membrane microdomains16.
The S. aureus T7SS is pivotal for bacterial virulence. Indeed, S. aureus mutants lacking the entire T7SS7 or specific T7SS components (EsxA, EssB, EssC, EsxC, EsxB, EsaB, EsaD or EsaE) were consistently shown to be less virulent and/or persistent in various mouse infection models11,17–21. EsxA is necessary to delay apoptosis of S. aureus-infected epithelial and dendritic cells, while other substrates modulate cytokine production18,22,23. Although the relevance of T7SS to S. aureus is less understood, a role for the toxin-antitoxin pair EsaD (or EssD) and EsaG (or EssI) was recently demonstrated in intraspecies competition9,15.
In the human host, S. aureus encounters fatty acids (FAs) in the blood, on the skin, the nasal mucosa and other lipid rich tissues24–26. Several unsaturated FAs, including palmitoleic acid (C16:1) and linoleic acid (C18:2) are known to inhibit S. aureus growth27,28. In mice, topical or intraperitoneal treatments with such antimicrobial FAs or diets rich in antimicrobial FAs decrease bacterial load and increase survival upon S. aureus infection29,30. Many saturated FAs like stearic acid, on the other hand, are non-toxic to this pathogen31. Irrespective of their saturation, host FAs can be incorporated into S. aureus phospholipids via fatty acid phosphorylation by a fatty acid kinase (Fak)32. It is thought that this incorporation may contribute to bacterial resistance against toxic host FAs31. Interestingly, S. aureus T7SS expression is induced in response to host-specific FAs20,21,33. The role of T7SS in bacterial resistance to antimicrobial FAs however remains unclear. In this study, we demonstrate that surprisingly, EsxC and other T7SS mutants were more sensitive to unsaturated FAs compared to the wild-type (WT). Although there were no differences in binding labelled linoleic acid (LA), LA induced a more leaky membrane in the T7SS mutants, and there was less incorporation of LA into EsxC mutant membrane phospholipids. Furthermore, cellular proteomics revealed that in addition to membrane discrepancies, T7SS mutants exhibited different redox and metabolic states, which likely resulted in a distinct response to LA.
Results
S. aureus esxC and essC mutants are more sensitive to antimicrobial fatty acids
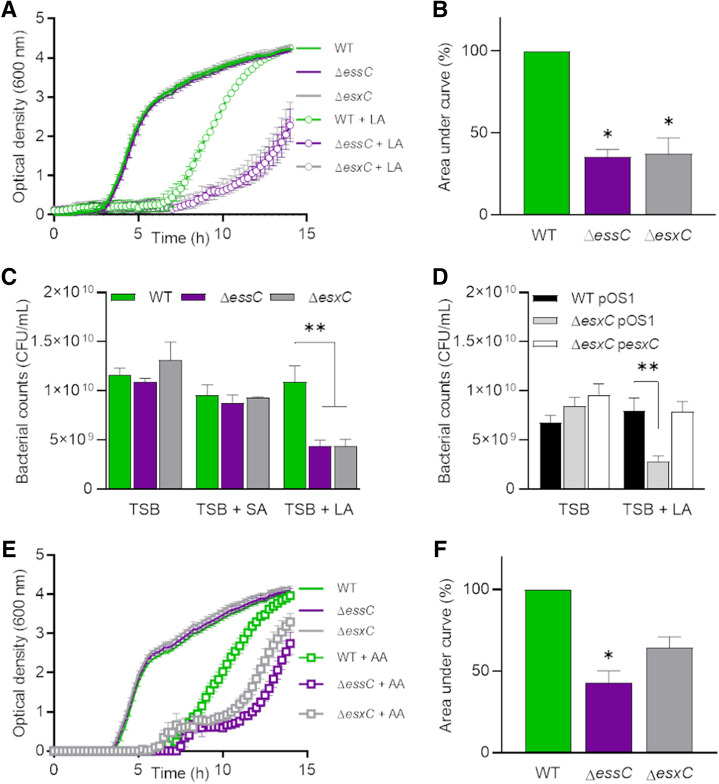
EsxC, a small 15-kDa protein secreted by the T7SS, is important for S. aureus persistence in mice19. However, mechanisms underlying EsxC- or T7SS-mediated bacterial survival are not known. In order to understand the role of EsxC, we generated an isogenic esxC mutant as described previously34, and the absence of any secondary site mutations was confirmed by whole genome sequencing. ΔesxC had a similar growth rate to the WT USA300 JE2 strain in standard rich medium (Supplementary Fig. S1). However, interestingly, when ΔesxC was cultured in the presence of an unsaturated FA (C18:2), linoleic acid (LA), at a concentration (80 µM) that still allows WT growth, it displayed significantly impaired growth, compared to the WT (Fig. 1A,B). ΔesxC did not show a growth defect when cultured in parallel in presence of stearic acid (SA), a saturated C18:0 FA (Supplementary Fig. S2A). A deletion mutant of the membrane-bound major ATPase EssC (a core T7SS component) also showed a similar growth defect in presence of LA but not SA. Both T7SS mutants displayed a decrease in optical density and colony forming units (CFU) in presence of LA but not SA (Fig. 1A,C, Supplementary Fig. S2A). Importantly, ΔesxC complemented with a plasmid containing the esxC gene reverted to the WT phenotype (Fig. 1D). The increased susceptibility of T7SS mutants to antimicrobial fatty acids was not restricted to linoleic acid as when cultured in the presence of arachidonic acid, another unsaturated FA (C20:4), growth of ΔesxC and ΔessC was inhibited more as compared to the WT (Fig. 1E, F).
Figure 1.
Enhanced S. aureus growth inhibition by antimicrobial fatty acids in esxC and essC mutants. (A) S. aureus WT USA300, ΔessC, and ΔesxC were grown in TSB or TSB supplemented with 80 µM linoleic (LA). Means ± standard error of the mean (SEM) are shown. n = 4. (B) The area under the curve (AUC) of biological replicates grown in TSB + LA in (A) were calculated and presented as % relative to the WT. Means ± SEM are shown. *Indicates P < 0.05 using a Kruskal–Wallis test with Dunn's multiple comparisons test. (C) After 14 h growth in TSB or TSB supplemented with 80 µM LA or stearic acid (SA), bacteria were serially diluted, and CFU were determined. Mean values are presented, and the error bars represent SEM. n = 3, **indicates P < 0.01 using one-way ANOVA with Dunnett’s test. (D) USA300 WT with the empty pOS1 plasmid (WT pOS1) and USA300 JE2 esxC mutant with either pOS1 (ΔesxC pOS1) or pOS1-esxC (ΔesxC pOS1-esxC) were grown in TSB or TSB + 80 µM LA as described in (A) followed by CFU estimation. Mean values are shown; error bars represent SEM. n = 5, **indicates P < 0.01 using one-way ANOVA with Dunnett’s test. (E) S. aureus WT USA300, ΔessC, and ΔesxC were grown in TSB or TSB supplemented with 80 µM arachidonic acid (AA). Means ± SEM are shown, n = 3. (F) AUCs of biological replicates grown in TSB + AA in (E) were calculated and presented as % relative to the WT. Means ± SEM are shown. *Indicates P < 0.05 using a Kruskal–Wallis test with Dunn's multiple comparisons test.
T7SS substrates contribute to S. aureus resistance to LA toxicity
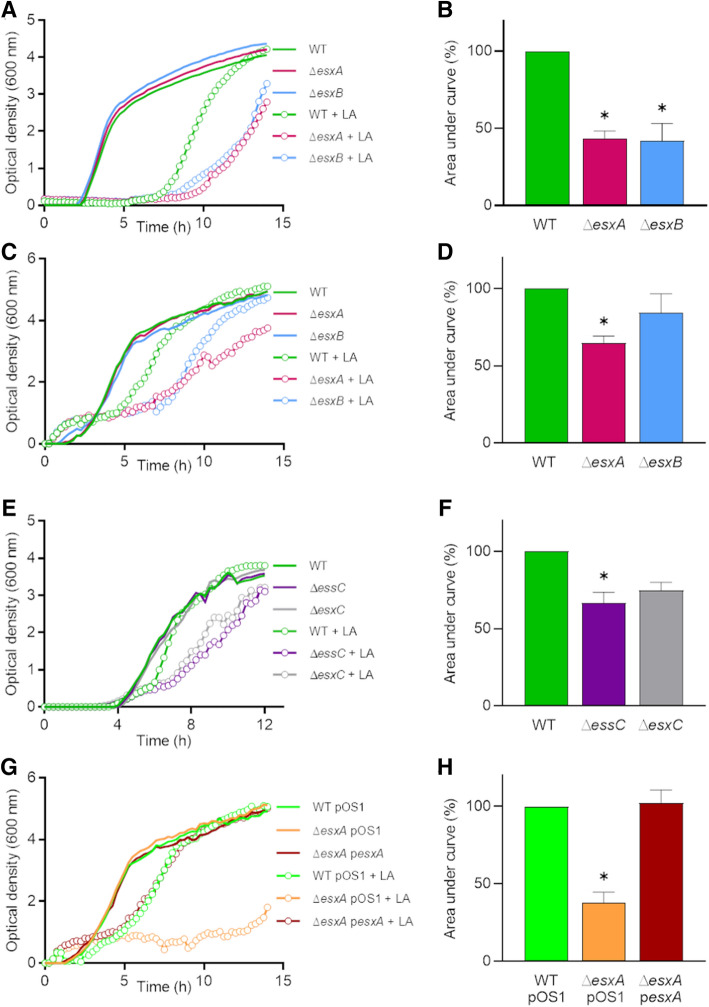
Next, we investigated whether T7SS proteins other than essC and esxC contributed to S. aureus growth in presence of LA. Mutants lacking two other substrates, ΔesxA and ΔesxB, were grown in presence of FAs. Both mutants grew significantly slower than the WT USA300 LAC (Fig. 2A,B), in line with previous studies demonstrating the inter-dependency of the different T7SS substrates7,14. To ensure that the increased sensitivity observed for the T7SS mutants was not strain specific, RN6390 ΔessC or ΔesxC and Newman ΔesxA or ΔesxB mutants were tested. Similar to the USA300 mutants, the growth of all these T7SS mutants was also impacted in the presence of LA (Fig. 2C–F). Newman ΔesxA, and RN6390 ΔessC showed significantly decreased growth, while Newman ΔesxB and RN6390 ΔesxC had a slight growth defect. The growth defect in Newman ΔesxA was abrogated upon complementation (Fig. 2G,H). None of the T7SS mutants grew differently compared with WT in presence of stearic acid (Supplementary Fig.S2B–E). Of note, the Newman WT was readily inhibited by a lower concentration of LA (40 µM), which is in agreement with the lower T7SS expression levels in this strain compared to USA3007,14. We conclude that a functional T7SS plays a role in S. aureus resistance to LA toxicity.
Figure 2.
T7SS substrates contribute to resistance to linoleic acid toxicity. (A) S. aureus USA300 wild-type (WT) and USA300 esxA (ΔesxA) or esxB (ΔesxB) deletion mutants were grown in TSB or TSB supplemented with 80 µM linoleic acid (LA). (B) AUCs of biological replicates grown in TSB + LA as in (A) were calculated and presented as % relative to the WT. Means ± SEM are shown. n = 4. (C) S. aureus Newman WT and Newman esxA (ΔesxA) or esxB (ΔesxB) deletion mutants were grown in TSB or TSB + 40 µM LA. (D) AUCs of biological replicates grown in TSB + LA as in (C) were calculated and presented as % relative to the WT. Means ± SEM are shown. n = 4. (E) Growth curves as described in (A) were done with RN6390 wild-type (WT) and RN6390 essC (ΔessC) or esxC (ΔesxC) deletion mutants. (F) AUCs of biological replicates grown in TSB + LA as in (E) were calculated and presented as % relative to the WT. Means ± SEM are shown. n = 3. (G) Newman WT with the empty pOS1 plasmid (WT pOS1) and Newman esxA mutant with either pOS1 (ΔesxA pOS1) or pOS1-esxA (ΔesxA pesxA) were grown in TSB or TSB + 40 µM LA. Data shown in (A,C,E,G) are representative of at least three independent experiments. (H) AUCs of biological replicates grown in TSB + LA as in (G) were calculated and presented as % relative to the WT. Means ± SEM are shown. n = 4. In (B,D,F,H) *indicates P < 0.05 using a Kruskal–Wallis test with Dunn's multiple comparisons test.
T7SS is required for maintaining membrane integrity in the presence of LA
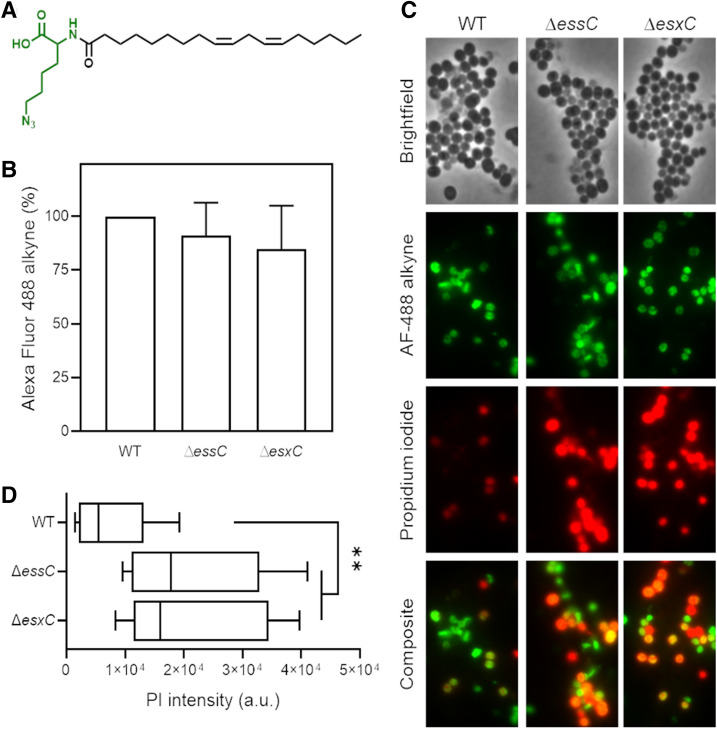
To study the mechanisms involved in T7SS mediated protection to LA toxicity, further studies were performed using mutants constructed in the USA300 JE2 strain lacking EsxC, a representative T7SS effector, or EssC the main T7SS transporter. To test if LA-mediated growth inhibition was due to an increased binding of LA to T7SS mutants, we chemically engineered LA to produce an azide functionalised LA (N6-diazo-N2-((9Z,12Z)-octadeca-9,12-dienoyl)lysine, N3-LA) or azide-LA (Fig. 3A). After incubating bacteria with azide-LA, click-chemistry with an alkyne dye (Click-iT Alexa Fluor 488 sDIBO alkyne) was used to stain azide-LA associated with bacteria. There were no obvious differences in the fluorescence from ΔessC and ΔesxC compared to the WT (Fig. 3B), suggesting that T7SS components are not involved in binding or sequestering LA.
Figure 3.
T7SS mutants display increased membrane permeability upon LA binding. (A) Chemical structure of azide functionalised linoleic acid (azide-LA; N6-diazo-N2-((9Z,12Z)-octadeca-9,12-dienoyl)lysine, N3-LA). Highlighted in green is the azido lysine. (B) S. aureus USA300 WT, ΔessC, and ΔesxC were grown with shaking in TSB to OD600 of 1.0. Bacteria were then stained for 15 min with 10 µM azide-LA prior to labelling for 1 h with alkyne Alexa Fluor 488. Mean percentage of fluorescence values relative to WT (100%) are presented; error bars represent SD, n = 5. (C) Micrographs of bacteria grown in TSB and treated as described in (B) and additionally stained with propidium iodide (PI). (D) ImageJ was used to quantitate PI fluorescence of bacterial clusters from 12 different fields per strain. Each box‐and‐whisker plot depicts the minimal and maximal PI intensities, the median is the vertical bar inside the box, which is delimited by the lower and upper quartiles. **Indicates P < 0.01 using one-way ANOVA with Dunnett’s test.
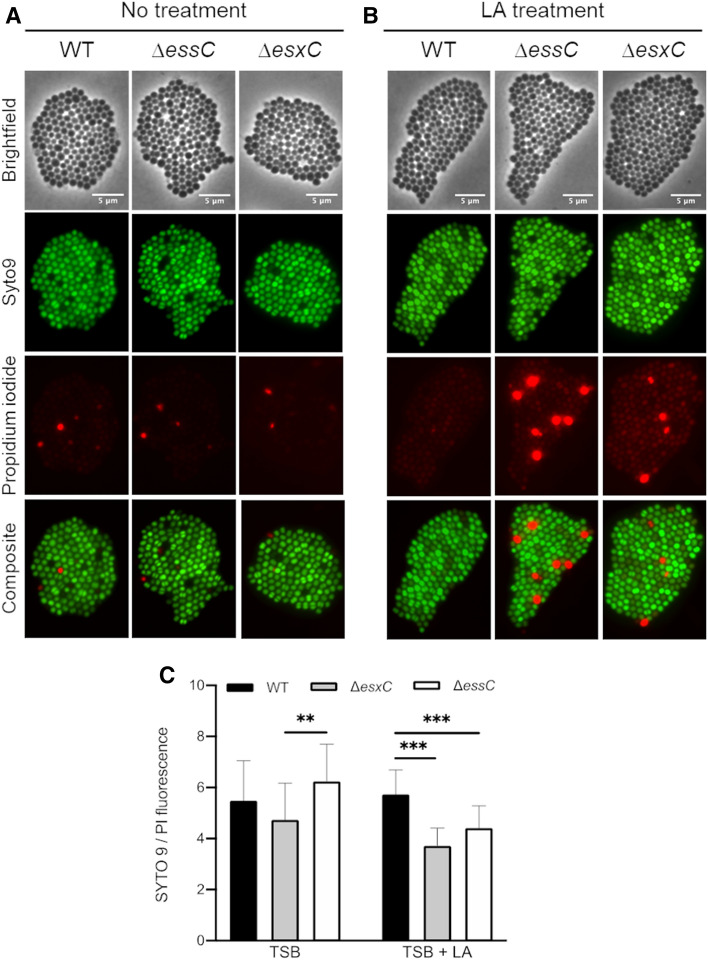
Unsaturated FAs have been well-documented to disrupt S. aureus membranes31,35. To study this, bacteria treated with azide-LA were also stained with propidium iodide (PI), a good indicator of membrane integrity. A more intense PI staining was observed for ΔessC and ΔesxC compared to the WT (Fig. 3C,D). Furthermore, to study the effects of unlabelled FA, WT and mutants were stained with PI and SYTO 9 after treatment with unlabelled LA. Again, an increased PI staining (Fig. 4A,B) and therefore lower SYTO 9/PI (Live/Dead) ratio was observed for both mutants (Fig. 4C). These data suggest that an intact T7SS helps S. aureus to maintain its membrane integrity when faced with the detergent-like effects of unsaturated FAs.
Figure 4.
T7SS mutants display increased PI staining when treated with LA. Live/Dead staining of S. aureus USA300 WT, ΔessC or ΔesxC mutants after growth to OD600 of 1.0, without (A) or with treatment with 80 µM (B) linoleic acid. Images are representative of 3 independent experiments. (C) The ratio of SYTO 9: PI fluorescence (live:dead cells) of 25 different fields per strain was quantitated with ImageJ. Means ± SD are shown, n = 3; ***Indicates P < 0.001, **Indicates P < 0.01 using a one-way ANOVA with Tukey’s multiple-comparison test.
LA-incorporation into membrane phospholipids is modulated by EsxC
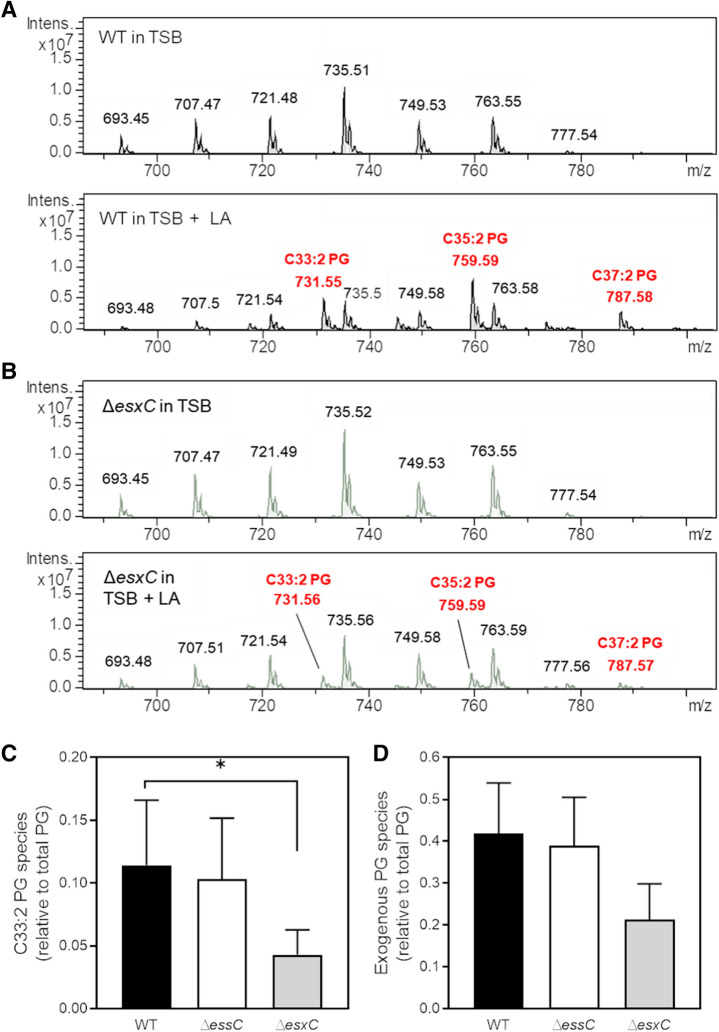
When grown in presence of unsaturated fatty acids, S. aureus has been shown to incorporate unsaturated FAs into its membrane31,36. To investigate if membrane lipids were altered in the T7SS mutants, lipids from WT USA300 and T7SS mutants were analysed by high-performance liquid chromatography (HPLC)-mass spectrometry (MS) in negative ionisation mode. As reported previously37,38, phosphatidylglycerol (PG) was the major phospholipid present in the membrane of WT grown in TSB (Supplementary Fig. S3A). ΔessC and ΔesxC grown with or without 10 µM LA (a concentration that has been previously shown to be sub-inhibitory for USA300)21 displayed lipid profiles similar to that of WT (Supplementary Fig. S3A,B). Notably, PG molecular species were significantly altered upon growth in LA-supplemented TSB for WT (Fig. 5A), ΔessC (Supplementary Fig. S3C) and ΔesxC (Fig. 5B). Three new LA-specific PG species with mass to charge ratios (m/z) 731 (C33:2), 759 (C35:2), and 787 (C37:2) appeared to contain LA (C18:2) or its elongated C20:2 or C22:2 versions, as revealed by their fragmentations (Supplementary Fig. S4A–C). PG species containing exogenous, unsaturated FAs were also present in ΔessC and ΔesxC. However, LA (C18:2)-containing PG species (C33:2) were less abundant in the esxC mutant compared to WT (Fig. 5C). A similar trend, although statistically non-significant (P > 0.05), was observed for C20:2- and C22:2-containing PG species (Supplementary Fig. S4D,E), and when all the unsaturated exogenous PG species were combined (Fig. 5D). However, there were no significant differences in the incorporation of LA for ΔessC compared to WT. A possible explanation is that although ΔessC is defective in the secretion of EsxC, EsxC that accumulates in the cytosol7,10 and membranes (as indicated by our initial studies, Supplementary Fig. S5) of the ΔessC mutant, may mediate FA incorporation. Our data suggest that lack of the T7SS component EsxC may compromise the elongation and incorporation of LA into S. aureus phospholipids.
Figure 5.
The esxC mutant is less able to incorporate LA into its phospholipids. Representative HPLC chromatograms of native phosphatidylglycerol (PG) species of S. aureus USA300 JE2 WT (A) or ΔesxC (B) grown in TSB (top panel) or in TSB supplemented with 10 µM LA (bottom panel), in negative ionisation mode. Relative quantification of the indicated PG species containing an unsaturated FA in LA-treated WT, ΔessC and ΔesxC. The C18:2-containing PG species, C33:2 (C) and total unsaturated fatty acid (C18:2, C20:2 and C22:2) containing exogenous PG species (C33:2, C35:2 and C37:2) (D) are presented as ratios of total PG species. Mean values are shown; error bars represent SD. n = 3, *indicates P < 0.05 using one-way ANOVA with Dunnett’s test.
T7SS mutations affect the total cellular content and S. aureus responses to LA
In order to gain further insight into T7SS-mediated modulation of proteins involved in FA incorporation and membrane homeostasis in presence of LA, we used an unbiased proteomic approach to study protein profiles of WT USA300, ΔessC, and ΔesxC grown exponentially with or without 10 µM LA. Of note, WT and both these T7SS mutants grew similarly in presence of up to 40 µM LA (Supplementary Fig. S6).
WT vs T7SS mutants in absence of LA treatment
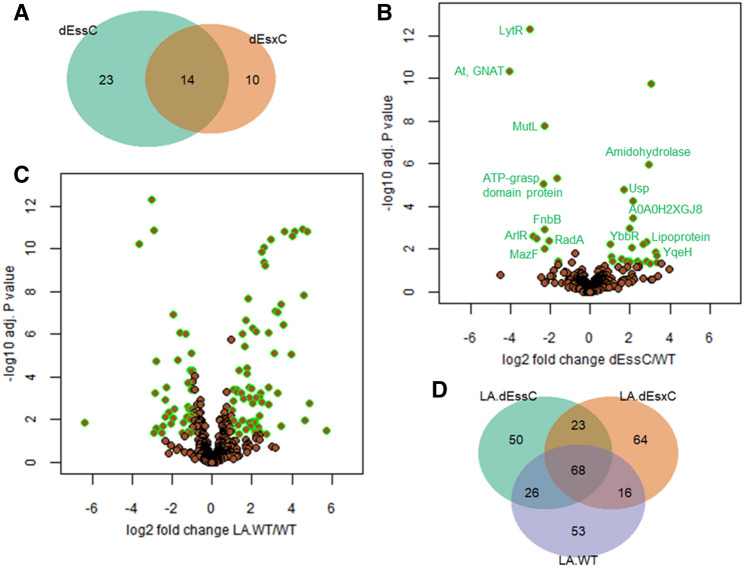
Interestingly, ΔessC or ΔesxC cultured in TSB readily displayed proteins with changed abundance when compared to the WT, with 37 and 24 proteins significantly (P < 0.05) altered in ΔessC and ΔesxC, respectively. Similarly, 14 proteins were differentially abundant in both ΔessC and ΔesxC (Fig. 6A,B). These included proteins associated with signal transduction (LytR and ArlR), the CW (acetyltransferase GNAT, FnbB and MazF), DNA repair (MutL and RadA), nucleotide binding (ATP-grasp domain protein and YqeH), hydrolysis (amidohydrolase), cell stress response [universal stress protein (Usp) family], or were uncharacterised (A0A0H2XGJ8, YbbR and lipoprotein) (Fig. 6B, Table 1). Of the 33 proteins changed only in ΔessC (23 proteins) or ΔesxC (10 proteins), nearly 40% (13 proteins) were associated with oxidation–reduction and other metabolic processes. Ten proteins which were annotated or reported to be membrane proteins, were more abundant in ΔessC (Table 1), which included SrrB, a membrane protein that is activated by impaired respiration39, and whose gene expression increased 6 times upon growth in presence of LA21. SrrB was also detected at higher levels in the esxC mutant although the increase was non-significant (P = 0.07).
Figure 6.
Quantitative proteomics shows altered cellular content and bacterial response to LA in T7SS mutants. S. aureus USA300 WT and mutants (ΔessC and ΔesxC) were grown in TSB or TSB supplemented with LA. (A) Venn diagram showing the number of proteins with altered abundance compared to WT specific to ΔessC (23) or ΔesxC (10), and common to ΔessC and ΔesxC (14). (B) The 14 proteins that are similarly changed in ΔessC and ΔesxC mutants are highlighted on a volcano plot. (C) Volcano plot showing the extensive change in the LA-treated WT compared to WT. (D) Venn diagram displaying the numbers of proteins with altered relative abundance upon LA challenge of WT (LA.WT), ΔessC (LA.dEssC) or ΔesxC (LA.dEsxC) compared to the respective untreated samples.
Table 1.
Proteins with changed abundance in ΔessC and ΔesxC mutants relative to the WT USA300 JE2.
| Functions | Uniprot ID | ΔessC/WT | ΔesxC/WT | Description | ||
|---|---|---|---|---|---|---|
| Log2 FC | Adjusted P value | Log2 FC | Adjusted P value | |||
| Signal transduction systems | Q2FK09 | − 3.0 | 4.90E−13 | − 3.1 | 4.90E−13 | Sensory transduction protein LytR |
| Q2FH23 | − 2.9 | 0.002527 | − 2.1 | 0.026184 | Response regulator ArlR | |
| Membrane proteins | A0A0H2XF42 | 3.0 | 1.75E−10 | 0 | 1 | Cytochrome D ubiquinol oxidase, subunit I |
| A0A0H2XDZ5 | 1.7 | 1.68E−05 | 0 | 1 | Uncharacterized membrane protein | |
| A0A0H2XFJ8 | 2.0 | 0.001077 | 0.5 | 0.883953 | Uracil permease | |
| A0A0H2XGW7 | 2.7 | 0.005757 | 2.5 | 0.010784 | Putative lipoprotein | |
| A0A0H2XIA9 | 1.0 | 0.006115 | 0 | 1 | Protein translocase subunit SecY | |
| Q2FIN2 | 1.6 | 0.029451 | 0.5 | 0.970614 | Prolipoprotein diacylglyceryl transferase LGT | |
| A0A0H2XKD9 | 2.0 | 0.038093 | 1.8 | 0.070347 | Staphylococcal respiratory response protein SrrB | |
| A0A0H2XFE1 | 3.4 | 0.039814 | 1.1 | 0.970614 | Peptidase | |
| A0A0H2XJV8 | 2.0 | 0.04444 | 1.5 | 0.231285 | Cyclic-di-AMP phosphodiesterase | |
| A0A0H2XGF4 | 3.0 | 0.044681 | 0.9 | 0.986458 | Sodium:dicarboxylate symporter family protein | |
| A0A0H2XHV2 | 2.5 | 0.049168 | 2.4 | 0.070182 | Glycine betaine transporter OpuD | |
| Stress response | A0A0H2XKH6 | 2.2 | 5.80E−05 | 2.5 | 1.07E−05 | Universal stress protein family |
| A0A0H2XIZ0 | 0.0 | 1 | − 3.4 | 1. 06E−10 | OsmC/Ohr family protein | |
| DNA repair | Q2FHE2 | − 2.3 | 1.79E−08 | − 2.3 | 1.19E-08 | DNA mismatch repair protein MutL |
| A0A0H2XI63 | − 2.0 | 0.004036 | − 2.0 | 0.004036 | DNA repair protein RadA | |
| A0A0H2XHT1 | − 0.3 | 0.708343 | − 1.9 | 0.008379 | Formamidopyrimidine-DNA glycosylase MutM | |
| Oxidation–reduction process | A0A0H2XJ90 | 1.1 | 0.038093 | 1.0 | 0.088334 | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| A0A0H2XHE0 | 2.4 | 0.039099 | − 0.1 | 1 | Thiol-disulphide oxidoreductase, DCC family protein | |
| A0A0H2XGR9 | 0.0 | 1 | 1.4 | 8.89E-08 | Oxidoreductase, Gfo/Idh/MocA family | |
| A0A0H2XK08 | 1.0 | 0.406975 | 2.9 | 0.00791 | Oxidoreductase, short chain dehydrogenase/reductase family | |
| A0A0H2XFZ3 | − 0.8 | 0.404225 | 2.1 | 0.016405 | Nitroreductase family protein | |
| Hydrolases | A0A0H2XE49 | 2.9 | 1.07E−06 | 2.9 | 5.99E−07 | Amidohydrolase |
| Q2FES9 | − 2.7 | 0.003385 | − 0.3 | 1 | Uncharacterized hydrolase | |
| A0A0H2XFF2 | − 0.8 | 0.016697 | − 0.5 | 0.157809 | Peptidase, U32 family | |
| A0A0H2XJH8 | 0.0 | 1 | 2.8 | 1.06E−10 | Peptidase M20 domain-containing protein 2 | |
| A0A0H2XJ54 | 0.0 | 1 | 2.0 | 0.000949 | Hydrolase (HAD superfamily) | |
| Q2FEG2 | − 0.2 | 0.854748 | − 2.9 | 0.004615 | Formimidoylglutamase | |
| Metabolism | A0A0H2XGU2 | − 1.6 | 4.99E−06 | − 0.1 | 1 | Pseudouridine synthase |
| A0A0H2XK15 | 2.8 | 0.004424 | 0.9 | 0.777138 | 1-phosphatidylinositol phosphodiesterase | |
| Q2FEK2 | − 1.6 | 0.038093 | − 0.3 | 1 | Urease accessory protein UreE | |
| Q2FI05 | 1.1 | 0.038093 | 0.0 | 1 | Bifunctional purine biosynthesis protein PurH | |
| Q2FIL2 | 2.9 | 0.038093 | 0.8 | 0.970614 | SsrA-binding protein | |
| A0A0H2XII6 | 1.8 | 0.038093 | 1.5 | 0.088334 | Orn/Lys/Arg decarboxylase | |
| A0A0H2XJR8 | − 0.9 | 0.04444 | − 0.5 | 0.61246 | RNA methyltransferase, RsmD family | |
| A0A0H2XKG7 | 0.0 | 1 | 1.4 | 8.18E−08 | Aspartokinase | |
| Cell wall composition | A0A0H2XJQ4 | − 4.0 | 4.92E−11 | − 4.0 | 3.28E−11 | Acetyltransferase, GNAT family |
| A0A0H2XKG3 | − 2.3 | 0.001238 | − 1.7 | 0.016405 | Fibronectin binding protein B | |
| A0A0H2XJC8 | − 2.3 | 0.009779 | − 3.0 | 0.000819 | Phi77 ORF017-like protein (Toxin MazF) | |
| Q2FE03 | 0.0 | 1 | 2.6 | 1.18E−12 | Fibronectin-binding protein A | |
| Nucleotide binding | A0A0H2XHY5 | − 2.3 | 8.58E−06 | − 2.3 | 6.01E−06 | ATP-grasp domain protein |
| A0A0H2XFA5 | 3.3 | 0.014259 | 3.5 | 0.008379 | Putative GTP-binding YqeH protein | |
| Uncharacterised proteins | A0A0H2XGJ8 | 2.2 | 0.000372 | 2.4 | 9.41E−05 | Uncharacterized protein |
| A0A0H2XE09 | 2.1 | 0.008231 | 1.9 | 0.016405 | Ybbr-like uncharacterized protein | |
| Q2FFI4 | 3.4 | 0.020136 | 0.9 | 0.970614 | UPF0316 membrane protein | |
| A0A0H2XG24 | 1.1 | 0.022345 | 0.8 | 0.088334 | Uncharacterized protein | |
WT vs T7SS mutants in presence of LA
We then compared the proteomic profiles of LA-treated strains (WT, ΔessC or ΔesxC) with their untreated counterparts. Clearly, the principal component analysis revealed that the differences due to the genetic makeup (WT or T7SS mutants) were less prominent than the dramatic changes induced by LA (Supplementary Fig. S7). These changes are exemplified for the WT; 163/1,132 proteins identified had an altered relative abundance upon growth with LA (Fig. 6C). 167 and 171 proteins were changed (P < 0.05) in ΔessC and ΔesxC, respectively, in response to LA, of which ~ 40% (68 proteins) were common to these mutants and their WT (Fig. 6D). At least 30% of proteins that were significantly different (P < 0.05) were unique to WT (53 proteins), ΔessC (50 proteins), or ΔesxC (64 proteins) (Fig. 6D), suggesting that each strain responds differently to LA. However, almost all proteins (13/14 proteins) that were similarly deregulated in ΔessC and ΔesxC grown without LA (Fig. 6B) were modulated in presence of LA (highlighted in bold in Dataset S1). Proteins that were less abundant in both mutants were, upon LA treatment, either increased to WT levels (MutL, acetyltransferase GNAT, Toxin MazF, and ATP-grasp domain protein), or were unchanged in the mutants and decreased in the LA-treated WT (LytR and FnbB) (Dataset S1). Likewise, proteins with increased abundance in ΔessC or ΔesxC were: (i) downregulated to WT levels (putative lipoprotein A0A0H2XGW7), (ii) unaltered in both mutants and upregulated in WT (Usp, amidohydrolase, and YbbR), (iii) or further increased in the essC mutant and strongly upregulated in WT (A0A0H2XGJ8) (Dataset S1). In sum, except for ArlR and RadA that were inversely regulated in all strains upon LA treatment, proteins similarly deregulated in ΔesxC and ΔessC were further modulated in response to LA, indicating that proteins altered by the lack of T7SS are important in the staphylococcal response to unsaturated fatty acids like LA.
Altered molecular functions in presence of LA
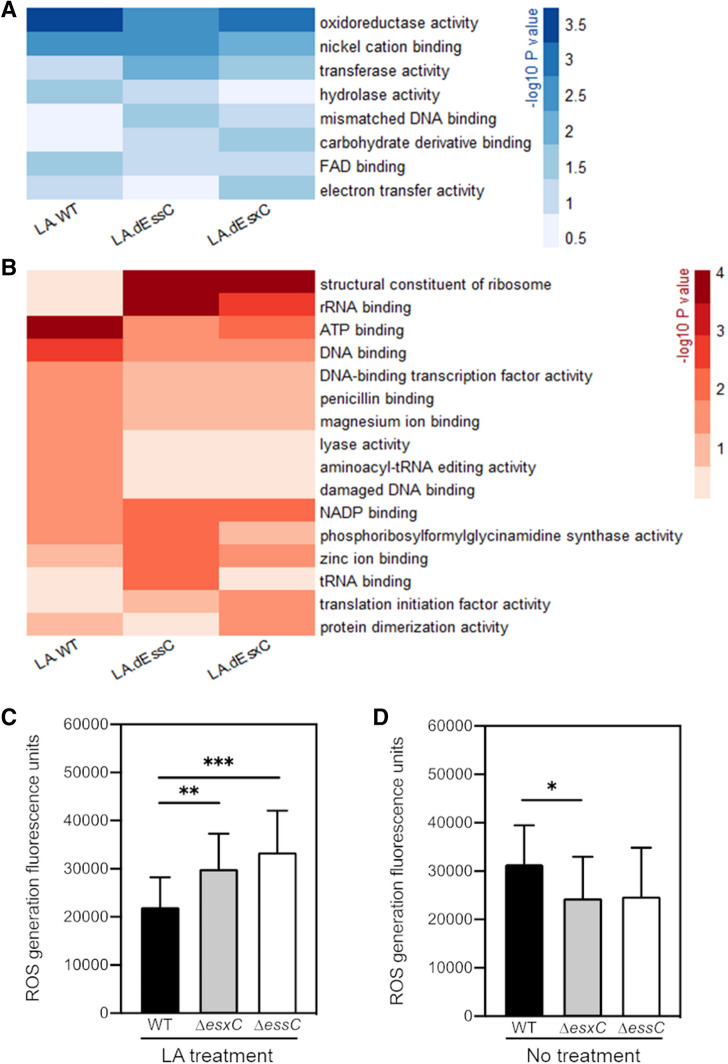
We then used QuickGO (a web-based tool for Gene Ontology searching)40 to retrieve GO terms associated with the ten most significantly upregulated proteins in LA-treated WT (Dataset S1). Strikingly, 9/10 proteins had a hydrolase or an oxidoreductase activity. A comprehensive, statistical analysis showed a clear enrichment of 8 specific molecular functions (P < 0.05) in at least one strain (WT or T7SS mutants) (Fig. 7A). Oxidoreductase and hydrolase activities were enhanced in LA-treated WT, while ΔessC and ΔesxC were less able to upregulate proteins with these molecular functions. Flavin adenine dinucleotide (FAD)-binding, which plays a role in oxidation–reduction and FA metabolic processes, was similarly more enriched in the LA-treated WT. In contrast, transferase activity, which is linked to CW synthesis, was induced more in T7SS mutants compared to the WT. Molecular functions that are decreased upon LA challenge were also determined (Fig. 7B). In agreement with reduced intracellular ATP levels following membrane damage by antimicrobial FAs35, genes with the ATP-binding function (mainly ATP-binding ABC transporters) were negatively impacted in the WT. ATP-dependent lyases were also repressed in the WT. On the contrary, T7SS mutants were less able to modulate ATP-binding proteins. Instead, a strong inhibition of ribosomal constituents and other translation-related components was seen (Fig. 7B).
Figure 7.
An altered oxidoreductive response in T7SS mutants in response to LA. Heatmaps depicting the P values of enriched (A) or diminished (B) molecular functions following a gene set analysis based on GO (gene ontology) annotations. Molecular functions that are changed in at least one strain (P < 0.05) following growth in presence of LA are shown. The shades of blue (A) or red (B) correspond to – log10 (P value). ROS levels were measured in cultures of S. aureus USA300 JE2 WT, ΔessC or ΔesxC grown to OD600 of 1.0 treated (C) with or without LA (D) using DCF reagent. Means ± SD are shown N = 5. *Indicates P < 0.05, **indicates P < 0.01, ***indicates P < 0.001 using the Kruskal–Wallis rank test.
To test the oxidoreductive states of the WT and the mutants, we stained bacteria with dichlorofluorescin (DCF), which detects reactive oxygen species41. Reflecting the changes seen in the proteomics data, when treated with 10 µM LA there is an increase in the ROS generated in the T7SS mutants compared to the WT (Fig. 7C). However, in bacteria grown without LA, the mutants have slightly less or no change in the ROS generated compared to WT (Fig. 7D). Taken together, our proteomic analyses reveal that the lack of T7SS induces altered membrane and metabolic states indicative of oxidative stress responses. While multiple pathways are modulated in the WT to mitigate LA-induced damage on the bacterial membrane, such responses are clearly altered in the absence of the T7SS.
Discussion
Host fatty acids (FAs) play a crucial role in the host defence to S. aureus infections. S. aureus is particularly sensitive to unsaturated FAs, which are abundant in the human skin27,29,31,33,42. We report here that the T7SS, an important component of S. aureus virulence arsenal, is critical in modulating the response to antimicrobial host FAs by maintaining the bacterial cell membrane integrity. A functional T7SS enables bacteria to mitigate LA-induced toxicity and grow better than mutants with a compromised T7SS. In the absence of T7SS components, LA is less incorporated into membrane phospholipids and enhances cell membrane damage. Furthermore, these bacteria are unable to activate adaptive mechanisms involved in LA resistance, as indicated by cellular proteomics.
T7SS loci in S. aureus vary in the organisation of four modules and their transcription across different strains. In this study, a protective role for the T7SS against FA toxicity was clear for S. aureus strains USA300, Newman, RN3690, which have a modular organisation representative of NCTC83258. However, we see a variation in the degree of inhibition by LA with certain substrates and strains (Newman ΔesxB and RN6390 ΔesxC), which may be due to differences in T7SS expression and regulation between strains7,14. It is noteworthy that strains with other T7SS modular configurations have been reported to activate T7SS in response to LA (MRSA252)33 or require a functional T7SS for infection of FA-rich mouse skin (ST398)43,44. Hence, although T7SS substrates in many staphylococcal strains are yet to be characterised, it is plausible that the protective role of T7SS against toxic FA is conserved in strains with different T7SS. Additionally, several studies on the USA300 T7SS have shown multiple interactions between staphylococcal T7SS components, although the precise molecular architecture of this system remains unclear. EsxC (previously EsaC) was first described as a secreted protein19. However, it has been subsequently shown to localize within staphylococcal membranes7,10. Based on the available data, EsxC is likely to be associated to EsxA, EsaD, or EsaE on the membrane9,14,18.
The cellular proteomics data reveal that the abundance of more proteins is altered in ΔessC (37) than esxC (24) in comparison to S. aureus WT, which is in keeping with the greater importance of EssC as the conserved driving force of the T7SS8. Importantly, almost 60% of proteins deregulated in ΔesxC are similarly affected in ΔessC, strongly suggesting that any modification of the T7SS core leads to a similar staphylococcal response. Surprisingly, proteins with altered abundance in USA300 ΔessC were distinct to the ferric uptake regulator (Fur)-controlled genes differentially expressed in RN6390 ΔessC45. This discrepancy might be due to strain differences, including rsbU defect in RN6390 that impairs SigB activity46,47. Nevertheless, given the known role of Fur in oxidative stress resistance48,49, both mutants may display an altered oxidative status following essC deletion. S aureus RN6390 also differentially expresses redox-sensitive genes in absence of EsaB50. Also, since the T7SS substrate EsxA is upregulated in response to hydrogen pyroxide45, one could speculate that lack of T7SS stimulates an oxidative stress response. A further indication of altered physiological states of ΔessC and ΔesxC was the decreased abundance of the two-component regulatory system proteins, LytSR, ArlSR and SrrAB, which was consistent with down-regulation of lytR transcription observed previously in the absence of arlR51. Importantly, the S. aureus response to antimicrobial FAs includes downregulation of lytRS33,52, and upregulation of srrB21. Given that LytSR is involved in bacterial surface and membrane potential modulation53,54, T7SS defects are likely to result in an altered cell envelope.
It is striking that the staphylococcal T7SS is strongly upregulated in presence of sub-inhibitory concentrations of LA21,33. FAs with more cis double bonds, which are more toxic toward S. aureus31, are also more potent T7SS activators21. Our current study interestingly suggests a protective role of T7SS against LA toxicity. Previously described S. aureus antimicrobial FA resistance mechanisms, including IsdA or wall teichoic acid-mediated modulation of cellular hydrophobicity29,31,55,56, and FA detoxification with the efflux pumps Tet38 and FarE57,58, do not appear to explain the increased susceptibility of T7SS mutants to LA, as indicated by cellular proteomics. In line with a role for T7SS in the oxidative stress response, T7SS mutants were less able to prime their redox-active proteins in response to LA-induced oxidative stress. Instead, to cope with LA, they appear to rely on strong inhibition of the protein synthesis machinery, which is reminiscent of the stringent response59.
FA can inhibit S. aureus growth by destabilising the cell membrane through several mechanisms including membrane permeabilisation31. However, both toxic and non-toxic FA can be incorporated into bacterial phospholipids31,60, and reduced incorporation at inhibitory concentrations correlated with an accumulation of free FAs31,36. The incorporation of exogenous FA into membrane phospholipids occurs via a two-component fatty acid kinase (Fak)31,32,60, which was reported to be important for T7SS activation by unsaturated FA21. FakB1 and FakB2, bind to FAs, and FakB-bound FAs are phosphorylated by FakA prior to their incorporation32. Our lipidomic analyses revealed that in the absence of EsxC, bacteria were less able to incorporate LA into their phospholipids (Fig. 5), and displayed an increased membrane permeability in presence of LA. However, as all the T7SS mutants showed increased sensitivity to FA, it seems counterintuitive that LA incorporation was impacted more in ΔesxC than in ΔessC given the central role of EssC in T7 secretion11–13. In the incorporation experiments which were performed in presence of low non-inhibitory LA concentration (10 µM), it is possible that EsxC that accumulates in the membrane (Supplementary Fig. S5) in the absence of protein secretion by EssC, mediates LA incorporation in the essC mutant. In higher concentrations of LA, however, any T7SS defects may affect incorporation and hence sensitivity to unsaturated FA. It is also worth noting that transcript levels of esxC, and not essC, were strongly upregulated in a S. aureus fakA mutant32, Proteomic analyses however showed that protein levels of Fak proteins in the T7SS mutants remained unaltered in presence or absence of LA, suggesting no T7SS-mediated regulatory control of the Fak pathway. Although we currently do not understand the precise mechanisms involved in T7SS-mediated protection, we speculate that EsxC and other interdependent T7SS substrates may play a role in facilitating Fak function in S. aureus membranes, either by mediating recruitment or targeting of Fak proteins to the membrane. Further investigations are necessary to clarify the molecular mechanisms underlying T7SS-mediated FA incorporation within staphylococcal membranes.
The increased susceptibility of T7SS mutants to LA might explain why they are less virulent in environments rich in LA and other antimicrobial FAs, like the mouse lungs (ΔessC)20, abscesses (ΔesxC and ΔesaB), liver and skin (ΔessB)21,44. Previous research showing T7SS induction by host-derived FAs further supports the importance of T7SS in such environments20,21. Taken together, we conclude that T7SS plays a key role in modulating the S. aureus cell membrane in response to toxic host FAs. Although at present, it is unclear how T7SS contributes to staphylococcal membrane architecture, T7SS interaction with the flotillin homolog FloA within functional membrane microdomains16 corroborates the idea that T7SS proteins interact with many other proteins to modulate S. aureus membranes. Indeed, our data also suggest that blocking T7SS activity would make S. aureus more vulnerable to antimicrobial FAs, a key anti-staphylococcal host defence, thus making T7SS a very attractive drug target.
Materials and methods
Bacterial strains and growth conditions
The plasmid cured USA300 LAC JE2 strain and its mutants (ΔessC and ΔesxC) were used for most parts of the study. All S. aureus strains used are listed in Table S1, and were grown aerobically in tryptic soy broth (TSB) overnight (O/N) at 37ºC for each experiment unless stated otherwise. For complemented S. aureus strains, TSB was supplemented with 10 µg/mL chloramphenicol.
Construction of bacterial mutants
The primers used are listed in Table S2. In-frame deletion of essC or esxC was performed as described previously34. Briefly, 1-kb DNA fragments up and downstream of the targeted gene sequence were PCR-amplified from USA300 LAC JE2 chromosomal DNA, and both PCR products fused via SOEing (splicing by overlap extension)-PCR. The 2-kb DNA fragment obtained was cloned into pKOR1, and used for in-frame deletion. Putative mutants were screened by PCR-amplification of a fragment including the gene of interest, whose deletion was confirmed by Sanger sequencing. Further, to confirm that successful mutants did not have any additional mutations, lllumina whole genome sequencing was performed on libraries prepared with the Nextera XT kit and an Illumina MiSeq instrument following manufacturers’ recommendations. For complementation, full-length esxC gene was cloned onto pOS1CK described previously23.
Growth curves
O/N bacterial cultures were diluted to an OD600 of 0.05 in plain TSB or TSB supplemented with fatty acids. Bacteria were then grown in a 96-well plate with shaking, and the OD600 was measured every 15 min with a FLUOstar OMEGA plate reader (BMG Labtech, UK). Areas under the curves were computed with GraphPad Prism 8.0.
Synthesis of azide functionalized linoleic acid
A 2-step synthesis was used to obtain N6-diazo-N2-((9Z,12Z)-octadeca-9,12-dienoyl)lysine, N3-LA (azide-LA). LA was first functionalized with N-hydroxysuccinimide (NHS) in anhydrous dimethyl formamide (DMF) in presence of N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride. The solvent was then removed and replaced by dicholoromethane (DCM), following which the reaction mixture was washed with water and dried over magnesium sulphate. The product, 2,5-dioxopyrrolidin-1-yl (9Z,12Z)-octadeca-9,12-dienoate (NHS-LA), was analysed using 1H nuclear magnetic resonance (NMR) spectroscopy (Supplementary Fig. S8A) and mass spectrometry (MS). MS: [M + Na]+ 400.5 (calculated), 400.5 (found).
NHS-LA was left O/N at room temperature (RT) to react with l-azidolysine hydrochloride in anhydrous DMF, and produce azide-LA. 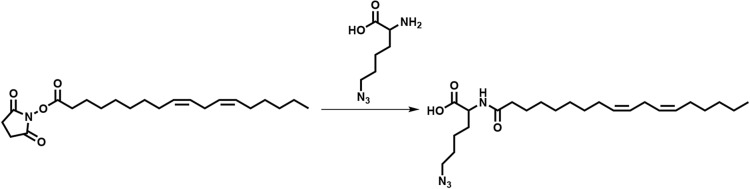
DMF was then removed, the reaction mixture precipitated in water, and dried under vacuum to obtain a clear oil. The composition of the oil was confirmed as being a mixture of azide-LA and unmodified LA (20% and 80%, respectively) based on 1H NMR (Supplementary Fig. S8B) and MS data. MS: [LA-H]− 279.5 (calculated), 279.2 (found), [M-H]− 433.3 (calculated), 433.6 (found).
Binding assays with azide-LA and click chemistry
S. aureus USA300 JE2 WT, ΔessC, and ΔesxC, grown to OD600 of 1.0, were treated with 10 µM azide-LA for 15 min at 37 °C with shaking. The samples were then centrifuged, and the bacterial pellets resuspended in PBS supplemented with 4 µg/mL Click-iT Alexa Fluor 488 sDIBO alkyne (Life Technologies LTD, UK). After incubation at 25 °C for 1 h with shaking, bacteria were washed with PBS, and binding to azide-LA was quantified by measuring fluorescence using a FLUOstar OMEGA plate reader (BMG Labtech, UK). The samples imaged with a microscope were additionally stained with 3 µM propidium iodide, following click chemistry. Bacteria stained with Click-iT Alexa Fluor 488 sDIBO alkyne and 3 µM propidium iodide were immobilized on agarose-covered glass slides, and viewed with a Leica DMi8 widefield microscope (Leica Microsystems LTD, UK). Images were analysed with the ImageJ processing package Fiji61.
Live/dead staining
Bacteria grown to OD600 of 1.0, were treated with 80 μM linoleic acid for 15 min at 37 °C with shaking. The samples were then centrifuged, and the bacterial pellets resuspended in PBS and supplemented with a 1:1 ratio of 2X LIVE/DEAD solution (6 μM SYTO-9 stain and 30 μM propidium iodide) from LIVE/DEAD BacLight kit (Invitrogen). After incubation in the dark for 15 min, bacteria were washed with PBS, spotted on to agarose pads and imaged using a Leica DMi8 widefield microscope (Leica Microsystems, UK). Acquired images were analysed with the ImageJ processing package, Fiji.
Lipid extraction and analyses
Lipids were extracted from bacterial cultures as described elsewhere62. Briefly, bacteria were grown to OD600 of 1.0 in TSB or TSB supplemented with 10 µM LA, centrifuged in a 2 mL glass Chromacol vial (Thermo Scientific), and resuspended in 0.5 mL MS grade methanol (Sigma-Aldrich). MS grade chloroform was then used to extract lipids. The extracted lipids were dried under nitrogen gas with a Techne sample concentrator (Staffordshire, UK), and the lipid pellets resuspended in 1 mL acetonitrile. The samples were then analysed by LC–MS with a Dionex 3400RS HPLC coupled to an amaZon SL quadrupole ion trap mass spectrometer (Bruker Scientific) via an electrospray ionisation interface. Both positive and negative ionisation modes were used for sample analyses. The Bruker Compass software package was utilized for data analyses, using DataAnalysis for peak identification and characterization of lipid class, and QuantAnalysis for quantification of the relative abundance of distinct PG species to total PG species.
Cellular proteomics
S. aureus strains were grown O/N at 37ºC on tryptic soy agar plates. The next day, single colonies were used to inoculate 10 mL plain TSB or TSB with 10 µM LA. Cultures were grown at 37ºC with 180-rpm shaking until an OD600 of 3.2 ± 0.2 was reached. The bacteria were then centrifuged, washed with PBS, and resuspended in lysis buffer (PBS, 250 mM sucrose, 1 mM EDTA, and 50 µg/mL lysostaphin) supplemented with cOmplete, mini EDTA-free protease inhibitor cocktail (Sigma-Aldrich, UK). After 15 min incubation at 37 °C, cells were lysed mechanically with silica spheres (Lysing Matrix B, Fischer Scientific, UK) in a fast-prep shaker as described previously16. Samples were then centrifuged, and the supernatants transferred to fresh tubes, where proteins were reduced and alkylated for 20 min at 70 °C with 10 mM TCEP (tris(2-carboxyethyl)phosphine) and 40 mM CAA (2-chloroacetamide), respectively. Next, the solvent was exchanged first to 8 M urea buffer then to 50 mM ammonium bicarbonate (ABC). Proteins were digested O/N at 37 °C with mass spectrometry grade lysyl endopeptidase LysC and sequencing grade modified trypsin (Promega LTD, UK).
Label-free protein quantification
Peptides prepared for proteome analyses were desalted and concentrated with a C18 cartridge in 40 µL MS buffer (2% acetonitrile plus 0.1% trifluoroacetic acid). For each sample, 20 µL were analysed by nanoLC-ESI–MS/MS using the Ultimate 3000/Orbitrap Fusion instrumentation (Thermo Scientific), and a 90-min LC separation on a 50 cm column. The data were used to interrogate the Uniprot Staphylococcus aureus USA300 database UP000001939, and the common contaminant database from MaxQuant63. MaxQuant software was used for protein identification and quantification using default settings. Intensities were log2-tansformed with the Perseus software, and proteins with one or no valid value for every sample in triplicate were filtered. Missing values in cellular proteomics data were imputed on R. Specifically, for each sample, the imputed value was either the lowest intensity across all samples if at least two biological replicates had missing values or the average of two valid values if only one was missing.
Immunoblotting
15 µg of proteins per sample (cell membrane and cell wall fractions) were loaded onto Mini-Protean TGX precast protein gels (Bio-Rad). After electrophoresis, proteins were transferred to PVDF membranes, which were cut and probed in parallel with anti-EsxC7 and anti-PBP2a64 antibodies.
Statistical analyses
Except for the proteomics results, the statistical tests were performed with GraphPad Prism 8.0 as indicated in the Figure legends, with P values < 0.05 considered significant. A paired two-tailed Student’s t-test or a paired Mann–Whitney U test was used for pairwise comparisons. An ordinary one-way analysis of variance (ANOVA) with Dunnett's multiple comparisons test or a Kruskal–Wallis test with Dunn's multiple comparisons test was applied to data form three or more groups. The fold changes and P values of the proteomics data were calculated with the R package limma66, with USA300 JE2 WT or bacteria grown without LA as references.
Supplementary information
Acknowledgements
This study was supported by a Medical Research Council (MRC) grant (MR/N010140/1) to M. U., an MRC Doctoral Training Partnership studentship in Interdisciplinary Biomedical Research (MR/J003964/1) awarded to R.A.J., a CSIRO (Commonwealth Scientific and Industrial Research Organisation) scholarship to A.K., and a Royal Society Wolfson Merit Award (WM130055) to S.P. We thank Professor Tracy Palmer (Newcastle University) and Professor Olaf Scheewind (University of Chicago) for providing us S. aureus strains and reagents. We thank GSK, Siena, Italy for providing the esxA, esxB mutant strains used in this study. We acknowledge the contribution of the Proteomics Research Technology Platform, University of Warwick.
Author contributions
A.K.T., K.W., R.J. and A.K. conducted experiments, analysed data and prepared figures, M.T.A. and A.K.T. analysed proteomics data, S.P.,Y.C. and M.U. contributed to experimental design and A.K.T and M.U. wrote the main manuscript text. All authors reviewed the manuscript.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE65 partner repository with the dataset identifier PXD013081 and 10.6019/PXD013081. Cellular proteomic samples are labelled MS18-193.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71653-z.
References
- 1.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study group. N. Engl. J. Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee AS, et al. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers. 2018;4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 4.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008;46(Suppl 5):S350–359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad WH, et al. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. U.S.A. 2017;114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The enigmatic Esx proteins: Looking beyond mycobacteria. Trends Microbiol. 2017;25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Kneuper H, et al. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol. Microbiol. 2014;93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warne B, et al. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17:222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2016;2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobrovskyy M, Willing SE, Schneewind O, Missiakas D. EssH peptidoglycan hydrolase enables Staphylococcus aureus type VII secretion across the bacterial cell wall envelope. J. Bacteriol. 2018 doi: 10.1128/JB.00268-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jager F, Kneuper H, Palmer T. EssC is a specificity determinant for Staphylococcus aureus type VII secretion. Microbiology. 2018;164:816–820. doi: 10.1099/mic.0.000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoltner M, et al. EssC: Domain structures inform on the elusive translocation channel in the Type VII secretion system. Biochem. J. 2016;473:1941–1952. doi: 10.1042/BCJ20160257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson M, Aly KA, Chen YH, Missiakas D. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol. Microbiol. 2013;90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohr RJ, Anderson M, Shi M, Schneewind O, Missiakas D. EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. J. Bacteriol. 2017 doi: 10.1128/JB.00528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mielich-Suss B, et al. Flotillin scaffold activity contributes to type VII secretion system assembly in Staphylococcus aureus. PLoS Pathog. 2017;13:e1006728. doi: 10.1371/journal.ppat.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson M, Chen YH, Butler EK, Missiakas DM. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J. Bacteriol. 2011;193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson M, et al. EssE promotes Staphylococcus aureus ESS-dependent protein secretion to modify host immune responses during infection. J. Bacteriol. 2017 doi: 10.1128/JB.00527-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burts ML, DeDent AC, Missiakas DM. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii K, et al. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect. Immunol. 2014;82:1500–1510. doi: 10.1128/IAI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez MS, et al. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 2017;114:11223–11228. doi: 10.1073/pnas.1700627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruciani M, et al. Staphylococcus aureus Esx factors control human dendritic cell functions conditioning Th1/Th17 response. Front. Cell Infect. Microbiol. 2017;7:330. doi: 10.3389/fcimb.2017.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korea CG, et al. Staphylococcal Esx proteins modulate apoptosis and release of intracellular Staphylococcus aureus during infection in epithelial cells. Infect. Immunol. 2014;82:4144–4153. doi: 10.1128/IAI.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Do TQ, et al. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J. Immunol. 2008;181:4177–4187. doi: 10.4049/jimmunol.181.6.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takigawa H, Nakagawa H, Kuzukawa M, Mori H, Imokawa G. Deficient production of hexadecenoic acid in the skin is associated in part with the vulnerability of atopic dermatitis patients to colonization by Staphylococcus aureus. Dermatology. 2005;211:240–248. doi: 10.1159/000087018. [DOI] [PubMed] [Google Scholar]
- 26.Abdelmagid SA, et al. Comprehensive profiling of plasma fatty acid concentrations in young healthy Canadian adults. PLoS ONE. 2015;10:e0116195. doi: 10.1371/journal.pone.0116195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arsic B, Zhu Y, Heinrichs DE, McGavin MJ. Induction of the staphylococcal proteolytic cascade by antimicrobial fatty acids in community acquired methicillin resistant Staphylococcus aureus. PLoS ONE. 2012;7:e45952. doi: 10.1371/journal.pone.0045952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian C, Frank MW, Batte JL, Whaley SG, Rock CO. Oleate hydratase from Staphylococcus aureus protects against palmitoleic acid, the major antimicrobial fatty acid produced by mammalian skin. J. Biol. Chem. 2019;294:9285–9294. doi: 10.1074/jbc.RA119.008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke SR, et al. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe. 2007;1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Svahn SL, et al. Dietary polyunsaturated fatty acids increase survival and decrease bacterial load during septic Staphylococcus aureus infection and improve neutrophil function in mice. Infect. Immunol. 2015;83:514–521. doi: 10.1128/IAI.02349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J. Bacteriol. 2012;194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons JB, et al. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenny JG, et al. The Staphylococcus aureus response to unsaturated long chain free fatty acids: Survival mechanisms and virulence implications. PLoS ONE. 2009;4:e4344. doi: 10.1371/journal.pone.0004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Cartron ML, et al. Bactericidal activity of the human skin fatty acid cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob. Agents Chemother. 2014;58:3599–3609. doi: 10.1128/AAC.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenway DL, Dyke KG. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J. Gen. Microbiol. 1979;115:233–245. doi: 10.1099/00221287-115-1-233. [DOI] [PubMed] [Google Scholar]
- 37.Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of exogenous fatty acids. J. Bacteriol. 2018 doi: 10.1128/JB.00728-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mashruwala AA, Guchte AV, Boyd JM. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife. 2017 doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binns D, et al. QuickGO: A web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George SE, et al. Oxidative stress drives the selection of quorum sensing mutants in the Staphylococcus aureus population. Proc. Natl. Acad. Sci. U.S.A. 2019;116:19145–19154. doi: 10.1073/pnas.1902752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelsey JA, Bayles KW, Shafii B, McGuire MA. Fatty acids and monoacylglycerols inhibit growth of Staphylococcus aureus. Lipids. 2006;41:951–961. doi: 10.1007/s11745-006-5048-z. [DOI] [PubMed] [Google Scholar]
- 43.Dai Y, et al. A novel ESAT-6 secretion system-secreted protein EsxX of community-associated Staphylococcus aureus lineage ST398 contributes to immune evasion and virulence. Front. Microbiol. 2017;8:819. doi: 10.3389/fmicb.2017.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, et al. Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci. Rep. 2016;6:25163. doi: 10.1038/srep25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casabona MG, et al. Haem-iron plays a key role in the regulation of the Ess/type VII secretion system of Staphylococcus aureus RN6390. Microbiology. 2017;163:1839–1850. doi: 10.1099/mic.0.000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassat J, et al. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology. 2006;152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 47.Giachino P, Engelmann S, Bischoff M. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 2001;183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson M, et al. Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int. J. Med. Microbiol. 2011;301:44–52. doi: 10.1016/j.ijmm.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casabona MG, et al. Functional analysis of the EsaB component of the Staphylococcus aureus type VII secretion system. Microbiology. 2017 doi: 10.1099/mic.0.000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang X, et al. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 2005;187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann Y, et al. The effect of skin fatty acids on Staphylococcus aureus. Arch. Microbiol. 2015;197:245–267. doi: 10.1007/s00203-014-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patton TG, Yang SJ, Bayles KW. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 2006;59:1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x. [DOI] [PubMed] [Google Scholar]
- 54.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2000;182:1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohler T, Weidenmaier C, Peschel A. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 2009;191:4482–4484. doi: 10.1128/JB.00221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moran JC, Alorabi JA, Horsburgh MJ. Comparative transcriptomics reveals discrete survival responses of S. aureus and S. epidermidis to sapienic acid. Front. Microbiol. 2017;8:33. doi: 10.3389/fmicb.2017.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alnaseri H, et al. Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J. Bacteriol. 2015;197:1893–1905. doi: 10.1128/JB.02607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Truong-Bolduc QC, Villet RA, Estabrooks ZA, Hooper DC. Native efflux pumps contribute resistance to antimicrobials of skin and the ability of Staphylococcus aureus to colonize skin. J. Infect. Dis. 2014;209:1485–1493. doi: 10.1093/infdis/jit660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geiger T, et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012;8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen MT, Hanzelmann D, Hartner T, Peschel A, Gotz F. Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect. Immunol. 2016;84:205–215. doi: 10.1128/IAI.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith AF, et al. Elucidation of glutamine lipid biosynthesis in marine bacteria reveals its importance under phosphorus deplete growth in Rhodobacteraceae. ISME J. 2019;13:39–49. doi: 10.1038/s41396-018-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Fernandez E, et al. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell. 2017;171:1354–1367. doi: 10.1016/j.cell.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE65 partner repository with the dataset identifier PXD013081 and 10.6019/PXD013081. Cellular proteomic samples are labelled MS18-193.