Abstract
Defects in epigenetic mechanisms are well-recognized in multiple neurodevelopmental disorders including Schizophrenia (SZ). In addition to aberrant epigenetic marks, dysregulated epigenetic machinery was also identified among the contributory factors in SZ patients. Among these, overexpression of DNA methyltransferase 1 (DNMT1) was the first to be identified. In this context, Dnmt1tet/tet (Tet/Tet), a mouse embryonic stem cell (ESC) line that overexpresses DNMT1 in ESCs and neurons, was developed to study abnormal neurogenesis. In an attempt to understand whether DNMT1 overexpression is associated with aberrant DNA methylation, we compared the genome-wide methylation levels of R1 (wild-type) and Tet/Tet ESCs and their neuronal derivatives by RRBS. The RRBS data (GSE152817) showed an average mappability of ∼59% and an average coverage of 40X per locus. The data was processed to determine the methylation percentages of target genes and was visualized using the UCSC genome browser. The observed methylation differences were validated by Combined Bisulfite Restriction Analysis (COBRA). The methylome data described here can be used to study the relationship between DNMT1 overexpression, alterations in methylation levels and dysregulation of SZ-associated genes.
Keywords: Epigenetics, Schizophrenia, Reduced representation bisulfite sequencing (RRBS), Next generation sequencing (NGS)
Specifications Table
| Subject | Neurogenetics |
|---|---|
| Specific Area | Psychiatric epigenetics |
| Type of data | Tables, Raw sequencing data |
| How data was acquired | Genomic DNAs were isolated, digested with MspI, gel-purified, treated with bisulfite and subjected to sequencing by RRBS method to obtain 40X coverage of the individual CpG islands |
| Data format | Raw (files): FASTQ files |
| Analyzed (files): bedGraph files | |
| Parameters for data collection | R1 and Dnmt1tet/tet (Tet/Tet) mouse embryonic stem cell lines, Reduced Representation Bisulfite Sequencing, bedGraph files for data visualization. |
| Description of data collection | Genomic DNAs from the cells above were digested with MspI, ligated to adapters, bisulfite converted and amplified to generate sequencing libraries by RRBS protocol. The libraries were sequenced by Next Generation Sequencing. |
| Data source location | Birla Institute of Technology and Science, Pilani Hyderabad Campus Hyderabad, India |
| Data accessibility | Repository name: Gene Expression OmnibusData identification number: GSE152817Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152817Raw data links for reviewer access: |
| Related research article | S. Saxena, P.A. Maroju, S. Choudhury, A. Anne, K.N. Mohan, Analysis of transcript levels of a few schizophrenia candidate genes in neurons from a transgenic mouse embryonic stem cell model overexpressing DNMT1, Gene. 757 (2020) 144934. https://doi.org/10.1016/j.gene.2020.144934. |
Value of the Data
-
•
The dataset described here provides genome-wide DNA methylation patterns at single nucleotide level among CpG islands located at the promoter regions, gene bodies and intergenic regions of R1 (wild-type) and the transgenic Dnmt1tet/tet (Tet/Tet) ESCs and their neuronal derivatives.
-
•
Since DNMT1 overexpression is one of the etiological factors for schizophrenia (SZ), the RRBS data generated helps in studying the epigenetic basis of DNMT1 overexpression in abnormal neurogenesis.
-
•
The bedGraph files allow visualization of the methylation levels of desired sites in the genome and their comparison across different cell types.
1. Data Description
This manuscript describes methylome datasets generated using Reduced Representational Bisulfite Sequencing (RRBS) of genomic DNAs from mouse embryonic stem cells (ESCs) and neurons obtained through their differentiation. The data can be accessed from the Gene Expression Omnibus database with accession number GSE152817. These data were obtained for R1 (wild-type) ESCs and the transgenic Dnmt1tet/tet (Tet/Tet) ESCs and their respective neuronal derivatives. A total of four RRBS data files, viz., GSM4626846 (R1 ESCs), GSM4626847 (Tet/Tet ESCs), GSM4626848 (R1 neurons) and GSM4626849 (Tet/Tet neurons) were deposited. The data includes the raw reads of all the samples for both paired-end read orientations (read 1 and read 2) in FASTQ format. The sequenced data were deposited at NCBI Sequence Read Archive with accession number SRP267969 under the BioProject accession number PRJNA640537. In order to confirm the integrity of the uploaded datafiles, their md5checksum values were examined (Table 1).
Table 1.
Details of the files included in the dataset.
| Accession Number | Description | File Type | File Names | File Checksum |
|---|---|---|---|---|
| GSM4626846 | R1 ESCs | FASTQ | zr2736_1_R1.fq | b74d3a9436fa4992572be1b228084e8b |
| FASTQ | zr2736_1_R2.fq | 7dcc6fe3e612bfed313c1495af9fd30c | ||
| bedGraph | R1 ES RRBS meth calling new.txt | ccf1fee1702c5976894e0c349086f3b4 | ||
| GSM4626847 | Tet/Tet ESCs | FASTQ | zr2736_2_R1.fq | 0178f0a3d64550dd430b8dfec82912c1 |
| FASTQ | zr2736_2_R2.fq | e0306f9cbba075f6778b2a0ec94b4781 | ||
| bedGraph | Tet ES RRBS meth calling new.txt | 1d83c72cc6d335dfe2dac393b3c78e58 | ||
| GSM4626848 | R1 neurons | FASTQ | zr2544_1_R1.fq | 07091d6141e21f079032097199887fa5 |
| FASTQ | zr2544_1_R2.fq | 71aa2e738e249c86639a768bd3ccb468 | ||
| bedGraph | R1 neu RRBS meth calling new.txt | 7d8e6f71c8c8c2370fbf52946c2e245e | ||
| GSM4626849 | Tet/Tet neurons | FASTQ | zr2544_2_R1.fq | 0720d0140ca114e712fdaf55553a2027 |
| FASTQ | zr2544_2_R2.fq | 282d3954c0a397f5ddd16de18d9d7b9f | ||
| bedGraph | Tet neu RRBS meth calling new.txt | e015fcd49cae521b8d5923ea46cfede0 |
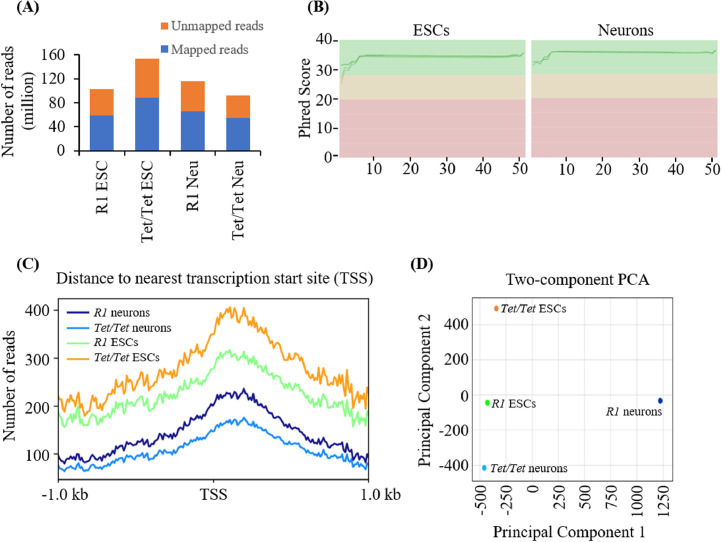
The present dataset was obtained by classic RRBS which covers ≥ 70% of the CpG islands (CGIs) and > 75% of all gene promoters and 1.5 – 2.0 million unique CpG sites that can be identified [1]. A total number of 92.6 – 153.7 million reads were generated from all four samples with 99% conversion rate and 56.8 – 58.7% mappable reads (Fig 1A). Overall, an average coverage of 40X per locus was achieved with Phred Scores > 20 corresponding to > 99% accuracy (Fig 1B). The distribution of RRBS-sequences containing converted and unconverted cytosines for the four cell types suggested that the qualitative distribution of the sequences from 1 kb upstream to 1 kb downstream of the transcription start sites (TSS) is similar (Fig. 1C). Principal component analysis revealed that methylation differences exist between R1 and Tet/Tet ESCs and, between R1 and Tet/Tet neurons (Fig. 1D).
Fig. 1.
Broad features of RRBS sequencing data from R1 and Tet/Tet ESCs and their corresponding neurons. (A) Number of reads obtained and the proportion of mappable reads in the four cell types. Millions of reads are shown in Y axis against the different cell types on the X-axis. (B) Phred Scores of the RRBS data from the four samples. (C) Distribution of the identified CpG sites with respect to the transcription start sites. (D) Two-component Principal Component Analysis (PCA) of R1 ESCs, Tet/Tet ESCs, R1 neurons and Tet/Tet neurons.
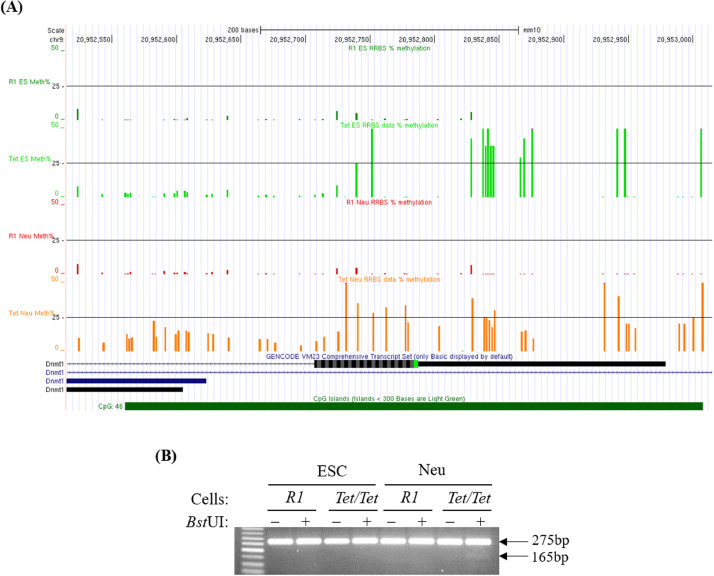
Processed data in the form of bedGraph files were provided as supplementary data associated with the accession number GSE152817 (Table 1, GSE152817_RAW.tar). These files were uploaded in the UCSC genome browser for visualization of the RRBS data. As an example, Fig. 2A shows the methylation levels of the CpG sites visualized by querying for the gene Dnmt1. Upon focusing on the promoter region, we observed ∼25% increase in methylation of a 288 bp region downstream to the Dnmt1 promoter in Tet/Tet neurons (Fig 2A). Using the same procedure, genome-wide DNA methylation levels can be studied and target genes with dysregulated methylation patterns can be identified.
Fig. 2.
Use of RRBS data in identifying DNA methylation in promoters of genes of interest. (A) DNA methylation data for Dnmt1 promoter in ESCs and neurons from R1 and Tet/Tet cells viewed in UCSC browser. (B) Validation of DNA methylation data by Combined Bisulfite Restriction Analysis (COBRA). Agarose gel electrophoresis of bisulfite PCR products obtained from biological replicates of ESCs and neurons from R1 and Tet/Tet cells using primers for the indicated 288 bp region. +: reaction mixture was incubated with BstUI enzyme, -: no enzyme control. The presence of a faint 165 bp fragment confirms presence of methylated CpGs in the analyzed genomic region in the DNA from Tet/Tet neurons.
2. Experimental design, materials and methods
The Dnmt1tet/tet ESC line is a transgenic cell line obtained by knocking in tet-off constructs downstream to the Dnmt1 (DNA methyltransferase 1) promoter of the R1 ESCs (wild-type) [2]. The cassettes were upstream to the translation start site located in the exon1 of Dnmt1. As a result, the Tet/Tet ESCs overexpressed DNMT1 and the resultant neurons also showed increased levels of the enzyme [3]. The main purpose of the study was to generate nucleotide level information on the cytosine methylation levels of the genomes of R1 ESCs, Tet/Tet ESCs, R1 neurons and Tet/Tet neurons. DNAs from all the four cell types were isolated by standard procedures and subjected to RRBS analysis (Section 2.2). The RRBS data described here enables analysis of DNA methylation levels of target sequences in R1 and Tet/Tet ESCs and neurons. In future, the data enables identification of target DNA sequences subject to differential methylation due to increased DNMT1 levels. Such information is important to study the role of DNMT1 overexpression in abnormal neurogenesis.
2.1. Cell culture
All reagents were of tissue culture or molecular biology grade and obtained from vendors such as ThermoFisher (USA), Merck (Germany), Takara Biotech (USA), HiMedia (India) and New England BioLabs (USA). ESCs were grown in ES media (Knockout DMEM/F-12 with 10% ESC-grade FBS, 1X Non-Essential Amino acids, 1X Glutamax, 1X Penicillin/Streptomycin, 100 µM β-meracptoethanol, 10 ng/ml Leukemia Inhibitory factor) till confluence in 0.1% Gelatin coated cell culture dishes. In case of Tet/Tet ESCs, the medium was supplemented with G418 (150 µg/ml) and puromycin (1 µg/ml). After reaching confluence, the cells were treated with 1X Trypsin-EDTA for ten minutes to obtain single cell suspensions. The cells were plated onto non-adherent dishes containing embryoid body medium (ES medium without Leukemia Inhibitory Factor) to obtain EBs after two days. The EBs were differentiated using NDiff 227 medium on cell culture-treated dishes for 10–12 days. At the end of 12 days, a majority of the cells differentiated into neurons with well-branched neurites. A more detailed description of the differentiation process is given in Saxena et al. [3,4].
2.2. Extraction of DNA and reduced representational bisulfite sequencing (RRBS)
The RRBS was performed at Zymo Research (USA) using genomic DNAs from R1 and Tet/Tet ESCs and their neuronal derivatives. Genomic DNAs were isolated using SDS-proteinase K digestion and phenol: chloroform extraction method and quantified using Nanodrop® [5]. The method followed for generating RRBS data was adapted from Meissner et al. [1]. About 200 ng each of the DNA samples were digested with 30 units MspI and purified using DNA Clean and Concentrator™-5 (Zymo Research). The fragments were ligated to pre-annealed adapters containing 5′-methylcytosine instead of cytosine as per Illumina's specified guidelines. Adapter -ligated fragments were purified using DNA Clean and Concentrator™-5 and then treated with bisulfite using EZ DNA methylation Lightening Kit (Zymo Research). The converted DNAs were amplified with adapter-specific primers. The amplified products were analyzed by bioanalyzer to ensure correct size distribution and the PCR products were sequenced using next generation sequencing technology using Illumina HighSeq 1500 platform.
Sequence reads were identified using Illumina base calling software and the sequences corresponding to the adapters were removed using TrimGalore 0.6.4 software [6]. The effect of trimming and overall quality distributions of the data were assessed by FASTQC 0.11.8 software [7]. The sequences were mapped to mm10 mouse genome using Bismark 0.19.0 software [8]. Methylated and unmethylated read totals for each CpG site were called using MethylDackel 0.3.0 software [9]. Methylation level of each sampled cytosine was estimated as the number of reads reporting a cytosine divided by the total number of reads reporting cytosine or thymine. In order to identify significant differentially methylated cytosines, Fisher's exact test was performed on sequences with a minimum read coverage of five in each sample with at least 10% difference in the level of methylation. Genes with significant differences in the level of methylation with a p-value < 0.05 were identified by summing up all the CpG sites in one gene and the value was used to determine if there is hypomethylation or hypermethylation.
2.3. Visualization of methylation data in UCSC browser
The sequence data was converted to bedGraph files and were uploaded as custom tracks in the UCSC genome browser [10]. The methylation levels were displayed in the UCSC browser as vertical bars. For display of methylation levels, the Y-axis in the UCSC settings can be set from 0 to 100 or a desired range, with 100 representing complete methylation (100%) and zero representing no methylation. In order to distinguish the individual tracks, the track files were coded in different colours. The level of methylation for the individual CpG sites for a given genomic region were visualized in the UCSC genome browser by entering the chromosome coordinates or gene name in the query box.
Validation of methylation differences by Combined Bisulfite Restriction Analysis (COBRA)
The RRBS data generated here and the procedure in general provides an average of 10X - 40X coverage of the target sequences. However, an independent validation of differences in methylation identified by RRBS data is needed as confirmatory evidence. Combined Bisulfite Restriction Analysis (COBRA) is a method that provides an overall estimate of the level of methylation using a larger number of molecules than sampled by RRBS or sequencing of cloned PCR products obtained from bisulfite-treated genomic DNA [11]. In COBRA, specific primers for target sequences under investigation are used to amplify bisulfite-treated DNA. The obtained PCR products are gel-purified for incubation with restriction enzymes containing one or more CpGs in their recognition sites. Unmethylated sequences are converted by bisulfite into uracils and are observed as thymines in the amplified products. Methylated sequences on the other hand are resistant to the conversion and are retained as cytosines and CpGs in the amplified products. Therefore, the extent of digestion of the PCR products by the restriction enzymes mentioned above allows an approximation of methylation levels from a large number of target sequences in the sample. Fig. 2B shows validation of the methylation difference identified in the 500 bp region downstream to the Dnmt1 promoter in Tet/Tet neurons. Agarose gel electrophoresis of samples incubated with the enzyme BstUI (CGCG) suggested digestion of a small proportion of the PCR products obtained from bisulfite treated DNA from Tet/Tet neurons but not from the other three cell types. Therefore, the COBRA results confirmed the DNA methylation difference downstream to Dnmt1 promoter region identified by RRBS (Fig. 2A).
Declaration of Competing Interest
The authors declare no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Ethics statement
This work was approved by the Institutional Biosafety Committee of BITS Pilani, Hyderabad Campus.
Acknowledgements
This work is supported by grants from Scientific and Engineering Research Board and Department of Biotechnology in the initial stages and by OPERA (BITS Pilani) at later stages. Sumana Choudhury is supported by fellowship from Centre for Human Disease Research, BITS Pilani Hyderabad Campus. Sonal Saxena is supported by fellowships from Department of Biotechnology and for a brief period from OPERA.
References
- 1.Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucl. Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Aiuto L., Di Maio R., Mohan K.N., Minervini C., Saporiti F., Soreca I., Greenamyre J.T., Chaillet J.R. Mouse ES cells overexpressing DNMT1 produce abnormal neurons with upregulated NMDA/NR1 subunit. Differentiation. 2011;82:9–17. doi: 10.1016/j.diff.2011.03.003. http:// doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena S., Maroju P.A., Choudhury S., Anne A., Mohan K.N. Analysis of transcript levels of a few schizophrenia candidate genes in neurons from a transgenic mouse embryonic stem cell model overexpressing DNA Methyltransferase 1 (DNMT1) Gene. 2020;757 doi: 10.1016/j.gene.2020.144934. [DOI] [PubMed] [Google Scholar]
- 4.Saxena S., Choudhury S., Mohan K.N. Reproducible differentiation and characterization of neurons from mouse embryonic stem cells. MethodsX. 2020 doi: 10.1016/j.mex.2020.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green M.R., Sambrook J. Isolation of high-molecular-weight DNA using organic solvents. Cold Spring Harb. Protoc. 2017;2017:356–359. doi: 10.1101/pdb.prot093450. [DOI] [PubMed] [Google Scholar]
- 6.F. Krueger, "Trim galore." A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter trimming to FastQ files, 516 (2015) 517. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 7.Andrews S. 2010. FastQC: A Quality Control Tool For High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk?/projects/fastqc/ [Google Scholar]
- 8.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan D.P. 2019. MethylDackel.https://github.com/dpryan79/methyldackel [Google Scholar]
- 10.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong Z., Laird P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucl. Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]