Abstract
Emerging viruses are a major public health problem. Most zoonotic pathogens originate in wildlife, including human immunodeficiency virus (HIV), influenza, Ebola, and coronavirus. Severe acute respiratory syndrome (SARS) is a viral respiratory illness caused by a coronavirus called SARS-associated coronavirus (SARS-CoV). Viruses are charged colloidal particles that have the ability to adsorb on surfaces depending on pH. Their sorptive interaction with solid particles has important implications for their behavior in aquatic environments, soils, sewage sludge, and other solid materials and their removal or concentration by water treatment processes.
Current state of knowledge on the potential of wastewater surveillance to understand the COVID-19 pandemic is reviewed. This study also identified wastewater irrigation systems with a higher risk of COVID-19 transmission. Emphasis was placed on methodologies for the detection and quantification of SARS-CoV-2 in wastewater.
Keywords: COVID-19 pandemic, Wastewater surveillance, Treatment methods, Wastewater treatment plants
Graphical abstract
1. Introduction
In light of recent epidemics around the world, there is increasing awareness of the risk of exposure to emerging pathogens during wastewater collection and treatment. Emerging pathogens may enter wastewater systems from pathogen shedding in human waste, introduction of decontamination wastewater, illicit activity, animal farming, and hospital effluents, or surface water runoff following a wide-area biological incident. Some emerging pathogens (e.g., Ebola virus [EBOV] and SARS-CoV-2) pose a significant health threat and their exposure in a wastewater system could have potentially serious health consequences. The need to assess the potential exposure and transmission of disease through sanitation systems is therefore necessary. Excretion of SARS-CoV-2 and its RNA from the body in saliva, sputum, and feces can be found in sewage. The main route of transmission of this virus would be inhalation by person-to-person transmission and aerosol/droplet, as well as fomite and hand contamination. However, currently available data indicate that there is a need to better understand the role of wastewater as a potential source of epidemiological data and a risk factor for public health. The detection of SARS-CoV-2 in feces has prompted several groups around the world to promote the analysis of wastewater to assess its circulation in populations (Mallapaty, 2020; Lodder and de Roda Husman, 2020).
Wastewater is all the water from homes and urban public facilities (hospitals, schools, etc.) as well as from certain industries (if it does not require specific treatment). This water is conveyed, via the “sewerage system”, to treatment plants where it is treated and then discharged into the environment. Wastewater and wastewater-based epidemiology (WBE) monitoring is a tool to guide epidemiological surveillance and mitigation efforts for infectious diseases, such as the Global Polio Eradication Initiative (Asghar et al., 2014). It is essential to continuously monitor the prevalence of SARS-CoV-2 and take appropriate measures to prevent and control the spread of the disease in the community. However, it is very difficult to track the virus because most people are asymptomatic; and further, it is not possible to do active clinical testing of all individuals, due to resource and cost constraints. Furthermore, COVID-19 may also display second or more waves. Under these circumstances, the passive, but effective, method of sewage or wastewater monitoring can be used to trace and track the presence of SARS-CoV-2, through their genetic material RNA, and screen entire community. The presence of SARS-CoV-2 in wastewater is predictable because it can infect the gastrointestinal tract and are shed through the stools of the patients (Xiao et al., 2020; Leung et al., 2003; Zumla et al., 2015; Gu et al., 2020).
With the objective of keeping the water community informed of COVID-19 related findings, this review highlights the recent scientific evidence, as well as topics not previously discussed. Emphasis was placed on their presence and persistence, as well as detection methods in different water matrices. Virus-infected wastewater treatment techniques are also highlighted.
2. Persistence of SARS-CoV-2 on surfaces and environment
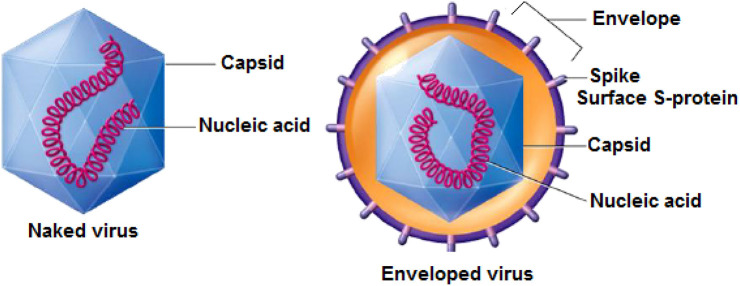
Environmental persistence refers to the length of time a pathogen, such as the SARS CoV-2, is able to survive outside the human body; the longer it survives, the more likely it is to cause infection. For the most part, investigations on the fate of viruses in the aquatic environment have focused on non-enveloped enteric viruses, given that these viruses are characterized by high resistance under a variety of environmental conditions (Annalaura et al., 2020). As for the number of studies concerning the fate of enveloped viruses in aquatic compartments, it is rather limited, because enveloped viruses are predisposed to deactivate in water (Wigginton et al., 2015). In fact, although enveloped viruses are inactivated more quickly than non-enveloped viruses, the survival time of enveloped viruses can still be very long depending on the specific environmental conditions. This is explained by the fact that the surface S-proteins (Fig. 1 ) are deeply anchored and only pass through the envelope: if the envelope is altered, but the surface S-proteins are preserved, infectivity is maintained.
Fig. 1.
Virus structure.
The persistence of viruses can be affected both by the type of environment (e.g., surface, water, wastewater) and by the physical and chemical properties of the environment (e.g., temperature, pH, humidity, exposure to sunlight and the type of surface (Rzeżutka and Cook, 2004; Thevenin et al., 2013). Furthermore, it is also possible to be influenced by the composition of the media. This is consistent with a recent study published detailing the survival rates of SARS-CoV-2 on different non-living surfaces: 72 h on stainless steel and plastic, 24 h on cardboard, and 4 h on copper (Van Doremalen et al., 2020).
Studies to verify the survival of SAR-CoV and SAR-CoV-2 have a strong phylogenetic similarity (Forster et al., 2020), showing the survival rate of a pathogen with a viral titer of 105 on aluminum, plastic, metal, wood and paper of 4–5 days at room temperature (Kampf et al., 2020). Unfortunately, data on the environmental sustainability of SARS-CoV-2 is limited to date. However, the basic understanding of its persistence can be explained by the results of studies on other coronaviruses such as SARS CoV-1 and MERS CoV.
3. SARS-CoV-2 in wastewater
Generally speaking, viruses are microscopic pathogenic agents ranging in size from 18 to 1500 nm (Hurst and Gerba, 1989) and are abundant in water and wastewater (Gantzer et al., 1998), especially domestic and urban effluents, which are particularly laden with animal and bacterial viruses. These two groups are of particular interest for environmental and human health studies.
Infected subjects are considerable and can have up to 1010 per gram of stool (Masson, 1996). The transmission of these viruses to humans generates several diseases and epidemics that affect all age groups. In some cases, they can be fatal for children (e.g., gastroenteritis leading to morbidity and mortality during the first five years of life) (Parashar et al., 2013). Humans are considered to be the primary contaminator, and secondary receptor (Gantzer et al., 1998). Several potential routes ensure viral transmission to humans: domestic use of the contaminated surface, or groundwater, bathing in contaminated water, consumption of infected seafood, or crops grown in contaminated soil.
There are many different types of viruses to include, for example, Picornaviridae (e.g. polioviruses, echoviruses, and hepatitis A), Reoviridae (e.g. reoviruses, rotaviruses), Adenoviridae (e.g. adenovirus A.), Coronaviridae (e.g. coronaviruses), Caliciviridae (e.g. caliciviruses), small round viruses (e.g. astroviruses, Norwalk), and bacteriophages (Gantzer et al., 1998; Girard and Hirth, 1989; Havelaar, 1991; Grabow et al., 1995; Havelaar et al., 1986; Havelaar et al., 1990; Joffre, 1991).
Bacterial viruses or bacteriophages (or phages) are present everywhere there is bacterial life. In every environment, there are phages from native bacteria and phages from elsewhere. The abundance of phages in water environments, especially those polluted by fecal matter, is great. They come from various ecological niches, including the digestive tract (Grabow et al., 1995; Havelaar et al., 1986; Havelaar et al., 1990; Joffre, 1991). According to Kai et al. (1985), their concentration in the feces is less than l05 particles per gram.
Owing to their characteristics, bacteriophages are considered by researchers as indicators of fecal pollution and as models for monitoring the fate of viruses in different water environments and WWTP (Havelaar, 1991). Bacteriophages have a morphology, structure, and composition similar to those of enteric viruses. Resistance and behavior, especially of F-specified RNA phages, are comparable to those of enteric viruses (Havelaar, 1991; Havelaar and Pot-Hogeboom, 1988; Lewis, 1995). Concretely, on the one hand, phages are not involved in human pathology.
One of the main problems is the spread of viruses between polluted environmental waters and populations. It happens that water is contaminated by humans on the one hand, and water becomes a means of infection for humans on the other hand. Since wastewater contains viruses that are repelled by everyone, regardless of their health, monitoring for viruses in wastewater and environmental waters that receive effluent from wastewater treatment plants (WWTPs) can determine the true prevalence and molecular epidemiology of gastroenteritis viruses and the risks to human health (Guan et al., 2020; Huang et al., 2020; Wang et al., 2020) in a given geographical area rather than clinical research (Prevost et al., 2015; Kazama et al., 2017). Such data would be a useful indicator, especially for poor countries, as it would allow simple, reliable and inexpensive epidemiological surveillance. In such a situation, the two fundamental factors that require attention to prevent the spread of infection, especially in high-risk locations such as hospitals, are the interconnection of the plumbing system and its condition (Gormley et al., 2020). The wastewater piping system is designed to be a precursor to pathogenic microorganisms that, in some cases, can cause airborne spread of viruses such as SARS-CoV-2.
Several studies in different countries have detected the genetic material of SARS-CoV-2 in the human feces of patients with COVID-19 (Wölfel et al., 2020; Lescure et al., 2020; Holshue et al., 2020; Xiao et al., 2020). The genetic material of SARS-CoV-2 has been detected in the feces of COVID-19 patients with or without gastrointestinal symptoms (Wölfel et al., 2020; Lescure et al., 2020) and in cured individuals who no longer have symptoms (Wölfel et al., 2020; Lescure et al., 2020; Holshue et al., 2020). However, the presence of SARS-CoV-2 genetic material in the stool does not necessarily indicate infection or disease. A few studies that have attempted to detect a viable infectious virus in the stool have produced conflicting results. Three studies reported the detection of live virus (Wölfel et al., 2020; Lescure et al., 2020; Holshue et al., 2020) in the stool and one study reported no detection of live virus despite the detection of the SARS-CoV-2 genetic material (Holshue et al., 2020).
Researchers in Netherlands were the first to successfully isolate and detect SARS-CoV-2 in wastewater (Medema et al., 2020). They demonstrated that the coronavirus genome can be detected at several wastewater collection sites within days of the identification of the first human case of COVID-19. This hypothesis was confirmed by Wurtzer and his colleagues (https://www.sciencemag.org/news/2020/04/coronavirus-found-paris-sewage-points-early warning-system). It also demonstrates, for the first time, that the quantities of viral genomes detected in wastewater are increasing in line with the number of COVID-19-related hospitalizations at the regional level. Preliminary results obtained even more recently at the same sites show a very significant reduction in the viral load in wastewater, an expected consequence of containment measures on the circulation of the virus.
Very recently, researchers from Australia have also advocated sewage monitoring for SARS-CoV-2 surveillance in the community (Ahmed et al., 2020). Researchers are now developing techniques to track the spread of new coronaviruses in water. The early warning and monitoring system will recognize the virus by its genetic material or RNA (ribonucleic acid). The presence of coronavirus in untreated wastewater has been proven by researchers at the University of Queensland and the Australian National Scientific Agency CSIRO that took samples from a suburban pumping station and WWTP. They analyzed the wastewater samples using RT-PCR tests, which helps identify gene fragments of the SARS-CoV-2 virus. This is also the method used by hospitals to test for the virus in human samples.
Passive surveillance through wastewater mining and monitoring of SARS-CoV-2, as a subset of the National Water Quality Monitoring Network, therefore, can be utilized to assess community or public health in COVID-19 pandemic as well as post-pandemic scenarios. The monitoring should commence in red, orange, as well as green zones to alert health officials in advance about the possible second wave of COVID-19.
Culture techniques can detect viruses and provide information on the viability of the virus. However, these methods remain more difficult, especially for SARS-CoV-2, and take more time than most molecular techniques, which explains why they are less frequently used. In order to develop and test methods for monitoring SARS-CoV-2 genetic material in sewage, a study is currently underway (Kitajima et al., 2020). With this approach, it is possible to estimate the frequency of the disease in communities, identify areas where few tests are performed, predict a possible second wave of infection, and monitor vaccine results. The approach is not yet ready and is not an alternative to human testing.
4. Ecological and health impacts of COVID-19 contaminated wastewater
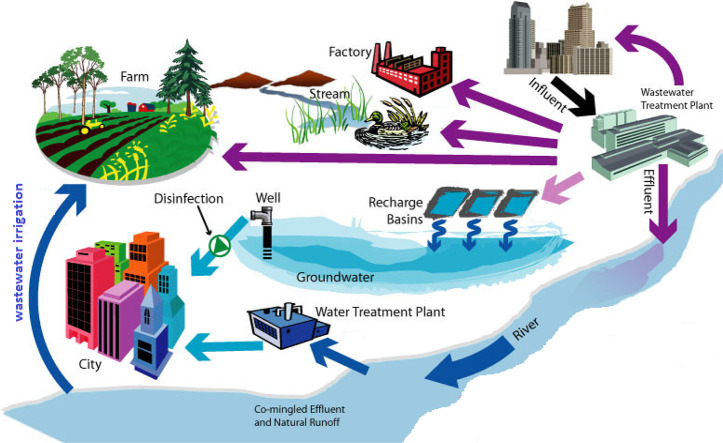
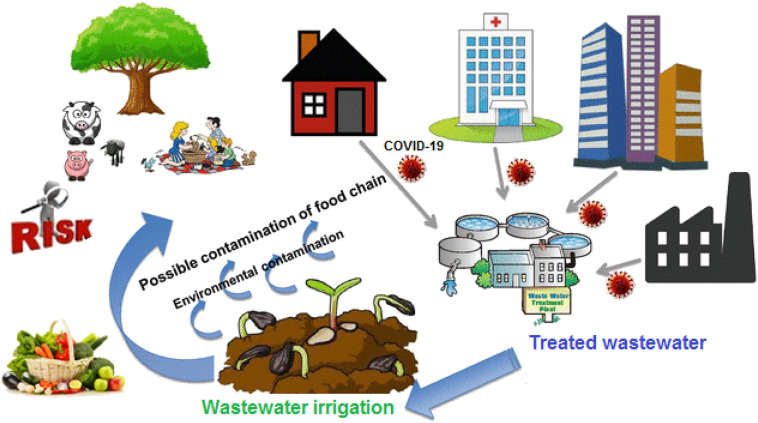
In many countries, due to the lack of water resources, the following are used to wastewater for irrigation of agricultural land. In addition, sludge from the treated wastewater is excellent fertilizer; it is increasingly used in agricultural amendment. The viruses contained in this wastewater and sludge is thus deposited on crops and on the soil where they are likely to survive. There are then creations of a risk to public health either as a result of the contamination of water bodies by migration of viruses through the soil or the consumption of contaminated market garden produce.
The reuse of treated and untreated wastewater in agriculture is a practice that is steadily increasing. It is therefore important to know the fate of these viruses, particularly on plants. This can only be done with enteric viruses that can multiply on cell cultures. Indeed, the presence of viral genome does not indicate the presence of infectious viral particles.
Studies reported in the literature show that viruses can survive on plants irrigated with sewage for variable periods ranging from a few days to nearly 4 weeks depending on conditions (Badawy et al., 1990; Tierney et al., 1977). In addition to the nature of the vegetable under consideration, it appears that two factors have a fundamental influence on the survival time of viruses on plants after irrigation. These are the initial level of contamination and the temperature associated with sunlight. It must be fully aware that even in regions with a very hot climate it takes more than 6 h to achieve a 2-log reduction in virus contamination. This means, for example, that in case of irrigating a golf course with wastewater in the morning, enteric viruses are likely to be still present when the players will be on the course during the day. The same situation can be found on irrigated lawns in public parks. In both cases, players or children are likely to come into contact with enteric viruses.
On the other hand, with regard to vegetables, it is clear, and this has been well confirmed by Croci et al. (1991), that plants irrigated during cultivation with contaminated water retain significant amounts of viruses on their surface. In addition, storage at +4 °C not only does not accelerate viral inactivation but slows it down. It is therefore extremely important to have virus-free vegetable crops at harvest time. Fig. 2 illustrates the potential impacts of reusing recycled wastewater for agricultural irrigation.
Fig. 2.
The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment.
In addition, since the onset of the crisis, the impact of the COVID-19 pandemic on the environment has attracted attention, including observation and analysis of recent impacts, and estimates related to long-term changes. Qualitative assumptions prevail, while consistent quantitative research must await relevant data sets and additional knowledge. Most aspects of the environmental impact of the COVID-19 pandemic do not come directly from the virus itself. Sudden restrictions or closures of economic sectors (such as heavy industry, transportation, or hotel businesses) had a direct impact on the environment. From an anthropocentric point of view, the pandemic may lead to a more sustainable future, and particularly to more resilient socio-ecological systems or shorter supply chains, which is a positive development.
However, countries can still choose less sustainability by pursuing rapid economic growth and reducing environmental concerns. Although the overall negative impact on the economy and society can be enormous, it is likely that the reduction in global economic activity due to the COVID-19 crisis will trigger many major improvements in the quality of service of the environment and climate systems. However, not all the environmental consequences of the crisis are or will be positive. Some of these include the increase in the amount of non-recyclable waste, the large amount of organic waste generated due to declining agricultural and fisheries exports, and the failure to maintain and monitor natural ecosystems. https://unctad.org/en/pages/newsdetails.aspx?OriginalVersionID=2333.
Anecdotal evidence indicates reduced biochemical oxygen demand and coliform levels in rivers, improved air quality by reducing nitrous oxide (NOx) loading, particulate matter (PM) and the formation of ozone (O3) associated with other pollutants (UNCTAD, n.d). https://unctad.org/en/pages/newsdetails.aspx?OriginalVersionID=2333
Many rural and coastal populations depend on the sustainable use of the local environment and its natural resources, whether they are small farmers, small and medium enterprises (SMEs) or micro, small and medium enterprises (MSMEs) involved in the production of organic trade, forest and fisheries products and ecotourism services. Because the crisis has disrupted their links to national and international markets on the demand side, rural producers (many of whom are women supporting entire families) can no longer fully maintain their business models and livelihood resources. Attention needs to be paid to the threats to the environment and natural resources posed by the coronavirus pandemic, and the social and economic consequences that result.
As in many countries in the world, wastewater offers Morocco an alternative solution as the pressure on freshwater is constantly increasing, particularly due to repeated droughts. In the country, wastewater reuse is intended for industry, particularly phosphate washing, but also for golf course irrigation. A letter from the Ministry of the Interior addressed to the local authorities prohibits the use of wastewater before it has been treated due to the possible presence of traces of genomes from the stools of affected persons (https://www.leconomiste.com/article/1061485-les-eaux-usees-un-moyen-de-tracage-du-covid-19). The letter mentions that the use of this wastewater is set by the laws and regulations in force.
5. SARS-CoV-2 detection in wastewater
The detection of viruses in wastewater and drinking water requires the following detection methods: sensitive, resistant to false-positive results, and enabling full automation. In addition, the method used must be fast and inexpensive. A method, which fulfills all needed requirements, as yet was not worked out.
In addition to economic agents, the implementation of methods for the detection of viruses in the aquatic environment can come up against several obstacles, including: the considerable dilution of the sample, the influence of the environmental matrix on the analytical results and the mutagenic variability of viruses. In faecally contaminated water, viruses are present in relatively small amounts. It is therefore often necessary to concentrate the samples before determining the exact virus content (Marzouk et al., 1979). These include, for example: Polymerase chain reaction (PCR) (Li et al., 2002; Pina et al., 1998; Lee et al., 2005; Cho et al., 2000; Fout et al., 2003; Schvoerer et al., 2000), nucleic acid sequence-based amplification (NASBA) (Rutjes et al., 2006; Jean et al., 2002; Jean et al., 2001), DNA chip technique (Nettikadan et al., 2003; Alhamlan et al., 2013), atomic force microscope (AFM) (Nettikadan et al., 2003), fluorescence microscopy (Hara et al., 1991; Hennes and Suttle, 1995; Noble and Fuhrman, 1998; Shibata et al., 2006; Wen et al., 2004; Weinbauer and Suttle, 1997), electron microscopy (Weinbauer and Suttle, 1997; Bettarel et al., 2000; Alonso et al., 1999; Wommack et al., 1992), biosensor application (Liu and Zhu, 2005; Rengevych et al., 1999), enzyme-linked immunosorbent assay (ELISA) (Kittigul et al., 2000; Nasser and Metcalf, 1987; El-Esnawy, 2000; Nishida et al., 2009) and flow cytometry (Marie et al., 1999; Abad et al., 1998; Brussaard, 2004; Chen et al., 2001). Some of these methods have been modified by: i) concentration (ELISA tests, PCR and NASBA reactions, application of microarrays) (Li et al., 2002; Alhamlan et al., 2013; Kittigul et al., 2000), ii) combination of different methods (PCR reaction combined with plate-forming tests, atomic force microscopy combined with protein microarray technology) (Straub et al., 1995; Haab et al., 2001; Zhu and Snyder, 2001), iii) change in the pore size of the filter (epifluorescence microscopy) (Weinbauer and Suttle, 1997), iv) dilution of the sample (flow cytometry) (Nettikadan et al., 2003). Combined methods can also be used. For example, the combination of PCR with plate formation tests is possible. Polymerase chain reaction is used to amplify the specific sequence of deoxyribonucleic acid (DNA). In this reaction, double-stranded DNA, called template DNA, is amplified. The PCR technique, because of its high specificity, has been adopted for several years for the detection of viruses in the environment, in particular enteroviruses and hepatitis A virus (HAV) (Egger et al., 1995; Gantzer et al., 1997). PCR is a technique that amplifies (i.e. synthesizes many copies) a specific segment of DNA of interest using short sequences of synthetic nucleotides called primers that bind to unique regions of the target genome for organism-specific identification.
While much simpler than cell culture, molecular methods also present many challenges. The degree of methodological sensitivity and specificity depends both on the efficiency of virus recovery from samples by concentration and purification processes and on the final degree of purity of the virus recovered by removal of PCR inhibitory substances. The system still does not meet the established standards for virus detection. It is not possible to determine unequivocally whether the virus detected is infectious or even if the PCR result is positive. In addition, the PCR reaction can only detect one type of virus at a time and there are many different types of viruses in water (Li et al., 2002). Molecular methods pose another problem when it comes to translating nucleic acid test results in public health risks. Using nucleic acid-based analysis, the result is the expression of all viral genetic material present, without distinguishing between infectious and non-infectious particles (Metcalf et al., 1995). It is beyond the scope of PCR to measure the extent to which viruses are affected by different disinfectants. In fact, PCR targets specific parts or fragments of RNA, regardless of the type and severity of damage caused by the disinfectant to the genome (e.g., RNA structural modification, phosphate backbone scission, etc.) (Torrey et al., 2019). Recent efforts to monitor COVID-19 virus in wastewater in the Netherlands, Australia, and the United States have relied on RT-PCR as a detection tool. These studies had the primary objective of both supporting public health surveillance and estimating the number of infections in the community.
Promising method which may be used in the near future is a device which is called laboratory-on-a chip (LOC). It is kinds of biosensors which can respond to certain properties of analyte and converts these responses into detectable signal; the most common kind of signal is electrical. These are complex devices with a network of fluid microchannels, valves, mixers, pumps, reaction chambers, and interconnected detectors, and they can perform many laborious procedures without manual intervention (Jung et al., 2015). These features make the LOCs suitable for clinical diagnostic applications and immediate care tests (Wang et al., 2017), allowing mass production at low cost. The LOC-based technology has been widely used for virus detection, and research has been published in the past, such as a comprehensive description of pathogen detection using microfluidic systems (Mairhofer et al., 2009). Other reviews on viral infection and microfluidic technology are specifically used to diagnose a single virus, including methods for sample preparation and detection, such as Ebola virus (Coarsey et al., 2017), dengue fever (Darwish et al., 2018), hepatitis (Duchesne and Lacombe, 2018) and human immunodeficiency virus (HIV) (Mauk et al., 2017).
With LOC technology, the detection of microbiological pathogens in environmental samples is fast and sensitive. Its use in environmental microbiology can be done in several ways. LOC-based detection system is cost-effective, rapid, sensitive, specific, and has relatively high throughput for parallel identification, which is especially suitable for resource-limited facilities/areas and POC testing (Huang et al., 2017). Biosensors or methods based on connection between plaque-forming tests and PCR reactions seem to be the right alternatives. The application of biosensors has the capability to detect viruses for 7–16 min (Liu and Zhu, 2005). In addition, the technology is highly sensitive and allows observation of the interaction between molecules in real-time (Liu and Zhu, 2005; Keusgen, 2002). Considering the need for rapid information on the infection status of the population, and the presence of the virus in the environment, biosensors can play an essential role in the diagnosis and surveillance of the disease in the current global pandemic. Qiu et al. (2020) were able to demonstrate a dual-function plasmonic biosensor that combines plasmonic photothermal effect (PPT) and localized surface plasmon resonance (LSPR) to detect SARS-CoV-2 RNA transduction without the use of RT-PCR. The biosensor demonstrated high specificity and a low detection limit of SARS-COV-2 sequences down to 0.22 pM (Qiu et al., 2020). As the viral concentration is relatively low in a patient sample, the detection limit is of paramount importance. LOC devices appear to have the lowest detection limits, making them most immediately relevant for this application (Qiu et al., 2020; Peng et al., 2019; Kaarj et al., 2018; Seo et al., 2020).
Plaque forming tests connected with PCR reaction enable detection of viruses present in water in time of 2 to 4 days (Straub et al., 1995). In this method, the detection period is considerably shorter than that of the traditional plate-forming test, as well as a shorter incubation period. Techniques based on biosensors in terms of time-saving are much better (Liu and Zhu, 2005). Comparison of described methods is presented in Table 1 .
Table 1.
Advantages and limitations of virus detection methods for environmental sample application.
| Method for virus detection | Limitations | Advantages |
|---|---|---|
| PCR |
|
|
| Plaque forming test |
|
|
| Plaque forming test combined with PCR |
|
|
| NASBA |
|
|
| ELISA |
|
|
| Biosensors |
|
|
6. Treatment of virus-contaminated wastewater
Wastewater treatment is designed to reduce or eliminate suspended solids, dissolved and particulate organic matter, nutrients, and heavy metals. The degree of wastewater treatment is often indicated by the effluent standards prescribed by regulatory agencies, and by the end-use of the effluent. The sanitation and health guidelines of the World Health Organization (WHO) must always be followed (WHO, 2018). At present, no additional measures specific to COVID-19 are recommended by WHO, the US Centers for Disease Control and Prevention (CDC), or the Occupational Safety and Health Administration (OSHA).
With well-designed and well-functioning wastewater treatment plants and on-site sanitation systems with reliable in-situ disposal or a network for emptying and discharge to a sewage sludge treatment plant, the risk posed by fecal pathogens, including SARS-CoV-2, should be limited.
As a further precaution, WWTPs may consider adding a final disinfection step (often called tertiary treatment) to further reduce the risk posed by viral pathogens, such as SARS-CoV-2, before spilling. The methods are available for the inactivation of viruses in wastewater effluents and water range from the purely physical (ionizing radiation by gamma rays, non-ionizing radiation by ultraviolet light, photodynamic oxidation and heat) to the purely chemical (chlorine, chlorine dioxide, ozone, iodine, bromine, and bromine chloride). Two treatments such as the addition of chlorine to provide a residue after ozonation can be used to produce finished water free of toxic residues.
Chlorine is the most widely used disinfectant because it is effective at low concentrations, relatively inexpensive, and forms a residue if applied in sufficient doses. It can be applied as gas or hypochlorite, the gaseous form being the most common. Chlorine gas reacts readily with water to form hypochlorous acid and hydrochloric acid; the hypochlorous acid produced then dissociates to give the hypochlorite ion:
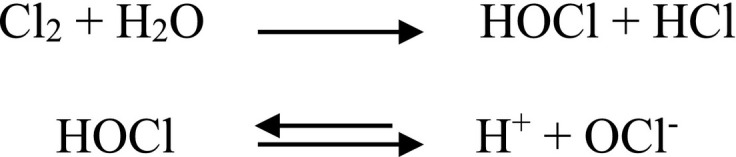
Hypochlorous acid (HOCl) form is the main form of chlorine responsible for its disinfecting properties. At neutral and lower pH levels, more HOCl is formed, resulting in greater disinfection capacity at these pH levels. Chlorine in the form of HOCl or OCl− is the available free chlorine. The combination of HOCl with ammonia and organic compounds in wastewater produces combined chlorine in the form of chloramines. Low-level wastewater disinfection can effectively inactivate SARS-CoV-1 (0.5 mg L−1 free chlorine residual), although standard dosage recommendations must be followed (Wang et al., 2005). Therefore, the safety of drinking water and wastewater depends on the appropriate selection of the disinfectant dose and contact time in the treated environment, which are very important analytical techniques and methods that can detect viruses.
Viruses are charged colloidal particles that have the ability to adsorb on surfaces. Their adsorption interaction with solid particles is very important for their behavior in aquatic and soil environments and for their elimination or concentration by water treatment processes (Bitton, 1975). To control the adsorption of viruses on surfaces, factors such as the type of virus and the relevant surface, pH, electrolytes, and the presence or absence of interfering substances in the suspension medium must be considered (Bitton et al., 1976).
As with all other treatment systems capable of removing viruses from water and effluents, carbon requires continuing study. Although it is not an exciting system for this purpose, carbon does retain viruses. The extent to which it acts as a filter by adsorbing or filtering other materials that adsorb viruses should be investigated.
Carbon, of course, is important in the disinfection process because it removes organic compounds that interfere with disinfectants. Carbon may also alter effluent quality in a way that adversely affects disinfection. For example, iron used for coagulation may react in carbon columns and subsequently reduce iodine making it unavailable for disinfection. Problems of this kind must always be monitored.
Treatment processes are generally unable to remove all viruses from sewage. Research must therefore focus on how best to remove substances that interfere with the disinfection process from effluents and raw waters. Techniques that lead to an increase in pH are of particular interest because strong alkalinity has a destructive effect on viruses. The concentration of viruses entering treatment facilities varies greatly from season to season, from one location to another, and even within 24-h (Bitton et al., 1976). Management of virus removal by treatment procedures requires coordination and synchronization of sampling. Quantitative methods of virus concentration must be developed to accurately assess the effectiveness of treatments.
The ability of some viruses to survive in disinfected effluent can be problematic (Cromeans et al., 2010; Li and Mitch, 2018). To overcome this dilemma, one solution is to adopt wastewater treatment technologies that achieve high levels of pathogen inactivation while removing carbon and nutrients. For example, the removal of pathogens (as well as biochemical oxygen demand (BOD) and nutrient removal) can be achieved by size exclusion using membrane bioreactors (Purnell et al., 2016). But high membrane costs and frequent fouling limit their large-scale application. The efficient operation of membrane bioreactors is suggested in this regard by filtering coronaviruses attached to suspended solids (Naddeo and Liu, 2020). Use of electrospun nanofibre membranes, which have the property of attracting the genetic material of viruses, is a potential tool for WBE surveillance (Venugopal et al., 2020). In addition, pathogens can be reduced by bioabsorption, photodegradation, gravity settling or chemical lysis (Curtis et al., 1992; Curtis, 2003).
The use of Algal-based WWT should also be evaluated. They were introduced in the 1950s to minimize the energy requirements of the activated sludge (AS) process and/or improve secondary effluent to meet nutrient discharge standards. In recent years, algae-based wastewater treatment systems have evolved to become sustainable and cost-effective alternatives to conventional, energy-intensive wastewater treatment systems (Rawat et al., 2011; Oswald and Gotaas, 1957). These systems have been shown to inactivate pathogens at high levels while removing carbon and nutrients (Young et al., 2016; Buchanan, 2014).
7. Conclusions and future perspectives
The presence of SARS-CoV-2 genetic material in wastewater can be used to monitor the spread of COVID-19 in a community. The use of wastewater as a disease surveillance tool is not widespread, but it is beginning to grow. Lack of a standardized protocol for monitoring SARS-Cov-2 in wastewater is a major challenge. Detecting viral genetic material in wastewater requires a virus concentration step to enable extraction and detection, but there is limited knowledge on how to do this efficiently for SARS-CoV-2. Understanding how the virus breaks down in the aquatic environment is also critical to assessing risks to human health at present; the stability of the SARS-CoV-2 genome in wastewater is unclear. It is possible to use an effective surveillance tool to provide an alert if viral particles rise above thresholds, allowing for quicker action and containing the infection before it spreads at an alarming rate.
Further research is also needed in other fields: methods for the quantitative detection of viruses adsorbed on solids; suitability of soils to eliminate viruses; search for better indicators for the presence of viruses; viral concentrations in the shellfish; frequency of human infections caused by shellfish by ingestion of small amounts of viruses contained in the water; epidemiology of waterborne human viral infections; effects of non-human viruses on people; presence of oncogenic agents in humans; presence of oncogenic agents in the water.
During this period, the authorities may reconsider their recommendations for local irrigation of wastewater. This also requires a renewed effort to promote micro-irrigation technology that can safely irrigate the soil without leaving the farmers and bring fresh produce into direct contact with the wastewater.
As the latest data on SARS-CoV-2 and other coronaviruses show, they can survive on different environmental surfaces for hours or days. Accordingly, further research is needed to better understand such differences in persistence duration. In this manner, supports-based these materials can be developed for the treatment of COVID-19 in wastewater.
CRediT authorship contribution statement
S. Lahrich: Conceptualization, Methodology, Supervision, Visualization, Writing - original draft, Writing - review & editing. F. Laghrib: Methodology, Writing - original draft. A. Farahi: Writing - original draft, Supervision. M. Bakasse: Conceptualization, Writing - review & editing. S. Saqrane: Conceptualization, Methodology. M.A. El Mhammedi: Conceptualization, Methodology, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abad F.X., Pintó R.M., Bosch A. Flow cytometry detection of infectious rotaviruses in environmental and clinical samples. Appl. Environ. Microbiol. 1998;64(7):2392–2396. doi: 10.1128/aem.64.7.2392-2396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W.…Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. Article 138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhamlan F.S., Ederer M.M., Green T.L., Brinkman C.K., Coats E.R., Crawford R.L. A novel screening tool using microarray and PCR to detect pathogens in agriculturally impacted waters. International Journal of Environmental Protection. 2013;3(3):17. [Google Scholar]
- Alonso M.C., Rodríguez J., Borrego J.J. Enumeration and isolation of viral particles from oligotrophic marine environments by tangential flow filtration. Int. Microbiol. 1999;2(4):227–232. [PubMed] [Google Scholar]
- Annalaura C., Ileana F., Dasheng L., Marco V. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L.…Burns C.C. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. The Journal of infectious diseases. 2014;210(suppl_1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawy A.S., Rose J.B., Gerba C.P. Comparative survival of enteric viruses and coliphage on sewage irrigated grass. Journal of Environmental Science & Health Part A. 1990;25(8):937–952. [Google Scholar]
- Bettarel Y., Sime-Ngando T., Amblard C., Laveran H. A comparison of methods for counting viruses in aquatic systems. Appl. Environ. Microbiol. 2000;66(6):2283–2289. doi: 10.1128/aem.66.6.2283-2289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton G. Adsorption of viruses onto surfaces in soil and water. Water Res. 1975;9(5–6):473–484. [Google Scholar]
- Bitton G., Pancorbo O., Gifford G.E. Factors affecting the adsorption of polio virus to magnetite in water and wastewater. Water Res. 1976;10(11):973–980. [Google Scholar]
- Brussaard C.P. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004;70(3):1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan A.N. Flinders University, School of the Environment; 2014. Comparing the Performance of a High Rate Algal Pond With a Waste Stabilisation Pond in Rural South Australia. Doctoral dissertation. [Google Scholar]
- Chen F., Lu J.R., Binder B.J., Liu Y.C., Hodson R.E. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 2001;67(2):539–545. doi: 10.1128/AEM.67.2.539-545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.B., Lee S.H., Cho J.C., Kim S.J. Detection of adenoviruses and enteroviruses in tap water and river water by reverse transcription multiplex PCR. Can. J. Microbiol. 2000;46(5):417–424. [PubMed] [Google Scholar]
- Coarsey C.T., Esiobu N., Narayanan R., Pavlovic M., Shafiee H., Asghar W. Strategies in Ebola virus disease (EVD) diagnostics at the point of care. Crit. Rev. Microbiol. 2017;43(6):779–798. doi: 10.1080/1040841X.2017.1313814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci L., Fiore A., De Medici D., Toti L. Persistence of Escherichia coli and poliovirus 1 in contaminated vegetables. MAN Microbiologie, Aliments, Nutrition. 1991;9(3):257–262. [Google Scholar]
- Cromeans T.L., Kahler A.M., Hill V.R. Inactivation of adenoviruses, enteroviruses, and murine norovirus in water by free chlorine and monochloramine. Appl. Environ. Microbiol. 2010;76(4):1028–1033. doi: 10.1128/AEM.01342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis T. The Handbook of Water and Wastewater Microbiology. 2003. Bacterial pathogen removal in wastewater treatment plants; pp. 477–490. [Google Scholar]
- Curtis T.P., Mara D.D., Silva S.A. The effect of sunlight on faecal coliforms in ponds: implications for research and design. Water Sci. Technol. 1992;26(7–8):1729–1738. [Google Scholar]
- Darwish N.T., Sekaran S.D., Khor S.M. Point-of-care tests: a review of advances in the emerging diagnostic tools for dengue virus infection. Sensors Actuators B Chem. 2018;255:3316–3331. [Google Scholar]
- Duchesne L., Lacombe K. Innovative technologies for point-of-care testing of viral hepatitis in low-resource and decentralized settings. J. Viral Hepat. 2018;25(2):108–117. doi: 10.1111/jvh.12827. [DOI] [PubMed] [Google Scholar]
- Egger D., Pasamontes L., Ostermayer M., Bienz K. Reverse transcription multiplex PCR for differentiation between polio-and enteroviruses from clinical and environmental samples. J. Clin. Microbiol. 1995;33(6):1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Esnawy N.A. Examination for hepatitis E virus in wastewater treatment plants and workers by nested RT-PCR and ELISA. The Journal of the Egyptian Public Health Association. 2000;75(1–2):219–231. [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fout G.S., Martinson B.C., Moyer M.W., Dahling D.R. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 2003;69(6):3158–3164. doi: 10.1128/AEM.69.6.3158-3164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantzer C., Senouci S., Maul A., Levi Y., Schwartzbrod L. Enterovirus genomes in wastewater: concentration on glass wool and glass powder and detection by RT-PCR. J. Virol. Methods. 1997;65(2):265–271. doi: 10.1016/s0166-0934(97)02193-9. [DOI] [PubMed] [Google Scholar]
- Gantzer C., Dubois É., Crance J.M., Billaudel S., Kopecka H., Schwartzbrod L.…Le Guyader F. Devenir des virus entériques en mer et influence des facteurs environnementaux. Oceanologica acta. 1998;21(6):983–992. [Google Scholar]
- Girard M., Hirth L. Virologie moleculaire Doin Cd. Paris. Appl. Enriron. Microbial. 1989;53 [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob. Health. 2020;8(5):e643. doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow W.O.K., Neubrech T.E., Holtzhausen C.S., Jofre J. Bacteroides fragilis and Escherichia coli bacteriophages: excretion by humans and animals. Water Sci. Technol. 1995;31(5–6):223–230. [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020;158(6):1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X.…Du B. Clinical characteristics of coronavirus disease 2019 in China. New England journal of medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab B.B., Dunham M.J., Brown P.O. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2(2) doi: 10.1186/gb-2001-2-2-research0004. (research0004-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S., Terauchi K., Koike I. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl. Environ. Microbiol. 1991;57(9):2731–2734. doi: 10.1128/aem.57.9.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar A.H. Bacteriophages as model viruses in water quality control. Water Research (Oxford) 1991;25(5):529–541. [Google Scholar]
- Havelaar A.H., Pot-Hogeboom W.M. F-specific RNA-bacteriophages as model viruses in water hygiene: ecological aspects. Water Sci. Technol. 1988;20(11−12):399–407. [Google Scholar]
- Havelaar A.H., Furuse K., Hogeboom W.M. Bacteriophages and indicator bacteria in human and animal faeces. J. Appl. Bacteriol. 1986;60(3):255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]
- Havelaar A.H., Pot-Hogeboom W.M., Furuse K., Pot R., Hormann M.P. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 1990;69(1):30–37. doi: 10.1111/j.1365-2672.1990.tb02908.x. [DOI] [PubMed] [Google Scholar]
- Hennes K.P., Suttle C.A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr. 1995;40(6):1050–1055. [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H.…Tong S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Huang Q., Xie L., Xiang G., Wang L., Xu H.…Jin X. A rapid, low-cost, and microfluidic chip-based system for parallel identification of multiple pathogens related to clinical pneumonia. Scientific reports. 2017;7(1):1–10. doi: 10.1038/s41598-017-06739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.…Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst C.J., Gerba C.P. Fate of viruses during wastewater sludge treatment processes. Crit. Rev. Environ. Sci. Technol. 1989;18(4):317–343. [Google Scholar]
- Jean J., Blais B., Darveau A., Fliss I. Detection of hepatitis A virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 2001;67(12):5593–5600. doi: 10.1128/AEM.67.12.5593-5600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J., Blais B., Darveau A., Fliss I. Simultaneous detection and identification of hepatitis A virus and rotavirus by multiplex nucleic acid sequence-based amplification (NASBA) and microtiter plate hybridization system. J. Virol. Methods. 2002;105(1):123–132. doi: 10.1016/s0166-0934(02)00096-4. [DOI] [PubMed] [Google Scholar]
- Joffre J. Tee Dot-Lavoisier; Paris: 1991. Les bacteriophages dans les milieux hydriques. Schwartzbrod L.(Cd), Virologie des milieux hydriques; pp. 253–274. [Google Scholar]
- Jung W., Han J., Choi J.W., Ahn C.H. Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies. Microelectron. Eng. 2015;132:46–57. [Google Scholar]
- Kaarj K., Akarapipad P., Yoon J.Y. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M., Watanabe S., Furuse K., Ozawa A. Bacteroides bacteriophages isolated from human feces. Microbiol. Immunol. 1985;29(9):895–899. doi: 10.1111/j.1348-0421.1985.tb00891.x. [DOI] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama S., Miura T., Masago Y., Konta Y., Tohma K., Manaka T.…Oshitani H. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Applied and environmental microbiology. 2017;83(9) doi: 10.1128/AEM.03406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusgen M. Biosensors: new approaches in drug discovery. Naturwissenschaften. 2002;89(10):433–444. doi: 10.1007/s00114-002-0358-3. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A.…Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittigul L., Raengsakulrach B., Siritantikorn S., Kanyok R., Utrarachkij F., Diraphat P.…Chaichantanakit N. Detection of poliovirus, hepatitis A virus and rotavirus from sewage and water samples. Southeast Asian journal of tropical medicine and public health. 2000;31(1):41–46. [PubMed] [Google Scholar]
- Lee S.H., Lee C., Lee K.W., Cho H.B., Kim S.J. The simultaneous detection of both enteroviruses and adenoviruses in environmental water samples including tap water with an integrated cell culture–multiplex-nested PCR procedure. J. Appl. Microbiol. 2005;98(5):1020–1029. doi: 10.1111/j.1365-2672.2004.02496.x. [DOI] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S.…Enouf V. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. The Lancet Infectious Diseases. 2020:S1473–3099. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N.…Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D. F-specific bacteriophage as an indicator of human viruses in natural waters and sewage effluents in Northern New Zealand. Water Sci. Technol. 1995;31(5–6):231–234. [Google Scholar]
- Li X.F., Mitch W.A. Drinking water disinfection byproducts (DBPs) and human health effects: Multidisciplinary challenges and pportunities. Environ. Sci. Technol. 2018;52(4):1681–1689. doi: 10.1021/acs.est.7b05440. [DOI] [PubMed] [Google Scholar]
- Li J.W., Wang X.W., Yuan C.Q., Zheng J.L., Jin M., Song N.…Chao F.H. Detection of enteroviruses and hepatitis A virus in water by consensus primer multiplex RT-PCR. World journal of gastroenterology. 2002;8(4):699–702. doi: 10.3748/wjg.v8.i4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.T., Zhu L. Environmental microbiology-on-a-chip and its future impacts. Trends Biotechnol. 2005;23(4):174–179. doi: 10.1016/j.tibtech.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. The Lancet Gastroenterology & Hepatology. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer J., Roppert K., Ertl P. Microfluidic systems for pathogen sensing: a review. Sensors. 2009;9(6):4804–4823. doi: 10.3390/s90604804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Marie D., Brussaard C.P., Thyrhaug R., Bratbak G., Vaulot D. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 1999;65(1):45–52. doi: 10.1128/aem.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzouk Y., Goyal S.M., Gerba C.P. Prevalence of enteroviruses in ground water of Israel. Groundwater. 1979;17(5):487–491. doi: 10.1111/j.1745-6584.1979.tb03345.x. [DOI] [PubMed] [Google Scholar]
- Masson C.F. 3rd ed. Longman; 1996. Biology of Freshwater Pollution. (356 p) [Google Scholar]
- Mauk M., Song J., Bau H.H., Gross R., Bushman F.D., Collman R.G., Liu C. Miniaturized devices for point of care molecular detection of HIV. Lab Chip. 2017;17(3):382–394. doi: 10.1039/c6lc01239f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Metcalf T.G., Melnick J.L., Estes M.K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 1995;49(1):461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environmental Science: Water Research & Technology. 2020;6(5):1213–1216. [Google Scholar]
- Nasser A.M., Metcalf T.G. An A-ELISA to detect hepatitis A virus in estuarine samples. Appl. Environ. Microbiol. 1987;53(5):1192–1195. doi: 10.1128/aem.53.5.1192-1195.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettikadan S.R., Johnson J.C., Mosher C., Henderson E. Virus particle detection by solid phase immunocapture and atomic force microscopy. Biochem. Biophys. Res. Commun. 2003;311(2):540–545. doi: 10.1016/j.bbrc.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Nishida M.K., Ruiz V.L.A., Gregori F. Detection of rotavirus from pig livestock wastewater of São Paulo State, Brazil. Ars Veterinaria. 2009;25(3):136–141. [Google Scholar]
- Noble R.T., Fuhrman J.A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 1998;14(2):113–118. [Google Scholar]
- Oswald W.J., Gotaas H.B. Photosynthesis in sewage treatment. Trans. Am. Soc. Civ. Eng. 1957;122(1):73–105. [Google Scholar]
- Parashar U.D., Nelson E.A., Kang G. Diagnosis, management, and prevention of rotavirus gastroenteritis in children. BMJ (Clinical Research Ed.) 2013;347:f7204. doi: 10.1136/bmj.f7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Luo G., Wu Z., Wen W., Zhang X., Wang S. Fluorescent-magnetic-catalytic nanospheres for dual-modality detection of H9N2 avian influenza virus. ACS Appl. Mater. Interfaces. 2019;11(44):41148–41156. doi: 10.1021/acsami.9b16718. [DOI] [PubMed] [Google Scholar]
- Pina S., Puig M., Lucena F., Jofre J., Girones R. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 1998;64(9):3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S., Ambert-Balay K., Pothier P., Moulin L., Wurtzer S. Deciphering the diversities of astroviruses and noroviruses in wastewater treatment plant effluents by a high-throughput sequencing method. Appl. Environ. Microbiol. 2015;81(20):7215–7222. doi: 10.1128/AEM.02076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell S., Ebdon J., Buck A., Tupper M., Taylor H. Removal of phages and viral pathogens in a full-scale MBR: implications for wastewater reuse and potable water. Water Res. 2016;100:20–27. doi: 10.1016/j.watres.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Rawat I., Kumar R.R., Mutanda T., Bux F. Dual role of microalgae: phycoremediation of domestic wastewater and biomass production for sustainable biofuels production. Appl. Energy. 2011;88(10):3411–3424. [Google Scholar]
- Rengevych O.V., Shirshov Y.M., Ushenin Y.V., Beketov A.G. Separate determination of thickness and optical parameters by surface plasmon resonance: accuracy consideration. Semiconductor Physics Quantum Electronics & Optoelectronics. 1999;2:28–35. [Google Scholar]
- Rutjes S.A., van den Berg H.H., Lodder W.J., de Roda Husman A.M. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 2006;72(8):5349–5358. doi: 10.1128/AEM.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeżutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004;28(4):441–453. doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Schvoerer E., Bonnet F., Dubois V., Cazaux G., Serceau R., Fleury H.J., Lafon M.E. PCR detection of human enteric viruses in bathing areas, waste waters and human stools in Southwestern France. Res. Microbiol. 2000;151(8):693–701. doi: 10.1016/s0923-2508(00)90132-3. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B.…Kim S.J. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shibata A., Goto Y., Saito H., Kikuchi T., Toda T., Taguchi S. Comparison of SYBR Green I and SYBR Gold stains for enumerating bacteria and viruses by epifluorescence microscopy. Aquat. Microb. Ecol. 2006;43(3):223–231. [Google Scholar]
- Straub T.M., Pepper I.L., Gerba C.P. Comparison of PCR and cell culture for detection of enteroviruses in sludge-amended field soils and determination of their transport. Appl. Environ. Microbiol. 1995;61(5):2066–2068. doi: 10.1128/aem.61.5.2066-2068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenin T., Lobert P.E., Hober D. Inactivation of coxsackievirus B4, feline calicivirus and herpes simplex virus type 1: unexpected virucidal effect of a disinfectant on a non-enveloped virus applied onto a surface. Intervirology. 2013;56(4):224–230. doi: 10.1159/000350556. [DOI] [PubMed] [Google Scholar]
- Tierney J.T., Sullivan R., Larkin E.P. Persistence of poliovirus 1 in soil and on vegetables grown in soil previously flooded with inoculated sewage sludge or effluent. Appl. Environ. Microbiol. 1977;33(1):109–113. doi: 10.1128/aem.33.1.109-113.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey J., von Gunten U., Kohn T. Differences in viral disinfection mechanisms as revealed by quantitative transfection of Echovirus 11 genomes. Appl. Environ. Microbiol. 2019;85(14) doi: 10.1128/AEM.00961-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNCTAD News details. https://unctad.org/en/pages/newsdetails.aspx?OriginalVersionID=2333 Available online.
- Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N.…Lloyd-Smith J.O. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New England Journal of Medicine. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., SS S.R., Govindasamy V., Arunachalam M., Narayanasamy A.…Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Current Opinion in Environmental Science & Health. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N.…Si B.Y. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. Journal of virological methods. 2005;126(1‐2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Maw M.M., Yu X., Dai B., Wang G., Jiang Z. Applications and perspectives on microfluidic technologies in ships and marine engineering: a review. Microfluid. Nanofluid. 2017;21(3):39. [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. 2020. A Novel Coronavirus Outbreak of. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M.G., Suttle C.A. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat. Microb. Ecol. 1997;13(3):225–232. [Google Scholar]
- Wen K., Ortmann A.C., Suttle C.A. Accurate estimation of viral abundance by epifluorescence microscopy. Appl. Environ. Microbiol. 2004;70(7):3862–3867. doi: 10.1128/AEM.70.7.3862-3867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environmental Science: Water Research & Technology. 2015;1(6):735–746. [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A.…Hoelscher M. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wommack K.E., Hill R.T., Kessel M., Russek-Cohen E., Colwell R.R. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 1992;58(9):2965–2970. doi: 10.1128/aem.58.9.2965-2970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2018. Guidelines on Sanitation and Health. [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Buchanan N., Fallowfield H.J. Inactivation of indicator organisms in wastewater treated by a high rate algal pond system. J. Appl. Microbiol. 2016;121(2):577–586. doi: 10.1111/jam.13180. [DOI] [PubMed] [Google Scholar]
- Zhu H., Snyder M. Protein arrays and microarrays. Curr. Opin. Chem. Biol. 2001;5(1):40–45. doi: 10.1016/s1367-5931(00)00170-8. [DOI] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]