Highlights
-
•
Abrogating AGTR1 expression in cancer cells lead to reduced oncogenicity.
-
•
AGTR1 is post-transcriptionally regulated by miRNA-155.
-
•
MiR-155 mitigates AGTR1-mediated angiogenesis, EMT and stemness.
-
•
MiR-155 disrupts oncogenic AGTR1/NF-κB /CXCR4 axis.
-
•
IKK inhibitors or miR-155 therapies might prove beneficial for AGTR1-positive cancers.
Keywords: Glioblastoma, AGTR1, miR-155, CXCR4, NF-κB signaling
Abstract
Glioblastoma (GBM) represents the most aggressive malignancy of the central nervous system. Increased expression of Angiotensin II Receptor Type 1 (AGTR1) has been associated with proliferative and infiltrative properties of glioma cells. However, the underlying mechanism of AGTR1 upregulation in GBM is still unexplored. To understand the post-transcriptional regulation of AGTR1 in GBM, we screened 3′untranslated region (3′UTR) of AGTR1 for putative miRNA binding by using prediction algorithms. Interestingly, miR-155 showed conserved binding on the 3′UTR of AGTR1, subsequently confirmed by luciferase reporter assay. Furthermore, miR-155 overexpressing GBM cells show decrease in AGTR1 expression accompanied with reduced cell proliferation, invasion, foci formation and anchorage-independent growth. Strikingly, immunodeficient mice implanted with stable miR-155 overexpressing SNB19 cells show negligible tumor growth. Notably, miR-155 attenuates NF-κB signaling downstream of AGTR1 leading to reduced CXCR4 as well as AGTR1 levels. Mechanistically, miR-155 mitigates AGTR1-mediated angiogenesis, epithelial-to-mesenchymal transition, stemness, and MAPK signaling. Similar effects were observed by using pharmacological inhibitor of IκB Kinase (IKK) complex in multiple cell-based assays. Taken together, we established that miRNA-155 post-transcriptionally regulates AGTR1 expression, abrogates AGTR1/NF-κB/CXCR4 signaling axis and elicits pleiotropic anticancer effects in GBM. This study opens new avenues for using IKK inhibitors and miRNA-155 replacement therapies for the treatment of AGTR1-positive malignancies.
Introduction
Glioblastoma (GBM) is the leading cause of cancer-related deaths among all adult primary malignant brain tumors. Regardless of the recent developments in GBM treatment and therapeutics comprising maximum safe resection, followed by radio-chemotherapy, the average patient survival time is less than 15 months [1], [2]. Surgical treatments present many limitations as the tumor infiltrates and diffuses in the surrounding parenchyma leading to disease relapse [3]. GBM has been categorized into proneural, neural, classical and mesenchymal subtypes; the mesenchymal subgroup has been linked to the worst prognosis [4]. The intratumoral heterogeneity of GBM poses several challenges such as post-therapy resistance and compromised responses to radio-chemotherapy [5]. Therefore, it’s imperative to investigate the molecular mechanisms underlying poor prognosis of GBM, and develop novel therapeutic approaches based on targeting specific molecular subtypes.
Increased levels of Angiotensin II Receptor Type 1 (AGTR1), a G-protein coupled receptor has been linked to high grade astrocytomas and poor patient prognosis [6]. Moreover, GBM cell lines overexpressing AGTR1 and Angiotensin II Receptor Type 2 (AGTR2) exhibit a mitogenic response upon treatment with angiotensin peptides [7]. Furthermore, AGTR1 overexpression has also been linked to multiple human malignancies including ovarian, renal, pancreatic, breast, and brain [8], [9], [10], [11]. While in breast cancer (BCa), significant overexpression of AGTR1 represents a subpopulation of Human epidermal growth factor (HER2)-negative and estrogen receptor (ER) positive patients [12], in ovarian cancer its high expression has been linked to metastasis by promoting multicellular spheroid formation [13]. Mechanistically, overexpression of AGTR1 has been observed to inhibit apoptosis while promoting angiogenesis, cell proliferation and inflammation [14], [15]. Conversely, blockage of AGTR1 in C6 rat glioma cells results in attenuated angiogenesis, cell proliferation and tumor progression. [11]. Although, AGTR1 has been deemed as an oncogene in various cancers, its role in GBM is yet to be explored completely. Further, the underlying mechanism of its overexpression in GBM is not well understood and needs to be elucidated.
One of the mechanisms for AGTR1-mediated tumorigenesis has been attributed to activation of multiple signaling pathways downstream of AGTR1, for instance, nuclear factor-κB (NF-κB) signaling in BCa [16]. Moreover, angiotensin II (ATII), the ligand of AGTR1 is known to activate NF-κB signaling downstream of AGTR1 in endothelial and vascular smooth muscle cells [17]. The IκB Kinase (IKK) complex is important for the positive regulation of NF-κB, which by phosphorylation-induced, proteasome-mediated degradation of IκB results in nuclear accumulation of NF-κB, wherein it binds to its target genes involved in cell survival, growth, immune response and inflammation [18]. Activation of NF-κB signaling in GBM often triggers multiple downstream targets, such as activation of GPCR named C-X-C chemokine receptor type 4 (CXCR4) [19], which promotes cell migration and metastases [20], [21]. CXCR4 is a major chemokine receptor, and a known marker for GBM metastasis. It mediates the cell survival, invasion [22], [23] and proliferation of GBM progenitor cells [24], [25], deeming it as a promising therapeutic target in GBM.
In this study, we discovered a mechanism underlying AGTR1 overexpression in GBM and its downstream signaling pathways associated with AGTR1-mediated oncogenesis. Further, we established the role of miR-155 in the post-transcriptional regulation of AGTR1 in GBM. We also showed that activation of AGTR1 via ATII stimulation in GBM cells instigates NF-κB signaling leading to CXCR4 overexpression as well as AGTR1 upregulation, thereby forming a positive feedback regulatory loop. Moreover, ectopic expression of miR-155 in GBM cells attenuates AGTR1 downstream signaling thereby disrupting this regulatory loop. Alternatively, targeting NF-κB signaling by an IKK complex inhibitor, results in downregulation of AGTR1 and CXCR4 expression, leading to reduced AGTR1-mediated oncogenicity. Conclusively, this study reveals a novel regulatory mechanism involving miR-155, which targets AGTR1/NF-κB/CXCR4 axis and abrogates GBM progression.
Materials and methods
Xenograft model
NOD/SCID (NOD.CB17-Prkdcscid/J) mice of about five to six weeks old were randomly placed in two groups. The mice were subjected to anaesthesia with a cocktail of xylazine/ketamine (5 and 50 mg/kg, respectively) through the intraperitoneal route. Thereafter, SNB19-CTL and SNB19-miR-155 cells (5 × 106 cells for each condition), suspended in 100 µl saline and mixed with 20% Matrigel were injected into dorsal flank of mice on both the sides. Digital Vernier’s calipers were used to measure tumor growth, twice a week, in a blinded assessment, and the formula (π/6) (L × W2), (L = length; W = width) was used to calculate the tumor volume. All procedures involving animals were approved by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) and were in accordance with the guidelines of the Institutional Animal Ethics Committee at Indian Institute of Technology Kanpur.
Luciferase promoter reporter assay
Dual-Luciferase reporter vector pEZX-MT01 (Renilla/Firefly) from GeneCopoeia was used to clone a 525 bp region of AGTR1 3′UTR from human genomic DNA. Another similar region with mutated residues in the binding site of miR-155 was also cloned in the luciferase vector. SNB19 cells at a confluency of ∼30–40% were co-transfected with 25 ng pEZX-MT01 wild type and mutant constructs and 30 pmol of miR-155 mimics using Lipofectamine RNAiMax (Invitrogen) for two consecutive days. Thereafter, the luciferase assay was terminated using the Dual-Glo Luciferase assay kit (Promega) following the manufacturer’s instructions. Normalization of Firefly Luciferase activity to Renilla luciferase activity was carried out for every sample analyzed [26].
Gene expression array analysis
For gene expression profiling studies, RNA extracted from stable SNB19-CTL and SNB19-miR-155 cells was subjected to Whole Human Genome Oligo Microarray profiling (dual color) using Agilent Platform (8 × 60 k format) in accordance with the manufacturer’s protocol. Two separate microarray hybridizations were performed using SNB19-miR-155 cells against the SNB19-CTL cells. Locally weighted linear regression (Lowess normalization) was used to normalize the microarray data. To recognize significant gene expression patterns for differentially regulated genes, Pearson correlation coefficient-based hierarchical clustering algorithm was utilized. To identify differentially expressed genes, Benjamini and Hochberg procedure was used to calculate FDR- corrected P-values (FDR < 0.05). Genes with differential expression (log2 (fold change) > 0.6 or <−0.6, P < 0.05) were further analyzed using Database for Annotation, Visualization and Integrated Discovery (DAVID) for major deregulated pathways [27]. Gene set enrichment analysis (GSEA) was used for the identification of enriched molecular signatures in control with respect to miRNA overexpression [28]. Enrichment Map for critical biological pathways and processes was generated using Enrichment Map, a plug-in for Cytoscape network visualization software (http://baderlab.org/Software/EnrichmentMap/). Heatmap.2 function of ‘gplots’ in R was used to generate heatmaps.
Western blot analysis
Protein samples were analyzed by estimating the cell lysates using the BCA protein estimation kit (G-Biosciences), sample preparation in SDS loading dye and subjecting them to SDS-PAGE followed by transfer onto polyvinylidine difluoride (PVDF) membrane (GE Healthcare). Blocking was done in 2% bovine serum albumin (BSA) or 5% non-fat dry milk (NFDM) prepared in 1X tris-buffered saline with 0.1% Tween 20 (TBS-T) for an hour at room temperature (RT). The membrane was incubated overnight at 4 °C with the following primary antibodies: anti-AGTR1 (Abcam, ab124734, ab9391), p-Akt (CST, 2965), total Akt (CST, 9272), p-ERK (CST, 4377), ERK (CST, 4695), Bcl-xl (CST, 2764), Bcl2 (CST, 762870), E-cadherin (CST, 3195), N-cadherin (CST, 4061), Vimentin (CST, 3932), β-actin (Abcam, ab6276), phospho-NF-κB p65 (CST, 3033), NF-κB p65 (CST, 4764), CXCR4 (ab181020). Consequently, the blots were washed with 1X TBS-T followed by incubation with secondary anti-rabbit or anti-mouse antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch, USA) for each respective antibody in 2% NFDM or BSA at RT for 2 hours. Finally, the membrane was washed and subjected to Pierce™ ECL Western Blotting Substrate (ThermoFisher) to visualize signals as per the manufacturer’s protocol.
Chick chorioallantoic membrane (CAM) angiogenesis assay
Fertilized eggs were incubated at 37 °C with 70% humidity. A small hole was made on the blunt side of each egg and approximately 3 ml of albumin was aspirated out from each egg with a needle, followed by forming a small window on top of each egg which was later secured by scotch tape and returned to the incubator. On day six [29], SNB19-CTL (3 × 106 cells) were implanted onto the CAM of a batch of 8 eggs and SNB19-miR-155 (3 × 106 cells) on another batch of 8 eggs. The cells were suspended in saline and mixed with 20% Matrigel. Subsequently, the windows were sealed properly before returning the eggs to the incubator. Three days post implantation, the eggs were taken out and the seal was carefully removed. The cells were visualised in stereomicroscope under fluorescent light to detect their location as the cells have red fluorescence and then the image was taken in bright field via the microscope. Following this, the CAM was moistened with saline and fixed with 4% paraformaldehyde (PFA). The fixed CAM was then resected out of the egg and imaged under the stereomicroscope at 1.8X. The images obtained from both control (n = 8) and experimental (n = 8) set of eggs were quantified by the Angiogenesis analyzer for ImageJ [30].
Statistics
Two-tailed Student’s t-test was employed to assess the statistical significance of two distinct samples or indicated otherwise. The experimental groups were ascertained to be significantly different based on the P-value. Significance was determined as follows: *P ≤ 0.05, **P ≤ 0.001. Error bars depict the standard error of mean (SEM) acquired from three independent experiments.
Supplementary methods and tables
Further methodologic details can be found in Supplementary Methods. Primers can be found in Supplementary Table S1.
Data availability
The gene expression microarray data from this study has been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nih.gov/geo/) under the accession number GSE138737.
Results
AGTR1 is highly expressed in glioblastoma and silencing its expression leads to reduced oncogenicity
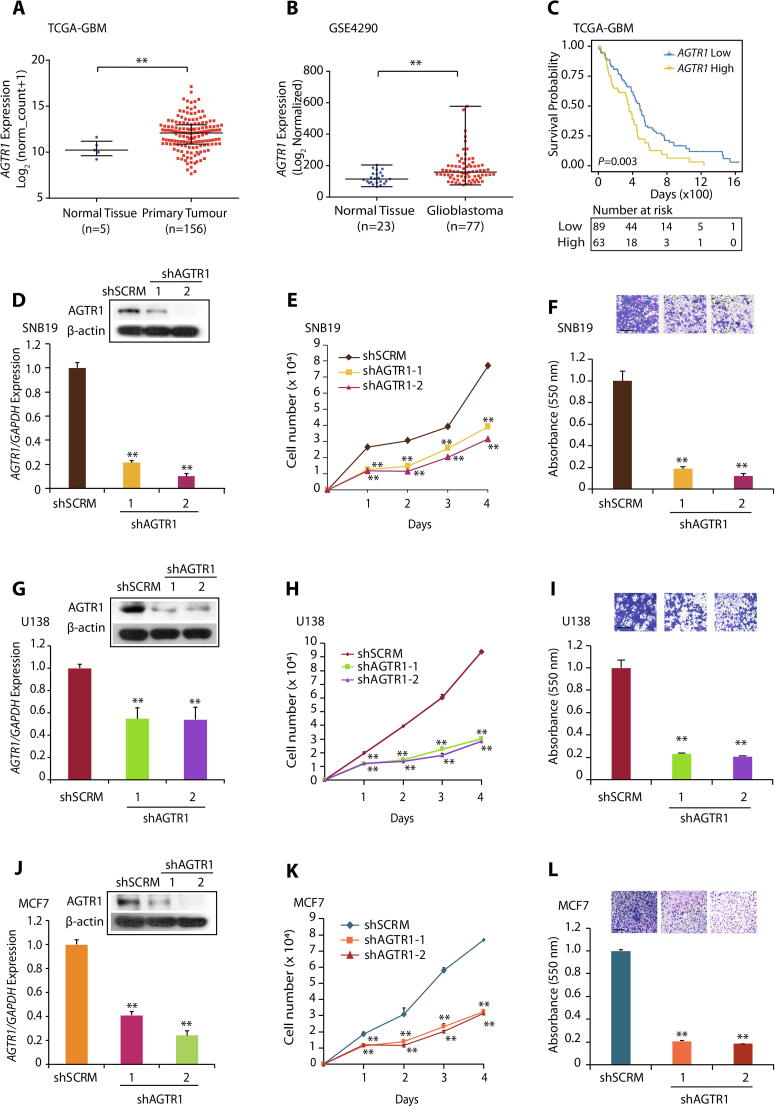
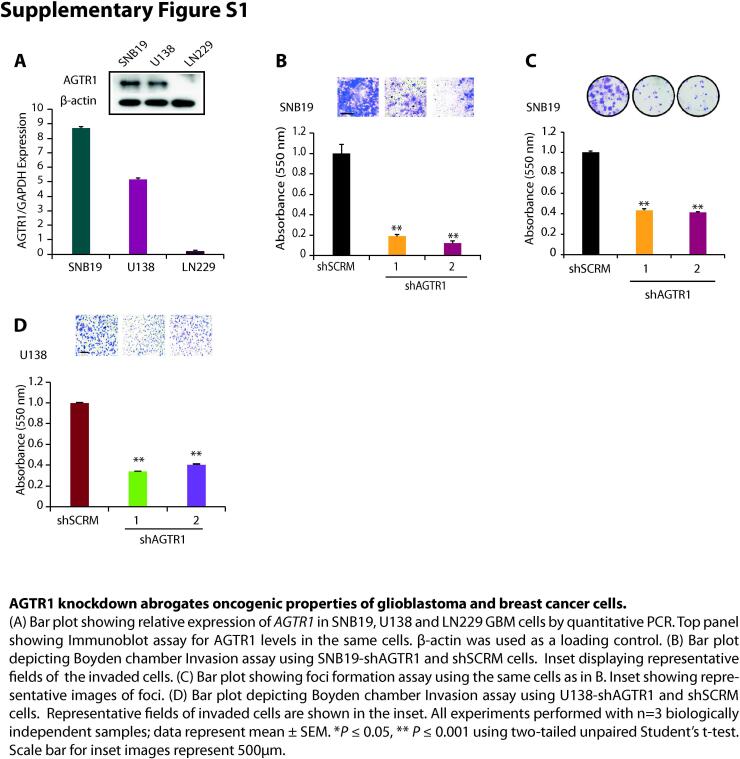
To understand the functional significance of AGTR1 we first examined its expression in TCGA-GBM (The Cancer Genome Atlas-Glioblastoma) and Sun Brain (GSE4290) datasets. In both cohorts, we observed increased expression of AGTR1 in GBM tumors with respect to normal tissue (Fig. 1A and B). We next evaluated the overall survival probability of GBM patients (TCGA-GBM) with high versus low AGTR1 expression. Interestingly, patients with high AGTR1 expression show overall low survival probability compared to the patients with low AGTR1 levels (Fig. 1C), indicating an association between elevated AGTR1 levels and poor survival of the clinically advanced GBM patients. Several independent studies implicated AGTR1 upregulation in cell proliferation, invasion and distant metastases in multiple malignancies [6], [12], [16]. Therefore, to ascertain the role of AGTR1 in GBM oncogenesis, we examined the expression of AGTR1 in GBM cell lines, namely SNB19, U138 and LN229, and found relatively higher expression of AGTR1 in SNB19 and U-138 cells (Supplementary Fig. S1A). We therefore performed stable shRNA-mediated knockdown of AGTR1 in SNB19 (Fig. 1D) and U-138 cells (Fig. 1G) followed by characterization of their oncogenic properties. Importantly, a significant decrease in proliferation of SNB19-shAGTR1 cells was observed with respect to control (Fig. 1E). Similarly, a marked decrease in the migratory as well as invasive potential was also observed in SNB19-shAGTR1 cells (∼80% and 85% respectively) as compared to control (Fig. 1F and Supplementary Fig. S1B). Moreover, a significant reduction (∼60%) in the foci forming ability of these cells was also noted (Supplementary Fig. S1C). On a similar note, a marked reduction in the cell proliferation of U138-shAGTR1 cells (Fig. 1H) along with decrease in the migration (∼80%) (Fig. 1I) and invasion (∼60%) (Supplementary Fig. S1D) was observed compared to control, signifying the oncogenic role of AGTR1 in GBM. Since, elevated level of AGTR1 is already known to exert oncogenic properties and lymph nodes metastases in breast cancer [12], [31], therefore we also silenced AGTR1 in MCF7 BCa cells (Fig. 1J). As anticipated a significant decrease in proliferation and invasion (Fig. 1K and L) in MCF7-shAGTR1 cells was recorded. Collectively, our findings confirm the oncogenic role of AGTR1 in GBM, and overall poor survival probability of GBM patients with elevated AGTR1 expression.
Fig. 1.
AGTR1 is highly expressed in glioblastoma and silencing its expression leads to reduced oncogenicity. (A) Scatter plot showing expression of AGTR1 in normal tissue (n = 5) and primary tumors (n = 156) in the TCGA-GBM cohort. (B) Scatter plot showing expression of AGTR1 in normal tissue (n = 23) and glioblastoma patients (n = 77) in the GSE4290 cohort. (C) Kaplan-Meier curve showing survival probability of GBM patients with high versus low AGTR1 expression in the TCGA-GBM cohort. (D) Bar plot showing expression of AGTR1 in SNB19-shAGTR1 and SNB19-shSCRM cells by quantitative PCR. Immunoblot assay for AGTR1 levels in same cells. β-actin was used as a loading control. (E) Line graph showing cell proliferation assay using same cells as in D. (F) Bar plot depicting Boyden chamber migration assay using same cells as in D. Inset displaying representative fields of migrated cells. (G) Bar plot showing expression of AGTR1 in U138-shAGTR1 and U138-shSCRM cells by quantitative PCR. Immunoblot assay for AGTR1 levels in same cells. β-actin was used as a loading control. (H) Line graph showing cell proliferation assay using same cells as in G. (I) Bar plot depicting Boyden chamber migration assay using same cells as in G. Inset displaying representative fields of migrated cells. (J) Bar plot showing expression of AGTR1 in MCF7-shAGTR1 and MCF7-shSCRM cells by quantitative PCR. Immunoblot assay for AGTR1 levels in same cells. β-actin was used as a loading control. (K) Line graph showing cell proliferation assay using same cells as in J. (L) Bar plot depicting Boyden chamber invasion assay using same cells as in J. Inset displaying representative fields of invaded cells. All experiments performed with n = 3 biologically independent samples; data represent mean ± SEM. *P ≤ 0.05, ** P ≤ 0.001 using two-tailed unpaired Student’s t-test. Scale bar for inset images represent 500 μm.
Ectopic expression of miR-155 in GBM cells attenuates AGTR1-mediated oncogenesis
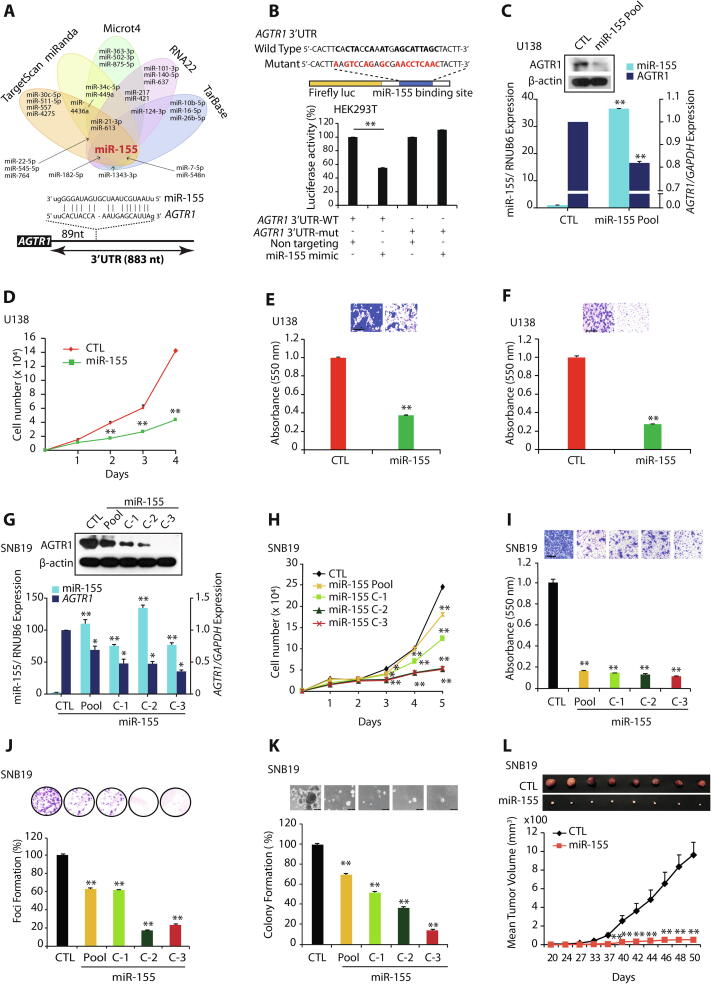
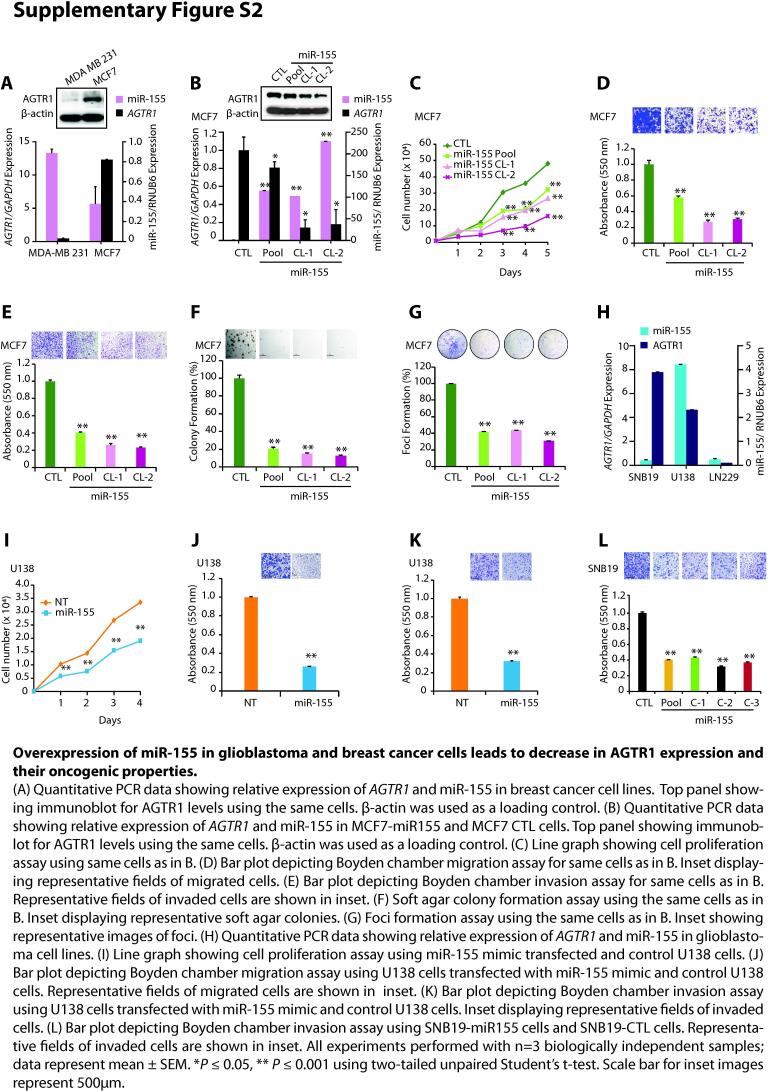
To examine the role of miRNAs in the post-transcriptional regulation of AGTR1, we screened putative miRNA binding sites on the 3′-UTR of AGTR1 transcript by employing miRNA prediction algorithms, namely TargetScan, miRanda (microRNA.org), MicroT4 (DIANA tools), RNA22 and TarBase. Interestingly, all the five algorithms independently predicted consensus binding site of miR-155 on the 3′UTR of AGTR1 (Fig. 2A). Next, to validate the miR-155 binding to the 3′-UTR of AGTR1, we generated Firefly/Renilla dual-luciferase reporter constructs by cloning wild type (3′-UTR-WT) and mutant (3′-UTR-mut) 3′-UTR of AGTR1, and co-transfected them along with synthetic miR-155 mimic in HEK293T cells. A significant decrease in the luciferase reporter activity was recorded in 3′-UTR-WT transfected HEK293T cells, while no change in luciferase activity was observed with 3′-UTR-mut construct (Fig. 2B). As proof-of-principal, we stably overexpressed miR-155 in AGTR1-positive MCF7 BCa cells, which show lower expression of miR-155 (Supplementary Fig. S2A). As anticipated, ectopically overexpressing miR-155 in MCF7 (MCF7-miR-155) cells show significant decrease in both transcript and protein levels of AGTR1 (Supplementary Fig. S2B). Next, we characterized these cells for any change in oncogenic properties, and as compared to control, MCF7-miR-155 cells exhibit a significant reduction in the cell proliferation, migration, invasion, as well as attenuated growth of soft agar colonies and foci formation (Supplementary Fig. S2C, D, E, F and G). Next to ascertain role of miR-155 in GBM, we examined the miR-155 expression in GBM cell lines, namely SNB19, U138 and LN229, and minimal expression of miR-155 accompanied with elevated AGTR1 levels was detected in SNB19 cells (Supplementary Fig. S2H). Therefore, to establish miR-155-mediated negative regulation of AGTR1, we stably overexpressed miR-155 in SNB19 and U138 cells, and examined the expression of AGTR1, a significant reduction in the AGTR1 levels was observed both at transcript and protein levels in miR-155 overexpressing GBM cells (Fig. 2C and G). Collectively, these findings confirm miR-155 mediated post-transcriptional repression of AGTR1 in GBM. We also examined any change in the oncogenic properties of stable miR-155 overexpressing U138 cells (U138-miR-155). Notably, a significant decrease in cell proliferation was observed in U138-miR-155 cells compared to control (Fig. 2D). Moreover, a significant reduction in cell migration (∼60%) and invasive potential (∼70%) of U138-miR-155 cells was observed (Fig. 2E and F). Similar results were obtained upon transfecting U138 cells with synthetic miR-155 mimic (Supplementary Fig. S2I, J and K). Likewise, stable overexpression of miR-155 in SNB19 cells also resulted in significant decrease in the cell proliferation (Fig. 2H), cell migration (∼80%) and invasion (∼60%) (Fig. 2I and Supplementary Fig. S2L). Notably, this change in migration and invasion of U138-miR-155 or SNB19-miR-155 cells was not due to a decrease in the proliferation rate of these cells, as invasion and migration assays were terminated at 24 and 16 hours, respectively. Furthermore, oncogenic transformation of SNB19-miR-155 cells was examined by foci formation assay, and a significant reduction in the number of foci was observed (Fig. 2J). Moreover, a striking decrease (∼50–80%) in the number and size of soft agar colonies was also recorded in SNB19-miR-155 overexpressing cells, indicating compromised anchorage-independent growth (Fig. 2K). To further examine the effect of miR-155 on tumorigenesis, we subcutaneously implanted SNB19-miR-155 and SNB19-CTL cells in the flank region of immunodeficient NOD/SCID mice and monitored tumor growth. Intriguingly, the mice implanted with SNB19-miR-155 cells exhibited negligible tumor growth compared to the mice implanted with SNB19 CTL cells (Fig. 2L).
Fig. 2.
MiR-155 post-transcriptionally regulates AGTR1 and its overexpression abrogates oncogenic properties of glioblastoma cells. (A) Venn diagram displaying computationally predicted miRNAs targeting AGTR1 by TargetScan, miRanda, Microt4, RNA22, and TarBase. Schema showing predicted binding site of miR-155 on the 3′UTR of AGTR1. (B) Schema showing the luciferase reporter assay constructs with wild-type or mutated (red residues with alteration) AGTR1 untranslated region (3′UTR) downstream of firefly luciferase reporter gene (top panel). Bar plot showing luciferase reporter activity in HEK293T cells co-transfected with miR-155 mimic and mutant or wild-type 3′UTR AGTR1 constructs. (C) Quantitative PCR data showing relative expression of AGTR1 and miR-155 in U138-miR155 and U138-CTL cells. Top panel showing immunoblot for AGTR1 levels using same cells. β-actin was used as a loading control. (D) Line graph showing cell proliferation assay using same cells as in C. (E) Bar plot depicting Boyden chamber migration assay using same cells as in C. Inset displaying representative fields of migrated cells. (F) Bar plot depicting Boyden chamber invasion assay using same cells as in C. Inset displaying representative fields of invaded cells. (G) Quantitative PCR data showing relative expression of AGTR1 and miR-155 in SNB19-miR155 and SNB19-CTL cells. Top panel showing immunoblot for AGTR1 levels using same cells. β-actin was used as a loading control. (H) Line graph showing cell proliferation assay using same cells as G. (I) Bar plot depicting Boyden chamber migration assay using same cells as in G. Inset displaying representative fields of migrated cells. (J) Foci formation assay using same cells as in I. Inset showing representative images of foci. (K) Soft agar colony formation assay using same cells as in I. Inset displaying representative soft agar colonies. (L) Line graph showing mean tumor growth in NOD/SCID mice (n = 8 per group) subcutaneously implanted with SNB19-CTL and SNB19-miR-155 cells. All experiments performed with n = 3 biologically independent samples; data represent mean ± SEM. *P ≤ 0.05, ** P ≤ 0.001 using two-tailed unpaired Student’s t-test. Scale bar for all inset images represent 500 μm.
Collectively, our findings implicate the tumor suppressive role of miR-155 via modulating AGTR1 expression in GBM and abrogating AGTR1-mediated tumorigenesis. Thus, our findings suggest the tumor suppressive role of miR-155, which could be achieved by miRNA restoration therapy in AGTR1-positive GBM cases.
Global gene expression profile of miR-155 overexpressing GBM cells reveal multiple deregulated oncogenic biological processes
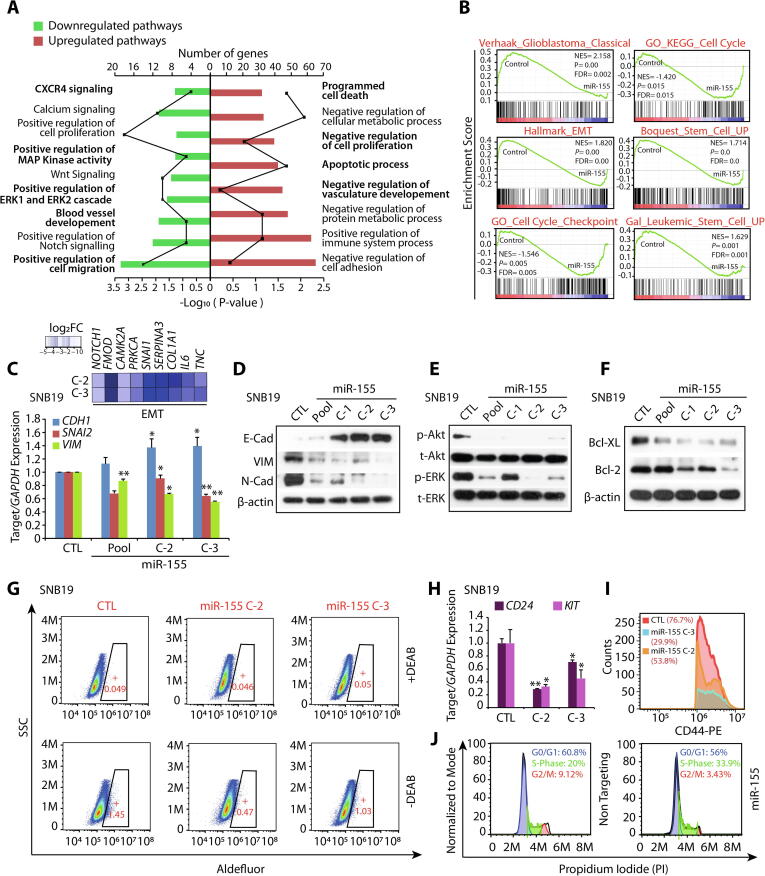
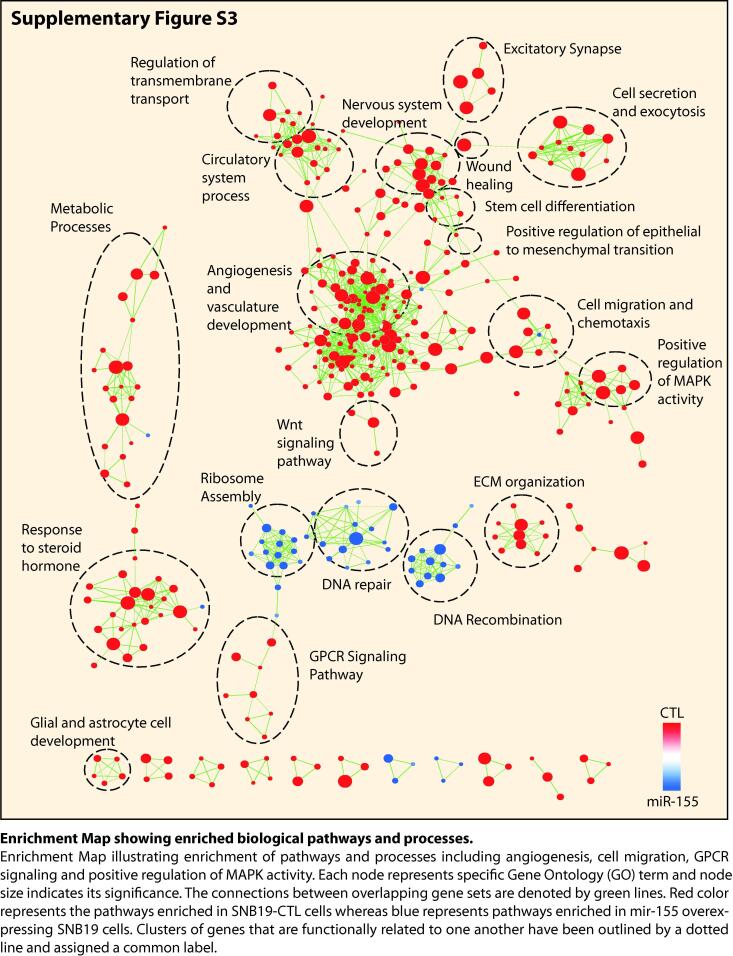
To investigate the biological pathways involved in miR-155 mediated reduction in oncogenic potential of GBM cells, we performed global gene expression profiling of SNB19-miR-155 and SNB19-CTL cells. Next, to examine the biological processes that were differentially regulated in GBM cells with miR-155 overexpression, we employed DAVID (Database for Annotation, Visualization and Integrated Discovery), GSEA (Gene Set Enrichment Analysis), and Enrichment Map analysis (Supplementary Fig. S3). Interestingly, majority of genes downregulated upon miR-155 overexpression were associated with CXCR4 signaling pathway, positive regulation of MAPK, ERK1/2 cascade, development of blood vessel and positive regulation of cell migration. While, genes associated with programmed cell death, apoptotic processes, negative regulation of cell proliferation and negative regulation of vasculature development were upregulated in miR-155 overexpressing cells (Fig. 3A). Additionally, GSEA of SNB-19-miR-155 cells showed a significant reduction in the enrichment of genes associated to the classical subtype of GBM, and features associated with cell-cycle checkpoint and stemness (Fig. 3B). Furthermore, our GSEA output shows a significant reduction in the expression of genes related to epithelial-to-mesenchymal transition (EMT) (Fig. 3B). Next, we validated this data, and a diminished expression of genes associated with EMT in GBM was observed (Fig. 3C). Notably, a significant increase in E-Cadherin (CDH1), an epithelial marker with a concomitant decrease in mesenchymal markers namely Vimentin (VIM) and SLUG (SNAI2) was observed (Fig. 3C). Similarly, a significant increase in the E-Cadherin and a decrease in Vimentin and N-cadherin at the protein level was also noted (Fig. 3D). Since, both MAPK and Akt signaling pathways play a key role in regulating cell proliferation and survival, we therefore examined the phosphorylation status of ERK (p-ERK) and Akt (p-Akt) as a readout of these pathways. As apparent in the DAVID analysis, a significant reduction of the p-ERK and p-Akt levels was detected in SNB19-miR-155 cells with respect to control (Fig. 3E). Furthermore, apoptosis was another key pathway delineated by our gene expression analysis, hence we explored the expression of anti-apoptotic Bcl-2 and Bcl-xL proteins, which are critical in inhibiting mitochondria-dependent extrinsic and intrinsic cell death pathways [32]. Intriguingly, we found a remarkable decrease in Bcl-2 and Bcl-XL in SNB19-miR-155 cells compared to control (Fig. 3F). Moreover, Aldehyde dehydrogenase (ALDH) is a known stem cell marker for GBM [33], therefore we performed ALDH assay, and a significant decrease (∼67%) in its activity in SNB19-miR-155 cells was recorded (Fig. 3G). Further, we analyzed the expression of CD117 (KIT), a type III receptor tyrosine kinase, known to play important role in GBM cell viability [34], as well as CD24 and CD44 expression, known to be present in treatment resistant tumor initiating and glioma stem cells [35]. Intriguingly, a significant decrease in the expression of CD24 (∼30–70%), KIT (∼45–30%) (Fig. 3H) and CD44 positive cells (∼60%) in miR-155-SNB19 cells was observed compared to control (Fig. 3I). As prominent in our GSEA analysis, we also carried out cell cycle analysis of SNB19 cells transiently transfected with miRNA mimics, and a marked increase in S phase cell population, indicative of S phase arrest was observed (Fig. 3J). Conclusively, our findings show that miR-155 downregulates the expression of genes associated with various oncogenic pathways namely, ERK/MAPK signaling, EMT, stemness, promotes apoptosis and induces cell cycle arrest, signifying that miR-155 replacement strategies may offer an alternative therapeutic intervention for the glioblastoma patients.
Fig. 3.
Overexpression of miR-155 in glioblastoma cells abrogates multiple oncogenic pathways. (A) Bar plot showing DAVID analysis of gene expression profile data of SNB19-miR-155 cells compared to SNB19-CTL cells, showing downregulated (green) and upregulated (red) pathways. Frequency polygon (line in black) represents the number of genes and the bars represent –Log10 (P value). (B) Gene Set Enrichment Analysis (GSEA) plots depicting deregulated processes using same gene expression profile data as in A. (C) Heatmap depicting the relative expression of genes related to EMT in same cells as A (top panel). Shades of blue represent log2 fold change in gene expression. Bar plot showing quantitative PCR data for the relative expression of EMT markers in same cells as A. (D) Immunoblots for E-Cadherin, Vimentin, and N-Cadherin using same cells as in A. β-actin was used as a loading control. (E) Immunoblots for phospho (p), total (t) Akt and ERK1/2 using same cells as in A. (F) Same as in E, except immunoblots for anti-apoptotic proteins Bcl-2 and Bcl-XL. (G) Aldeflour assay showing aldehyde dehydrogenases (ALDH) expression using SNB19-miR-155 (C-2 and C-3) and SNB19-CTL cells. The graphs depict the fluorescence intensity of the catalysed ALDH substrate in the presence and absence of DEAB (ALDH activity inhibitor). Windows marked in graphs represent the percentage of ALDH positive cell population. (H) Bar plot showing quantitative PCR data for the relative expression of CD24 and KIT in same cells as in G. (I) Flow cytometry analysis for CD44 expression using same cells as in G. (J) Flow cytometry analysis depicting S-phase cell cycle arrest of SNB19 cells transfected with miR-155 mimics stained with propidium iodide (PI). Data represent mean ± SEM. *P ≤ 0.05, ** P ≤ 0.001 using two-tailed unpaired Student’s t-test.
MiR-155 mediates its tumor-suppressive effects by downregulating angiogenesis
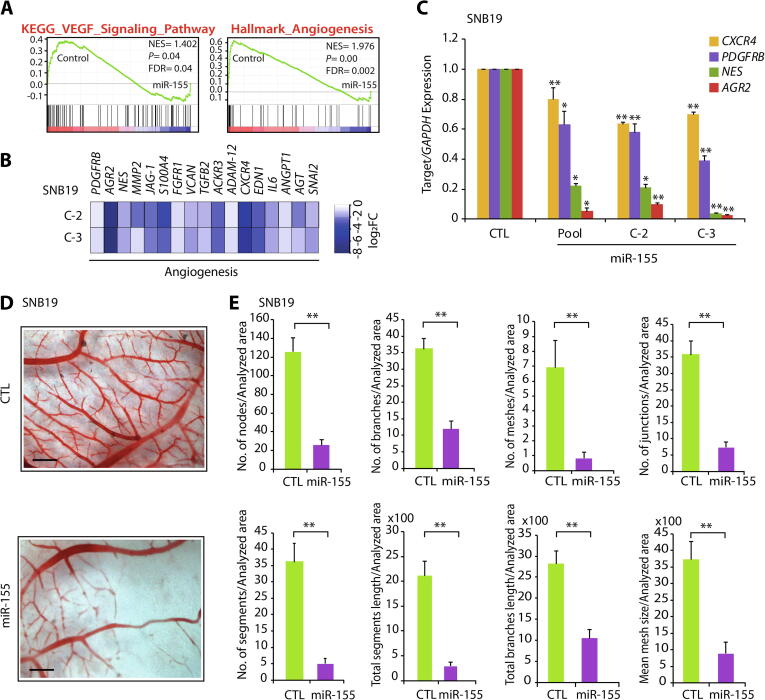
Angiogenesis or formation of new blood vessels from the pre-existing vasculature is crucial for the growth and survival of all cancers including GBM. It has been known that administering bevacizumab, an angiogenesis inhibitor, in GBM patients result in an increase in progression-free survival in phase III clinical trials [36], [37]. Notably, our enrichment map as well as pathway analysis shows a decrease in the expression of genes involved in angiogenesis and Vascular endothelial growth factor (VEGF) signaling in miR-155 overexpressing cells (Supplementary Fig. S3; Fig. 4A and B). Therefore, we sought to investigate the effect of miR-155 on angiogenesis and examined the expression of genes associated with angiogenesis in GBM. Interestingly, a significant reduction in the expression of established angiogenesis markers, namely, Nestin (NES) [38], Atypical Chemokine Receptor 3 (ACKR3) [39], Platelet Derived Growth Factor β (PDGFRB) [40], and Anterior Grade Protein 2 (AGR2) [41] was observed (Fig. 4C). To investigate the angiogenic potential of miR-155 in GBM, we implanted SNB19-miR-155 or SNB19-CTL cells onto chick chorioallantoic membrane (CAM) of the fertilized eggs (n = 8 in each group), and performed CAM assay, a popular in vivo model for studying angiogenesis. Intriguingly, a remarkable reduction in angiogenesis was observed in the CAMs implanted with SNB19-miR155 cells compared to SNB19-CTL cells (Fig. 4D). Measurement of the features of angiogenic vessels by using ImageJ angiogenesis analyzer [30] showed a significant reduction in the number of nodes, branches, meshes, junctions, segments along with mean mesh size, total segments’ length and total branches’ length in SNB19-miR-155 implanted CAM group compared to control (Fig. 4E). Taken together, our data establishes that high level of miR-155 attenuates angiogenesis in AGTR1-positive SNB19 GBM cells, thus ascertaining the importance of miR-155 as a potential therapeutic intervention against neovascularization and angiogenesis in glioblastoma.
Fig. 4.
MiR-155 attenuates neovascularization in chorioallantoic membrane (CAM) assay. (A) The Gene Set Enrichment Analysis (GSEA) plots depicting angiogenesis and VEGF signaling to be enriched and their corresponding statistical metrics in SNB19-miR155 and control cells. (B) Heatmap depicting the relative expression of genes related to angiogenesis using same cells as in A. Shades of blue represent log2 fold change in gene expression. (C) Bar plots showing quantitative PCR data for the relative expression of angiogenesis markers using same cells as in A. (D) Representative images of chick CAM assay using same cells as in A. Scale bar represents 1 cm. (E) Bar plots representing angiogenic properties of the CAM assay using microscopic images analyzed by Angiogenesis analyzer in Image J. Data represent mean ± SEM. *P ≤ 0.05, **P ≤ 0.001 using two-tailed unpaired Student’s t-test.
Disrupting the AGTR1/NF-κB/CXCR4 axis attenuates oncogenesis in glioblastoma
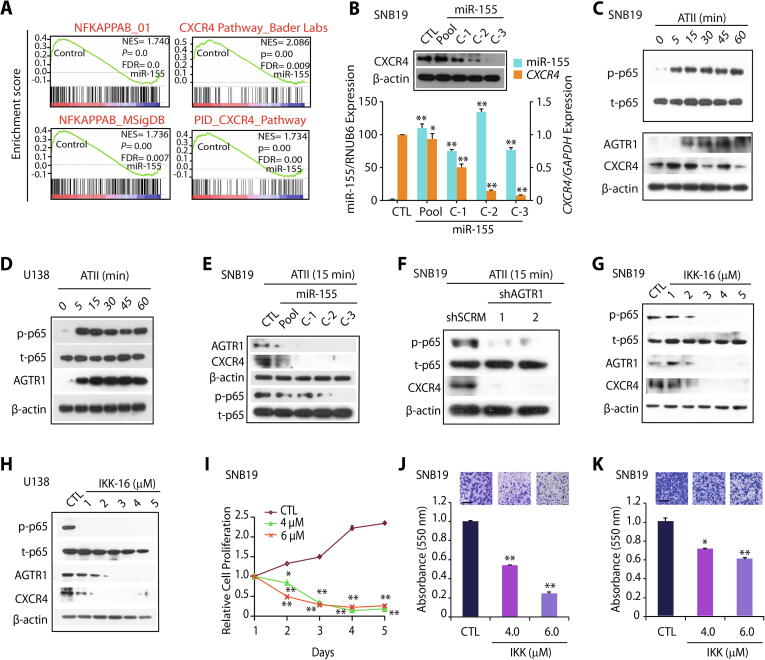
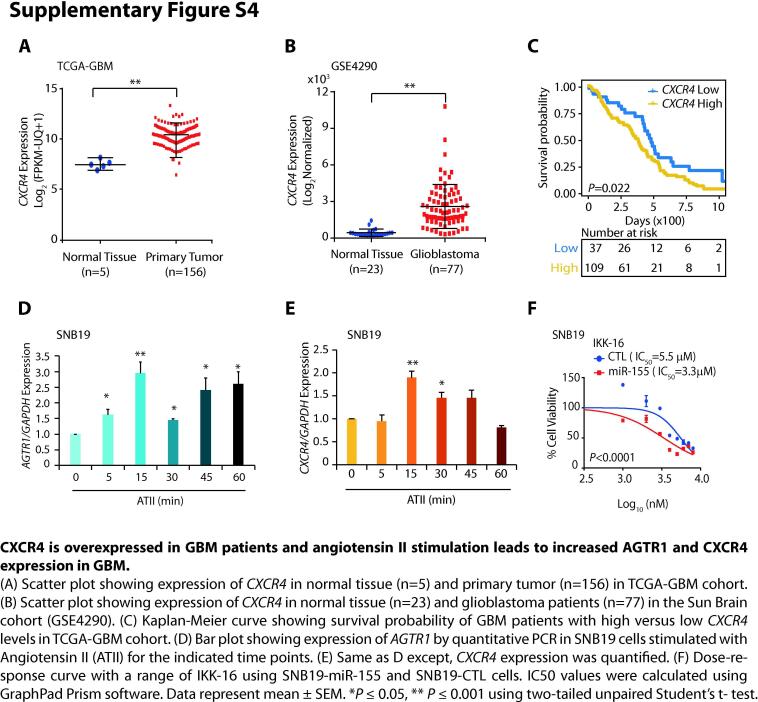
The expression of C-X-C chemokine receptor type 4 (CXCR4) is known to mediate tumor invasion, and targeting this receptor has been shown to reduce the invasive properties of GBM cells, sensitizing them to radiation-induced apoptosis [42]. Importantly, as evident by our DAVID and GSEA analyses, CXCR4 signaling pathway emerged as one of the most significant biological processes downregulated upon miR-155 overexpression in SNB19 cells (Figs. 3A and 5A). Hence, we next analyzed TCGA-GBM and Sun Brain (GSE4290) cohorts for the expression of CXCR4 and found it to be significantly overexpressed in both cohorts (Supplementary Fig. S4A and B). Moreover, TCGA-GBM patients with higher expression of CXCR4 show reduced overall survival probability compared to patients with low CXCR4 expression (Supplementary Fig. S4C), suggesting that its higher expression is associated with poor patient survival. Hence, we next examined the expression of CXCR4 in SNB19-miR-155 cells, and as expected, a remarkable decrease in CXCR4 expression was observed with respect to control (Fig. 5B). To pinpoint the exact mechanism involved in reduced CXCR4 expression, we examined whether miR-155 binds on the 3′UTR of the CXCR4 by using miRNA prediction tools; nonetheless absence of miR-155 binding on its 3′-UTR indicates that some alternate regulatory mechanism might be involved. Interestingly, our GSEA data revealed both CXCR4 and NF-κB signaling pathways to be significantly downregulated in SNB19-miR155 cells (Fig. 5A). NF-κB signaling being a master regulator of cell survival, immunity, inflammation [19], as well as considering its critical role in the maintenance of cancer stem-like cells and resistance to radiotherapy in GBM [19], we sought to investigate the possible link between miR-155 and NF-κB signaling. Previously, NF-κB signaling is known to induce CXCR4 expression in BCa cells [20] as well as through hepatocyte growth factor stimulation in glioma cells [43], hence we conjectured that the decrease in CXCR4 expression in miR-155 overexpressing cells might be influenced by NF-κB signaling. Furthermore, it has been shown that ATII activates NF-κB and CREB signaling via p38 MAPK leading to the upregulation of AGTR1 [44], hence we speculated that NF-κB signaling might be involved in upregulation of AGTR1 in GBM. To confirm this, we examined the phosphorylation level of p65 subunit (p-p65) upon stimulation with ATII which is a bonafide ligand for AGTR1. As expected, higher p-p65 level with a concomitant increase in AGTR1 and CXCR4 expression was observed in ATII stimulated SNB19 cells (Supplementary Fig. S4D and E; Fig. 5C). Likewise, ATII stimulation in U138 cells also led to an increase in p-p65 and AGTR1 levels (Fig. 5D). Conversely, stimulating SNB19-miR-155 cells with ATII resulted in a robust decrease in the p-p65 levels compared to SNB19-CTL cells (Fig. 5E). Similarly, a marked decrease in AGTR1 and CXCR4 levels was also noted in ATII stimulated SNB19-miR-155 cells (Fig. 5E). Taken together, our data suggest that ATII stimulation activates NF-κB signaling, which in turn results in increased expression of CXCR4 and AGTR1 in GBM cells, while the presence of miR-155 disrupts this feedback regulatory loop.
Fig. 5.
Targeting AGTR1 in glioblastoma results in reduced NF-κB signaling and CXCR4 expression. (A) Gene Set Enrichment Analysis (GSEA) for NF-κB and CXCR4 pathway and their associated statistical metrics in SNB19-miR-155 and SNB19-CTL cells. (B) Bar plot showing relative expression of CXCR4 and miR-155 by quantitative PCR using the same cells as in A. Immunoblot assay for CXCR4 levels using same cells. β-actin was used as a loading control. (C) Immunoblot showing phospho (p) and total (t) p65, AGTR1 and CXCR4 levels using ATII (1 mM) stimulated SNB19 cells at indicated time points. β-actin was used as a loading control. (D) Immunoblot for phospho (p), total (t) p65, AGTR1 levels in ATII (1 mM) stimulated U138 cells at indicated time points. β-actin was used as a loading control. (E) Immunoblot assays for AGTR1, CXCR4, phospho (p) and total (t) p65 levels using SNB19-miR-155 and SNB19-CTL cells stimulated with ATII (1 mM) for 15 minutes. β-actin was used as a loading control. (F) Immunoblots for phospho (p) and total (t) p65 and CXCR4 levels using SNB19-shAGTR1 and SNB19-shSCRM cells stimulated with ATII (1 mM) for 15 minutes. β-actin was used as a loading control. (G) Immunoblots for phospho (p) p65, total (t) p65, AGTR1 and CXCR4 levels using ATII (1 mM) stimulated and IKK-16 treated SNB19 cells. β-actin was used as a loading control. (H) Same as in G except U138 cells were used. β-actin was used as a loading control. (I) Line graph showing cell proliferation assay for IKK-16 treated SNB-19 cells at the indicated concentration. (J) Bar plot depicting Boyden chamber invasion assay using same cells as in I. Inset showing representative fields of invaded cells (n = 3 biologically independent samples; data represent mean ± SEM). (K) Bar plot depicting Boyden chamber migration assay using same cells as in I. Inset showing representative fields of migrated cells (n = 3 biologically independent samples; data represent mean ± SEM). Data represent mean ± SEM. *P ≤ 0.05, ** P ≤ 0.001 using two-tailed unpaired Student’s t- test.
To further corroborate that miR-155 disrupts AGTR1/NF-κB /CXCR4 axis in GBM, we stimulated SNB19-shAGTR1 cells with ATII and examined for NF-κB signaling. Interestingly, ATII stimulation in SNB19-shAGTR1 cells show decreased p-p65 levels accompanied with a notable reduction in CXCR4 expression compared to control SNB19-shSCRM cells (Fig. 5F). Collectively, silencing AGTR1 in GBM cells demonstrate similar outcome as achieved via miR-155 overexpression, thereby providing compelling evidence that the downregulation of AGTR1 results in reduced NF-κB signaling. Finally, our findings suggest that targeting AGTR1 either by miR-155 or RNA-interference could abrogate NF-κB downstream signaling in AGTR1-positive GBM subtype.
Next, we sought to examine whether abrogating NF-κB signaling by specific pharmacological inhibitor, could lead to reduction in the expression of AGTR1 and CXCR4, as well as associated oncogenic properties. To inhibit NF-κB signaling, we used IKK-16, a selective IκB kinase (IKK) inhibitor for IKK-1, IKK-2 and IKK complex [45]. Interestingly, a robust decrease in p-p65 levels was observed in ATII stimulated SNB19 cells upon treatment with IKK-16 (Fig. 5G). Moreover, a marked decrease in both AGTR1 and CXCR4 expression was also noted in these cells (Fig. 5G). Likewise, a remarkable decrease in p-p65 levels, AGTR1, and CXCR4 expression was observed in U138 GBM cells (Fig. 5H).
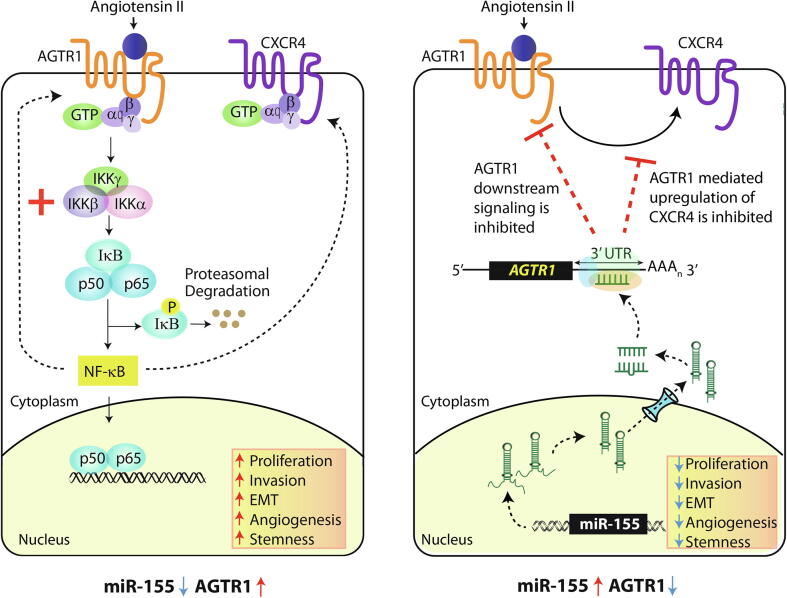
Next, we also investigated any alteration in oncogenic properties by abrogating the NF-κB signaling pathway via IKK-16, and a significant decrease in cell proliferation was observed in ATII stimulated and IKK-16 treated SNB19 cells compared to vehicle control (Fig. 5I). Additionally, a marked reduction in invasion and migration was recorded in IKK-16 treated SNB19 cells post ATII stimulation (Fig. 5J and K). Furthermore, we measured any change in drug-sensitivity in miR-155 overexpressing GBM cells, and found that SNB19-miR-155 cells show enhanced chemosensitivity towards IKK-16 compared to SNB19-CTL cells (Supplementary Fig. S4F), which suggests that replenishing miR-155 via miRNA replacement therapy, might be beneficial in overcoming drug resistance in GBM. Conclusively, our findings implicate the role of miR-155 in abrogating NF-κB signaling downstream of AGTR1, thereby leading to inhibition of CXCR4 and AGTR1 expression via feedback loop (Fig. 6). Collectively, our findings nominate miR-155 as a key player in modulating AGTR1 expression and suggest miR-155 replacement strategy for AGTR1-positive GBM cases. Moreover, disrupting AGTR1/NF-κB/CXCR4 axis by miR-155 or using pharmacological inhibitors against NF-κB signaling might be beneficial for treating AGTR1-positive subset of GBM patients.
Fig. 6.
MiR-155 targets AGTR1 resulting in reduced NF-κB signaling and CXCR4 levels thereby attenuating their associated oncogenic properties. Schematic representation of AGTR1 downstream signaling leading to activation of NF-κB pathway which in turn increases CXCR4 expression and mediates feedback upregulation of AGTR1 (left panel). Right panel depicts post-transcriptional regulation of AGTR1 via miR-155 which leads to reduced NF-κB signaling, decrease in CXCR4 expression and inhibition of feedback regulatory loop. Collectively, miR-155 disrupts the oncogenic NF-κB/CXCR4/AGTR1 signaling axis in GBM thereby decreasing AGTR1-mediated tumorigenesis.
Discussion
Elevated levels of AGTR1 have been associated with high grade cancer, angiogenesis, and poor survival in glioma [6], [11]. However, the underlying molecular mechanism involved in AGTR1 upregulation in GBM still needs to be understood. Here, we uncovered a regulatory mechanism involved in AGTR1 overexpression, wherein miR-155 post-transcriptionally regulates AGTR1 expression in GBM and elicits pleiotropic anticancer effects as evident by inhibition of angiogenesis in CAM assay and arrested tumor growth in mice xenograft model. Although, miR-155 has been implicated as a probable oncogenic miRNA in multiple cancers [46], [47], its tumor-suppressive role has also been reported, wherein it inhibits the invasive as well as metastatic potential in colorectal carcinoma [48]. In BCa, miR-155 directly attenuates the expression of TCF4 transcription factor, an important regulator of EMT [49]. Moreover, miR-155 is also known to inhibit HER2 expression by post-transcriptional regulation as well as by targeting Histone Deacetylase 2 (HDAC2), its transcriptional activator [50]. In triple negative breast cancer (TNBC), high miR-155 levels are known to be associated with low recombinase RAD51 levels and better patients’ survival [51]. Futhermore, miR-155 is also known to reduce the CD44+/CD117+ ovarian cancer-initiating cells which display stem cell-like properties by targeting CLDN1 [52] and inhibit cancer stem cell like properties in colorectal cancer by targeting TGFβ/SMAD signaling pathway [53]. In conjunction with these studies, we observed that miR-155 inhibits the oncogenic potential of GBM by inhibiting EMT, stemness and promoting apoptosis.
Interestingly, miR-155 is shown to target AGTR1 in human umbilical vein endothelial cells, leading to decreased ATII induced ERK1/2 phosphorylation [54]. In corroboration to this, we observed a decrease in ERK1/2, MAPK signaling, mesenchymal markers along with an increase in apoptosis in miR-155 overexpressing GBM cells. Earlier, it has been known that miR-155 exerts anti-angiogenic effects by suppression of AGTR1 and Suppressor Of Cytokine Signaling 1 (SOCS-1) [55], which supports our findings wherein miR-155 overexpressing GBM cells fail to induce angiogenesis in chick CAM angiogenesis assay. Of late, it has been reported that miR-155 mediated gene regulation is dependent on the cellular context [56], and the phenotype associated with miR-155 targeting the same transcript could vary depending on the cell type [57]. Taken together, our study established the tumor-suppressive role of miR-155 in AGTR1-positive subtype of GBM, wherein, its overexpression results in inhibition of multiple oncogenic properties and signaling pathways.
The NF-κB plays a major role in cancer development by dysregulation of IKK activity leading to cell proliferation, angiogenesis, migration, and metastasis in multiple cancers including GBM [58], [59], [60]. Moreover, AGTR1 is known to mediate its oncogenic effects in BCa through activation of NF-κB signaling [16]. It has also been established that NF-κB subunits p65 and p50 directly bind to the CXCR4 promoter, and activate its expression, leading to increased cell migration and metastasis [20]. Interestingly, ATII stimulated murine neuronal cells also exhibit a positive feedback mechanism of AGTR1 upregulation via NF-κB and CREB activation [44]. Furthermore, inhibiting NF-κB activity by using a NEMO (NF-κB essential modifier)–binding domain (NBD) peptide, depletion of IκB kinase 2 (IKK2), expression of a IκBαM super repressor, or by targeting inducible NF-κB genes are considered as attractive therapeutic strategies for treatment of GBM patients [61]. Similarly, it has been shown that overexpression of IκB, an inhibitor of κB in TNBC cells, led to reduced expression of CXCR4 [20], and mutant IκBα super-repressor mediated repression of NF-κB in prostate cancer results in the decrease of CXCR4 expression, resulting in reduced cell adhesion and transendothelial migration [62].
Recently, it has been shown that AGTR1 overexpression in BCa engages both ligand-independent and -dependent activation of NF-κB signaling through a triad of CARMA3, Bcl10, and MALT1, known as CBM signalosome [16]. Moreover, it has been demonstrated that CBM-dependent activation of NF-κB promotes tumor angiogenesis by inducing cancer cell-extrinsic effects. In the current study, we found that ATII stimulation results in the activation of NF-κB signaling pathway, subsequently resulting in increased CXCR4 and AGTR1 levels. Similarly, overexpression of miR-155 in GBM cells also results in the downregulation of AGTR1 via post-transcriptional regulation, which in turn attenuates the NF-κB signaling pathway thereby leading to reduced CXCR4 and AGTR1 levels. Alternatively, we also demonstrated that pharmacological inhibition of NF-κB signaling pathway by an IKK inhibitor attenuates the expression of CXCR4 and AGTR1 in GBM. Taken together, our study highlights that disrupting the AGTR1/NF-κB/CXCR4 axis via miR-155 and/or IKK inhibitor would be an effective therapeutic approach for the AGTR1-positive GBM patients.
In view of the pleiotropic anticancer effects displayed by miR-155 associated with attenuation of the oncogenic AGTR1/NF-κB/CXCR4 axis, we propose that miRNA-155 replacement therapy could be explored for AGTR1-positive subtype of GBM. Nevertheless, the stability and accurate in vivo delivery of miRNAs still present challenges to translate it to the clinics [63]. Although, magnetic resonance guided focused ultrasound has been used to deliver miRNAs across blood brain barrier as an effective treatment modality for GBM [64], hence well-planned clinical trials with miR-155 are highly warranted for GBM patient with higher AGTR1 levels. Besides, several strategies including small molecule inhibitors against IKK and its related kinases have been tested in preclinical studies [65], [66] as well as in clinical trials for other diseases (NCT00883584), which might be introduced for GBM after complete evaluation in clinical trials.
Financial support
This work is supported by the extra-mural research grant from the Department of Biotechnology, Ministry of Science and Technology, Government of India [BT/PR8675/GET/119/1/2015 to BA].
Conflict of interest
The authors declare no potential conflicts of interest.
CRediT authorship contribution statement
Anukriti Singh: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Nidhi Srivastava: Formal analysis, Validation. Anjali Yadav: Methodology, Software. Bushra Ateeq: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Visualization, Project administration, Resources, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was supported by a research funding from the Department of Biotechnology (BT/PR8675/GET/119/1/2015). Financial supports from the Wellcome Trust/ DBT India Alliance (IA/I(S)/12/2/500635) and Science and Engineering Research Board, Government of India, (EMR/2016/005273) are also acknowledged. AS thanks Indian Council of Medical Research, India for the Senior Research fellowship (ICMR number: 3/1/3/JRF-2011/HRD-88). We thank Prof. K. Somsundaram for GBM cell lines, Prof. S. Ganesh for extending the use of microscopy facility, Prof. Jonaki Sen for the fertilized eggs for CAM assay and imaging facility. We are also thankful to Ayush Praveen, Vipul Bhatia, Nishat Manzar and Ritika Tiwari for insightful discussion and critically reading the manuscript.
Authors’ contributions
AS and BA designed and directed the experimental studies. AS performed the experiments and interpreted the results. AS, AY and BA performed the mice xenograft studies. AS, NS, and BA interpreted and analyzed the data. AS and BA wrote the manuscript. Overall supervision of the study was provided by BA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.08.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
References
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro-oncology 2015;17(suppl_4):iv1-iv62. [DOI] [PMC free article] [PubMed]
- 2.Stupp R., Mason W.P., Van Den Bent M.J., Weller M., Fisher B., Taphoorn M.J. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Wick W, Osswald M, Wick A, Winkler F. Treatment of glioblastoma in adults. Therapeutic advances in neurological disorders 2018;11:1756286418790452. [DOI] [PMC free article] [PubMed]
- 4.Behnan J., Finocchiaro G., Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142(4):847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh D., Nandi S., Bhattacharjee S. Combination therapy to checkmate glioblastoma: clinical challenges and advances. Clin Transl Med. 2018;7(1):33. doi: 10.1186/s40169-018-0211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrieta O., Pineda-Olvera B., Guevara-Salazar P., Hernández-Pedro N., Morales-Espinosa D., Cerón-Lizarraga T. Expression of AT1 and AT2 angiotensin receptors in astrocytomas is associated with poor prognosis. Br J Cancer. 2008;99(1):160. doi: 10.1038/sj.bjc.6604431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogarty D.J., Sánchez-Gómez M.V., Matute C. Multiple angiotensin receptor subtypes in normal and tumor astrocytes in vitro. Glia. 2002;39(3):304–313. doi: 10.1002/glia.10117. [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto Y., Sasaki T., Tsuchida A., Chayama K. Angiotensin II type 1 receptor expression in human pancreatic cancer and growth inhibition by angiotensin II type 1 receptor antagonist. FEBS Lett. 2001;495(3):197–200. doi: 10.1016/s0014-5793(01)02377-8. [DOI] [PubMed] [Google Scholar]
- 9.Miyajima A., Kosaka T., Asano T., Asano T., Seta K., Kawai T. Angiotensin II type I antagonist prevents pulmonary metastasis of murine renal cancer by inhibiting tumor angiogenesis. Cancer Res. 2002;62(15):4176–4179. [PubMed] [Google Scholar]
- 10.Suganuma T., Ino K., Shibata K., Kajiyama H., Nagasaka T., Mizutani S. Functional expression of the angiotensin II type 1 receptor in human ovarian carcinoma cells and its blockade therapy resulting in suppression of tumor invasion, angiogenesis, and peritoneal dissemination. Clin Cancer Res. 2005;11(7):2686–2694. doi: 10.1158/1078-0432.CCR-04-1946. [DOI] [PubMed] [Google Scholar]
- 11.Arrieta O., Guevara P., Escobar E., Garcia-Navarrete R., Pineda B., Sotelo J. Blockage of angiotensin II type I receptor decreases the synthesis of growth factors and induces apoptosis in C6 cultured cells and C6 rat glioma. Br J Cancer. 2005;92(7):1247–1252. doi: 10.1038/sj.bjc.6602483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes D.R., Ateeq B., Cao Q., Tomlins S.A., Mehra R., Laxman B. AGTR1 overexpression defines a subset of breast cancer and confers sensitivity to losartan, an AGTR1 antagonist. Proc Natl Acad Sci. 2009;106(25):10284–10289. doi: 10.1073/pnas.0900351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q., Yu S., Lam M.M.T., Poon T.C.W., Sun L., Jiao Y. Angiotensin II promotes ovarian cancer spheroid formation and metastasis by upregulation of lipid desaturation and suppression of endoplasmic reticulum stress. J Exp Clin Cancer Res. 2019;38(1):116. doi: 10.1186/s13046-019-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egami K., Murohara T., Shimada T., Sasaki K.-i., Shintani S., Sugaya T. Role of host angiotensin II type 1 receptor in tumor angiogenesis and growth. J Clin Investig. 2003;112(1):67–75. doi: 10.1172/JCI16645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A., Srivastava N., Amit S., Prasad S., Misra M., Ateeq B. Association of AGTR1 (A1166C) and ACE (I/D) polymorphisms with breast cancer risk in North Indian population. Transl Oncol. 2018;11(2):233–242. doi: 10.1016/j.tranon.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekambaram P., Lee J.-Y.-L., Hubel N.E., Hu D., Yerneni S., Campbell P.G. The CARMA3–Bcl10–MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin ii receptor-positive breast cancer. Cancer Res. 2018;78(5):1225–1240. doi: 10.1158/0008-5472.CAN-17-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAllister-Lucas L.M., Jin X., Gu S., Siu K., McDonnell S., Ruland J. The CARMA3-Bcl10-MALT1 signalosome promotes angiotensin II-dependent vascular inflammation and atherogenesis. J Biol Chem. 2010;285(34):25880–25884. doi: 10.1074/jbc.C110.109421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.J., Hawke N., Baldwin A.S. NF-κB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006;13(5):738. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- 19.Soubannier V., Stifani S. NF-κB signalling in glioblastoma. Biomedicines. 2017;5(2):29. doi: 10.3390/biomedicines5020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helbig G., Christopherson K.W., Bhat-Nakshatri P., Kumar S., Kishimoto H., Miller K.D. NF-κ B promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278(24):21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 21.Schimanski C.C., Schwald S., Simiantonaki N., Jayasinghe C., Gönner U., Wilsberg V. Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res. 2005;11(5):1743–1750. doi: 10.1158/1078-0432.CCR-04-1195. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., Larsen P.H., Hao C., Yong V.W. CXCR4 is a major chemokine receptor on glioma cells and mediates their survival. J Biol Chem. 2002;277(51):49481–49487. doi: 10.1074/jbc.M206222200. [DOI] [PubMed] [Google Scholar]
- 23.Ehtesham M., Winston J., Kabos P., Thompson R. CXCR4 expression mediates glioma cell invasiveness. Oncogene. 2006;25(19):2801. doi: 10.1038/sj.onc.1209302. [DOI] [PubMed] [Google Scholar]
- 24.Ehtesham M., Mapara K.Y., Stevenson C.B., Thompson R.C. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274(2):305–312. doi: 10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson C.B., Ehtesham M., McMillan K.M., Valadez J.G., Edgeworth M.L., Price R.R. CXCR4 expression is elevated in glioblastoma multiforme and correlates with an increase in intensity and extent of peritumoral T2-weighted magnetic resonance imaging signal abnormalities. Neurosurgery. 2008;63(3):560–570. doi: 10.1227/01.NEU.0000324896.26088.EF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatia V., Yadav A., Tiwari R., Nigam S., Goel S., Carskadon S. Epigenetic silencing of miRNA-338-5p and miRNA-421 drives SPINK1-positive prostate cancer. Clin Cancer Res. 2019;25(9):2755–2768. doi: 10.1158/1078-0432.CCR-18-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deryugina E.I., Quigley J.P. Chick embryo chorioallantoic membrane models to quantify angiogenesis induced by inflammatory and tumor cells or purified effector molecules. Methods Enzymol. 2008;444:21–41. doi: 10.1016/S0076-6879(08)02802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpentier G, Martinelli M, Courty J, Cascone I. Angiogenesis analyzer for ImageJ. 2012. p 198-201.
- 31.Ma Y., Xia Z., Ye C., Lu C., Zhou S., Pan J. AGTR1 promotes lymph node metastasis in breast cancer by upregulating CXCR4/SDF-1α and inducing cell migration and invasion. Aging (Albany Ny) 2019;11(12):3969. doi: 10.18632/aging.102032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Z., Zheng X., Rich K.M. Down-regulation of Bcl-2 and Bcl-xL expression with bispecific antisense treatment in glioblastoma cell lines induce cell death. J Neurochem. 2003;84(2):273–281. doi: 10.1046/j.1471-4159.2003.01522.x. [DOI] [PubMed] [Google Scholar]
- 33.Rasper M., Schäfer A., Piontek G., Teufel J., Brockhoff G., Ringel F. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro-oncology. 2010;12(10):1024–1033. doi: 10.1093/neuonc/noq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson J.R., Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduction Targeted Ther. 2017;2:17040. doi: 10.1038/sigtrans.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palanichamy K., Jacob J.R., Litzenberg K.T., Ray-Chaudhury A., Chakravarti A. Cells isolated from residual intracranial tumors after treatment express iPSC genes and possess neural lineage differentiation plasticity. EBioMedicine. 2018;36:281–292. doi: 10.1016/j.ebiom.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick W., Brandes A.A., Gorlia T., Bendszus M., Sahm F., Taal W. American Society of Clinical Oncology; 2016. EORTC 26101 phase III trial exploring the combination of bevacizumab and lomustine in patients with first progression of a glioblastoma. [Google Scholar]
- 37.Wen P.Y., Macdonald D.R., Reardon D.A., Cloughesy T.F., Sorensen A.G., Galanis E. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda Y., Hagio M., Ishiwata T. Nestin: a novel angiogenesis marker and possible target for tumor angiogenesis. World J Gastroentero: WJG. 2013;19(1):42. doi: 10.3748/wjg.v19.i1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neves M., Fumagalli A., van den Bor J., Marin P., Smit M.J., Mayor F. The role of ACKR3 in breast, lung and brain cancer. Mol Pharmacol. 2019;118 doi: 10.1124/mol.118.115279. mol. 118.115279. [DOI] [PubMed] [Google Scholar]
- 40.Roberts W.G., Whalen P.M., Soderstrom E., Moraski G., Lyssikatos J.P., Wang H.-F. Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 2005;65(3):957–966. [PubMed] [Google Scholar]
- 41.Hong X.-Y., Wang J., Li Z. AGR2 expression is regulated by HIF-1 and contributes to growth and angiogenesis of glioblastoma. Cell Biochem Biophys. 2013;67(3):1487–1495. doi: 10.1007/s12013-013-9650-4. [DOI] [PubMed] [Google Scholar]
- 42.Yadav V.N., Zamler D., Baker G.J., Kadiyala P., Erdreich-Epstein A., DeCarvalho A.C. CXCR4 increases in-vivo glioma perivascular invasion, and reduces radiation induced apoptosis: a genetic knockdown study. Oncotarget. 2016;7(50):83701. doi: 10.18632/oncotarget.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esencay M., Newcomb E.W., Zagzag D. HGF upregulates CXCR4 expression in gliomas via NF-κB: implications for glioma cell migration. J Neurooncol. 2010;99(1):33–40. doi: 10.1007/s11060-010-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haack K.K., Mitra A.K., Zucker I.H. NF-κB and CREB are required for angiotensin II type 1 receptor upregulation in neurons. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0078695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waelchli R., Bollbuck B., Bruns C., Buhl T., Eder J., Feifel R. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg Med Chem Lett. 2006;16(1):108–112. doi: 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 46.Witten L.W., Cheng C.J., Slack F.J. miR-155 drives oncogenesis by promoting and cooperating with mutations in the c-Kit oncogene. Oncogene. 2019;38(12):2151. doi: 10.1038/s41388-018-0571-y. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J., Wang W., Gao Z., Peng X., Chen X., Chen W. MicroRNA-155 promotes glioma cell proliferation via the regulation of MXI1. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Chen Z., Xiang J., Gu X. MicroRNA-155 acts as a tumor suppressor in colorectal cancer by targeting CTHRC1 in vitro. Oncol Lett. 2018;15(4):5561–5568. doi: 10.3892/ol.2018.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang X., Zhuang X., Ju S., Zhang S., Jiang H., Mu J. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30(31):3440. doi: 10.1038/onc.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He X., Zhu W., Yuan P., Jiang S., Li D., Zhang H. miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene. 2016;35(46):6015. doi: 10.1038/onc.2016.132. [DOI] [PubMed] [Google Scholar]
- 51.Gasparini P., Lovat F., Fassan M., Casadei L., Cascione L., Jacob N.K. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci. 2014;111(12):4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin W., Ren Q., Liu T., Huang Y., Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587(9):1434–1439. doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 53.Miao L. Mo1432–inhibition of the cancer stem cells-like properties by Mir-155, involved in the targeting of transforming growth factor beta/Smad2 signal. Gastroenterology. 2019;156(6) S-1306. [Google Scholar]
- 54.Cheng W., Liu T., Jiang F., Liu C., Zhao X., Gao Y. microRNA-155 regulates angiotensin II type 1 receptor expression in umbilical vein endothelial cells from severely pre-eclamptic pregnant women. Int J Mol Med. 2011;27(3):393–399. doi: 10.3892/ijmm.2011.598. [DOI] [PubMed] [Google Scholar]
- 55.Pankratz F., Bemtgen X., Zeiser R., Leonhardt F., Kreuzaler S., Hilgendorf I. MicroRNA-155 exerts cell-specific antiangiogenic but proarteriogenic effects during adaptive neovascularization. Circulation. 2015;131(18):1575–1589. doi: 10.1161/CIRCULATIONAHA.114.014579. [DOI] [PubMed] [Google Scholar]
- 56.Hsin J.-P., Lu Y., Loeb G.B., Leslie C.S., Rudensky A.Y. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol. 2018;19(10):1137. doi: 10.1038/s41590-018-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michaille J.J., Awad H., Fortman E.C., Efanov A.A., Tili E. miR-155 expression in antitumor immunity: the higher the better? Genes Chromosom Cancer. 2019;58(4):208–218. doi: 10.1002/gcc.22698. [DOI] [PubMed] [Google Scholar]
- 58.Lee D.-F., Hung M.-C. Advances in targeting IKK and IKK-related kinases for cancer therapy. Clin Cancer Res. 2008;14(18):5656–5662. doi: 10.1158/1078-0432.CCR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia Y., Shen S., Verma I.M. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2(9):823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robe P.A., Bentires-Alj M., Bonif M., Rogister B., Deprez M., Haddada H. In vitro and in vivo activity of the nuclear factor-κB inhibitor sulfasalazine in human glioblastomas. Clin Cancer Res. 2004;10(16):5595–5603. doi: 10.1158/1078-0432.CCR-03-0392. [DOI] [PubMed] [Google Scholar]
- 61.Friedmann-Morvinski D., Narasimamurthy R., Xia Y., Myskiw C., Soda Y., Verma I.M. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci Adv. 2016;2(1) doi: 10.1126/sciadv.1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kukreja P., Abdel-Mageed A.B., Mondal D., Liu K., Agrawal K.C. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1α (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway–dependent NF-κB activation. Cancer Res. 2005;65(21):9891–9898. doi: 10.1158/0008-5472.CAN-05-1293. [DOI] [PubMed] [Google Scholar]
- 63.Adams B.D., Parsons C., Walker L., Zhang W.C., Slack F.J. Targeting noncoding RNAs in disease. J Clin Investig. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vega R.A., Zhang Y., Curley C., Price R.L., Abounader R. 370 magnetic resonance-guided focused ultrasound delivery of polymeric brain-penetrating nanoparticle microRNA conjugates in glioblastoma. Neurosurgery. 2016;63(CN_suppl_1):210. [Google Scholar]
- 65.Chang J., Liu F., Lee M., Wu B., Ting K., Zara J.N. NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proceedings of the National Academy of Sciences. 2013;110(23):9469–9474. doi: 10.1073/pnas.1300532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yemelyanov A., Gasparian A., Lindholm P., Dang L., Pierce J., Kisseljov F. Effects of IKK inhibitor PS1145 on NF-κB function, proliferation, apoptosis and invasion activity in prostate carcinoma cells. Oncogene. 2006;25(3):387. doi: 10.1038/sj.onc.1209066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression microarray data from this study has been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nih.gov/geo/) under the accession number GSE138737.