Abstract
Hypertension has been described as a condition of premature vascular aging, relative to actual chronological age. In fact, many factors that contribute to the deterioration of vascular function as we age are accelerated and exacerbated in hypertension. Nonetheless, the precise mechanisms that underlie the aged phenotype of arteries from hypertensive patients and animals remain elusive. Classically, the aged phenotype is the buildup of cellular debris and dysfunctional organelles. One means by which this can occur is insufficient degradation and cellular recycling. Mitophagy is the selective catabolism of damaged mitochondria. Mitochondria are organelles that contribute importantly to the determination of cellular age via their production of reactive oxygen species (ROS; Harman’s free radical theory of aging). Therefore, the accumulation of dysfunctional and ROS-producing mitochondria could contribute to the acceleration of vascular age in hypertension. This review will address and critically evaluate the current literature on mitophagy in vascular physiology and hypertension.
Keywords: blood pressure, hypertension, mitophagy, premature vascular aging
While the increased incidence of cardiovascular events as we age is generally attributable to the natural decline in organ function, in hypertension, dysfunction is premature in its onset and particularly pronounced. As a result, hypertension has been classified as a condition of premature aging.1 In particular, arteries from hypertensive patients and animals present a range of phenotypes including, hypercontractility, stiffening and remodeling, inflammation, oxidative stress, and cellular senescence which are all relatively early in their onset compared with age-matched normotensive controls.2 Clinically, vascular age determination, as opposed to chronological age per se, has now been introduced into guidelines for cardiovascular disease prevention.3 Nonetheless, a critical barrier to our progress in reducing the morbidity and mortality of hypertensive patients is our lack of understanding of the mechanisms that cause arteries to prematurely age.
The aged phenotype is classically viewed as the accumulation of cellular debris and dysfunctional organelles. Normally, endogenous cellular recycling mechanisms function to repair or clear damaged macromolecules. The two major pathways by which eukaryotic cells perform degradation and cellular recycling are autophagy and the ubiquitin–proteasome system.4 While both systems function to eliminate cellular debris and dysfunctional organelles, there are mechanistic differences between the two proteolytic pathways. For example, the proteasome system functions primarily to degrade single proteins, while autophagy specializes in the degradation of larger cellular materials indiscriminately (e.g., protein aggregates, organelles, and pathogens).4 However, if either one of these systems is dysregulated, cellular debris can accumulate and confer the classical “aged” phenotype. Given that several recent publications have revealed that autophagy is downregulated in the vasculature of old animals,5–8 new and exciting questions have emerged regarding the connection between impaired autophagic mechanisms and premature vascular aging observed in hypertension.9 Therefore, the broad focus of the following review is on the emerging role of autophagy in the etiology of premature vascular aging in hypertension, with a particular emphasis on the organelle-specific autophagic mechanism, mitophagy.
AUTOPHAGY AND VASCULAR AGING
Autophagy is the evolutionarily conserved catabolic process essential for maintaining homeostasis via the removal of cellular debris and dysfunction organelles, to provide micro- and macronutrients during times of stress (e.g., prolonged fasting or extreme exercise), and to initiate cell death pathways (e.g., apoptosis or necrosis). Broadly, autophagy can be classified by three unique subclasses: microautophagy, chaperone-mediated autophagy, and macroautophagy, and it can be further refined based on the constituents being degraded.10 These classes have been shown to include different initiation markers, materials being catabolized, and the cell type in which the degradation occurs.10 Microautophagy involves the lysosomal membrane folding inwards to directly internalize cytosolic constituents to be degraded.11 In contrast, the chaperone-mediated autophagy mechanism includes the chaperone protein heat shock cognate 71 kDa protein (Hsc70), a member of the heat shock protein 70 family. Hsc70, along with cochaperones, can recognize and unfurl substrate proteins containing the KFERQ amino acid sequence.12 After unfolding, substrates adhere to lysosome-associated membrane protein-2 isoform A (Lamp-2A) and are subsequently translocated across the lysosomal membrane for hydrolysis.12 Perhaps the most described type of autophagy is macroautophagy, which is evolutionarily conserved from single-cell organisms to whole animals. Macroautophagy is further divided into bulk autophagy or selective autophagy, each with different initiating conditions.13 For example, starvation of the cell and a lack of vital nutrients initiates bulk autophagy; whereas selective autophagy occurs to clear damaged, dysfunctional, or otherwise extraneous organelles, including mitochondria. Macroautophagy commences when an isolation membrane termed the phagophore engulfs a portion of the cytosol or an entire organelle. This subsequently forms a double membrane structure termed an autophagosome and the autophagosome then merges with the lysosome to form an autolysosome. The autolysosome is the structure within which the hydrolytic degradation of the contents of the autophagosome occurs, completing the macroautophagy mechanism.11 Uniquely, mitochondria are removed by a special form of macroautophagy called mitophagy, which will be discussed more below.
Autophagy has long been associated with longevity for multiple, compelling reasons,14 including the lengthening of lifespan,15 and it has been well established that induction of autophagy reduces the “aged” vascular phenotype.16,17 Currently, our understanding of how autophagy exerts a beneficial effect on the vasculature centered on the premise that reduced autophagy leads to the accumulation of damaged cellular debris and dysfunctional organelles. If undegraded, this buildup results in a proinflammatory/prooxidative milieu that promotes the generation of vasocontracting factors,9 quenching of nitric oxide bioavailability,6,7 and uncoupling of endothelial nitric oxide synthase.18 Therefore, upregulation or reconstitution of autophagy decreases these vascular dysfunctions.
MITOPHAGY IN PHYSIOLOGY
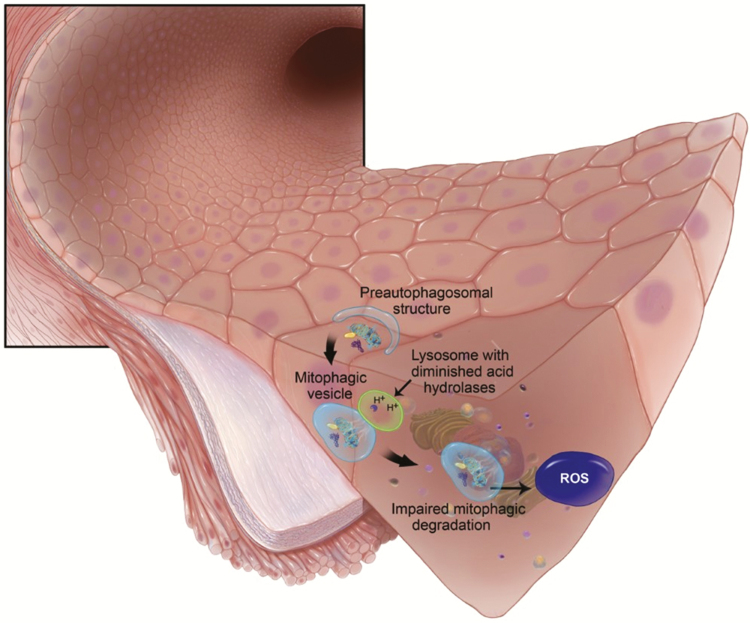
Mitophagy is the selective elimination of dysfunctional, damaged, or superfluous mitochondria, requiring the two major degradation systems: autophagy and the ubiquitin–proteasome system, working separately or in concert.19 It has been well established that disruptions to mitophagy contribute to disease, including age-associated pathologies.20 Therefore, it is tempting to rationalize that decreased mitophagy, resulting in oxidative stress and inflammation from residual and dysfunctional mitochondria, may be a novel mechanism of premature vascular aging in hypertension (Figure 1). We briefly introduce some of the normal molecular pathways of mitophagy to provide context for which dysfunctional mitophagy may promote pathophysiology.
Figure 1.
Impaired mitophagic degradation of damaged mitochondria leads to oxidative stress. In endothelial cells this prooxidative milieu promotes the release of contractile factors, quenches nitric oxide bioavailability, and uncouples endothelial nitric oxide synthase (eNOS). Abbreviation: ROS, reactive oxygen species.
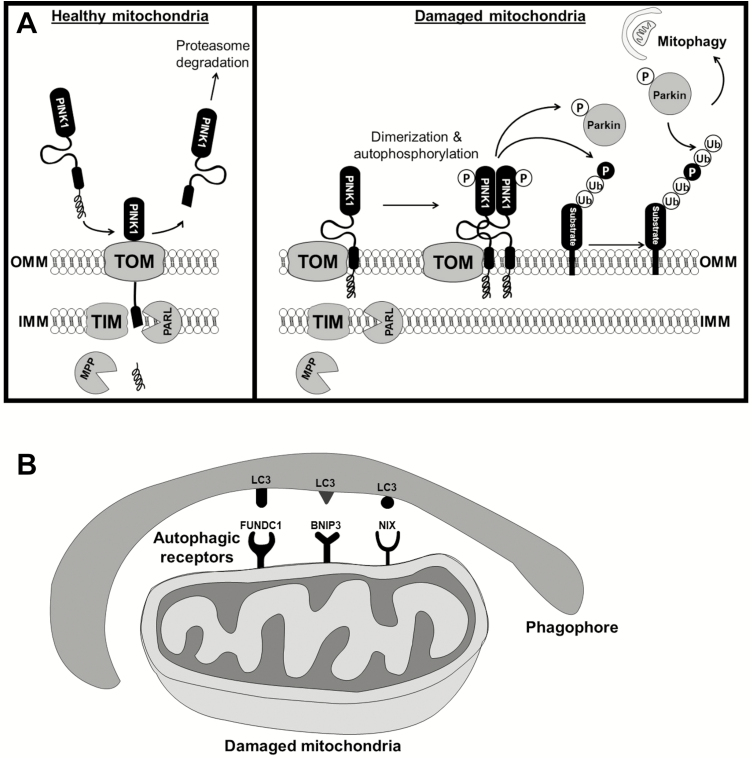
Phosphatase and tensin homolog-induced putative protein kinase 1 (PINK1) is a nuclear encoded, mitochondrial targeted serine/threonine-protein kinase. PINK1 is involved in mitochondrial quality control by identifying dysfunctional mitochondria and targeting these mitochondria for degradation. Healthy mitochondria maintain a membrane potential that can be used to import PINK1 via the translocase of the outer membrane and translocase of the inner membrane complexes at the outer and inner mitochondrial membrane, respectively. The mitochondrial targeting sequence is then cleaved off by the mitochondrial processing peptidase located in the matrix. Subsequently, the inner mitochondrial membrane protease presenilin-associated rhomboid-like protease (PARL) cleaves PINK1. The resulting peptide is then retrotranslocated to the cytosol, where it is subjected to constitutive degradation via the proteasome through the N-end rule pathway21 (Figure 2a).
Figure 2.
Mitophagy signaling pathways. (a) Parkin dependent mitophagy: healthy mitochondria import PINK1 via the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) complexes. The mitochondrial targeting sequence is then cleaved off by the mitochondrial processing peptidase (MPP) and the inner mitochondrial membrane protease presenilin-associated rhomboid-like protease (PARL) cleaves PINK1. The resulting peptide is then retrotranslocated to the cytosol, where it is subjected to degradation via the proteasome. However, when mitochondria are damaged, PINK1 accumulates at the outer mitochondrial membrane bound to the TOM complex. As a result, PINK1 dimerizes and is activated by autophosphorylation. PINK1 subsequently phosphorylates Parkin and ubiquitin chains, resulting in Parkin activation and relocation to the mitochondria where it further ubiquitinates mitochondrial substrates and signals the removal of the damaged organelle. (b) Parkin-independent mitophagy: microtubule-associated protein 1A/1B light chain 3 (LC3) proteins anchored in the membrane of the phagophore can bind to LC3-interacting region (LIR) containing autophagic receptors (e.g., FUNDC1, BNIP3, and NIX) that are constitutively expressed on the outer membrane of mitochondria. Subsequently, the autophagosome can engulf the damaged mitochondria for degradation.
On the other hand, in severely damaged mitochondria that lack sufficient membrane potential, PINK1 accumulates on the outer membrane. As a result, PINK1 interacts with the translocase of the outer membrane complex, dimerizes, and PINK1 kinase activity becomes activated through autophosphorylation.21 PINK1 phosphorylates ubiquitin, which triggers recruitment of Parkin to the outer mitochondrial membrane and activation of its E3 ligase activity. At the same time, phosphoubiquitin recruits autophagy receptors to initiate autophagosome formation. Parkin acts as an enhancer of this signaling through further ubiquitination of mitochondrial proteins22 (Figure 2a). This mitochondrial ubiquitination acts as the autophagic signal and adaptor proteins, such as adaptor p62/SQSTM1 via its ubiquitin-binding domain, recognizes and initiates autophagosome formation.23 If all components are functional, the PINK1-Parkin-mediated mitophagic pathway is the major mechanism by which damaged mitochondria are marked for degradation and cleared from cells before causing deleterious downstream effects.
Another major pathway by which mitochondria are cleared after being damaged is Parkin-independent mitophagy.24 In contrast to PINK1-Parkin-mediated mitophagy, which requires the translocation of Parkin to the damaged mitochondria followed by recruitment of autophagic receptors, there exist several light chain 3 (LC3)-interacting region containing autophagic receptors (e.g., FUN14 domain-containing protein 1 (FUNDC1), BCL2-interacting protein 3 (BNIP3), and NIP3-like protein X (NIX)) that are constitutively expressed on the outer membrane of mitochondria and can bind to microtubule-associated protein 1A/1B LC3 proteins anchored in the membrane of the phagophore. As a result, the autophagosome engulfs the mitochondria for degradation and recycling24 (Figure 2b). Interestingly, it has been proposed that basal mitophagy occurs in a Parkin-independent manner within tissues of high metabolic demand, including the vasculature.25
DYSFUNCTIONAL MITOPHAGY AND HYPERTENSION
It is clear that dysfunction in the mitophagy process can cause the accumulation of dysfunctional mitochondria that can contribute to a diverse range of pathologies. However, much of the breadth of pathophysiological research on mitophagy pertains to (age-related) neurodegenerative diseases.20,26 Conditions, such as Huntington’s disease, Parkinson’s disease, and Charcot–Marie–Tooth 2A, have been linked to altered, damaged, or absent mitophagy mechanisms, usually due to genetic mutations. For example, deleterious mutations in Parkin and PINK1 are linked with familial forms of Parkinson’s disease.27 Additional pathologies that are linked to dysfunctional mitophagy include cancers,28 lysosomal storage disorders (e.g., Pompe disease),29 Duchenne muscular dystrophy,30 and innate immune defense.31
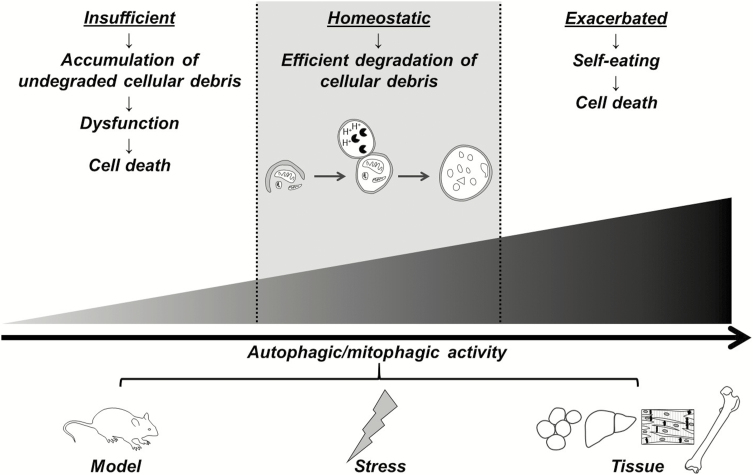
Although mitophagy has been measured in vivo in several different hypertensive models including, spontaneously hypertensive rats,32 Goldblatt two-kidney, one-clip (2K1C) hypertension,33 unilateral renal artery stenosis,34 and in vitro after cellular exposure to hypertensive stimuli (e.g., elevated angiotensin II,35–39 a high fat diet,40 reactive oxygen species (ROS),41 pressure overload,42,43 ischemia,44,45 oxidized low-density lipoprotein,46 and high glucose and free fatty acids47,48), our understanding of mitophagy in hypertension is still nascent. Indeed most of this literature has focused on cardiomyocytes,32,34,37–40,42–45 and only a few studies have investigated mitophagy in response to prohypertensive factors in arteries,49 vascular smooth muscle cells,35,46 or endothelial cells36,44,47,48 (Table 1). Moreover, the direction of change in mitophagy activity varies between these studies, and this may be model-, stressor-, or tissue-dependent (e.g., in the heart, inhibition of mitophagy ameliorates pressure overload induced heart failure43 and conversely, mitophagic activity is protective against angiotensin II-induced cardiac injury38) (Figure 3). Therefore, it is still premature to conclude whether mitophagy is in fact a cause or an effect of hypertension, as no one has directly manipulated mitophagy status in hypertensive animals in vivo.
Table 1.
Investigations that have reported changes in mitophagy in vascular cells, including the model (or stressor to induce mitophagy) and the direction of mitophagy change (increased or decreased)
| Model | Tissue/cell type | Mitophagy status | Reference |
|---|---|---|---|
| Angiotensin II | Endothelial cells | ↓ | 36 |
| Vascular smooth muscle cells | ↓ | 35 | |
| Glucose and palmitate | Endothelial cells | ↓ | 48 |
| Ischemia | Endothelial cells | ↓ | 44 |
| Oxidized low-density lipoprotein | Vascular smooth muscle cells | ↑ | 46 |
| Palmitic acid | Endothelial cells | ↑ | 47 |
Figure 3.
Autophagy and mitophagy activity spans a continuum, where too much or too little is detrimental to homeostasis and health. Differences between studies indicate that increases or decreases in activity are model-, stressor-, or tissue-dependent. We hypothesize that decreases in mitophagy confer an aged phenotype in the vasculature of hypertensive patients and animals.
It has also been well established that mitochondrial dynamics (fusion, fission, and biogenesis) regulates mitophagy status. It is widely considered that mitochondrial fission precedes and facilitates mitophagy by dividing elongated mitochondria into a manageable size for autophagosome encapsulation,50 while fusion protect elongated mitochondria from mitophagy.51 In relation to hypertension, it has been reported that prohypertensive stimuli such as elevated levels of aldosterone,52 ROS,53 angiotensin II,54 dietary salt,55 and calcium56 can mediate mitochondrial fission, and inhibition of fission is cardioprotective.57,58 Interestingly, acute inhibition of mitochondrial fission in vascular smooth muscle cells can antagonize contractile responses, and vasoconstriction stimuli induce mitochondrial fission.59 Moreover, hyperhomocysteinemia increased mitochondrial fission, causing endothelial cell damage, and collagen deposition in the mesenteric arteries.49 Nonetheless, the cause-and-effect relationship between mitochondrial dynamics and mitophagy is difficult to discern because fission is not only essential for the removal of damaged mitochondria, it can facilitate cell death during high levels of stress, and it can also stimulate creation of de novo mitochondria.60
Given the importance of mitophagy in maintaining cellular homeostasis and it participation in a number of pathophysiological conditions, there are numerous clinical and experimental drugs that have shown efficacy at activating and inhibiting mitophagy (Table 2). These drugs include classical autophagy activators such as mammalian target of rapamycin inhibitors (e.g., rapamycin), inositol monophosphatase inhibitors (e.g., carbamazepine), and epigenetic mediators (e.g., spermidine) and classical autophagy inhibitors such as lysosomal alkalizers (e.g., chloroquine) and phosphatidylinositol 3-kinase inhibitors (e.g., 3-methyladenine). Table 2 also contains a number of novel mitophagy drugs. These drugs offer the opportunity to therapeutically manipulate the mitophagy pathway a number of different ways and depending on the conditions. However, caution needs to be observed when proposing the use of these drugs as mediators and modulators of mitophagy (and autophagy), as these drugs have a number of known and unknown pleiotropic effects.
Table 2.
Clinical and experimental drugs that that shown efficacy at activating and inhibiting autophagy and mitophagy, including the putative mechanism of active, and mitophagy-specific reference
| Drug | Putative autophagic action | Reference |
|---|---|---|
| Autophagy/mitophagy activators | ||
| AICAR | AMPK-dependent inhibition of mTOR | 61 |
| Betulinic acid | AKT-mTOR inhibitor | 62 |
| BEZ235 | mTOR inhibitor | 63 |
| Carbamazepine | IMPase inhibitor and mTOR-independent activator | 64 |
| 3-Carboxyl proxyl nitroxide | AMPK-dependent inhibition of mTOR | 65 |
| CCI-779 | mTOR inhibitor | 66 |
| Ceramide | (i) AKT-mTOR inhibitor | 67 |
| (ii) Dissociation of the Beclin 1:Bcl-2 complex | ||
| Lithium chloride | IMPase inhibitor and mTOR-independent activator | 68 |
| Metformin | AMPK-dependent inhibition of mTOR | 69 |
| MDL-28170 | Inhibition of calpains I and II and subsequent cleavage of autophagic machinery | 70 |
| Mitochondrial toxins | ROS-induced translocation of Parkin to mitochondria | 71,72 |
| Mito-metformin | AMPK-dependent inhibition of mTOR | 65 |
| Nicotinamide derivatives | SIRT1 activators | 73 |
| Olanzapine | (i) ROS-induction of FoxO transcription factor | 74 |
| (ii) AMPK-dependent inhibition of mTOR | ||
| p62-mediated mitophagy inducer (PMI) | Parkin-dependent and -independent mitophagy | 75 |
| Phenanthroline | Mitochondrial fission | 76 |
| Rapamycin | mTOR inhibitor | 77 |
| RAD001 | mTOR inhibitor | 63 |
| Resveratrol | NAD+-dependent deacetylase and mTOR-independent activator | 78 |
| Rilmenidine | mTOR inhibitor | 79 |
| Selenite | Superoxide-induced mitochondrial damage | 80,81 |
| Spermidine | Acetyltransferase inhibitor and mTOR-independent activator | 82 |
| Trehalose | mTOR-independent activator | 83 |
| Urolithin A | (i) Mitochondrial fission | 84 |
| (ii) AMPK-dependent inhibition of mTOR | ||
| Autophagy/mitophagy inhibitors | ||
| Acid protease inhibitors | Lysosomal alkalizers | 85 |
| Ammonium chloride | Lysosomal alkalizer | 86 |
| Antioxidants (butylated hydroxyanisole, N-acetylcysteine) | (i) Protection of mitochondrial from ROS-mediated damage | 87 |
| (ii) Inhibition of the MPTP | ||
| Bafilomycin A1 | Vacuolar-type H+-ATPase inhibitor | 88 |
| Brefeldin A | Inhibitor of intracellular protein transport and alternative (Atg5/Atg7-independent) autophagy | 89 |
| Chloroquine | Lysosomal alkalizer | 88 |
| Cyclosporine A | Inhibition of the MPTP | 85,87 |
| Idebenone | (i) Protection of mitochondrial from ROS-mediated damage | 90 |
| (ii) Inhibition of the MPTP | ||
| Liensinine diperchlorate | Inhibition of autophagosome–lysosome fusion | 91 |
| LY294002 | Class III PI3K inhibitor | 85 |
| 3-Methyladenine | Class III PI3K inhibitor | 77,85 |
| Mitochondrial division inhibitor 1 (Mdivi-1) | Inhibition of mitochondrial fission | 43 |
| Wortmannin | Nonspecific PI3K inhibitor | 85 |
Abbreviations: AKT, protein kinase B; AMPK, 5′AMP-activated protein kinase; Atg, autophagy-related gene; Bcl-2, B-cell lymphoma 2; IMPase, inositol monophosphatase; MPTP, mitochondrial permeability transition pore; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; ROS, reactive oxygen species; SIRT, sirtuin.
HARMAN’S FREE RADICAL THEORY OF AGING: A POTENTIAL LINK BETWEEN MITOPHAGY AND VASCULAR AGE
First described over 60 years ago, one of the most prominent theories on aging is Harman’s free radical theory of aging. This theory postulated that decreased cellular longevity is caused by increased ROS.92,93 ROS are highly reactive and short lived due to their unpaired valence shell electron. While ROS occur as a consequence of normal cellular metabolism, when ROS formation overwhelms antioxidant defenses, it is defined as oxidative stress. Oxidative stress can cause damage to cellular constituents (e.g., lipids, proteins, and nucleic acids), and has been suggested to lead to a variety of pathophysiological conditions. Hence, an ideal balance between ROS production and antioxidant defense is paramount for maintenance of physiological homeostasis. Harman’s theory held that these nonspecific, essentially irreversible oxidative reactions with cellular macromolecules over time are likely involved in the chronological aging process, and conversely, the longevity of an organism can be increased by slowing the incidence of ROS.92,93 Progressively, the understanding of ROS reactions has evolved and extended Harman’s initial theory94,95; however, the basis of the senescence-promoting nature of ROS, remains consistent.
More recent work has implicated the role of mitochondria as a primary target of ROS-mediated damage.96 It is well known that mitochondria are a major source of intracellular ROS as a natural byproduct of mitochondrial respiration and energy production. Exacerbated ROS production causes damage to certain macromolecules, most notably mitochondrial DNA, which can then induce an amplification and further buildup of ROS within cells.96 Recently, it was confirmed that vascular mitochondrial respiratory capacity significantly deteriorates with advancing age as a result of declining mitochondrial content.97 Despite this decline, aging also resulted in greater mitochondrial-derived ROS.97 These findings are a direct application of Harman’s theory to the aging vasculature and support the idea that increasing the degradation of damaged mitochondria can prevent age-related vascular dysfunction.8 Nonetheless, if decreased mitophagy is also involved in exacerbated mitochondrial ROS production in the vasculature of hypertensive patients or animals is yet to be determined.
Mitophagy is the selective catabolic process for removing damaged mitochondrial. Insufficient mitophagy would lead to the accumulation of dysfunctional mitochondria. Given that mitochondria are a prominent source of intracellular ROS in all cell types, including cells of the vasculature, and our knowledge of Harman’s free radical theory of aging,92,93 we hypothesize that diminished mitophagy in the vasculature contributes to oxidative stress and the vascular aging phenotype associated with hypertension. This hypothesis is supported by studies which showed an upregulation of autophagy and mitophagy reversed several phenotypes of vascular aging in old mice6–8 and premature vascular aging in spontaneously hypertensive rats.9
ACKNOWLEDGMENTS
We would like to thank Roy E. Schneider (University of Toledo Medical Illustration) for his artwork in Figure 1.
FUNDING
This work was supported by the American Heart Association (18POST34060003) and National Institutes of Health (R00GM118885 and R01HL143082).
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1. Guzik TJ, Touyz RM. Oxidative stress, inflammation, and vascular aging in hypertension. Hypertension 2017; 70:660–667. [DOI] [PubMed] [Google Scholar]
- 2. McCarthy CG, Wenceslau CF, Webb RC, Joe B. Novel contributors and mechanisms of cellular senescence in hypertension-associated premature vascular aging. Am J Hypertens 2019; 32:709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perk J, De Backer G, Gohlke H, Graham I, Reiner Z, Verschuren WM, Albus C, Benlian P, Boysen G, Cifkova R, Deaton C, Ebrahim S, Fisher M, Germano G, Hobbs R, Hoes A, Karadeniz S, Mezzani A, Prescott E, Ryden L, Scherer M, Syvänne M, Scholte Op Reimer WJ, Vrints C, Wood D, Zamorano JL, Zannad F; Fifth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice ; European Association for Cardiovascular Prevention and Rehabilitation. European Guidelines on cardiovascular disease prevention in clinical practice (version 2012): the fifth joint task force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of nine societies and by invited experts). Atherosclerosis 2012; 223:1–68. [DOI] [PubMed] [Google Scholar]
- 4. Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol 2013; 4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 5. Kaplon RE, Hill SD, Bispham NZ, Santos-Parker JR, Nowlan MJ, Snyder LL, Chonchol M, LaRocca TJ, McQueen MB, Seals DR. Oral trehalose supplementation improves resistance artery endothelial function in healthy middle-aged and older adults. Aging (Albany NY) 2016; 8:1167–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. LaRocca TJ, Gioscia-Ryan RA, Hearon CM Jr, Seals DR. The autophagy enhancer spermidine reverses arterial aging. Mech Ageing Dev 2013; 134:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol 2012; 590:3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LaRocca TJ, Hearon CM Jr, Henson GD, Seals DR. Mitochondrial quality control and age-associated arterial stiffening. Exp Gerontol 2014; 58:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCarthy CG, Wenceslau CF, Calmasini FB, Klee NS, Brands MW, Joe B, Webb RC. Reconstitution of autophagy ameliorates vascular function and arterial stiffening in spontaneously hypertensive rats. Am J Physiol Heart Circ Physiol 2019; 317:H1013–H1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J 2017; 36:1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol 2010; 21:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol 2016; 428:1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yen WL, Klionsky DJ. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda) 2008; 23:248–262. [DOI] [PubMed] [Google Scholar]
- 15. Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 2013; 4:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abdellatif M, Sedej S, Carmona-Gutierrez D, Madeo F, Kroemer G. Autophagy in cardiovascular aging. Circ Res 2018; 123:803–824. [DOI] [PubMed] [Google Scholar]
- 17. Nussenzweig SC, Verma S, Finkel T. The role of autophagy in vascular biology. Circ Res 2015; 116:480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang JX, Qu XL, Chu P, Xie DJ, Zhu LL, Chao YL, Li L, Zhang JJ, Chen SL. Low shear stress induces vascular eNOS uncoupling via autophagy-mediated eNOS phosphorylation. Biochim Biophys Acta Mol Cell Res 2018; 1865:709–720. [DOI] [PubMed] [Google Scholar]
- 19. Song WH, Yi YJ, Sutovsky M, Meyers S, Sutovsky P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc Natl Acad Sci USA 2016; 113:E5261–E5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palikaras K, Daskalaki I, Markaki M, Tavernarakis N. Mitophagy and age-related pathologies: development of new therapeutics by targeting mitochondrial turnover. Pharmacol Ther 2017; 178:157–174. [DOI] [PubMed] [Google Scholar]
- 21. Voigt A, Berlemann LA, Winklhofer KF. The mitochondrial kinase PINK1: functions beyond mitophagy. J Neurochem 2016; 139(Suppl 1):232–239. [DOI] [PubMed] [Google Scholar]
- 22. Sekine S, Youle RJ. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 2018; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippai M, Lőw P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int 2014; 2014:832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Villa E, Marchetti S, Ricci JE. No parkin zone: mitophagy without parkin. Trends Cell Biol 2018; 28:882–895. [DOI] [PubMed] [Google Scholar]
- 25. McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab 2018; 27:439–449.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodolfo C, Campello S, Cecconi F. Mitophagy in neurodegenerative diseases. Neurochem Int 2018; 117:156–166. [DOI] [PubMed] [Google Scholar]
- 27. Liu J, Liu W, Li R, Yang H. Mitophagy in Parkinson’s disease: from pathogenesis to treatment. Cells 2019; 8:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab 2015; 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim JA, Kakhlon O, Li L, Myerowitz R, Raben N. Pompe disease: shared and unshared features of lysosomal storage disorders. Rare Dis 2015; 3:e1068978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang C, Badr MA, Kyrychenko V, Eskelinen EL, Shirokova N. Deficit in PINK1/PARKIN-mediated mitochondrial autophagy at late stages of dystrophic cardiomyopathy. Cardiovasc Res 2018; 114:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gkikas I, Palikaras K, Tavernarakis N. The role of mitophagy in innate immunity. Front Immunol 2018; 9:1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Y, Mi C, Liu J, Gao F, Long J. Compromised mitochondrial remodeling in compensatory hypertrophied myocardium of spontaneously hypertensive rat. Cardiovasc Pathol 2014; 23:101–106. [DOI] [PubMed] [Google Scholar]
- 33. Fedorova LV, Sodhi K, Gatto-Weis C, Puri N, Hinds TD Jr, Shapiro JI, Malhotra D. Peroxisome proliferator-activated receptor δ agonist, HPP593, prevents renal necrosis under chronic ischemia. PLoS One 2013; 8:e64436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X, Li ZL, Crane JA, Jordan KL, Pawar AS, Textor SC, Lerman A, Lerman LO. Valsartan regulates myocardial autophagy and mitochondrial turnover in experimental hypertension. Hypertension 2014; 64:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu Y, Li S, Wu H, Bian Z, Xu J, Gu C, Chen X, Yang D. Beneficial effects of astragaloside IV against angiotensin II-induced mitochondrial dysfunction in rat vascular smooth muscle cells. Int J Mol Med 2015; 36:1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei T, Huang G, Gao J, Huang C, Sun M, Wu J, Bu J, Shen W. Sirtuin 3 deficiency accelerates hypertensive cardiac remodeling by impairing angiogenesis. J Am Heart Assoc 2017; 6:e006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiong W, Hua J, Liu Z, Cai W, Bai Y, Zhan Q, Lai W, Zeng Q, Ren H, Xu D. PTEN induced putative kinase 1 (PINK1) alleviates angiotensin II-induced cardiac injury by ameliorating mitochondrial dysfunction. Int J Cardiol 2018; 266:198–205. [DOI] [PubMed] [Google Scholar]
- 38. Xiong W, Ma Z, An D, Liu Z, Cai W, Bai Y, Zhan Q, Lai W, Zeng Q, Ren H, Xu D. Mitofusin 2 participates in mitophagy and mitochondrial fusion against angiotensin II-induced cardiomyocyte injury. Front Physiol 2019; 10:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao W, Li Y, Jia L, Pan L, Li H, Du J. Atg5 deficiency-mediated mitophagy aggravates cardiac inflammation and injury in response to angiotensin II. Free Radic Biol Med 2014; 69:108–115. [DOI] [PubMed] [Google Scholar]
- 40. Zhang X, Li ZL, Eirin A, Ebrahimi B, Pawar AS, Zhu XY, Lerman A, Lerman LO. Cardiac metabolic alterations in hypertensive obese pigs. Hypertension 2015; 66:430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8:1462–1476. [DOI] [PubMed] [Google Scholar]
- 42. Chaanine AH, Jeong D, Liang L, Chemaly ER, Fish K, Gordon RE, Hajjar RJ. JNK modulates FOXO3a for the expression of the mitochondrial death and mitophagy marker BNIP3 in pathological hypertrophy and in heart failure. Cell Death Dis 2012; 3:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PLoS One 2012; 7:e32388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou H, Zhu P, Guo J, Hu N, Wang S, Li D, Hu S, Ren J, Cao F, Chen Y. Ripk3 induces mitochondrial apoptosis via inhibition of FUNDC1 mitophagy in cardiac IR injury. Redox Biol 2017; 13:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One 2011; 6:e20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Swiader A, Nahapetyan H, Faccini J, D’Angelo R, Mucher E, Elbaz M, Boya P, Vindis C. Mitophagy acts as a safeguard mechanism against human vascular smooth muscle cell apoptosis induced by atherogenic lipids. Oncotarget 2016; 7:28821–28835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu W, Xu H, Wang Z, Mao Y, Yuan L, Luo W, Cui Z, Cui T, Wang XL, Shen YH. PINK1-Parkin-mediated mitophagy protects mitochondrial integrity and prevents metabolic stress-induced endothelial injury. PLoS One 2015; 10:e0132499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu N, Wu J, Zhang L, Gao Z, Sun Y, Yu M, Zhao Y, Dong S, Lu F, Zhang W. Hydrogen Sulphide modulating mitochondrial morphology to promote mitophagy in endothelial cells under high-glucose and high-palmitate. J Cell Mol Med 2017; 21:3190–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Familtseva A, Kalani A, Chaturvedi P, Tyagi N, Metreveli N, Tyagi SC. Mitochondrial mitophagy in mesenteric artery remodeling in hyperhomocysteinemia. Physiol Rep 2014; 2:e00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011; 13:589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuan Y, Zhang A, Qi J, Wang H, Liu X, Zhao M, Duan S, Huang Z, Zhang C, Wu L, Zhang B, Zhang A, Xing C. p53/Drp1-dependent mitochondrial fission mediates aldosterone-induced podocyte injury and mitochondrial dysfunction. Am J Physiol Renal Physiol 2018; 314:F798–F808. [DOI] [PubMed] [Google Scholar]
- 53. Wu S, Zhou F, Zhang Z, Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J 2011; 278:941–954. [DOI] [PubMed] [Google Scholar]
- 54. Qi J, Wang F, Yang P, Wang X, Xu R, Chen J, Yuan Y, Lu Z, Duan J. Mitochondrial fission is required for angiotensin II-induced cardiomyocyte apoptosis mediated by a Sirt1-p53 signaling pathway. Front Pharmacol 2018; 9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qi X, Disatnik MH, Shen N, Sobel RA, Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase C{delta} under oxidative stress conditions in vivo. Mol Biol Cell 2011; 22:256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hom JR, Gewandter JS, Michael L, Sheu SS, Yoon Y. Thapsigargin induces biphasic fragmentation of mitochondria through calcium-mediated mitochondrial fission and apoptosis. J Cell Physiol 2007; 212:498–508. [DOI] [PubMed] [Google Scholar]
- 57. Disatnik MH, Ferreira JC, Campos JC, Gomes KS, Dourado PM, Qi X, Mochly-Rosen D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J Am Heart Assoc 2013; 2:e000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 2010; 121:2012–2022. [DOI] [PubMed] [Google Scholar]
- 59. Liu MY, Jin J, Li SL, Yan J, Zhen CL, Gao JL, Zhang YH, Zhang YQ, Shen X, Zhang LS, Wei YY, Zhao Y, Wang CG, Bai YL, Dong DL. Mitochondrial fission of smooth muscle cells is involved in artery constriction. Hypertension 2016; 68:1245–1254. [DOI] [PubMed] [Google Scholar]
- 60. Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012; 337:1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yao N, Wang C, Hu N, Li Y, Liu M, Lei Y, Chen M, Chen L, Chen C, Lan P, Chen W, Chen Z, Fu D, Ye W, Zhang D. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis 2019; 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, Tanaka K, Matsutani T, Iwanami A, Aronow BJ, Manway L, Maira SM, Thorgeirsson SS, Mischel PS, Thomas G, Kozma SC. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med 2012; 4:139ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL Jr, Conrad C, Rothberg JM, Lipton JO, Kölker S, Hoffmann GF, Han MJ, Polleux F, Sahin M. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep 2016; 17:1053–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boyle KA, Van Wickle J, Hill RB, Marchese A, Kalyanaraman B, Dwinell MB. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J Biol Chem 2018; 293:14891–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, Chen HY, Ghavami A, Stein M, DiPaola RS, Zhang D, Rabinowitz JD, White E. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One 2012; 7:e41831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol 2012; 8:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Natale G, Lenzi P, Lazzeri G, Falleni A, Biagioni F, Ryskalin L, Fornai F. Compartment-dependent mitochondrial alterations in experimental ALS, the effects of mitophagy and mitochondriogenesis. Front Cell Neurosci 2015; 9:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Song YM, Lee WK, Lee YH, Kang ES, Cha BS, Lee BW. Metformin restores parkin-mediated mitophagy, suppressed by cytosolic p53. Int J Mol Sci 2016; 17 (doi: 10.3390/ijms17010122). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen Q, Thompson J, Hu Y, Dean J, Lesnefsky EJ. Inhibition of the ubiquitous calpains protects complex I activity and enables improved mitophagy in the heart following ischemia-reperfusion. Am J Physiol Cell Physiol 2019; 317:C910–C921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen Y, Gibson SB. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy 2008; 4:246–248. [DOI] [PubMed] [Google Scholar]
- 72. Xiao B, Goh JY, Xiao L, Xian H, Lim KL, Liou YC. Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J Biol Chem 2017; 292:16697–16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jang SY, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J Biol Chem 2012; 287:19304–19314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vucicevic L, Misirkic-Marjanovic M, Paunovic V, Kravic-Stevovic T, Martinovic T, Ciric D, Maric N, Petricevic S, Harhaji-Trajkovic L, Bumbasirevic V, Trajkovic V. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy 2014; 10:2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. East DA, Fagiani F, Crosby J, Georgakopoulos ND, Bertrand H, Schaap M, Fowkes A, Wells G, Campanella M. PMI: a ΔΨm independent pharmacological regulator of mitophagy. Chem Biol 2014; 21:1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park SJ, Shin JH, Kim ES, Jo YK, Kim JH, Hwang JJ, Kim JC, Cho DH. Mitochondrial fragmentation caused by phenanthroline promotes mitophagy. FEBS Lett 2012; 586:4303–4310. [DOI] [PubMed] [Google Scholar]
- 77. Li Q, Zhang T, Wang J, Zhang Z, Zhai Y, Yang GY, Sun X. Rapamycin attenuates mitochondrial dysfunction via activation of mitophagy in experimental ischemic stroke. Biochem Biophys Res Commun 2014; 444:182–188. [DOI] [PubMed] [Google Scholar]
- 78. Das S, Mitrovsky G, Vasanthi HR, Das DK. Antiaging properties of a grape-derived antioxidant are regulated by mitochondrial balance of fusion and fission leading to mitophagy triggered by a signaling network of Sirt1-Sirt3-Foxo3-PINK1-PARKIN. Oxid Med Cell Longev 2014; 2014:345105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79. Perera ND, Sheean RK, Lau CL, Shin YS, Beart PM, Horne MK, Turner BJ. Rilmenidine promotes MTOR-independent autophagy in the mutant SOD1 mouse model of amyotrophic lateral sclerosis without slowing disease progression. Autophagy 2018; 14:534–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim EH, Sohn S, Kwon HJ, Kim SU, Kim MJ, Lee SJ, Choi KS. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res 2007; 67:6314–6324. [DOI] [PubMed] [Google Scholar]
- 81. Kim EH, Choi KS. A critical role of superoxide anion in selenite-induced mitophagic cell death. Autophagy 2008; 4:76–78. [DOI] [PubMed] [Google Scholar]
- 82. Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Büttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Mühlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med 2016; 22:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rodríguez-Navarro JA, Rodríguez L, Casarejos MJ, Solano RM, Gómez A, Perucho J, Cuervo AM, García de Yébenes J, Mena MA. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis 2010; 39:423–438. [DOI] [PubMed] [Google Scholar]
- 84. Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med 2016; 22:879–888. [DOI] [PubMed] [Google Scholar]
- 85. Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2006; 2:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu L, Xie R, Nguyen S, Ye M, McKeehan WL. Robust autophagy/mitophagy persists during mitosis. Cell Cycle 2009; 8:1616–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cotán D, Cordero MD, Garrido-Maraver J, Oropesa-Ávila M, Rodríguez-Hernández A, Gómez Izquierdo L, De la Mata M, De Miguel M, Lorite JB, Infante ER, Jackson S, Navas P, Sánchez-Alcázar JA. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB J 2011; 25:2669–2687. [DOI] [PubMed] [Google Scholar]
- 88. Redmann M, Benavides GA, Berryhill TF, Wani WY, Ouyang X, Johnson MS, Ravi S, Barnes S, Darley-Usmar VM, Zhang J. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol 2017; 11:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang J, Fang Y, Yan L, Yuan N, Zhang S, Xu L, Nie M, Zhang X, Wang J. Erythroleukemia cells acquire an alternative mitophagy capability. Sci Rep 2016; 6:24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dombi E, Diot A, Morten K, Carver J, Lodge T, Fratter C, Ng YS, Liao C, Muir R, Blakely EL, Hargreaves I, Al-Dosary M, Sarkar G, Hickman SJ, Downes SM, Jayawant S, Yu-Wai-Man P, Taylor RW, Poulton J. The m.13051G>A mitochondrial DNA mutation results in variable neurology and activated mitophagy. Neurology 2016; 86:1921–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL, Huang C, Li P, Gao N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 2015; 11:1259–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol 1956; 11:298–300. [DOI] [PubMed] [Google Scholar]
- 93. Harman D. Free radical theory of aging: an update: increasing the functional life span. Ann NY Acad Sci 2006; 1067:10–21. [DOI] [PubMed] [Google Scholar]
- 94. Afanas’ev I. Signaling and damaging functions of free radicals in aging-free radical theory, hormesis, and TOR. Aging Dis 2010; 1:75–88. [PMC free article] [PubMed] [Google Scholar]
- 95. Pérez VI, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta 2009; 1790:1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 2014; 94:909–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Park SH, Kwon OS, Park SY, Weavil JC, Andtbacka RHI, Hyngstrom JR, Reese V, Richardson RS. Vascular mitochondrial respiratory function: the impact of advancing age. Am J Physiol Heart Circ Physiol 2018; 315:H1660–H1669. [DOI] [PMC free article] [PubMed] [Google Scholar]