We assessed the impact on Salmonella physiology of reciprocally translocating the genes encoding the Fis and Dps nucleoid-associated proteins (NAPs) and of inverting their growth-phase production patterns such that Fis was produced in stationary phase (like Dps) and Dps was produced in exponential phase (like Fis). Changes to peak binding of Fis were detected by ChIP-seq on the chromosome, as were widespread impacts on the transcriptome, especially when Fis production mimicked Dps production. Virulence gene expression and the expression of a virulence phenotype were altered. Overall, these radical changes to NAP gene expression were well tolerated, revealing the robust and well-buffered nature of global gene regulation networks in the bacterium.
KEYWORDS: nucleoid-associated protein, Fis, Dps, ChIP-seq, RNA-seq, transcriptome, Salmonella enterica serovar Typhimurium, virulence
ABSTRACT
The Fis nucleoid-associated protein controls the expression of a large and diverse regulon of genes in Gram-negative bacteria. Fis production is normally maximal in bacteria during the early exponential phase of batch culture growth, becoming almost undetectable by the onset of stationary phase. We tested the effect on the Fis regulatory network in Salmonella of moving the complete fis gene from its usual location near the origin of chromosomal replication to the position normally occupied by the dps gene in the right macrodomain of the chromosome, and vice versa, creating the gene exchange (GX) strain. In a parallel experiment, we tested the effect of rewiring the Fis regulatory network by placing the fis open reading frame under the control of the stationary-phase-activated dps promoter at the dps genetic location within the right macrodomain, and vice versa, creating the open reading frame exchange (OX) strain. Chromatin immunoprecipitation sequencing (ChIP-seq) was used to measure global Fis protein binding levels and to determine gene expression patterns. Strain GX showed few changes compared with the wild type, although we did detect increased Fis binding at Ter, accompanied by reduced binding at Ori. Strain OX displayed a more pronounced version of this distorted Fis protein-binding pattern together with numerous alterations in the expression of genes in the Fis regulon. OX, but not GX, had a reduced ability to infect cultured mammalian cells. These findings illustrate the inherent robustness of the Fis regulatory network with respect to the effects of rewiring based on gene repositioning alone and emphasize the importance of fis expression signals in phenotypic determination.
INTRODUCTION
trans-Acting regulatory proteins play a prominent role in controlling the expression of bacterial genes at the level of transcription. Whether acting positively or negatively, these proteins typically influence their target genes by binding at, or close to, transcriptional promoters (1). Transcription factors differ in the number of genes that are under their control (2), and a majority of these proteins exist with moderate copy numbers of between 1 and 100 monomers (3). Nucleoid-associated proteins (NAPs) play a role in the architecture of the genome, and many contribute to the regulation of transcription (4). NAPs share many of the features of transcription factors, but many are present in thousands of copies. The distinction between “transcription factor” and “NAP” is quite blurry; these are operational descriptions for proteins that lie along a continuum extending from factors with pervasive influences on gene expression to those that have just a few gene targets (5).
Transcription factors must make the journey from the gene that encodes them to the target promoter. This process seems to involve a combination of sliding along DNA and movements between DNA segments in which the interactions with DNA consist of nonspecific and specific binding events (6, 7). In the case of a low-copy-number transcription factor such as the Lac repressor protein, LacI (40 monomers per cell), the addition of an inducer (IPTG [isopropyl-β-d-thiogalactopyranoside]) reduces specific but not nonspecific binding; the protein explores the nucleoid as it searches for its specific binding sites (6). Many NAPs rely on indirect readout to identify their DNA targets, making their binding site selections on the basis of the DNA conformation rather than on the basis of the sequence alone (5). This, combined with their high copy numbers, might allow NAPs to spread rapidly through the nucleoid to locate their DNA targets. Alternatively, their weak requirement for specific base sequences at their DNA targets might increase their dwell time at the many nonspecific sites that they encounter as they migrate through the nucleoid.
The significance of the genomic positions of genes encoding transcription factors or NAPs has been investigated, principally in the model bacterium Escherichia coli. There, a correlation is seen between gene position along the replichore extending from the origin of chromosome replication, oriC, to the terminus and the period in the growth cycle when the gene product is most required (8). Gene position also correlates with gene copy number in fast-growing bacteria, when replication recommences before cell division is complete. This correlation is conserved across the Gammaproteobacteria (8) suggesting that it is biologically meaningful and may have consequences for the operation of bacterial gene control networks. This hypothesis is supported by previously reported observations suggesting that gene relocations that occur naturally, e.g., through inversions of chromosomal segments, tend to preserve the distance of the affected gene(s) from oriC (9–11). It suggests that moving regulatory genes, including genes that encode NAPs, to novel chromosomal locations could result in changes to bacterial physiology. Consistent with this proposal, repositioning the gene encoding the factor for inversion stimulation (FIS) NAP in E. coli alters the cell’s capacity to manage its global DNA topology (12).
Fis is a prominent member of the family of NAPs in Gram-negative bacteria (13, 14) that alters the transcription of hundreds of genes, directly or indirectly and positively or negatively (15–19), and contributes architecturally to site-specific recombination systems (20–22), chromosome replication initiation (23–26), transposon activity (27, 28), and bacteriophage life cycles (22, 27, 29, 30). Fis is not essential, despite its pervasive influence on cell biology, but it enhances the fitness of a wild-type (WT) bacterium when competing with an otherwise isogenic fis knockout mutant (31). Among the genes that are regulated by Fis are those encoding components of the translational apparatus of the bacterium, its chemotaxis and motility functions, and many metabolic pathway proteins and (in the case of pathogens) numerous virulence genes (17, 19, 32–34). The association of high Fis concentrations with the early exponential (EE) phase of growth is thought to be indicative of a role for Fis in signaling growth-cycle-related information to the global gene expression program of the cell (31, 35, 36).
The Fis protein influences the topology of DNA both directly, through DNA binding (31), and indirectly, through its influence on the expression of the genes encoding DNA gyrase (gyrA and gyrB) and DNA topoisomerase I (topA) (37–39). The fis gene is part of the bicistronic dusB-fis operon (40, 41), whose stringently controlled promoter is stimulated by negatively supercoiled DNA, creating a regulatory connection between the global supercoiling level in bacterial DNA, the physiology of the bacterium, and the initiation of fis transcription (42). The single dusB-fis promoter is autorepressed by the Fis protein (43), with translation of the Fis protein relying on mRNA secondary structure and nucleotide sequence motifs in the upstream message (44).
The production of high levels of Fis protein is associated with the early exponential phase of bacterial growth, with Fis being present at very low levels in the stationary phase (35, 37, 45, 46). This pattern is sensitive to aeration; cultures of Salmonella enterica serovar Typhimurium (S. Typhimurium) exhibit sustained, low-level production of Fis into the stationary phase of growth in the absence of aeration (47, 48).
We explored the subtleties of Fis biology by testing the significance of the geographical location of the fis gene in the nonstructured left (NS-Left) region of the bacterial chromosome and the physiological significance of the characteristic early-growth-phase-dependent fis expression profile. This was achieved by exploiting the chromosome location, promoter, and expression pattern of the dps gene. This gene is located in the right macrodomain, where it encodes a ferritin-like, DNA-protecting protein, Dps, which is produced in high concentrations in stationary-phase cells (49–54). A comparison of the proteomes of wild-type E. coli and an isogenic dps knockout mutant revealed many differences in protein production, suggesting a role for Dps in the control of protein production at some level (49). Although Dps is a DNA binding protein, it is thought not to influence transcription (51, 55). The dps gene is transcribed on entry into stationary phase using RpoS, the stress-and-stationary-phase sigma factor of RNA polymerase, although this can be overridden during oxidative stress when dps is transcribed using RpoD in an OxyR- and IHF-dependent mechanism (56, 57). Fis represses dps transcription by RNA polymerase containing the RpoD sigma factor, but not RpoS (58). Thus, Fis and Dps production is normally associated with opposite ends of the growth cycle: the early exponential and the stationary phases, respectively (46). The fis and dps genes are both transcribed in the orientation opposite the direction of DNA replication in the left and right replichores, respectively (Fig. 1A).
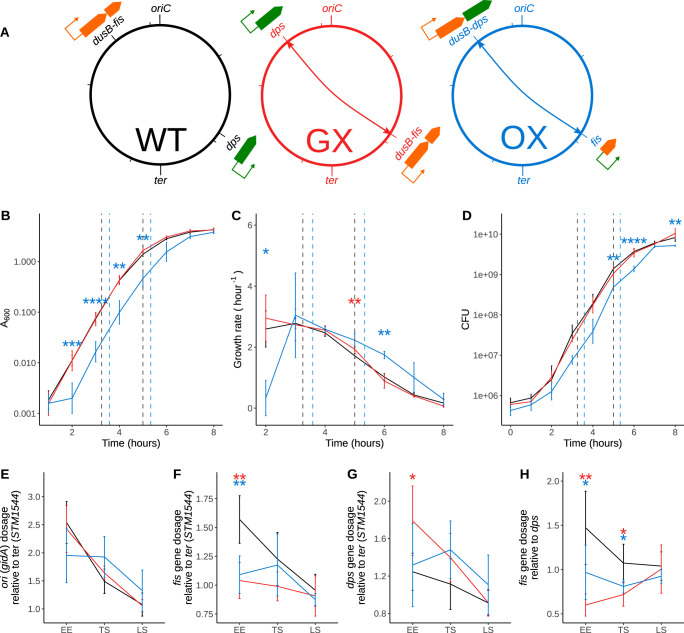
FIG 1.
Genomic exchanges of fis and dps transcription units or ORFs. (A) Schematic depicting wild-type (WT) SL1344 strain with the original locations of dusB-fis and dps transcription units shown, the constructed gene exchange (GX) strain with the transcriptional units exchanged, and the ORF exchange (OX) strain with only the fis-dps ORFs exchanged. (B) Optical density-based growth curves showing the impact of these genetic changes on growth. Early exponential (EE), transition to stationary (TS), and late stationary (LS) time points selected for other analyses are shown. Significance was determined relative to the wild type using Welch's t test, and P values were adjusted using the Benjamini Hochberg (BH) method. Adjusted P values are indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C) Growth rate curve derived from the OD-based growth curves. (D) CFU-based growth curves. (E) Origin-to-terminus gradients in these strains during the course of the growth curve analysis calculated by assaying the abundance of gidA (oriC proximal gene) DNA and STM1544 (ter proximal gene) DNA using qPCR. The late stationary (LS) phase is the 24-h time point in the growth curve not shown in the earlier plots. (F) Dosages of the fis gene relative to the terminus during the course of the growth curve analysis. (G) Dosages of the dps gene relative to the terminus. (H) Dosages of the fis gene relative to dps analyzed to understand the impact of the gene position swap.
We anticipated that altering the expression pattern of the fis gene, in a radical manner that fell short of eliminating Fis protein production completely, would provide valuable insights into (i) the robustness or the vulnerability of bacterial physiology in the face of changes to the expression of a prominent NAP gene, (ii) the impact on the operations of the cell of expressing the fis gene from a position in the replichore opposite the normal location, and (iii) the significance of the temporal expression pattern of the fis gene with respect to the in vivo role of the Fis protein. To test our ideas, we relocated the fis gene, with its native control elements, from the NS-Left region of the chromosome of S. Typhimurium to the dps locus in the right macrodomain. In a second genetic exchange, just the protein-encoding, open reading frame (ORF) of fis was substituted for that of dps, giving fis the stationary-phase-specific expression profile of the dps gene. Reciprocally exchanging the positions of the entire dusB-fis operon and the entire dps gene had a relatively modest impact on the physiology of the bacterium. On the other hand, placing fis under the control of the dps regulatory sequences at the dps locus, while driving dps gene expression with the dusB-fis regulatory sequences at the dusB-fis locus, had profound effects on the physiology of the bacterium at the levels of global gene expression, Fis binding patterns around the chromosome, and the ability of the bacterium to infect human cells.
RESULTS
Reciprocally exchanging the fis and dps genes.
The fis gene is the downstream component of the bicistronic dusB-fis operon (Fig. 1A). We moved the complete dusB-fis operon from its usual location near the origin of chromosomal replication to the position normally occupied by the dps gene in the right macrodomain of the chromosome, and vice versa, creating the strain GX (gene exchange strain). In a parallel strain construction, we placed the fis open reading frame under the control of the stationary-phase-activated dps promoter at the dps genetic location within the right macrodomain, and vice versa, creating strain OX (open reading frame exchange strain) (Fig. 1A). The construction of the GX and OX strains is described in Materials and Methods (see also Fig. S1 in the supplemental material). The genetic map locations of fis and dps in the wild-type (WT), GX, and OX strains are summarized in Fig. 1A. We used whole-genome sequencing to rule out the presence of mutations that might have been introduced during strain construction (Fig. S2).
Schematic representation of the strain construction process. For details, see Materials and Methods. Download FIG S1, EPS file, 0.7 MB (722.8KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of SNP analysis of constructed strains. Heat maps depict summarized Breseq analysis results. (A) Predicted mutations. (B) Unassigned missing coverage evidence. (C) Unassigned new junction evidence. In panels A and B, the color scale indicates the presence (1) or absence (0) of a mutation. In panel C, the color scale indicates the frequency of the new junction among reads mapping to that locus. Download FIG S2, EPS file, 0.9 MB (914.9KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The GX and WT strains exhibited similar growth profiles in liquid medium measured using absorbance (Fig. 1B), growth rate (Fig. 1C), or colony forming units (CFU) counts (Fig. 1D). In contrast, the OX strain underperformed in comparison with the WT and GX strains in each of the three growth assessments, with evidence of an extended lag phase being detected (Fig. 1B to D). An exception to this was seen in the case of the growth rate of OX following the transition from exponential growth to stationary phase; here, the growth rate of OX declined more slowly than that of the other two strains (Fig. 1C).
The locations of the fis and dps genes on the wild-type chromosome are almost diametrically opposite, with fis being much closer to the origin of chromosome replication (oriC) and dps being nearer to the terminus gene, ter (Fig. 1A). The gene dosage gradients from oriC to ter were measured for each of the three strains using quantitative PCR (qPCR) of an origin-proximal gene (gidA) and a terminus-proximal gene (STM1544). The measurements were performed at three stages of growth: the early exponential (EE) phase, the transition from exponential growth to stationary phase (TS), and late stationary phase (LS) (for the values of absorbance at 600 nm [A600], see Materials and Methods). The WT and GX strains had almost identical profiles, while OX had a gradient with a higher variance at each stage of growth than the other two (Fig. 1E). The gene dosage of fis was lower in GX and OX than in WT in the EE phase, as expected given its greater proximity to the terminus in the two exchanged strains (Fig. 1F). At the EE phase of growth, the dps gene dosage was greater in the GX strain than in the WT, as expected, but was not greater in the OX strain (Fig. 1G), possibly due to the lower growth rate and gene dosage gradient in the OX strain (Fig. 1C) stemming from the lower growth rate of the OX strain (Fig. 1C) at the early time point (Fig. 1E). At the EE phase of growth, the fis gene in the GX strain had the lowest gene dosage relative to dps (Fig. 1H), thus clearly reflecting the fis-dps gene position exchange and associated fis-dps gene dosage exchange.
Effects of genetic exchanges on fis and dps gene expression and protein levels.
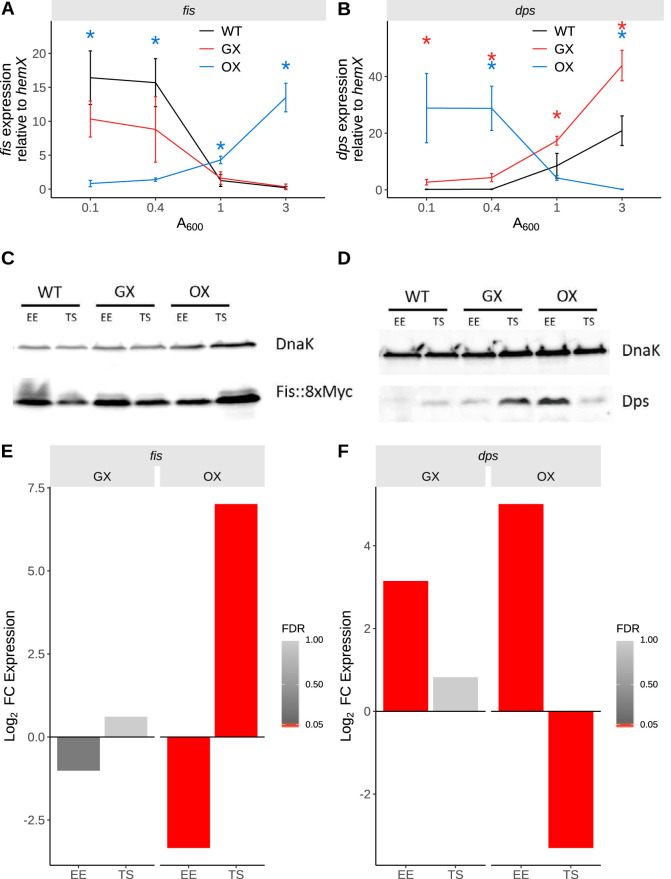
The transcription of the fis (Fig. 2A) and dps (Fig. 2B) genes was measured by qPCR using the hemX gene as a benchmark. In the WT, each gene was expressed as expected, with fis transcription being maximal in early exponential growth (Fig. 2A) and dps being transcribed maximally in the stationary phase (Fig. 2B). In the GX strain, both fis and dps retained their overall expression profiles as seen in the WT but the levels of the dps transcript were higher (Fig. 2A and B). In the OX strain, the transcription profile of fis was the reverse of that seen in the WT; fis transcript levels were highest in stationary phase and lowest in early exponential phase (Fig. 2A). In contrast, dps expression was highest in exponential growth and lowest in stationary-phase growth, the reverse of the WT pattern (Fig. 2B). Western blot analyses showed that the patterns of production of Fis (Fig. 2C) and Dps (Fig. 2D) resembled the transcription patterns for the corresponding genes; Dps production in OX mimicked Fis production in the WT, while Fis was produced in OX in a pattern that resembled that of Dps production in the WT. Notably, the expression of fis and dps genes determined using transcriptome sequencing (RNA-seq) (Fig. 2E and F) matched the expression determined using qPCR (Fig. 2A and B). These data showed that relocating the dusB-fis operon or the dps gene had a negligible effect on their expression patterns and on the production profiles of their products. However, exchanging just the fis and dps protein coding regions, with the concomitant switching of cognate regulatory elements, produced reciprocal changes in gene expression and protein production. We next examined the impacts of these changes on Fis protein binding patterns throughout the genome, on the downstream expression of the Fis regulon, and on the physiology of the bacterium.
FIG 2.
Gene expression changes. (A) Expression of fis transcript during the course of the growth curve assayed using RT-qPCR. Significance was determined relative to the wild type using Welch's t test, and P values were adjusted using the BH method. Adjusted P values are indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Black, WT; red, GX; blue, OX. (B) Expression of dps transcript during the growth curve. Black, WT; red, GX; blue, OX. (C) Fis protein levels determined by immunoblotting with anti-Fis monoclonal antibody. (D) Dps protein levels determined by immunoblotting with anti-Dps polyclonal serum. (E and F) Expression changes in fis and dps genes determined by RNA-seq in the GX and OX strains at the EE and TS phases of growth. The color bar indicates FDR, and bars representing genes with FDR values of <0.05 are red.
Fis binding to the genome in the GX and OX strains.
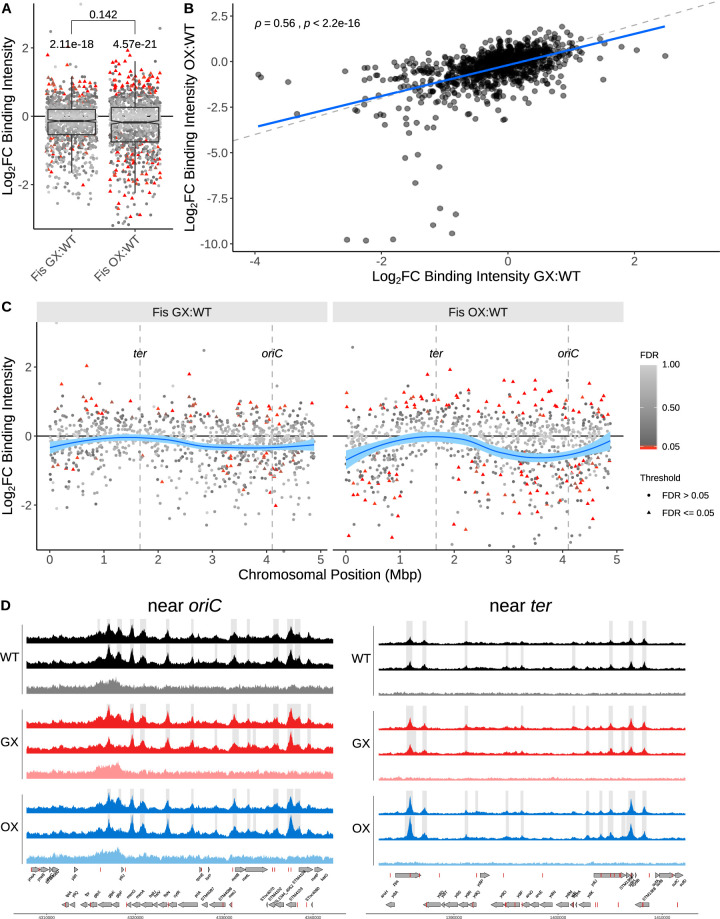
Chromatin immunoprecipitation sequencing (ChIP-seq) was used to examine the intensity and the distribution of Fis protein binding to the genomes in the GX and OX strains compared to the WT (Fig. 3 [see also Fig. S3]) (Fis motif, Fig. S4). The median Fis binding intensity was lower in both the GX and OX strains than in the WT (Fig. 3A). The intensity of Fis binding changed over a greater range, and was significantly changed at greater numbers of Fis peaks, in the OX strain than in the GX strain (Fig. 3A and C). Changes in Fis binding intensity in the GX strain positively correlated with those in the OX strain, suggesting that these two strains experienced similar changes in Fis binding across genomic loci (Fig. 3B and C). Binding intensity changes showed regional variations along the chromosomes in the OX-GX comparison (Fig. 3C). On average, the reduction in Fis binding was greater around the chromosome origin than around the terminus in both the OX and GX strains. However, the magnitude of the change was greater for OX versus WT than for GX versus WT. In both OX and GX, the Fis protein was being produced from a locus that was closer to the terminus than to the origin (i.e., the gene location was diametrically opposite that in the WT) (Fig. 1A). We also examined the binding patterns of Fis at the negatively autoregulated dusB-fis promoter in all three strains. OX and the WT had very similar patterns of Fis binding peaks, but the peaks showed reduced amplitudes in GX (Fig. S6). This may reflect the influence of the new chromosomal neighborhood following the relocation of those Fis binding sites together with dusB-fis to a novel chromosomal site (the dps locus) in GX.
FIG 3.
Fis binding changes assayed by ChIP-seq during EE phase. (A) ChIP-seq was used to identify Fis peaks in WT, GX, and OX. The R package DiffBind was then used to determined Log2 fold changes in binding intensities of Fis in these peaks in the GX and OX strains relative to the wild type. Significant peak intensity changes (FDR < 0.05) are shown as red filled triangles. To determine if the medians of the distributions of Log2 fold changes in binding intensities differed significantly from 0 (no change in Fis binding relative to WT), the Wilcoxon rank sum test was performed with μ = 0; the P values are displayed over the individual distributions. The difference between the medians of the GX:WT distribution and the OX:WT distribution was not significant with the Wilcoxon rank sum test as shown above the square bracket corresponding to the data from the single-sample tests (using the less conservative Welch's t test here gives a P value of 0.0105). (B) Spearman’s correlation of peak intensity changes in the GX in comparison with the OX strain. Dashed line has a slope of 1 and indicates the position where the fold changes in Fis binding intensity in GX would be equal to those in OX. The blue solid line is the regression linear fit to the data. (C) Peak intensity changes along the chromosomal position shown in millions of base pairs. The blue curve shows the local regression peak intensity change calculated using the R locally estimated scatterplot smoothing (LOESS) function. In calculating these intensity changes, the appropriate mock data (WT mock for WT peaks, GX mock for GX peaks, OX mock for OX peaks) were used to control for ori-ter gradient changes (Fig. S5). (D) Fis ChIP-seq coverage of the WT, GX, and OX strains (two biological replicates and mock) of example regions near oriC and ter loci. The ranges of the data indicated on the y-axis scale were the same for all the coverage traces and have been omitted for brevity. For each strain, the two biological replicates are represented by the first two coverage traces in the darker color shading. The third, lighter color trace represents the mock coverage for that strain. Gray-shaded regions show peaks called in each sample relative to the mock. Genes (gray arrows) and Fis binding motifs (red blocks) are shown on the positive and negative reference strands. ChIP-seq data for plasmids can be found in Fig. S3.
Fis binding regions on the chromosome and plasmids. For the SL1344 chromosome, predicted Fis binding regions are shown for the two biological replicates of the WT, GX, and OX strains in early exponential phase. Increased Fis binding around the ter gene in the GX and OX strains resulted in peak calling in this region that was slightly better than that seen with the WT. For plasmids, levels of coverage corresponding to the ChIP-seq experiment (solid traces) and predicted Fis binding regions (shaded boxes) are shown for the two biological replicates of the WT, GX, and OX strains in early exponential phase. For comparison, the wild-type mock coverage is also shown as a solid gray trace. Predicted Fis binding motifs are shown in orange, and the motif used for the prediction is briefly mentioned in the legend. This motif is described in detail in the Fig. S4 legend. Chromosomal positions are indicated in millions of base pairs, and plasmid positions are in thousands of base pairs. Download FIG S3, TIF file, 2.9 MB (2.8MB, tif) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding motif obtained from ChIP-seq data. A 15-nucleotide-long sequence logo represents the Fis binding motif discovered using MEME on the set of Fis binding regions in the wild-type strain identified using ChIP-seq. Each position in the 15-nucleotide-long palindromic motif is indicated on the x axis. The stack of letters at each position represents the most common nucleotides found at that position. The height of each letter represents the relative frequency of that nucleotide at that position in the motif. The overall height of the stack of letters at each position represents the degree of conservation at that position in the motif (if the stack is very short, that position is highly variable). Various statistics reported by MEME are presented below the logo. This motif is similar to the E. coli Fis binding motif. Download FIG S4, EPS file, 0.1 MB (159.5KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene dosage gradient differences in the GX and OX strains compared to the WT visualized using coverage data from the ChIP-seq mock experiments performed with these strains. For clarity, the coverage data at each position are not shown; only the local average calculated using the R LOESS function along the chromosomal position in millions of base pairs is shown. There were only slight gene dosage gradient differences in the GX and OX strains at the early exponential time point at which chromatin was collected for ChIP-seq. These differences are considered in the calculation of differentially bound regions. Download FIG S5, EPS file, 0.05 MB (54.1KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding around fis gene. (A) Log2 fold changes of Fis binding upstream of the dusB-fis transcription unit in GX and OX strains relative to the WT. (B) Fis ChIP-seq coverage of the WT, GX, and OX strains (two biological replicates and mock) of the region around the fis gene. The ranges of the y-axis scale are the same for all the coverage traces and have been omitted for brevity. For each strain, the two biological replicates are represented by the first two coverage traces in the darker color shade. The third, lighter color trace represents the mock coverage for that strain. Gray-shaded regions show the peaks called in each sample relative to the mock. Genes (gray arrows) and Fis binding motifs (red blocks) are shown on the positive and negative reference strands. The fis gene is a part of the dusB-fis transcription unit. The promoter of this transcription unit contains two Fis binding motifs, as predicted by FIMO from the MEME suite, and the ChIP-seq data show substantial Fis binding. ChIP-seq data were analyzed using the wild-type reference sequence; thus, the coverage reported here is plotted relative to wild-type gene locations, even though those locations had been altered in the GX and OX strains. In the GX strain, the entire region that included dusB, fis, and 315 bp of its upstream promoter region was relocated to the dps locus. In the OX strain, only the fis ORF was relocated to the dps locus. In the GX and OX strains, due the strain construction process, an rrnB terminator was inserted 63 bp downstream of the yhdG-fis operon. This is represented as a dip in the coverage and was expected. Download FIG S6, EPS file, 0.1 MB (158.1KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Salmonella Typhimurium SL1344 harbors two large plasmids, pSLT (the virulence plasmid) and pCol1B9, both of which are bound by Fis (Fig. S7). As with the chromosome, both plasmids displayed reduced Fis binding at most loci in the GX and OX strains (Fig. S7A) and these changes were positively correlated between the two strains (Fig. S7B).
Fis binding changes on the Salmonella megaplasmids assayed by ChIP-seq during the EE phase. (A) Peak intensity changes along the plasmid position shown in base pairs. In calculating these intensity changes, the appropriate mock data (WT mock for WT peaks, GX mock for GX peaks, OX mock for OX peaks) were used as controls. Significantly differentially bound peaks are roughly annotated with genes that are located in their vicinity. (B) Spearman’s correlation of peak intensity changes in the GX strain in comparison with the OX strain. The dashed line has a slope of 1 and indicates the position where the fold changes in Fis binding intensity in GX would be equal to those in OX. The blue solid line represents the regression linear fit to the data. Download FIG S7, EPS file, 0.4 MB (431.2KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The impact of the genetic exchanges on Fis target gene expression.
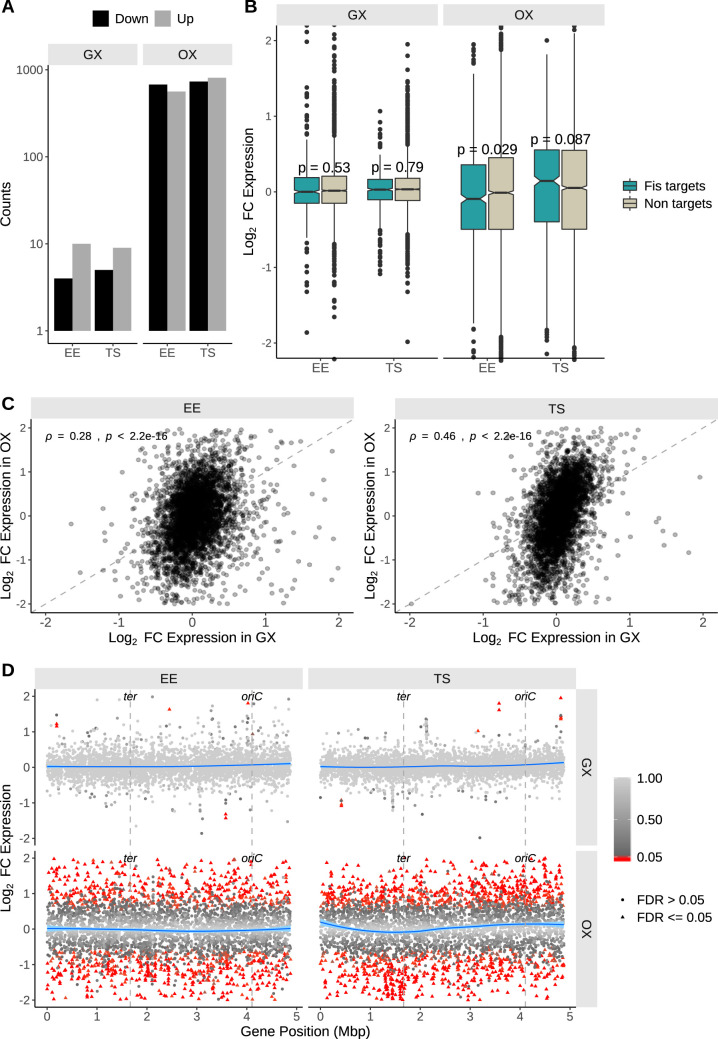
RNA-seq was used to compare the transcriptomes of the GX and the OX strains with that of the WT in cultures at the early exponential phase and at the exponential-phase-to-stationary-phase transition of the growth cycle (Fig. 4). The OX strain showed a higher number of genes with altered expression than the GX strain at both phases of growth (Fig. 4A), and the changes in expression were also greater in the OX strain than in the GX strain (Fig. 4B). Furthermore, the differences between the median values of log2 fold change in expression (log2FC) of the Fis target genes and the nontarget genes were more significant in the OX strain than in the GX strain. In the OX strain, the median log2FC of the Fis target genes was lower in the early exponential phase, and higher in the transition to stationary phase, than that of the nontarget genes. This difference in the levels of Fis target and nontarget gene expression mirrors the greater Fis levels seen during the exponential phase and the lower Fis levels seen in the transition to stationary phase in the OX strain. As with the Fis binding data, we saw slight positive correlations between the GX and OX strains at both the EE and TS phases (Fig. 4C), thus highlighting similarities in the Fis binding changes and associated gene expression changes in the two strains. Plotting the data as a function of position along the chromosome, the OX strain was again found to show a far greater degree of disturbance of the normal expression of its transcriptome than the GX strain compared to the WT, and differentially expressed genes were scattered throughout the chromosome (Fig. 4D). These global gene expression patterns were consistent with the much greater changes seen in fis expression, Fis protein production (Fig. 2), and Fis protein binding (Fig. 3) in the OX strain.
FIG 4.
Transcriptomic changes assayed by RNA-seq. (A) Counts of differentially expressed genes. (B) Box plot distribution representations of Log2 fold change in expression of genes in the GX and OX strains. The genes have been split into two groups based on whether or not they are predicted to be Fis targets. P values obtained using the two-sample Wilcoxon test for the comparison between Fis targets and nontargets are indicated over each pair of box plots. (C) Spearman’s correlation of Log2 fold changes in expression of genes in the GX with the OX strain. The dashed line has a slope of 1 and indicates the position where the fold changes are equal. (D) Log2 fold change in expression of genes along their position on the chromosome in millions of base pairs. The blue curve shows the local regression peak intensity change calculated using the R LOESS function. Significant gene expression changes (FDR < 0.05) are shown as red filled triangles.
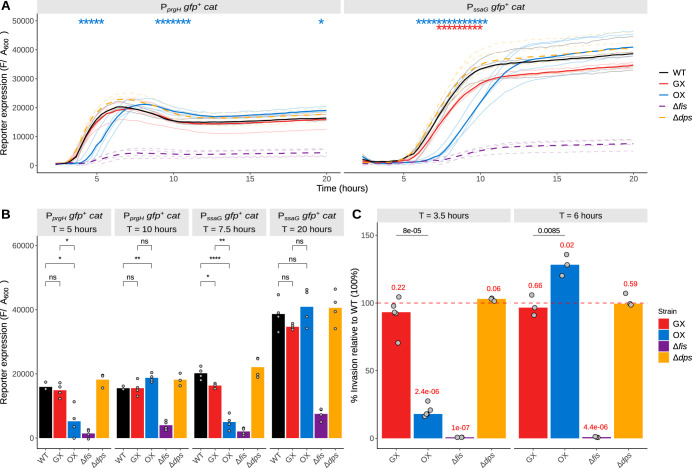
Among the known Fis-dependent genes, those encoding the three type 3 secretion systems found in Salmonella exhibited changes in expression (Fig. S8). These genes encode the flagellar apparatus and the Salmonella pathogenicity island 1 (SPI-1) and SPI-2 injectisomes. In each case, the largest, most statistically significant effects were seen in the OX strain (Fig. S8). The altered patterns of transcription detected in SPI-1 and SPI-2 in the RNA-seq experiments were investigated in more detail using the following gfp reporter gene fusions to representative promoters from each pathogenicity island: PprgH (SPI-1) and PssaG (SPI-2) (Fig. 5A and B). These experiments confirmed that both promoters were dependent on Fis because their full activities were lost in a fis knockout mutant. In contrast, the complete removal of Dps from the cell by introducing a dps knockout mutation had no effect on either promoter (Fig. 5B). In the OX strain, the SPI-1 PprgH promoter had reduced activity compared to the WT at 5 h in batch culture but was significantly more active at 10 h (Fig. 5A and B). PssaG from SPI-2 was also less active in the OX strain at earlier stages of growth but showed no significant differences from its activity in the WT at 20 h (Fig. 5A and B). The PprgH promoter was as active in the GX strain as it was in the WT at all stages of growth; the PssaG promoter differed from the WT by being less active at the 7.5-h time point.
FIG 5.
Pathogenicity island expression changes assayed using chromosomal Pgfp+ reporters. (A) SPI-1 expression corresponding to the PprgH gfp+ chromosomal fusions and SPI-2 expression corresponding to the PssaG gfp+ chromosomal fusions are reported. Reporter expression is measured by determining the fluorescence (F) of gfp+ expression divided by A600. Means of results from biological replicates are shown as solid lines, and data from the individual biological replicates are shown in the background as lines with lighter shading. Significance was determined relative to the wild type using Welch's t test, and P values were adjusted using the BH method. Adjusted P values (WT versus GX and WT versus OX comparisons only) are indicated with asterisks (*, P < 0.05). (B) A slice of the data in panel A showing reporter expression at early time points (SPI-1, 5 h; SPI-2, 15 h) versus later time points (SPI-1, 15 h; SPI-2, 20 h). Bars represent median values, and the individual biological replicates are represented by the scatter. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Results of epithelial cell invasion assay carried out with subcultures grown to two different time points (early, 3.5 h; late, 6 h). Plotted are percent invasions of the strains relative to the WT (100% [not shown], depicted by the red dashed line) in HeLa cells determined 90 min postinfection. Median values of percent invasion are represented by the bars, and values from biological replicates are represented by the scatter. P values obtained from one-sample t tests (μ = 100) are shown in red above each strain designation. P values obtained from Welch’s two-sample t tests between GX and OX are indicated over these comparisons in black.
Fold changes of some gene sets in the GX and OX strains. (A and B) Motility genes (A) and SPI-1 and SPI-2 genes (B). Expression changes in early exponential (EE) phase and in the transition to the stationary (TS) phase are shown. None of these gene sets were significantly differentially expressed in the GX strain. Note that some of the class 3 motility genes (fliC, fljA, and fljB) were differentially expressed due to phase switching in the GX and OX strains during their construction. The orientations of these genes in the constructed strains were different from those seen in the wild-type strain against which the gene expression changes were calculated, thus resulting in phase-variation-related gene expression changes. It is unlikely that these changes were related to our study. A general pattern of reversal in gene expression between the early exponential phase and the transition to the stationary phase was seen in the OX strain (this might have been related to Fis levels being higher in the stationary phase in this strain). The color bar indicates FDR, and bars representing genes with FDR values of <0.05 are red. Download FIG S8, EPS file, 0.7 MB (739.7KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Given the changes to SPI-1 and SPI-2 gene expression detected in the OX strain, this strain and the GX and WT strains were compared in an in vitro infection assay using cultured HeLa cells. The OX strain exhibited a strong reduction in infectivity at the 3.5-h time point, although not as strong as that seen with the fis knockout mutant control (Fig. 5C). In contrast, the GX strain showed a modest reduction in invasiveness compared to the WT at both the 3.5-h and the 6-h time points. The OX strain showed enhanced invasiveness at 6 h (Fig. 5C). The dps knockout mutant was indistinguishable from the wild type in this respect at either time point.
We also examined the transcriptomes of the two large plasmids in the GX, OX, and WT strains (Fig. S9). As with the chromosome, genes on the pSLT and pCol1B9 plasmids had shown changes in expression in the OX strain, but not in the GX strain, compared with the WT, indicating that the changes made to Fis production in OX had effects beyond the chromosome (Fig. S9). Notably, in the OX strain at the TS phase, we saw an upregulation of the tra genes on pCol1B9.
Transcriptomic changes on the Salmonella megaplasmids assayed by RNA-seq. Data represent Log2 fold changes in expression of genes along their positions on the plasmid in base pairs. The blue curve shows the local regression peak intensity change calculated using the R LOESS function. Significant gene expression changes (FDR < 0.05) are shown as red filled triangles. Download FIG S9, EPS file, 0.5 MB (500.6KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We have investigated the effect of repositioning the fis gene on the S. Typhimurium chromosome. This gene encodes the conditionally abundant nucleoid-associated protein Fis, which is present in over 50,000 copies in rapidly growing bacteria but in very few copies in stationary-phase cells (35). Previous work investigating the effects of regulatory gene translocation had used lacI, the gene encoding the transcriptional repressor, LacI, present in approximately 40 monomers per cell (6, 59). Fis has a much larger regulon than LacI, influencing the transcription of hundreds of genes, positively or negatively (19), and it plays a role in organizing the local structure of the nucleoid (60, 61). Its influence extends beyond transcription and includes effects on chromosome replication (26), transposition (28), and site-specific recombination (20, 22). We sought to investigate if, due to the pervasive influence of Fis, altering the location of the fis gene might produce significant changes to bacterial physiology. We were encouraged in our investigation by a previous study in which the translocation of the dusB-fis operon to the Ter region of the E. coli chromosome (without repositioning the dps gene) correlated with changes to the topology of a reporter plasmid and stress resistance, albeit without alterations to Fis protein levels in the cell (12). The lack of alteration in Fis protein levels was a consequence of an increase in fis expression, which compensated for the reduction in the fis gene copy number (12). In our study, the same effect was not observed in the GX strain: fis expression levels and Fis concentrations remained similar to those of the WT strain in the EE phase, despite the reduced fis gene dosage at this point in growth. Furthermore, the GX strain did not exhibit a growth defect whereas the E. coli strain with a translocated dusB-fis operon, described previously by Gerganova et al. (12), did. Together, these data indicate that other factors (including bacterial species differences) affect fis expression at a given location on the bacterial chromosome. Among these species-specific factors may be the known differences in DNA supercoiling set points that exist between E. coli and S. Typhimurium (62–64). The impact of Fis on the homeostatic control of DNA supercoiling, either through its influence on topoisomerase gene expression (36–39) or via direct Fis-mediated buffering of local DNA topology (31), may account for the changes to the transcriptome that we detected. In addition to dusB-fis translocation, in our experiments we also studied the effects of rewiring the fis gene so that it had the expression profile of dps, a gene with a growth-phase expression profile that is diametrically opposite that of fis (46, 58). By doing so, we were able to saturate the cell with Fis during stationary-phase growth, a period when Fis is normally undetectable. Similarly, we were able to reduce the production of Fis to a very low level in EE (early exponential) phase, an interval when the protein is normally very abundant.
DNA-binding regulators of transcription and of other molecular processes must communicate with their genomic targets by translocation through the cell, and some studies based on the LacI protein have suggested that the distance to be travelled influences the efficiency of the regulatory connection (65–67). There is some evidence that translated mRNA is spatially constrained in bacteria, remaining close to the site of transcription (68), while other work has suggested that the localization of mRNA is driven by the nature of the protein product and its cellular destination (69, 70). Collectively, these studies suggest that the physical location of a gene in the folded chromosome may determine the influence exerted by its protein product in the cell. A corollary to this proposition is that the repositioning of a gene may alter the sphere of influence of its product.
Our results obtained with the GX strain show that moving the entire dusB-fis operon to the right macrodomain site normally occupied by the dps gene had relatively modest effects on Salmonella physiology. This was true in terms of the growth kinetics of GX compared with the growth kinetics of the wild type (Fig. 1B to D), in the transcriptomic comparison (Fig. 4), and in comparisons of the GX strain and the wild type in terms of SPI-1 prgH virulence gene expression (Fig. 5A and B). In contrast, the activity of the SPI-2 virulence gene promoter PssaG was significantly reduced in GX compared to the wild type and GX exhibited a reduction in HeLa cell invasion (Fig. 5). Both the PprgH and the PssaG promoters were shown previously to have reduced activity in a fis knockout mutant of Salmonella, indicating a dependence on this NAP for normal function (19). Our new data show that altering the physical location of the fis gene influences PssaG negatively and is associated with a reduction in a virulence phenotype. In GX, the dusB-fis operon is moved from the nonstructured left region of the chromosome, where SPI-1 is also located, to the right macrodomain, adjacent to the right-hand segment of the Ter macrodomain where SPI-2 is found. The production of the Fis protein in GX increased at the transition to stationary-phase growth compared to the wild type (Fig. 2C), perhaps reflecting the homeostatic resetting of Fis levels reported previously when the dusB-fis operon was moved to a Ter-proximal site in E. coli (12). Looking in detail at the SPI-1 and SPI-2 pathogenicity islands, the SPI-2 gene cluster showed the greatest decrease in expression in the OX strain at the onset of stationary phase (see Fig. S8 in the supplemental material), which is consistent with the impact on virulence (Fig. 5C).
Overall, moving the fis gene to the native chromosomal location of dps and expressing fis using the regulatory elements normally used by dps in the OX strain had much more profound effects on Salmonella physiology than were seen in the GX strain. These effects included an extended lag phase in the OX strain during growth in batch culture (Fig. 1); a much more profound impact on the transcriptome, with many more genes in OX being affected than in GX and with a greater amplitude of positive and negative changes in transcription (Fig. 4); and a greater influence of the expression of key virulence genes and of a virulent phenotype (Fig. 5). The OX strain produced the Fis protein with a profile equivalent to that of Dps in the wild type and expressed Dps in a Fis-like manner (Fig. 2). In E. coli, producing Fis at high levels late in the growth cycle slows growth (71). In contrast, the Salmonella OX strain, with its extended lag phase, did not exhibit a growth defect later in the growth cycle (Fig. 1), indicating another point of difference with E. coli. Dps has a negligible impact on transcription (51), so the impact on gene expression in OX is likely to reflect differences in Fis protein levels, a proposition that is supported by the finding that many known Fis targets are among the genes showing altered expression.
Can the changes to Salmonella physiology in the GX and OX strains be explained by novel patterns of Fis distribution along the chromosome? The ensemble experiments conducted in this study relied on ChIP-seq to monitor Fis protein distributions during the early exponential phase of growth in living cells. The intensities of the protein binding peaks did change in the two rewired strains, especially in the vicinity of Ter, with OX showing by far the greater change compared to the wild type (Fig. 3C and D). This likely reflects the more radically altered pattern of Fis production in OX, where the transcription signals of the dps gene were driving the expression of fis (Fig. 1A). Changes to DNA topology caused by altered Fis production may also influence the Fis binding patterns that we detect in our strains (72), but the relative contributions of Fis abundance and target site topology are very difficult to tease apart in global surveys using ensemble experimental methods. The bacterium used in this study, S. Typhimurium strain SL1344, also harbors the large plasmids pSLT (a virulence plasmid) and pCol1B9. These autonomously replicating circular DNA molecules are independent of the chromosome, and they also contain Fis binding sites. The patterns of Fis binding to the two plasmids were generally similar in the GX, OX, and wild-type strains (Fig. S3). Plasmid gene expression was unaffected in the GX strain compared to the wild type, but statistically significant differences were detected in the OX strain (Fig. S9).
With the exception of H-NS (73, 74), no NAP in Salmonella is essential for the life of the bacterium, and this includes Fis and Dps. The contributions made by NAPs seem to be auxiliary to the functions that they influence. Our findings show that the Salmonella genome is well buffered against even quite radical changes to the production of Fis and Dps. The reciprocal inversion of their growth-phase-dependent production profiles, such that Fis assumes the pattern that is characteristic of Dps, and vice versa, does not seriously perturb the life of the cell, although the OX strain does exhibit the many physiological changes that have been documented here. Prominent among these are the effects on Salmonella plasmid and virulence gene expression and pathogenesis that we detected. The inversion of the SPI expression pattern in the OX strain relative to the WT (Fig. S8B) and the inversion of the pattern of invasion of this strain relative to the WT (Fig. 5C) demonstrate the role of the timing of Fis expression in controlling the normal timing of expression of these islands. These changes are likely to have their most telling effects when the bacterium is experiencing stress or is trying to compete with the wild type, compromising its ability to compete with the wild-type strain.
MATERIALS AND METHODS
Bacterial strains, growth media, and chemicals.
The bacterial strains used in this study and their genotypes and sources (where applicable) are listed in Table 1; all strains were derivatives of Salmonella enterica serovar Typhimurium strain SL1344. Unless otherwise stated, the bacterial strains were cultured in Miller lysogeny broth (LB; 1% NaCl). Kanamycin, carbenicillin, ampicillin, and chloramphenicol were used at concentrations of 50 μg/ml, 100 μg/ml, 100 μg/ml, and 35 μg/ml, respectively.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference(s) and/or description |
|---|---|---|
| SL1344 | rpsL hisG46 manXE95V menCL148L | 85, 101 |
| SL1344 fis::kan | fis::kan | Kanamycin resistance cassette inserted 63 bp downstream of fis ORF |
| SL1344 dps::cat | Pdps::cat | Chloramphenicol resistance cassette inserted 149 bp upstream of dps ORF |
| SL1344 2xfG | ΔPdpsdps::PfisdusBfis |
dps ORF and upstream/downstream regulatory regions replaced with dusB-fis operon and regulatory regions |
| SL1344 2xdG | ΔPfisdusBfis::Pdpsdps |
dusB and fis ORFs and upstream/downstream regulatory regions replaced with dps ORF and regulatory regions |
| SL1344 2xfO | Δdps::fis | dps ORF replaced with fis ORF |
| SL1344 2xdO | Δfis::dps | fis ORF replaced with dps |
| SL1344 Δfis | Δfis::cat | 19 |
| SL1344 Δdps | Δdps | dps ORF deletion |
| SL1344 GX | ΔPdpsdps::PfisdusBfis ΔPfisdusBfis::Pdpsdps | Exchange of dusB-fis and dps and their cis regulatory regions |
| SL1344 OX | Δdps::fis Δfis::dps | Exchange of fis and dps ORFs |
| SL1344 fis::8xmyc | fis::8xmyc | Deletion of the fis stop codon and insertion of 8 copies of the Myc epitope tag separated by a 21-bp linker |
| SL1344 GX fis::8xmyc | GX fis::8xmyc | Deletion of the fis stop codon and insertion of 8 copies of the Myc epitope tag separated by a 21-bp linker |
| SL1344 OX fis::8xmyc | OX fis::8xmyc | Deletion of the fis stop codon and insertion of 8 copies of the Myc epitope tag separated by a 21-bp linker |
| SL1344 PprgH | PprgH::gfp+::cat | GFP+ fused to SPI-1 promoter, PprgH (99) |
| SL1344 GX PprgH | PprgH::gfp+::cat | GFP+ fused to SPI-1 promoter, PprgH |
| SL1344 OX PprgH | PprgH::gfp+::cat | GFP+ fused to SPI-1 promoter, PprgH |
| SL1344 Δfis PprgH | PprgH::gfp+::cat | GFP+ fused to SPI-1 promoter, PprgH |
| SL1344 Δdps PprgH | PprgH::gfp+::cat | GFP+ fused to SPI-1 promoter, PprgH |
| SL1344 PssaG | PssaG::gfp+::cat | GFP+ fused to SPI-2 promoter, PssaG (99) |
| SL1344 GX PssaG | PssaG::gfp+::cat | GFP+ fused to SPI-2 promoter, PssaG |
| SL1344 OX PssaG | PssaG::gfp+::cat | GFP+ fused to SPI-2 promoter, PssaG |
| SL1344 Δfis PssaG | PssaG::gfp+::cat | GFP+ fused to SPI-2 promoter, PssaG |
| SL1344 Δdps PssaG | PssaG::gfp+::cat | GFP+ fused to SPI-2 promoter, PssaG |
| DH5α |
fhuA2 lacΔU169 phoA glnV44 Φ80' lacZΔM15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 |
102 |
| DH5α pBLUESCRIPT- dusB-dps-cat |
pBLUESCRIPTSK(+)::dusB::dps::cat | dusB-dps operon with cat antibiotic resistance gene |
Strain construction.
All strains generated during this study were constructed using phage λ Red recombinase-mediated homologous recombination (75) and P22 transduction (see summary schematic in Fig. S1 in the supplemental material). To construct the gene exchange (GX) and open reading frame exchange (OX) strains, first, the FLP recombination target (FRT)-flanked kanamycin resistance gene (kan), amplified from pKD4, was inserted 62 bp downstream of the fis ORF (SL1344 fis::kan) and the FRT-flanked chloramphenicol resistance gene (cat), amplified from pKD3, was inserted 190 bp upstream of the dps ORF (SL1344 dps::cat). These strains were used as templates to amplify either fis::kan ORF or the dusB-fis::kan and dps::cat transcriptional units. SLiCE (76) was used to generate a dusB-dps-cat construct on the plasmid pBluescript SK(+). This plasmid was used as a template to amplify the dusB-dps-cat transcriptional unit (where the fis ORF is replaced by the dps ORF). As previously described, these amplicons were used to replace the reciprocal gene, generating the 2× intermediate strains in the process that harbored two copies of either the fis or dps transcriptional units or ORFs (Table 1; see also Fig. S1). P22 phage-mediated generalized transduction was used to generate the GX and OX strains. Finally, the FRT-flanked antibiotic resistance genes were eliminated using a helper plasmid (pCP20) encoding FLP recombinase.
Strain variants of SL1344 and the exchange strains expressing 8× Myc-epitope-tagged Fis were constructed using a modified version of phage λ Red recombinase-mediated recombination (75).
The DNA oligonucleotides used in this study are listed in Table S1 in the supplemental material.
List of oligonucleotides used in this study. The nucleotide sequences are written 5′ to 3′. Download Table S1, DOCX file, 0.1 MB (96.2KB, docx) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial growth measurement.
Bacterial cell density was measured using absorbance and viable counts.
Absorbance was measured, using a Thermo Scientific BioMate 3S spectrophotometer, at 600 nm (A600) using sterile LB for blank correction. Cell densities with A600 values greater than 1 were diluted 1:10 in sterile LB before measurement.
Viable counts were performed by plating serial 10-fold dilutions of the culture made in sterile phosphate-buffered saline (PBS) on sterile LB agar plates. Colonies obtained after overnight incubation at 37°C were counted and multiplied by the appropriate dilution factors to give the viable cell density in terms of CFU.
Overnight cultures were grown for at least 16 h in 4 ml LB in tubes maintained at an angle at 37°C with 200-rpm orbital shaking. For performing growth experiments, overnight cultures were diluted to an A600 of 0.003 in 25 ml LB in 250-ml conical flasks. These were incubated at 37°C with 200 rpm orbital shaking.
For all experiments, the bacterial cultures were harvested at the early exponential (EE; A600 of 0.4) phase, transition-to-stationary (TS; A600 of 3) phase, or late stationary (LS; A600 of 3.5, 24 h from inoculation) phase of growth.
Estimating gene dosages using quantitative reverse transcription-PCR (RT-qPCR).
Bacterial chromosomal DNA was isolated by harvesting 1 ml of cultures grown to the EE, TS, and LS phases of growth by centrifugation at 16,000 × g for 1 min. The cell pellet was resuspended in sterile distilled water, boiled at 100°C for 5 min, and subjected to vortex mixing, and cell debris was pelleted by centrifugation again. The supernatant was mixed with 1 volume of chloroform, and subjected to vortex mixing. The upper aqueous layer was isolated by centrifugation at 16,000 × g for 10 min. The concentration was determined using a DeNovix DS-11 spectrophotometer using A260 and adjusted to 100 ng/μl with nuclease-free water.
The genomic DNA (gDNA) was probed using primers specific to target genes in a StepOnePlus real-time PCR system. Each 20-μl reaction mixture, set up in the wells of a 96-well plate, contained 1× FastStart Universal SYBR green Master (Rox) (Merck, Wicklow, Ireland), 0.6 μM concentrations of a primer pair, 8 μl gDNA, and sterile distilled water added to reach a volume of 20 μl. For each primer pair, a standard curve of 10-fold serially diluted SL1344 gDNA was included. Threshold cycle (CT) values of gDNA samples were checked against the standard curves to ensure that they fell in the linear range for each primer pair, and the concentrations of gDNA in the samples were estimated from the standard curve. Cycle conditions for qPCRs were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Oligonucleotide primers were designed against an oriC (gidA) proximal gene and a Ter proximal gene (STM1554). To determine the gene dosage gradient, the ratio of oriC-proximal DNA to Ter-proximal DNA was identified. Specific gene dosages of fis and dps were determined by comparing the quantities of these genes to that of STM1554.
Estimating gene expression changes using quantitative reverse transcription-PCR (RT-qPCR).
At optical densities of 0.1, 0.4, 1, and 3, further growth was halted and intracellular RNA was stabilized by the addition of a 0.4 volume of stop solution (5% [vol/vol] phenol [pH 4.3]–ethanol) and incubation on ice for 30 min. A volume of cells equivalent to a cell density of 1 ml of culture at an A600 of 1 was harvested. Cells were pelleted by centrifugation at 3,220 × g for 10 min at 4°C. Cells were resuspended in Tris-EDTA buffer (TE; pH 8.0) containing 50 mg/ml lysozyme. RNA was extracted using an SV total RNA isolation kit (Promega, WI, USA). RNA was DNase treated using a Turbo DNase kit (Invitrogen). RNA integrity was assessed on a HT gel (77) and quantified using a DeNovix DS-11 spectrophotometer and A260.
RNA (400 ng) was reverse transcribed into cDNA using random oligonucleotides and a GoScript reverse transcriptase kit (Promega). The cDNA was probed using primers specific to target genes in the StepOnePlus real-time PCR system as described above. The hemX gene was used as a control gene (with unchanged expression assumed), and the changes in expression of the other genes were calculated against hemX expression.
The oligonucleotide primer pairs used in qPCR are listed in Table S1.
Estimating protein levels using immunoblotting.
At the EE phase or TS phase of growth, a volume of cells equivalent to a cell density of 1 ml of culture with an A600 of 1 was harvested and resuspended in 350 μl PBS and transferred to a sonication tube. Cells were lysed by sonication with 10 cycles, with each cycle consisting of 10-s bursts (10-μm amplitude) followed by 30 s of incubation on ice. Samples were diluted with equal volumes of 2× Laemmli sample buffer (final concentrations, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, and 0.125 M Tris HCl, pH 6.8) and heated at 100°C for 5 min prior to loading on a 12.5% SDS-PAGE gel.
Proteins were separated by electrophoresis in running buffer (25 mM Tris-HCl, 190 mM glycine, 0.1% [wt/vol] SDS) at 60 V through the stacking gel, which increased to 130 V as the dye moved through the resolving gel until the dye reached the base of the gel, and the proteins were then transferred onto methanol-activated polyvinylidene difluoride (PVDF) membranes (0.2-μm pore size) in transfer buffer (20% [vol/vol] methanol, 25 mM Tris, 190 mM glycine) at 300 mA for 1 h 45 min using a Trans-Blot apparatus (Bio-Rad).
Blocking of the membrane was performed for 1 h at room temperature with a blocking solution (5% skimmed milk powder, phosphate-buffered saline–0.1% Tween 20 [PBST]) and with rocking. Membranes were probed overnight at 4°C using the following primary antibody concentrations: 1:50,000 mouse anti-DnaK monoclonal antibody (Abcam; ab69617), 1:50,000 mouse anti-C-Myc monoclonal antibody (Sigma; M4439), and 1:2,000 rabbit anti-Dps polyclonal serum (Regine Hengge, Humboldt Universitat zu Berlin). All antibodies were diluted to the appropriate concentration in PBST containing 5% bovine serum albumin (BSA). Membranes were washed with PBST for 5 min each while rocking. Horseradish peroxidase (HRP)-conjugated secondary antibodies were diluted 1:10,000 and 1:2,000 for polyclonal goat anti-mouse (Bio-Rad; 170-6516) and goat anti-rabbit HRP (Dako; P0448), respectively. Secondary antibodies were diluted to the appropriate concentration in PBST containing 5% skimmed milk powder (Lab M). Membranes were incubated with secondary antibodies for 1 h 30 min at room temperature while rocking. Membranes were then washed three times with PBST and once with PBS. Blots were incubated in ECL reagent (Pierce) for 1 min, and bands were visualized using an ImageQuant LAS 4000 scanner (GE Healthcare).
Whole-genome sequencing (WGS) and single nucleotide polymorphism (SNP) analysis to check constructed strains.
(i) DNA extraction. Chromosomal DNA for whole-genome sequencing was prepared by a phenol-chloroform method which caused minimal shearing to DNA. Briefly, 2 ml of an overnight culture was harvested by centrifugation at 16,000 × g and the cell pellet was resuspended in TE (pH 8.0) (100 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0]). Lysis was performed by the addition of 1% (wt/vol) SDS and 2 mg/ml proteinase K and incubation at 37°C for 1 h. DNA was isolated by the addition of 1 volume of phenol (pH 8.0):chloroform:isoamyl alcohol (25:24:1), followed by centrifugation at 16,000 × g to separate the aqueous and organic layers. Following this, 1 volume of chloroform was added to the aqueous layer, and centrifugation was repeated. To remove contaminants, 0.3 M sodium acetate (pH 5.2) and 5 volumes of 100% (vol/vol) ethanol were added followed by precipitation at −20°C for 1 h. The DNA was pelleted by centrifugation, washed once with 70% (vol/vol) ethanol, dried at 65°C for up to 5 min, and resuspended in 50 μl sterile distilled water. Total gDNA was treated with 100 mg/ml RNase A at 37°C for 30 min. Following this, DNA was isolated by the addition of 1 volume of phenol (pH 8.0):chloroform:isoamyl alcohol as described above.
(ii) Library preparation. Library preparation and sequencing for all strains except OX and 2xdO were performed by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1). DNA was quantified in triplicate with a Quant-iT dsDNA HS (double-stranded DNA high-sensitivity) assay kit in an Eppendorf AF2200 plate reader. Genomic DNA libraries were prepared using a Nextera XT library prep kit (Illumina, San Diego, USA) following the manufacturer’s protocol with the following modifications: 2 ng of DNA was used as the input instead of 1 ng, and PCR elongation time was increased to 1 min from 30 s. DNA quantification and library preparation were carried out on a Hamilton Microlab Star automated liquid-handling system. Pooled libraries were quantified using a Kapa Biosystems library quantification kit for Illumina on a Roche LightCycler 96 qPCR machine. Libraries were sequenced on an Illumina HiSeq sequencer using a 250-bp paired-end protocol. Reads were subjected to adapter trimming using Trimmomatic 0.30 with a sliding-window quality cutoff of Q15 (78). De novo assembly was performed on samples using SPAdes version 3.7 (79), and contigs were annotated using Prokka 1.11 (80).
For the OX and 2xdO strains, library preparation was performed by the Sanger Institute (Hinxton, United Kingdom). Samples were quantified with a Biotium AccuClear ultra-high-sensitivity dsDNA quantitative kit using a Mosquito LV liquid-handling platform, an Agilent Bravo WS automation system, and a BMG FLUOstar Omega plate reader and were subjected to cherry picking to achieve 200 ng/120 μl using a Tecan liquid-handling platform. Cherry-picked plates were sheared to 450 bp using a Covaris LE220 instrument. After shearing, the samples were purified using Agencourt AMPure XP solid-phase reversible immobilization (SPRI) beads on an Agilent Bravo WS platform. Library construction (end repair [ER], A-tailing, and ligation) was performed using a NEB Ultra II custom kit on an Agilent Bravo WS automation system. The PCR system was set up using Kapa HiFi Hot Start mix and IDT 96 iPCR tag barcodes on Agilent Bravo WS automation system. The PCR cycles (6 standard cycles) were performed as follows: (i) 95°C for 5 min, (ii) 98°C for 30 s, (iii) 65°C for 30 s, (iv) 72°C for 1 min, (v) five repeats of cycles ii to iv, and (vi) incubation at 72°C for 10 min, followed by post-PCR plate purification using Agencourt AMPure XP SPRI beads on a Beckman BioMek NX96 liquid-handling platform. Libraries were quantified with a Biotium AccuClear ultra-high-sensitivity dsDNA quantitative kit using a Mosquito LV liquid-handling platform, a Bravo WS platform, and a BMG FLUOstar Omega plate reader. Libraries were pooled in equimolar amounts on a Beckman BioMek NX-8 liquid-handling platform. Libraries were normalized to 2.8 nM such that they were ready for cluster generation on a c-Bot cluster generation system and loading on an Illumina sequencing platform.
(iii) SNP analysis. SNP analysis was performed using Breseq (81–83). This is a pipeline that performs all necessary functions, including mapping reads using Bowtie2 (84) and calling SNPs and other genomic variations.
The SL1344 parental strain used in this study has two previously described SNPs, in manX (E95V) and menC (L148L), compared to the reference genome (85) (Fig. S2). While no additional SNPs were identified in the WT strain or the GX strain, the OX strain contained additional SNPs in ybiH (V173L) and SL1344_3357 (C260C) and in the intergenic region between SL1344_3765 and emrD (Fig. S2). None of the genes with SNPs were found to interact with either Fis or Dps by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis (86).
Revealing transcriptomic changes with RNA-seq.
(i) RNA extraction. At the EE phase or TS phase of growth, cells were harvested as described for RT-qPCR. The bacterial pellet was dissolved in TE buffer (pH 8.0) (100 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0]) containing 0.5 mg/ml lysozyme. Lysis was carried out with 1% SDS and 0.1 mg/ml proteinase K during incubation at 40°C for 20 min. RNA was isolated by the addition of 0.3 M sodium acetate (pH 6.5) and 1 volume of phenol (pH 4.3):chloroform (1:1), followed by centrifugation (16,000 × g, 4°C, 10 min) in a phase lock tube (Quantabio, VWR) to separate the aqueous and organic phases. Following this, 1 volume of chloroform was added to the aqueous layer and centrifugation (16,000 × g, 4°C, 10 min) was repeated in the same phase lock tube. RNA was precipitated by the addition of 5 volumes of 100% ethanol and incubation at −20°C for 1 h. RNA was pelleted by centrifugation (16,000 × g, 4°C, 10 min), washed once with 70% ethanol, and resuspended in 50 μl diethyl pyrocarbonate (DEPC)-treated water. RNA was diluted to 500 ng/μl, denatured at 65°C for 5 min, and treated with 10 U RNase-free DNase I (Thermo Fisher Scientific, Waltham, USA) in DNase 1 buffer at 37°C for 40 min. DNase-treated RNA was cleaned up using the phenol chloroform method again. RNA integrity was assessed as described in the paragraph under the RT-qPCR heading.
(ii) Library preparation. Strand-specific library preparation and sequencing of the DNase-treated RNA was performed by Vertis Biotechnologie AG (Freising-Weihenstefan, Germany). Briefly, samples were analyzed by capillary electrophoresis (Bioanalyzer, Agilent), and rRNA was depleted using a bacterial Ribo-Zero rRNA removal kit (Illumina). Ribodepleted RNA samples were fragmented using ultrasound (four 30-s pulses at 4°C). Oligonucleotide sequencing adapters were ligated to the 3′ end of each specific strand of the RNA molecules. First-strand cDNA synthesis was performed using Moloney murine leukemia virus (M-MLV) reverse transcriptase and the 3′ adapter as the primer. The first-strand cDNA was purified, and the 5′ Illumina TruSeq sequencing adapter was ligated to the 3′ end of the antisense cDNA in a strand-specific manner. The resulting cDNA was PCR amplified to about 10 to 20 ng/μl using a high-fidelity DNA polymerase. The cDNA was purified using an Agencourt AMPure XP kit (Beckman Coulter Genomics) and was analyzed by capillary electrophoresis (Bioanalyzer; Agilent). For Illumina NextSeq sequencing, the samples were pooled in approximately equimolar amounts. The cDNA pool was subjected to size fractionation in the size range of 200 to 550 bp using a differential cleanup method with the Agencourt AMPure kit. An aliquot of the size-fractionated pool was analyzed by capillary electrophoresis. Samples were PCR amplified for Truseq according to the instructions of Illumina. For the WT and GX strains, the cDNA pool was subjected to paired-end sequencing on an Illumina NextSeq 500 system using 2-by-75-bp read lengths. For the OX strain, the cDNA pool was subjected to single-end sequencing on an Illumina NextSeq 500 system using 1-by-75-bp read lengths.
(iii) Sequencing alignment. Raw read sequence qualities were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). BWA (87) was used to align reads to the Salmonella enterica subsp. enterica serovar Typhimurium strain SL1344 chromosome and plasmid reference sequences (chromosome, NC_016810.1; pCol1B9_SL1344, NC_017718.1; pRSF1010_SL1344, NC_017719.1, pSLT_SL1344, NC_017720.1). Aligned reads were sorted by the reference sequence coordinate, and low-mapping-quality reads (mapping quality < 30) were removed using SAMtools (88). SAMtools was also used to summarize mapping statistics. More than 90% of the reads were mapped, and >98% of the mapped reads were unique. Sequence coverages were calculated using Deeptools (89) and are available as BigWig (.bw) files that can be viewed using IGB (90).
(iv) Differential expression analysis. Counts of the number of reads mapping to genomic features (genes or small RNA [sRNA]) were obtained using the R package Rsubread (91). The R package EdgeR (92, 93) was used to identify differentially expressed genes. P values were adjusted using the Benjamini Hochberg method (false-discovery rate [FDR]), and a cutoff of 0.05 was used to call differentially expressed genes. These are available in an Excel file on GEO.
Revealing Fis binding changes with ChIP-seq.
(i) ChIP (chromatin immunoprecipitation). For all ChIP experiments, fis::8xmyc epitope-tagged variants of the WT, GX, and OX strains were used (72). Overnight cultures were diluted to a starting A600 of 0.003 in 25 ml LB broth and grown to an A600 of 0.4. Cells were harvested by centrifugation at 3,220 × g for 10 min at room temperature and resuspended in 50 ml PBS. Formaldehyde was added dropwise to reach a final concentration of 1% (vol/vol) while the cells were stirred continuously. After 10 min of cross-linking, the reaction was quenched with cold glycine (2 M) added dropwise to reach a final concentration of 0.125 M. Cells were stirred further for 5 min and then harvested by centrifugation at 3,220 × g for 10 min at 4°C. The pellet was suspended in 600 μl lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA [pH 8.0], 1% [wt/vol] SDS, Roche protease inhibitor cocktail) and incubated for 10 min on ice prior to the addition of 1.2 ml IP dilution buffer (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA [pH 8.0], 1% [vol/vol] Triton X-100, 0.01% [wt/vol] SDS, Roche protease inhibitor cocktail) and transferred to a sonication tube. Chromatin was sheared to an average length of 500 bp by sonication at an amplitude of 10 μm 12 times for 30 s each time, with 1 min of incubation on ice between bursts, using a Soniprep 150 instrument. Sheared chromatin was analyzed by agarose gel electrophoresis and stored at −80°C. To preclear chromatin for input samples, 50 μl normal rabbit IgG (Millipore, Cork, Ireland) was added and samples were incubated for 1 h on a model SB1 blood tube rotator at 4°C. Antibody was removed by the addition of 100 μl homogeneous protein G agarose (Roche) and further incubation for 3 h. The protein G agarose was removed by centrifugation at 3,220 × g for 2 min, and 200 μl was used as the input. For all ChIP experimentsm a mock-IP system and an experimental IP system were set up at the same time using 1,350 μl precleared chromatin and 10 μg of normal mouse IgG (Millipore) and monoclonal mouse anti-c-Myc, respectively. Samples were incubated at 4°C for 16 h on a model SB1 blood tube rotator. A 100-μl volume of homogeneous protein G agarose was added to each sample, and the samples were incubated for a further 3 h. Protein G agarose beads were pelleted by centrifugation at 5,200 × g and washed 4 times with high-salt IP wash buffer (50 mM HEPES [pH 7.9], 500 mM NaCl, 1 mM EDTA, 0.1% [wt/vol] SDS, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] deoxycholate) and twice with TE (pH 8.0). Protein-DNA complexes were eluted from the beads twice with IP elution buffer (100 mM NaHCO3, 1% [wt/vol] SDS).
DNA was purified and recovered by performing a standard phenol‐chloroform extraction, followed by ethanol precipitation with 5 μg of glycogen (Invitrogen; catalog no. 10814‐010). The DNA pellets of the IP samples were resuspended (by heating at 37°C) in 50 μl of sterile filtered water for experimental and mock IPs and in 100 μl for the input DNA samples.
(ii) Library preparation. ChIP sequencing was performed by Vertis Biotechnologie AG using Illumina NextSeq 500 technology. The DNA samples were first fragmented with ultrasound (2 pulses of 30 s at 4°C). After end repair was performed, TruSeq sequencing adapters were ligated to the DNA fragments. Finally, the DNA was PCR amplified to about 10 to 20 ng/μl using a high-fidelity DNA polymerase. Aliquots of the PCR-amplified libraries were examined by capillary electrophoresis (Bioanalyzer; Agilent). For Illumina NextSeq sequencing, the samples were pooled in approximately equimolar amounts. The DNA pool was eluted in the size range of 250 to 550 bp from a preparative agarose gel. An aliquot of the size-fractionated library pool was analyzed by capillary electrophoresis (Bioanalyzer; Agilent). The adapters were designed for TruSeq sequencing according to the instructions of Illumina. The shotgun library pool was sequenced on an Illumina NextSeq 500 system using 75-bp read length.
(iii) Sequencing alignment. See RNA-seq analysis above.
(iv) Peak calling. Since we were performing differential binding analysis, we did not exclude duplicate reads as they contain information regarding the binding intensity of the transcription factor being pulled down. MACS2 (94) was used to call peaks against the mock treatment. Each of the strains (WT, GX, and OX) had its own mock sample. We used a custom R script to roughly annotate peaks with the names of neighboring genes and sRNA. These lists are available as both Excel and .gff files. The sequences of Fis peaks from the WT were used to search for motifs using unbiased MEME (95). This Fis motif was then used to search for all possible motifs in the genome, bound or not in the ChIP, using FIMO software (96). This list is also available as a. gff file.
(v) Differential binding analysis. Differential binding analysis was performed in R (97) using the DiffBind package (98). To correct for differences in gene dosage gradient between strains, mock treatment samples derived from each strain were used as a control. The output of the differential binding analysis is available (both with and without applying the FDR cutoff) as Excel files.
Salmonella pathogenicity island expression analyses using Pgfp+ reporter fusions.
SPI-1 (PprgH-gfp+) and SPI-2 (PssaG-gfp+) promoter fusions were integrated into the SL1344 chromosome by transduction by bacteriophage P22 and were selected for on LB agar plates supplemented with 25 μg/ml chloramphenicol (99). Overnight cultures of SPI-1 and SPI-2 reporter fusion strains were diluted 1:100 in LB. These were transferred to the wells of a black 96-well plate with a flat and transparent bottom (Corning, Fisher Scientific) in 4 to 6 technical replicates. The plate was incubated at 37°C for 24 h with 300-rpm (5-mm) shaking in a Synergy H1 microplate reader (Biotek, VT, USA). A600 and fluorescence (excitation, 485-nm wavelength; emission, 528-nm wavelength) were measured every 20 min. Data were obtained from multiple biological replicates.
Epithelial cell invasion assay.
The gentamicin protection assay was used as described previously (100) to determine Salmonella invasion. HeLa cells (ATCC) were maintained by passaging in complete growth medium (CGM; minimum essential Eagle medium supplemented with 10% [vol/vol] fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate) every 3 to 4 days (37°C, 5% CO2). HeLa cells were seeded 20 to 24 h prior to invasion assay in the wells of a 24-well tissue culture plate at a density of ∼5 × 104 cells in 1 ml CGM.
Overnight cultures (16 h of growth at 37°C, 200 rpm) of the Salmonella WT, GX, OX, Δfis, and Δdps strains were diluted 1:33 in 10 ml LB in a 125-ml flask and grown for 3.5 h (early time point) or 6 h (late time point) at 37°C and 200 rpm. Cells were pelleted by centrifugation at 8,000 × g for 2 min. A 900-μl volume of the supernatant was discarded without disturbing the pellet and then resuspended in 900 μl Hanks’ buffered saline solution (HBSS−/−). Cells were diluted 1:10 in HBSS−/− and then diluted in CGM to form the infection mixture that was used to infect the HeLa cells at a multiplicity of infection (MOI) of 50 with two replicates per strain. No more than three strains were tested at a time, with the WT always being tested as the reference strain. The infection was allowed to proceed for 10 min at 37°C and 5% CO2, following which the infection mixture was discarded by rapid aspiration. Cells were washed rapidly with HBSS supplemented with calcium chloride and magnesium chloride (HBSS+/+) twice to get rid of extracellular bacteria and were covered with 1 ml CGM. These were incubated at 37°C and 5% CO2 for 20 min and then again washed rapidly with HBSS+/+ twice and covered with 1 ml CGM supplemented with 50 μg/ml gentamicin to kill any remaining extracellular bacteria. After 1 h of incubation at 37°C and 5% CO2, cells were washed rapidly with phosphate-buffered saline (PBS) twice and lysed with 1 ml 2% (wt/vol) sodium deoxycholate–PBS.
Viable counts of the subculture, infection mixture, and lysed cells were obtained. Percent invasion was obtained by calculating the percentage of bacteria in the infection mixture that could invade the cells relative to the WT (counts of intracellular bacteria were calculated relative to the counts of the infection mix to control for variations in infection load and then relative to the wild type to control for day-to-day variation). The one-sample t test was used to assess significance relative to the wild type (μ = 100%, 100% being the percentage of invasion of the WT), and the two-sample Welch t test was used to assess the significance of pairwise comparisons with α = 0.05. The P values from the two-sample tests were corrected for multiple testing using the BH method (FDR).
Data availability.
RNA-seq and ChIP-seq data can be found on the Gene Expression Omnibus (accession number GSE152228). WGS data can be found on the Sequence Read Archive (accession number PRJNA638833). WGS data for strains OX and 2xdO can be found on the European Nucleotide Archive (accession numbers ERS2515905 and ERS4653304, respectively). Scripts used in the analysis can be found on GitLab (project identifier 10206830 [https://gitlab.com/aalap.mogre/chip-seq-and-rna-seq-in-salmonella-swap-strains]).
ACKNOWLEDGMENTS
We thank Regine Hengge (Humboldt-Universität zu Berlin) for providing the rabbit anti-Dps polyclonal serum. We thank Carsten Kröger (Trinity College Dublin) and Jay Hinton (University of Liverpool) for providing the PprgH-gfp+ and PssaG-gfp+ reporter strains. We thank Ciaran Finn (NIH, Trinity College Dublin) and Olivia Steele-Mortimer (NIH) for providing the HeLa cells and helping us with the Salmonella invasion assay protocol. We are grateful to Matthew Dorman for assistance with depositing whole-genome sequence data in the European Nucleotide Archive.
Research in Charles J. Dorman’s laboratory is supported by Principal Investigator Award 13/1A/1875 from Science Foundation Ireland. Whole-genome sequencing at the Wellcome Sanger Institute was supported by grant 206194 from the Wellcome Trust.
We declare that we have no competing interests.
Footnotes
This article is a direct contribution from Charles J. Dorman, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by David Grainger, University of Birmingham, and Georgi Muskhelishvili, Agricultural University of Georgia.
Citation Bogue MM, Mogre A, Beckett MC, Thomson NR, Dorman CJ. 2020. Network rewiring: physiological consequences of reciprocally exchanging the physical locations and growth-phase-dependent expression patterns of the Salmonella fis and dps genes. mBio 11:e02128-20. https://doi.org/10.1128/mBio.02128-20.
REFERENCES
- 1.Browning DF, Butala M, Busby SJW. 2019. Bacterial transcription factors: regulation by Pick “N” Mix. J Mol Biol 431:4067–4077. doi: 10.1016/j.jmb.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Browning DF, Busby SJW. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 3.Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 5.Dorman CJ, Schumacher MA, Bush MJ, Brennan RG, Buttner MJ. 2020. When is a transcription factor a NAP? Curr Opin Microbiol 55:26–33. doi: 10.1016/j.mib.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garza de Leon F, Sellars L, Stracy M, Busby SJW, Kapanidis AN. 2017. Tracking low-copy transcription factors in living bacteria: the case of the lac repressor. Biophys J 112:1316–1327. doi: 10.1016/j.bpj.2017.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elf J, Li G-W, Xie XS. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci U S A 109:E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darling AE, Miklós I, Ragan MA. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet 4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campo N, Dias MJ, Daveran-Mingot M-L, Ritzenthaler P, Le Bourgeois P. 2004. Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol Microbiol 51:511–522. doi: 10.1046/j.1365-2958.2003.03847.x. [DOI] [PubMed] [Google Scholar]
- 11.Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S. 2001. Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biol 2:INTERACTIONS1004. doi: 10.1186/gb-2001-2-12-interactions1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerganova V, Berger M, Zaldastanishvili E, Sobetzko P, Lafon C, Mourez M, Travers A, Muskhelishvili G. 2015. Chromosomal position shift of a regulatory gene alters the bacterial phenotype. Nucleic Acids Res 43:8215–8226. doi: 10.1093/nar/gkv709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 14.Dorman CJ. 2014. Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation. J Mol Microbiol Biotechnol 24:316–331. doi: 10.1159/000368850. [DOI] [PubMed] [Google Scholar]
- 15.Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, Mavathur R, Muskhelishvili G, Pemberton I, Schneider R, Travers A. 2003. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J Mol Biol 331:331–344. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 16.Bokal AJ, Ross W, Gaal T, Johnson RC, Gourse RL. 1997. Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J 16:154–162. doi: 10.1093/emboj/16.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho B-K, Knight EM, Barrett CL, Palsson BØ. 2008. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res 18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahramanoglou C, Seshasayee ASN, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res 39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JCD, Dorman CJ. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RC, Bruist MF, Simon MI. 1986. Host protein requirements for in vitro site-specific DNA inversion. Cell 46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 21.Koch C, Kahmann R. 1986. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem 261:15673–15678. [PubMed] [Google Scholar]
- 22.Papagiannis CV, Sam MD, Abbani MA, Yoo D, Cascio D, Clubb RT, Johnson RC. 2007. Fis targets assembly of the Xis nucleoprotein filament to promote excisive recombination by phage lambda. J Mol Biol 367:328–343. doi: 10.1016/j.jmb.2006.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gille H, Egan JB, Roth A, Messer W. 1991. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res 19:4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasho K, Fujimitsu K, Matoba T, Oshima T, Katayama T. 2014. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res 42:13134–13149. doi: 10.1093/nar/gku1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao P, Rozgaja TA, Alqahtani A, Grimwade JE, Leonard AC. 2018. Low affinity DnaA-ATP recognition sites in E. coli oriC make non-equivalent and growth rate-dependent contributions to the regulated timing of chromosome replication. Front Microbiol 9:1673. doi: 10.3389/fmicb.2018.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC. 2004. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol Microbiol 51:1347–1359. doi: 10.1046/j.1365-2958.2003.03906.x. [DOI] [PubMed] [Google Scholar]
- 27.van Drunen CM, van Zuylen C, Mientjes EJ, Goosen N, van de Putte P. 1993. Inhibition of bacteriophage Mu transposition by Mu repressor and Fis. Mol Microbiol 10:293–298. doi: 10.1111/j.1365-2958.1993.tb01955.x. [DOI] [PubMed] [Google Scholar]
- 28.Weinreich MD, Reznikoff WS. 1992. Fis plays a role in Tn5 and IS50 transposition. J Bacteriol 174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbani MA, Papagiannis CV, Sam MD, Cascio D, Johnson RC, Clubb RT. 2007. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc Natl Acad Sci U S A 104:2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball CA, Johnson RC. 1991. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol 173:4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider R, Travers A, Muskhelishvili G. 1997. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol 26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 32.Duprey A, Muskhelishvili G, Reverchon S, Nasser W. 2016. Temporal control of Dickeya dadantii main virulence gene expression by growth phase-dependent alteration of regulatory nucleoprotein complexes. Biochim Biophys Acta 1859:1470–1480. doi: 10.1016/j.bbagrm.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol 51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 34.Lv M, Chen Y, Liao L, Liang Z, Shi Z, Tang Y, Ye S, Zhou J, Zhang L. 2018. Fis is a global regulator critical for modulation of virulence factor production and pathogenicity of Dickeya zeae. Sci Rep 8:341. doi: 10.1038/s41598-017-18578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ball CA, Osuna R, Ferguson KC, Johnson RC. 1992. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol 174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. 2006. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep 7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keane OM, Dorman CJ. 2003. The gyr genes of Salmonella enterica serovar Typhimurium are repressed by the factor for inversion stimulation, Fis. Mol Genet Genomics 270:56–65. doi: 10.1007/s00438-003-0896-1. [DOI] [PubMed] [Google Scholar]
- 38.Schneider R, Travers A, Kutateladze T, Muskhelishvili G. 1999. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol Microbiol 34:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein-Fischer D, Altuvia S. 2007. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol 63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 40.Crozat E, Winkworth C, Gaffé J, Hallin PF, Riley MA, Lenski RE, Schneider D. 2010. Parallel genetic and phenotypic evolution of DNA superhelicity in experimental populations of Escherichia coli. Mol Biol Evol 27:2113–2128. doi: 10.1093/molbev/msq099. [DOI] [PubMed] [Google Scholar]
- 41.Morett E, Bork P. 1998. Evolution of new protein function: recombinational enhancer Fis originated by horizontal gene transfer from the transcriptional regulator NtrC. FEBS Lett 433:108–112. doi: 10.1016/s0014-5793(98)00888-6. [DOI] [PubMed] [Google Scholar]
- 42.Schneider R, Travers A, Muskhelishvili G. 2000. The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol Microbiol 38:167–175. doi: 10.1046/j.1365-2958.2000.02129.x. [DOI] [PubMed] [Google Scholar]
- 43.Ninnemann O, Koch C, Kahmann R. 1992. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J 11:1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nafissi M, Chau J, Xu J, Johnson RC. 2012. Robust translation of the nucleoid protein Fis requires a remote upstream AU element and is enhanced by RNA secondary structure. J Bacteriol 194:2458–2469. doi: 10.1128/JB.00053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osuna R, Lienau D, Hughes KT, Johnson RC. 1995. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol 177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181:6361–6370. doi: 10.1128/JB.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron ADS, Kröger C, Quinn HJ, Scally IK, Daly AJ, Kary SC, Dorman CJ. 2013. Transmission of an oxygen availability signal at the Salmonella enterica serovar Typhimurium fis promoter. PLoS One 8:e84382. doi: 10.1371/journal.pone.0084382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O Cróinín T, Dorman CJ. 2007. Expression of the Fis protein is sustained in late-exponential- and stationary-phase cultures of Salmonella enterica serovar Typhimurium grown in the absence of aeration. Mol Microbiol 66:237–251. doi: 10.1111/j.1365-2958.2007.05916.x. [DOI] [PubMed] [Google Scholar]
- 49.Almirón M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 50.Antipov SS, Tutukina MN, Preobrazhenskaya EV, Kondrashov FA, Patrushev MV, Toshchakov SV, Dominova I, Shvyreva US, Vrublevskaya VV, Morenkov OS, Sukharicheva NA, Panyukov VV, Ozoline ON. 2017. The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner. PLoS One 12:e0182800. doi: 10.1371/journal.pone.0182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janissen R, Arens MMA, Vtyurina NN, Rivai Z, Sunday ND, Eslami-Mossallam B, Gritsenko AA, Laan L, de Ridder D, Artsimovitch I, Dekker NH, Abbondanzieri EA, Meyer AS. 2018. Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell 174:1188–1199.e14. doi: 10.1016/j.cell.2018.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karas VO, Westerlaken I, Meyer AS. 2015. The DNA-binding protein from starved cells (Dps) utilizes dual functions to defend cells against multiple stresses. J Bacteriol 197:3206–3215. doi: 10.1128/JB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer AS, Grainger DC. 2013. The Escherichia coli nucleoid in stationary phase. Adv Appl Microbiol 83:69–86. doi: 10.1016/B978-0-12-407678-5.00002-7. [DOI] [PubMed] [Google Scholar]
- 54.Nair S, Finkel SE. 2004. Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankey JT. 2008. The Dps protein of Salmonella enterica serovar Typhimurium. Thesis Trinity College, Dublin, Ireland. [Google Scholar]
- 56.Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 57.Jeong K-C, Baumler DJ, Kaspar CW. 2006. dps expression in Escherichia coli O157:H7 requires an extended −10 region and is affected by the cAMP receptor protein. Biochim Biophys Acta 1759:51–59. doi: 10.1016/j.bbaexp.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Grainger DC, Goldberg MD, Lee DJ, Busby SJW. 2008. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol Microbiol 68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert W, Müller-Hill B. 1966. Isolation of the lac repressor. Proc Natl Acad Sci U S A 56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 61.Schneider R, Lurz R, Lüder G, Tolksdorf C, Travers A, Muskhelishvili G. 2001. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res 29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Champion K, Higgins NP. 2007. Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J Bacteriol 189:5839–5849. doi: 10.1128/JB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cameron ADS, Stoebel DM, Dorman CJ. 2011. DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol 80:85–101. doi: 10.1111/j.1365-2958.2011.07560.x. [DOI] [PubMed] [Google Scholar]
- 64.Higgins NP. 2016. Species-specific supercoil dynamics of the bacterial nucleoid. Biophys Rev 8:113–121. doi: 10.1007/s12551-016-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pulkkinen O, Metzler R. 2013. Distance matters: the impact of gene proximity in bacterial gene regulation. Phys Rev Lett 110:198101. doi: 10.1103/PhysRevLett.110.198101. [DOI] [PubMed] [Google Scholar]
- 66.Kuhlman TE, Cox EC. 2013. DNA-binding-protein inhomogeneity in E. coli modeled as biphasic facilitated diffusion. Phys Rev E Stat Nonlin Soft Matter Phys 88:e022701. doi: 10.1103/PhysRevE.88.022701. [DOI] [PubMed] [Google Scholar]
- 67.Kuhlman TE, Cox EC. 2012. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol Syst Biol 8:610. doi: 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moffitt JR, Pandey S, Boettiger AN, Wang S, Zhuang X. 2016. Spatial organization shapes the turnover of a bacterial transcriptome. Elife 5:e13065. doi: 10.7554/eLife.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kannaiah S, Livny J, Amster-Choder O. 2019. Spatiotemporal organization of the E. coli transcriptome: translation independence and engagement in regulation. Mol Cell 76:574–589.e7. doi: 10.1016/j.molcel.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 71.Richins R, Chen W. 2001. Effects of FIS overexpression on cell growth, rRNA synthesis, and ribosome content in Escherichia coli. Biotechnol Prog 17:252–257. doi: 10.1021/bp000170f. [DOI] [PubMed] [Google Scholar]
- 72.Cameron ADS, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8:e1002615. doi: 10.1371/journal.pgen.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 74.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Werling U, Edelmann W. 2014. Seamless Ligation Cloning Extract (SLiCE) cloning method. Methods Mol Biol 1116:235–244. doi: 10.1007/978-1-62703-764-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mansour FH, Pestov DG. 2013. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem 441:18–20. doi: 10.1016/j.ab.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 81.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deatherage DE, Traverse CC, Wolf LN, Barrick JE. 2014. Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front Genet 5:468. doi: 10.3389/fgene.2014.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fitzgerald S, Dillon SC, Chao T-C, Wiencko HL, Hokamp K, Cameron ADS, Dorman CJ. 2015. Re-engineering cellular physiology by rewiring high-level global regulatory genes. Sci Rep 5:17653. doi: 10.1038/srep17653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freese NH, Norris DC, Loraine AE. 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 96.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 98.Stark R, Brown G. 2020. DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf.
- 99.Hautefort I, Proença MJ, Hinton JCD. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol 69:7480–7491. doi: 10.1128/aem.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cooper KG, Chong A, Starr T, Finn CE, Steele-Mortimer O. 2017. Predictable, tunable protein production in Salmonella for studying host-pathogen interactions. Front Cell Infect Microbiol 7:475. doi: 10.3389/fcimb.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 102.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the strain construction process. For details, see Materials and Methods. Download FIG S1, EPS file, 0.7 MB (722.8KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of SNP analysis of constructed strains. Heat maps depict summarized Breseq analysis results. (A) Predicted mutations. (B) Unassigned missing coverage evidence. (C) Unassigned new junction evidence. In panels A and B, the color scale indicates the presence (1) or absence (0) of a mutation. In panel C, the color scale indicates the frequency of the new junction among reads mapping to that locus. Download FIG S2, EPS file, 0.9 MB (914.9KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding regions on the chromosome and plasmids. For the SL1344 chromosome, predicted Fis binding regions are shown for the two biological replicates of the WT, GX, and OX strains in early exponential phase. Increased Fis binding around the ter gene in the GX and OX strains resulted in peak calling in this region that was slightly better than that seen with the WT. For plasmids, levels of coverage corresponding to the ChIP-seq experiment (solid traces) and predicted Fis binding regions (shaded boxes) are shown for the two biological replicates of the WT, GX, and OX strains in early exponential phase. For comparison, the wild-type mock coverage is also shown as a solid gray trace. Predicted Fis binding motifs are shown in orange, and the motif used for the prediction is briefly mentioned in the legend. This motif is described in detail in the Fig. S4 legend. Chromosomal positions are indicated in millions of base pairs, and plasmid positions are in thousands of base pairs. Download FIG S3, TIF file, 2.9 MB (2.8MB, tif) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding motif obtained from ChIP-seq data. A 15-nucleotide-long sequence logo represents the Fis binding motif discovered using MEME on the set of Fis binding regions in the wild-type strain identified using ChIP-seq. Each position in the 15-nucleotide-long palindromic motif is indicated on the x axis. The stack of letters at each position represents the most common nucleotides found at that position. The height of each letter represents the relative frequency of that nucleotide at that position in the motif. The overall height of the stack of letters at each position represents the degree of conservation at that position in the motif (if the stack is very short, that position is highly variable). Various statistics reported by MEME are presented below the logo. This motif is similar to the E. coli Fis binding motif. Download FIG S4, EPS file, 0.1 MB (159.5KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene dosage gradient differences in the GX and OX strains compared to the WT visualized using coverage data from the ChIP-seq mock experiments performed with these strains. For clarity, the coverage data at each position are not shown; only the local average calculated using the R LOESS function along the chromosomal position in millions of base pairs is shown. There were only slight gene dosage gradient differences in the GX and OX strains at the early exponential time point at which chromatin was collected for ChIP-seq. These differences are considered in the calculation of differentially bound regions. Download FIG S5, EPS file, 0.05 MB (54.1KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding around fis gene. (A) Log2 fold changes of Fis binding upstream of the dusB-fis transcription unit in GX and OX strains relative to the WT. (B) Fis ChIP-seq coverage of the WT, GX, and OX strains (two biological replicates and mock) of the region around the fis gene. The ranges of the y-axis scale are the same for all the coverage traces and have been omitted for brevity. For each strain, the two biological replicates are represented by the first two coverage traces in the darker color shade. The third, lighter color trace represents the mock coverage for that strain. Gray-shaded regions show the peaks called in each sample relative to the mock. Genes (gray arrows) and Fis binding motifs (red blocks) are shown on the positive and negative reference strands. The fis gene is a part of the dusB-fis transcription unit. The promoter of this transcription unit contains two Fis binding motifs, as predicted by FIMO from the MEME suite, and the ChIP-seq data show substantial Fis binding. ChIP-seq data were analyzed using the wild-type reference sequence; thus, the coverage reported here is plotted relative to wild-type gene locations, even though those locations had been altered in the GX and OX strains. In the GX strain, the entire region that included dusB, fis, and 315 bp of its upstream promoter region was relocated to the dps locus. In the OX strain, only the fis ORF was relocated to the dps locus. In the GX and OX strains, due the strain construction process, an rrnB terminator was inserted 63 bp downstream of the yhdG-fis operon. This is represented as a dip in the coverage and was expected. Download FIG S6, EPS file, 0.1 MB (158.1KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis binding changes on the Salmonella megaplasmids assayed by ChIP-seq during the EE phase. (A) Peak intensity changes along the plasmid position shown in base pairs. In calculating these intensity changes, the appropriate mock data (WT mock for WT peaks, GX mock for GX peaks, OX mock for OX peaks) were used as controls. Significantly differentially bound peaks are roughly annotated with genes that are located in their vicinity. (B) Spearman’s correlation of peak intensity changes in the GX strain in comparison with the OX strain. The dashed line has a slope of 1 and indicates the position where the fold changes in Fis binding intensity in GX would be equal to those in OX. The blue solid line represents the regression linear fit to the data. Download FIG S7, EPS file, 0.4 MB (431.2KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fold changes of some gene sets in the GX and OX strains. (A and B) Motility genes (A) and SPI-1 and SPI-2 genes (B). Expression changes in early exponential (EE) phase and in the transition to the stationary (TS) phase are shown. None of these gene sets were significantly differentially expressed in the GX strain. Note that some of the class 3 motility genes (fliC, fljA, and fljB) were differentially expressed due to phase switching in the GX and OX strains during their construction. The orientations of these genes in the constructed strains were different from those seen in the wild-type strain against which the gene expression changes were calculated, thus resulting in phase-variation-related gene expression changes. It is unlikely that these changes were related to our study. A general pattern of reversal in gene expression between the early exponential phase and the transition to the stationary phase was seen in the OX strain (this might have been related to Fis levels being higher in the stationary phase in this strain). The color bar indicates FDR, and bars representing genes with FDR values of <0.05 are red. Download FIG S8, EPS file, 0.7 MB (739.7KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptomic changes on the Salmonella megaplasmids assayed by RNA-seq. Data represent Log2 fold changes in expression of genes along their positions on the plasmid in base pairs. The blue curve shows the local regression peak intensity change calculated using the R LOESS function. Significant gene expression changes (FDR < 0.05) are shown as red filled triangles. Download FIG S9, EPS file, 0.5 MB (500.6KB, eps) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of oligonucleotides used in this study. The nucleotide sequences are written 5′ to 3′. Download Table S1, DOCX file, 0.1 MB (96.2KB, docx) .
Copyright © 2020 Bogue et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq and ChIP-seq data can be found on the Gene Expression Omnibus (accession number GSE152228). WGS data can be found on the Sequence Read Archive (accession number PRJNA638833). WGS data for strains OX and 2xdO can be found on the European Nucleotide Archive (accession numbers ERS2515905 and ERS4653304, respectively). Scripts used in the analysis can be found on GitLab (project identifier 10206830 [https://gitlab.com/aalap.mogre/chip-seq-and-rna-seq-in-salmonella-swap-strains]).