Abstract
Autophagy, a highly conserved cellular protein degradation process, has been involved in acute myeloid leukemia (AML). The present study aims to establish a novel, autophagy-related prognostic signature for prediction of AML prognosis. Differentially expressed autophagy-related genes in AML and healthy samples were screened using GSE1159. Univariate Cox regression analysis was applied to determine survival-associated autophagy-related genes in The Cancer Genome Atlas (TCGA) AML cohort. Lasso regression was performed to develop multiple-gene prognostic signatures. A novel six-gene signature (including CASP3, CHAF1B, KLHL24, OPTN, VEGFA, and VPS37C) DC was established for AML prognosis prediction. The Kaplan–Meier survival analysis revealed that patients in the high-risk score group had poorer overall survival (OS). The receiver operating characteristic (ROC) curve validated its good performance in survival prediction in TCGA AML cohort, and the area under the curve value was 0.817. Moreover, our signature could independently predict OS. A nomogram was constructed, including the six-gene signature and other clinical parameters, and predictive efficiency was confirmed using the ROC curve and calibration curve. Furthermore, gene set enrichment analyses identified several tumor-associated pathways that may contribute to explain the potential molecular mechanisms of our signature. Overall, we developed a new autophagy-associated gene signature and nomogram to predict OS of AML patients, which may help in clinical decision-making for AML treatment.
Keywords: acute myeloid leukemia, autophagy, signature, prognosis
Introduction
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous malignancy characterized by abnormal accumulation of immature hematopoietic progenitor cells in the bone marrow and peripheral blood (Dohner et al., 2015). Despite the advances in AML pathogenesis research and the occurrence of new drugs, the prognosis of AML remains poor. The 5-year overall survival (OS) of younger people with AML is <50% and only about 10% in AML patients older than 60 years (Estey, 2018; Shallis et al., 2019). Patients with AML showed a heterogeneous prognosis after receiving chemotherapy, partly depending on age, cytogenetic changes, and the karyotype (Slovak et al., 2000; Grimwade et al., 2001; Byrd et al., 2002; Dohner et al., 2010). Currently, molecular abnormalities and cytogenetic characteristics at diagnosis are considered the most crucial prognostic parameters and perform well in evaluating the complete remission rate, disease-free survival rate, and OS rate and in guiding rational AML management (Dohner et al., 2010; Rollig et al., 2011; Nebbioso et al., 2015; Prada-Arismendy et al., 2017). Thus, the genetic prognostic markers are crucial in evaluating patients with AML and in guiding rational management. Several markers have been proved to be associated with AML prognosis, including AML-ETO (Lin et al., 2017), FLT3 (Gilliland and Griffin, 2002), NPM1 (Heath et al., 2017), CEBPA (Green et al., 2010), and KIT (Qin et al., 2018) genes. However, since there is great heterogeneity among AML patients, the current markers may not apply to every patient. Therefore, there is an urgent need to screen novel and reliable prognostic biomarkers for the therapy and prognosis prediction of AML patients.
Autophagy is a highly conserved biological process that plays an important role in the maintenance of cellular homeostasis through degrading and recycling damaged cellular components and proteins to provide nutrition for cell growth (Choi et al., 2013). The dysregulation of autophagy is prevalent in tumor initiation, malignant progression, and chemotherapy resistance (Levy et al., 2017; Li et al., 2017; Mowers et al., 2018). The role of autophagy in AML has been previously reported (Auberger and Puissant, 2017). For instance, genetic silencing of ATG7 in AML could enhance chemotherapy sensitivity (Sumitomo et al., 2016); moreover, autophagy activated by ULK1 played a protective role in AML resistance to daunorubicin (Qiu et al., 2020), and LAMP2 deficiency in AML was reported to be associated with Aza resistance and hypersensitivity (Dubois et al., 2019). Autophagy was also shown to enhance chemoresistance and help in maintaining the stemness of leukemia stem cells (Jang et al., 2017).
Notably, most of the studies have just focused on investigating the relationship between autophagy and leukemia progression by individual genes; however, the prognosis role of global autophagy-related genes has not been extensively analyzed in patients with AML. In the present study, we identified an autophagy-related signature, including six genes, which could accurately and independently predict the OS for AML patients. Moreover, a novel prognostic nomogram, including the autophagy signature and clinical parameters, was established. Taken together, our study demonstrates the relationship between autophagy-related genes and the prognosis of AML patients and provides new insights for AML treatment.
Materials and Methods
Data acquisition and preparation
Raw microarray AML data were acquired from GSE1159. The robust multi-array average method was employed to normalize the raw microarray data. In addition, the RNA sequencing profiles of AML tissues with gene expression and clinicopathological information were downloaded from The Cancer Genome Atlas (TCGA) database. The ComBat method was applied to eliminate any batch effects so as to remove discrepancies among different datasets. GSE1159, including 285 AML samples and 8 healthy people, and TCGA, including 128 AML patients, were selected as the discovery cohort. GSE12417 containing 150 AML patients was used as our validation cohort. All patients without clinical information were initially excluded.
Identification of differentially expressed autophagy-related genes in AML
A list of 546 autophagy-related genes was obtained from the Human Autophagy Database and the Molecular Signatures Database, (GO_autophagy, M12441). edgeR (Robinson et al., 2010) was used to identify differentially expressed autophagy-related genes (DEARGs) in GSE1159. The cutoff values were demonstrated according to the false discovery rate (FDR) <0.05 and |log2-fold change (FC)| > 1. Gene expression of profile interactive analysis is a newly developed web server, which could analyze RNA expression data for tumor tissues and normal tissues in TCGA and Genotype-Tissue Expression (GTEx) projects (Tang et al., 2017). Since there were no normal AML samples in TCGA, the expression levels of DEARGs identified by GSE1159 were further validated by GEPIA using GDC TCGA Acute Myeloid Leukemia (LAML) tumor data and matched normal tissue data from GTEx. |Log2FC| > 1 and p < 0.05 were considered to be statistically significant.
Construction and evaluation of the prognostic risk score model based on DEARGs
TCGA LAML cohort was employed to screen survival-associated DEARGs; a univariate Cox proportional hazards regression analysis was performed and the genes with p value <0.05 were further analyzed by the Lasso–Cox regression. Then, a prognostic autophagy-related gene signature of AML patients was constructed according to a linear combination of regression coefficients (β) calculated by the Lasso–Cox regression based on the glmnet package in R (Candia and Tsang, 2019). The risk score was calculated using the following formula: risk score = expression of gene 1 × coefficient + expression of gene 2 × coefficient + … expression of gene n × coefficient (Wang et al., 2019). The risk score was calculated for each patient in TCGA cohort and validated set based on this model. Next, AML patients were divided into high- and low-risk groups according to the median value of the risk scores. The Kaplan–Meier survival analysis was used to assess the prognosis of AML patients in the high-risk score or low-risk score group using the survival package. To evaluate the prognostic capability of the autophagy-related risk signature, we calculated the area under the curve (AUC) with the R package survivalROC. Furthermore, to investigate whether the predictive power of the prognostic model could be independent of other clinicopathologic factors (including age, sex, risk group, FLT3 status, RAS status, and NPM1 status) for patients with AML, univariate and multivariate Cox proportional hazards regression analyses were conducted in TCGA LAML cohort.
Predictive nomogram construction and gene set enrichment analysis of function enrichment
The nomogram and calibration plot were generated using the rms R package. Gene set enrichment analysis (GSEA) was used to identify related pathways in AML patients. Enriched gene sets with a nominal p < 0.05 and FDR <0.25 were considered to be statistically significant.
Statistical analyses
All statistical analyses were conducted using the R software, and p < 0.05 was considered statistically significant. The distribution variables were analyzed using the chi-square test or Fisher's exact test. The Kaplan–Meier survival analysis and log-rank test were applied to analyze OS. A time-dependent receiver operating characteristic (ROC) curve was used to detect the accuracy of the models. ROC curve analysis was also used to estimate the diagnostic value of gene expression. Univariate and multivariate Cox regression analyses were performed to assess survival. The hazard ratio (HR) and 95% confidence interval (CI) were calculated to identify genes related with OS.
Results
Identification of DEARGs in AML patients
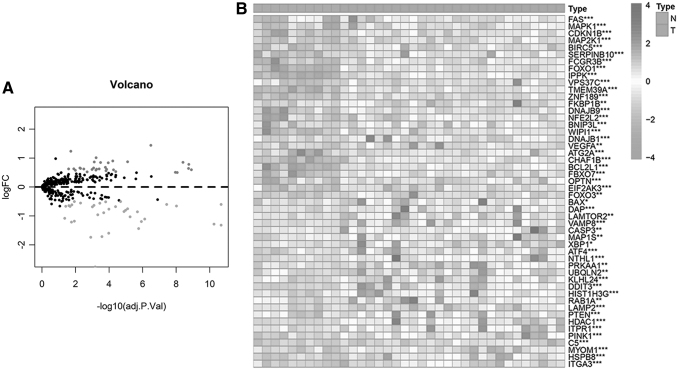
We analyzed 546 autophagy-related genes (ARGs) that were acquired from the Human Autophagy Database (232 genes) and the Molecular Signatures Database, v4.0. (GO_autophagy, M12441), (314 genes). Next, edgeR was used to analyze the expression of 546 ARGs in 285 AML and 8 healthy people from GSE1159, as shown in Figure 1A and B, a total of 66 DEARGs were identified, including 32 upregulated (red color) and 34 downregulated (green color) genes (|log2FC| > 1, FDR <0.05) (Supplementary Table S1).
FIG. 1.
Different expression levels of autophagy-related genes in AML patients. (A) Volcano plot of the differential expression of 546 autophagy-related genes in AML samples (n = 285) and healthy people (n = 8). Red dots represent upregulated genes, while green dots represent downregulated genes (p < 0.05 and |log2(FC)| > 1). (B) The heat map showing the expression of 66 DEARGs in AML samples and normal people (*p < 0.05, **p < 0.01, and ***p < 0.001). AML, acute myeloid leukemia; DEARGs, differentially expressed autophagy-related genes; FC, fold change.
Identification of prognostic-associated DEARGs in AML patients
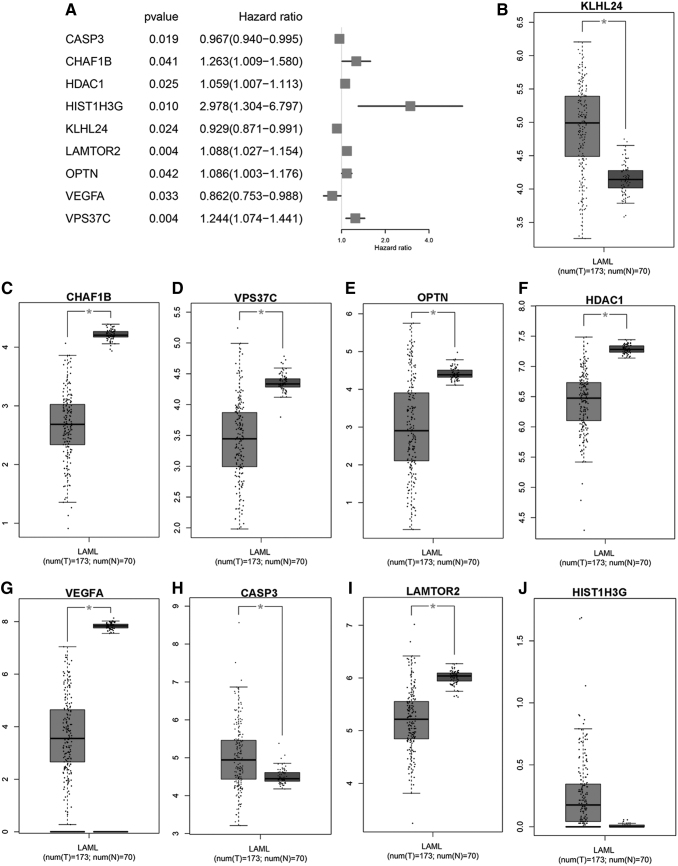
To explore the prognostic value of these 66 DEARGs in AML patients, univariate Cox regression analysis was applied according to the expression levels of the DEARGs from TCGA cohort consisting of 128 AML patients. As a result, 9 of 66 DEARGs were significantly correlated with OS (CASP3, CHAF1B, HDAC1, HIST1H3G, KLHL24, LAMTOR2, OPTN, VEGFA, and VPS37C) (p < 0.05, Fig. 2A) (Supplementary Table S2). Among these genes, three genes (VEGFA, KLHL24, and CASP3) were identified as protective factors (HR <1), while the other six genes (HDAC1, OPTN, LAMTOR2, VPS37C, CHAF1B, and HIST1H3G) were identified as risk factors (HR >1). The nine survival-associated genes in AML and healthy people were further confirmed in the GEPIA, which included 170 AML samples and 70 normal samples; the results revealed that except LAMTOR2 HIST1H3G and HDAC1, the expression levels of other genes were in keeping with GSE1159, therefore we chose these genes for further analysis (CASP3, CHAF1B, KLHL24, OPTN, VEGFA, and VPS37C).
FIG. 2.
Identification of survival-associated DEARGs and validation of their expression in TCGA cohort. (A) Forest plot illustrating the p value and HR of survival-associated DEARGs. (B–J) The bar graph showing the expression of survival-associated DEARGs in the GEPIA database (*p < 0.05). HR, hazard ratio; TCGA, The Cancer Genome Atlas.
Construction of a prognosis predicting the autophagy-related risk signature in AML
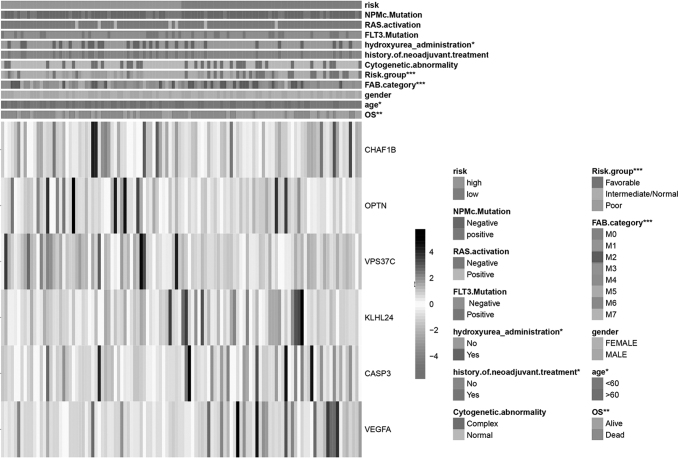
Next, the six selected DEARGs were employed to construct a signature; the Lasso–Cox regression analysis was performed to calculate the coefficients (Supplementary Fig. S1), and the risk score was calculated using the following formula based on the coefficient: (0.01532 × CASP3) + (0.0561 × CHAF1B) + (0.01959 × KLHL24) + (0.06514 × OPTN) + (−0.05621 × VEGFA) + (0.07141 × VPS37C), then AML patients were categorized into the high-risk group (n = 64) and low-risk group (n = 64) according to the median cutoff risk score (Supplementary Fig. S2). Interestingly, the expression of the protective and risk genes showed distinct patterns based on the risk score; the risk genes expressed higher levels in high-risk score AML patients, while protective genes expressed higher levels in low-risk score group patients (Fig. 3). Furthermore, there were significant clinical differences between the high- and low-risk groups (Fig. 3) (Table 1). In the high-risk group, patients tended to be older compared with those in the low-risk group (p < 0.05); moreover, more than half of the patients who were administered hydroxyurea were in the low-risk score group (67.8%) (p < 0.05). In addition, more poor-risk group AML patients were found in the high-risk group (70.8%) than in the low-risk group (29.2%), while more favorable-risk group patients were in the low-risk group (81.4%) compared with high-risk group (18.6%) (p < 0.001).
FIG. 3.
Construction of the autophagy-related risk signature in AML patients and correlation between the risk score and clinical characteristics. The heat map showing the expression of the six autophagy-related genes in high-risk and low-risk AML patients. The distribution of clinicopathological factors was compared between the high-risk and low-risk groups (*p < 0.05, **p < 0.01, and ***p < 0.001).
Table 1.
Demographic and Clinicopathologic Characteristics of Acute Myeloid Leukemia Patients in The Cancer Genome Atlas Cohort
| Variables | Total, N = 112 | Low risk (n = 56) | High risk (n = 56) | p |
|---|---|---|---|---|
| Age, years | ||||
| ≤60 | 44 | 16 (36.4%) | 28 (64.6%) | 0.032 |
| >60 | 68 | 40 (58.8) | 28 (41.2%) | |
| Sex | ||||
| Female | 55 | 29 (52.7%) | 26 (47.3%) | 0.23 |
| Male | 57 | 27 (47.3%) | 30 (52.6%) | |
| FAB category | ||||
| M0 | 11 | 7 (63.6%) | 4 (36.3%) | <0.001 |
| M1 | 26 | 11 (42.3%) | 15 (57.6) | |
| M2 | 26 | 17 (65.3) | 9 (34.6%) | |
| M3 | 13 | 13 (100%) | 0 | |
| M4 | 23 | 7 (30.4%) | 16 (69.6%) | |
| M5 | 10 | 0 | 10 (100%) | |
| M6 | 1 | 0 | 1 (100%) | |
| M7 | 1 | 1 (100%) | 0 | |
| Risk group | ||||
| Favorable | 27 | 22 (81.4%) | 5 (18.6%) | <0.001 |
| Intermediate/normal | 60 | 26 (43.3%) | 34 (56.7%) | |
| Poor | 24 | 7 (29.2%) | 17 (70.8%) | |
| Hydroxyurea administration | ||||
| Yes | 28 | 9 (32.1%) | 19 (67.8%) | 0.015 |
| No | 84 | 47 (55.9%) | 37 (44.1%) | |
| FLT3 status | ||||
| Mutated | 30 | 15 (50%) | 15 (50%) | 0.14 |
| Wild type | 82 | 41 (50%) | 41 (50%) | |
| NPM1 status | ||||
| Mutated | 28 | 10 (35.7%) | 18 (64.3%) | 0.57 |
| Wild type | 84 | 46 (54.8%) | 38 (45.2%) | |
Prognostic value of the six-autophagy-related gene signature in AML
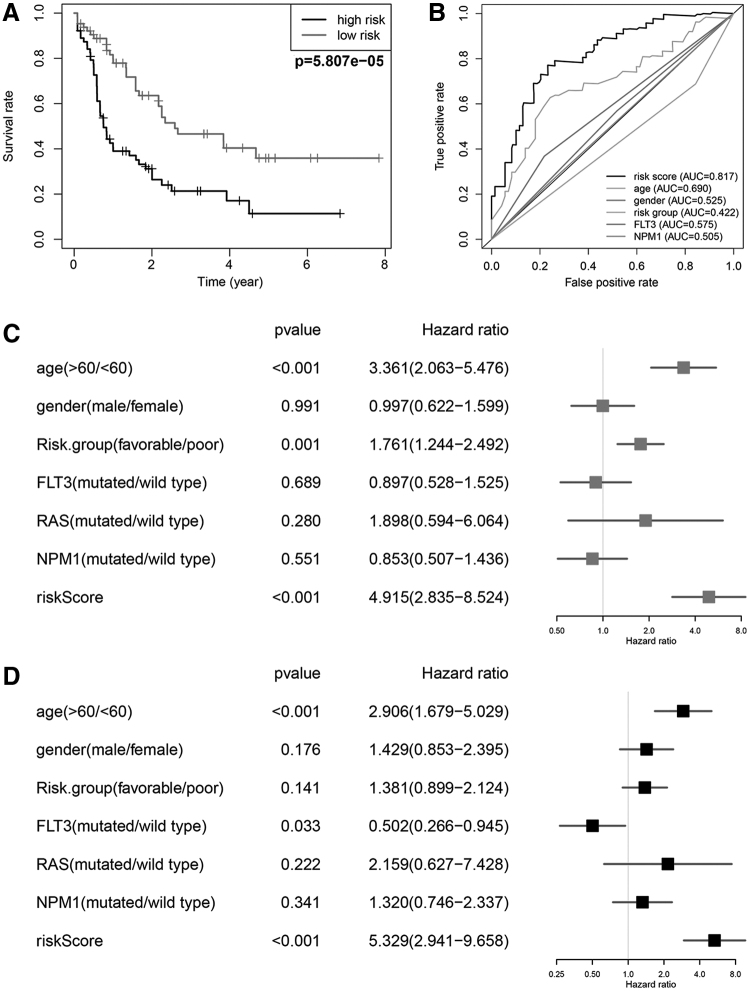
To better understand the prognostic role of our signature in AML patients, the Kaplan–Meier survival analysis was performed according to the median cutoff risk score, as a result, the high-risk score group had significantly shorter OS compared with the low-risk score group (Fig. 4A). The 5-year OS rate was 11.4% in the high-risk group and 35.9% in the low-risk group. A time-dependent ROC curve was employed to evaluate the specificity and sensitivity of the model, and results indicated that the AUC value of our model was 0.817, which was significantly higher compared with that of age (AUC = 0.768), gender (AUC = 0.448), risk group (AUC = 0.462), FLT3 mutation (AUC = 0.521), and NPM1 mutation (AUC = 0.583) (Fig. 4B). These results demonstrated that our six-gene risk signature performed well in survival prediction when compared with other clinical factors. To assess whether the six-gene signature was an independent prognostic factor, univariate and multivariate Cox regression analyses were conducted. As a consequence, age at diagnosis (p < 0.001, HR = 3.361, 95% CI = 2.063 − 5.476), risk group (p = 0.001, HR = 1.761, 95% CI = 1.244 − 2.492), and riskScore (p < 0.001, HR = 4.915, 95% CI = 2.835 − 8.524) were associated with OS in the univariate analysis (Fig. 4C) and only age at diagnosis (p < 0.001, HR = 2.906, 95% CI = 1.679 − 5.029) and riskScore (p < 0.001, HR = 5.329, 95% CI = 2.941 − 9.658) were still obviously related to OS (p < 0.05) in the multivariate Cox analysis (Fig. 4D). These findings demonstrated that the risk score retrieved from the six DEARGs could be regarded as an independent prognostic factor in AML.
FIG. 4.
Prognostic significance and performance of the autophagy-related gene signature in AML. (A) K–M survival curve for high- and low-risk patients. (B) Time-dependent ROC curve indicating the specificity and sensitivity for the risk score and other clinical factors. (C, D) Forest plot of the univariate and multivariate Cox regression analyses in AML. K–M, Kaplan–Meier; ROC, receiver operating characteristic.
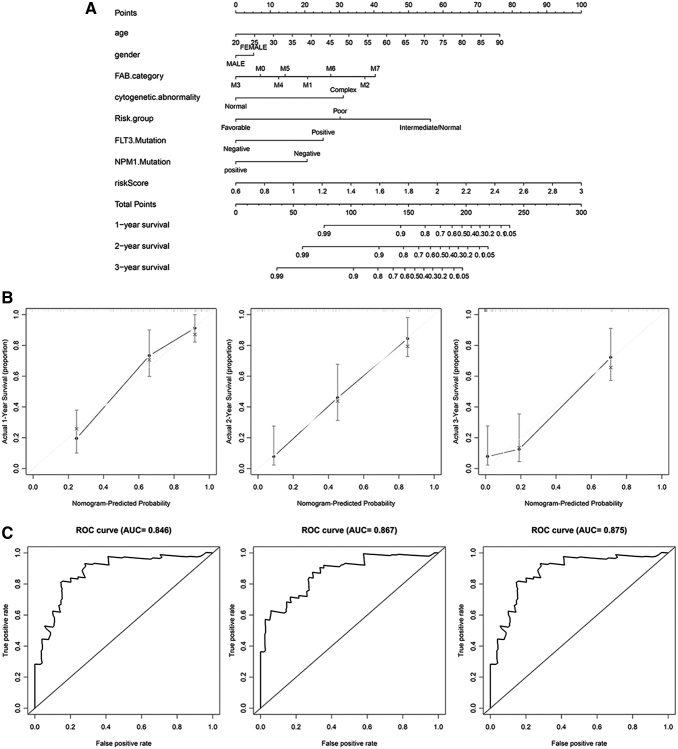
Nomogram development for prediction of prognostic risk in AML
To provide clinicians with a better quantitative way for predicting cancer prognosis, a nomogram was constructed using variables associated with OS (age, gender, FAB category, cytogenetic abnormality, risk group, FLT3 status, NPM1 status, and risk score) and it revealed that our signature risk score was the most important factor among the various clinical parameters (Fig. 5A). Calibration curves showed that the predicted and actual survival rates matched very well at 1, 2, and 3 years (Fig. 5B). Moreover, the time-dependent ROC curve was plotted to assess the efficiency of the nomogram, as shown in Figure, and AUC values of 1-, 2-, and 3-year OS were 0.846, 0.867, and 0.875, respectively (Fig. 5C). These findings suggest the appreciable accuracy of the nomogram.
FIG. 5.
Nomogram to predict the probability of patients with AML. (A) Prognostic nomogram predicting 1-, 2-, and 3-year OS for AML patients. (B) Calibration curves for the nomogram at 1, 2, and 3 years. (C) ROC curve analysis assessing the accuracy of the nomogram in predicting 1-, 2-, and 3-year OS. OS, overall survival.
Functional characteristics enrichment of the AML autophagy-related signature
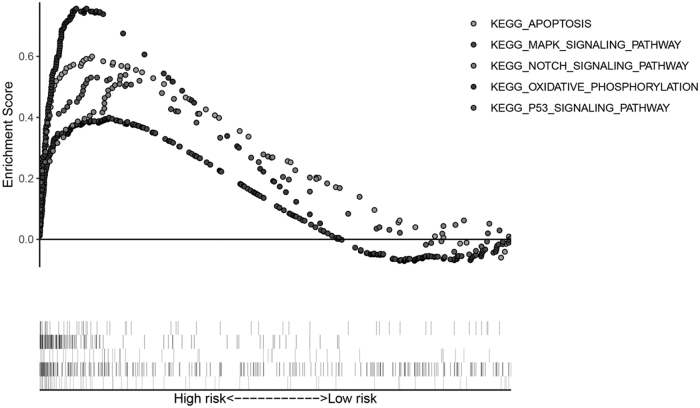
To better understand the molecular function of the six-autophagy-related gene signature, GSEA was performed in high-risk (n = 56) and low-risk (n = 56) AML patients based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways gene set from the MSigDB database; when p < 0.05 and FDR <0.25, the gene sets were thought to be significantly enriched. In our GSEA (enrichment) results, we observed that the pathways, KEGG_APOPTOSIS, NOTCH_SIGNALING_PATHWAY, MAPK_SIGNALING_PATHWAY, KEGG_OXIDATIVE_PHOSPHORYLATION, and KEGG_P53_SIGNALING_PATHWAY, were enriched in the high-risk score group (Fig. 6). Previous studies have reported that these pathways had a close relationship with AML. Briefly, the GSEA results revealed that the six-autophagy-related gene signature played a role in AML development and progress.
FIG. 6.
GSEA results in high-risk score AML patients. GSEA, gene set enrichment analysis.
Verification of the six-autophagy-related gene prognostic model in GSE12417
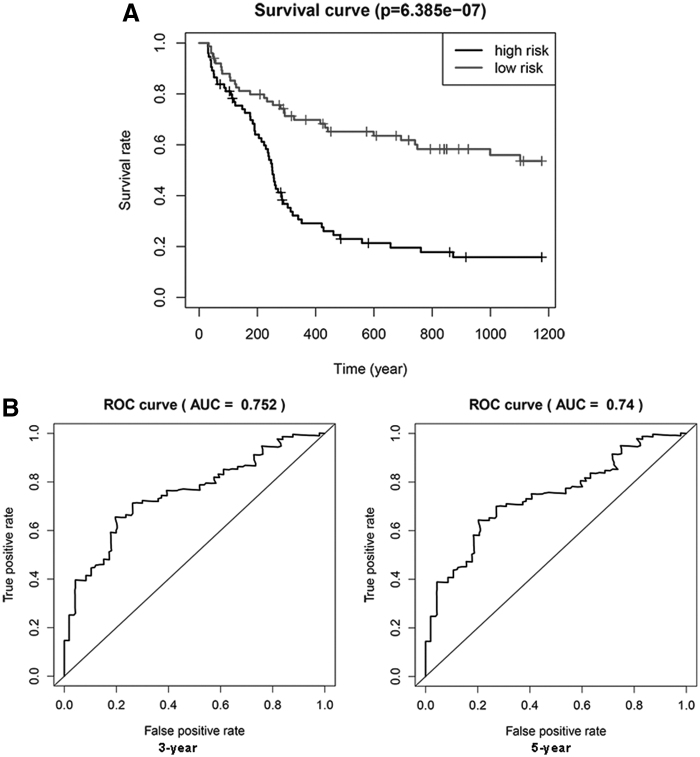
To further validate the accuracy of our prognostic model, we predict OS in an external Gene Expression Omnibus (GEO) cohort, including 150 AML patients (GSE12417, GPL 96). The patients in this dataset were divided into high-risk (n = 75) and low-risk (n = 75) groups based on the median risk score. As a result, patients in the high-risk score group had a poorer prognosis (p < 0.001) (Fig. 7A). In addition, the time-dependent ROC curve analysis was performed and AUC values for the OS model at 3 and 5 years were 0.752 and 0.74, respectively (Fig. 7B). Taken together, these results demonstrated that both the risk models accurately predicted the prognosis of AML patients.
FIG. 7.
Prognostic significance and performance of the autophagy-related gene signature in AML in a validated cohort. (A) K–M survival curve for high- and low-risk patients. (B) Time-dependent ROC curve for 3 and 5 years.
Discussion
Autophagy has been reported to play a crucial role in oncogenesis, progression aggressiveness, and chemotherapy resistance of various tumors, including hematologic malignancy (Auberger and Puissant, 2017). Piya et al. (2017) observed that high ATG7 levels in AML blasts are associated with shorter remission duration, and knockdown of ATG7 obviously enhanced the apoptosis induced by Ara-C. Nguyen et al. (2019) found that high p62 expression was correlated with poor prognosis in AML and silencing of p62 could suppress leukemia progression in a mouse. Heydt et al. (2018) reported that inhibition autophagy or ATF4 remarkably impaired FLT3-ITD-positive leukemic cell proliferation as well as tumor burden in a murine leukemia xenograft. Importantly, autophagy suppression also overcame FLT3 inhibitor resistance. Therefore, autophagy-related genes are promising prognostic predictors and therapeutic targets in AML. However, comprehensive expression patterns based on DEARGs have not been previously developed in AML.
In the current study, we first identified 66 DEARGs based on the GEO database (GSE1159), including 32 upregulated and 34 downregulated genes. Then, six survival-associated risk DEARGs (CASP3, CHAF1B, KLHL24, OPTN, VEGFA, and VPS37C) were identified through univariate Cox regression and Lasso–Cox regression. Several of them have been found to play an essential role in tumor development and progression. For instance, CASP3, a primary mediator of apoptosis (Yuan et al., 2016), has been reported to regulate invasion, migration, and metastasis of colon cancer cells (Zhou et al., 2018). VEGFA is an angiogenesis stimulator and overexpressed in majority malignancies with poor prognosis, which plays a significant role in tumor invasiveness, metastasis, increased vascular density, and recurrence (Nagy et al., 2009) (Kerbel, 2008; Lacal and Graziani, 2018). Several strategies that aim to target the VEGFA–VEGFR signaling pathway for treatment of neoplasm have been investigated (Kowanetz and Ferrara, 2006; Ellis and Hicklin, 2008). CHAF1B is a key facilitator in chromatin assembly in damaged DNA repair (Di et al., 2020). High expression of CHAF1B has been observed to be associated with poor prognosis in several solid tumors, including gliomas (de Tayrac et al., 2011), melanoma (Mascolo et al., 2010), and prostatic cancer (Staibano et al., 2009), and increased expression of CHAF1B also plays a role in tumor aggressiveness, cell cycle arrest, and apoptosis (Polo et al., 2010; Peng et al., 2018; Duan et al., 2019). Therefore, the identified DEARGs may also affect the prognosis and progression of AML.
Next, we constructed a novel prognostic risk signature based on the expression of six DEARGs. Based on the DEARG-based risk score model, patients with AML were categorized into the high-risk group and low-risk group. There were significant differences in prognosis and clinical characteristics between the two groups. Multivariate Cox regression analysis demonstrated that the prognostic models could independently predict the prognosis of AML patients.
The nomogram has been widely employed in clinical practice for its intuitive visual presentation (Wan et al., 2019). As far as we know, this nomogram is the first to combine an autophagy-related risk signature for predicting the survival of AML patients. In this study, we established a nomogram incorporating the autophagy risk signature, age, gender, FAB category, cytogenetic abnormality, risk group, FLT3 status, and NPM1 status. The calibration plot based on TCGA databases revealed that the actual survival rate was roughly in line with the predicted survival rate, suggesting the excellent predictive performance of our nomogram. This visual scoring system could help both physicians and patients perform individualized survival prediction, which would facilitate the selection of chemotherapy regimens.
Moreover, GSEA analyzed the differences between high-risk and low-risk groups, stratified by the autophagy-related signature. Several pathways that correlated with oncogenesis and progression in the high-risk score AML patients were identified, including the MAPK_SIGNALING_PATHWAY, KEGG_APOPTOSIS, KEGG_P53_SIGNALING_PATHWAY NOTCH_SIGNALING_PATHWAY, and KEGG_OXIDATIVE_PHOSPHORYLATION pathway. MAPK was found to be abnormally activated in leukemia cells and promoted cell proliferation by regulating the expression of oncogenic proteins (Rocca et al., 2018). It is known that dysregulation of apoptosis is the most obvious hallmark of various cancers (Pistritto et al., 2016). Moreover, the apoptosis arrest was one of the causes of leukemia chemoresistance (Valentin et al., 2018). P53 plays a central role in hematopoietic stem cell function, its aberrations could affect AML biology, progression, and even therapy response and usually predict poor prognosis of AML patients (Prokocimer et al., 2017). Increasing evidence indicates that dysregulation of oxidative phosphorylation was associated with leukemia stem cell chemoresistance (Kuntz et al., 2017). The mutations of Notch were proved to be involved in development of chronic lymphocytic leukemia or T cell acute lymphoblastic leukemia in several studies (Bellavia et al., 2018). Taken together, we revealed that our autophagy-related signature is involved in many oncogenic signaling pathways, suggesting their crucial roles in initiation and development of AML.
To our knowledge, this is the first study that makes a comprehensive and systematic exploration of the prognosis value of autophagy-related genes in AML. We acknowledge that there were some drawbacks and shortcomings in the current study. On the one hand, since the total number of AML patients involved in our study was limited, a larger dataset in the future study is urgently needed to further validate our prediction models. On the other hand, biological experiments and clinical trials are required to verify the function and clinical significance of the identified autophagy-related signatures.
Conclusion
In summary, in this study, we constructed a novel six-gene signature and nomogram to predict the OS of AML patients, which may contribute to the clinical decision-making for individual therapy.
Supplementary Material
Acknowledgments
The authors would like to thank the GEO and TCGA databases for the availability of data.
Disclosure Statement
No competing financial interests exist.
Funding Information
The present study was supported by the National Natural Science Foundation of China (NSFC) (No. 81170525 and No. 81470348).
Supplementary Material
References
- Auberger P., and Puissant A. (2017). Autophagy, a key mechanism of oncogenesis and resistance in leukemia. Blood 129, 547–552 [DOI] [PubMed] [Google Scholar]
- Bellavia D., Palermo R., Felli M.P., Screpanti I., and Checquolo S. (2018). Notch signaling as a therapeutic target for acute lymphoblastic leukemia. Expert Opin Ther Targets 22, 331–342 [DOI] [PubMed] [Google Scholar]
- Byrd J.C., Mrozek K., Dodge R.K., Carroll A.J., Edwards C.G., and Arthur D.C., et al. (2002). Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 100, 4325–4336 [DOI] [PubMed] [Google Scholar]
- Candia J., and Tsang J.S. (2019). eNetXplorer: an R package for the quantitative exploration of elastic net families for generalized linear models. BMC Bioinformatics 20, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A.M., Ryter S.W., and Levine B. (2013). Autophagy in human health and disease. N Engl J Med 368, 651–662 [DOI] [PubMed] [Google Scholar]
- de Tayrac M., Aubry M., Saikali S., Etcheverry A., Surbled C., and Guenot F., et al. (2011). A 4-gene signature associated with clinical outcome in high-grade gliomas. Clin Cancer Res 17, 317–327 [DOI] [PubMed] [Google Scholar]
- Di M, Wang M., Miao J., Chen B., Huang H., and Lin C., et al. (2020). CHAF1B induces radioresistance by promoting DNA damage repair in nasopharyngeal carcinoma. Biomed Pharmacother 123, 109748. [DOI] [PubMed]
- Dohner H., Estey E.H., Amadori S., Appelbaum F.R., Buchner T., and Burnett A.K., et al. (2010). Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115, 453–474 [DOI] [PubMed] [Google Scholar]
- Dohner H., Weisdorf D.J., and Bloomfield C.D. (2015). Acute myeloid leukemia. N Engl J Med 373, 1136–1152 [DOI] [PubMed] [Google Scholar]
- Duan Y., Liu T., Li S., Huang M., Li X., and Zhao H., et al. (2019). CHAF1B promotes proliferation and reduces apoptosis in 95D lung cancer cells and predicts a poor prognosis in nonsmall cell lung cancer. Oncol Rep 41, 2518–2528 [DOI] [PubMed] [Google Scholar]
- Dubois A., Furstoss N., Calleja A., Zerhouni M., Cluzeau T., and Savy C., et al. (2019). LAMP2 expression dictates azacytidine response and prognosis in MDS/AML. Leukemia 33, 1501–1513 [DOI] [PubMed] [Google Scholar]
- Ellis L.M., and Hicklin D.J. (2008). VEGF-targeted therapy: mechanisms of anti-tumour activity. Nat Rev Cancer 8, 579–591 [DOI] [PubMed] [Google Scholar]
- Estey E.H. (2018). Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol 93, 1267–1291 [DOI] [PubMed] [Google Scholar]
- Gilliland D.G., and Griffin J.D. (2002). The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532–1542 [DOI] [PubMed] [Google Scholar]
- Green C.L., Koo K.K., Hills R.K., Burnett A.K., Linch D.C., and Gale R.E. (2010). Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol 28, 2739–2747 [DOI] [PubMed] [Google Scholar]
- Grimwade D., Walker H., Harrison G., Oliver F., Chatters S., and Harrison C.J., et al. (2001). The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood 98, 1312–1320 [DOI] [PubMed] [Google Scholar]
- Heath E.M., Chan S.M., Minden M.D., Murphy T., Shlush L.I., and Schimmer A.D. (2017). Biological and clinical consequences of NPM1 mutations in AML. Leukemia 31, 798–807 [DOI] [PubMed] [Google Scholar]
- Heydt Q., Larrue C., Saland E., Bertoli S., Sarry J.E., and Besson A., et al. (2018). Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene 37, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.E., Eom J.I., Jeung H.K., Cheong J.W., Lee J.Y., and Kim J.S., et al. (2017). AMPK-ULK1-mediated autophagy confers resistance to BET inhibitor JQ1 in acute myeloid leukemia stem cells. Clin Cancer Res 23, 2781–2794 [DOI] [PubMed] [Google Scholar]
- Kerbel R.S. (2008). Tumor angiogenesis. N Engl J Med 358, 2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz M., and Ferrara N. (2006). Vascular endothelial growth factor signaling pathways: therapeutic perspective. Clin Cancer Res 12, 5018–5022 [DOI] [PubMed] [Google Scholar]
- Kuntz E.M., Baquero P., Michie A.M., Dunn K., Tardito S., and Holyoake T.L., et al. (2017). Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med 23, 1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal P.M., and Graziani G. (2018). Therapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacol Res 136, 97–107 [DOI] [PubMed] [Google Scholar]
- Levy J., Towers C.G., and Thorburn A. (2017). Targeting autophagy in cancer. Nat Rev Cancer 17, 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.J., Lei Y.H., Yao N., Wang C.R., Hu N., and Ye W.C., et al. (2017). Autophagy and multidrug resistance in cancer. Chin J Cancer 36, 52. [DOI] [PMC free article] [PubMed]
- Lin S., Mulloy J.C., and Goyama S. (2017). RUNX1-ETO leukemia. Adv Exp Med Biol 962, 151–173 [DOI] [PubMed] [Google Scholar]
- Mascolo M., Vecchione M.L., Ilardi G., Scalvenzi M., Molea G., and Di Benedetto M., et al. (2010). Overexpression of Chromatin Assembly Factor-1/p60 helps to predict the prognosis of melanoma patients. BMC Cancer 10, 63. [DOI] [PMC free article] [PubMed]
- Mowers E.E., Sharifi M.N., and Macleod K.F. (2018). Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J 285, 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J.A., Chang S.H., Dvorak A.M., and Dvorak H.F. (2009). Why are tumour blood vessels abnormal and why is it important to know? Br J Cancer 100, 865–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A., Benedetti R., Conte M., Iside C., and Altucci L. (2015). Genetic mutations in epigenetic modifiers as therapeutic targets in acute myeloid leukemia. Expert Opin Ther Targets 19, 1187–1202 [DOI] [PubMed] [Google Scholar]
- Nguyen T.D., Shaid S., Vakhrusheva O., Koschade S.E., Klann K., and Tholken M., et al. (2019). Loss of the selective autophagy receptor p62 impairs murine myeloid leukemia progression and mitophagy. Blood 133, 168–179 [DOI] [PubMed] [Google Scholar]
- Peng X., Fu H., Yin J., and Zhao Q. (2018). CHAF1B knockdown blocks migration in a hepatocellular carcinoma model. Oncol Rep 40, 405–413 [DOI] [PubMed] [Google Scholar]
- Pistritto G., Trisciuoglio D., Ceci C., Garufi A., and D'Orazi G. (2016). Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 8, 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S., Andreeff M., and Borthakur G. (2017). Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy 13, 214–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Theocharis S.E., Grandin L., Gambotti L., Antoni G., and Savignoni A., et al. (2010). Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology 57, 716–724 [DOI] [PubMed] [Google Scholar]
- Prada-Arismendy J., Arroyave J.C., and Rothlisberger S. (2017). Molecular biomarkers in acute myeloid leukemia. Blood Rev 31, 63–76 [DOI] [PubMed] [Google Scholar]
- Prokocimer M., Molchadsky A., and Rotter V. (2017). Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood 130, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.Z., Zhu H.H., Jiang Q., Xu L.P., Jiang H., and Wang Y., et al. (2018). Heterogeneous prognosis among KIT mutation types in adult acute myeloid leukemia patients with t(8;21). Blood Cancer J 8, 76. [DOI] [PMC free article] [PubMed]
- Qiu L., Zhou G., and Cao S. (2020). Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci 243, 117234. [DOI] [PubMed]
- Robinson M.D., McCarthy D.J., and Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca S., Carra G., Poggio P., Morotti A., and Brancaccio M. (2018). Targeting few to help hundreds: JAK, MAPK and ROCK pathways as druggable targets in atypical chronic myeloid leukemia. Mol Cancer 17, 40. [DOI] [PMC free article] [PubMed]
- Rollig C., Bornhauser M., Thiede C., Taube F., Kramer M., and Mohr B., et al. (2011). Long-term prognosis of acute myeloid leukemia according to the new genetic risk classification of the European LeukemiaNet recommendations: evaluation of the proposed reporting system. J Clin Oncol 29, 2758–2765 [DOI] [PubMed] [Google Scholar]
- Shallis R.M., Wang R., Davidoff A., Ma X., and Zeidan A.M. (2019). Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev 36, 70–87 [DOI] [PubMed] [Google Scholar]
- Slovak M.L., Kopecky K.J., Cassileth P.A., Harrington D.H., Theil K.S., and Mohamed A., et al. (2000). Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 96, 4075–4083 [PubMed] [Google Scholar]
- Staibano S., Mascolo M., Mancini F.P., Kisslinger A., Salvatore G., and Di Benedetto M., et al. (2009). Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology 54, 580–589 [DOI] [PubMed] [Google Scholar]
- Sumitomo Y., Koya J., Nakazaki K., Kataoka K., Tsuruta-Kishino T., and Morita K., et al. (2016). Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood 128, 1614–1624 [DOI] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., and Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1), W98–W102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin R., Grabow S., and Davids M.S. (2018). The rise of apoptosis: targeting apoptosis in hematologic malignancies. Blood 132, 1248–1264 [DOI] [PubMed] [Google Scholar]
- Wan B., Liu B., Yu G., Huang Y., and Lv C. (2019). Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma. Aging (Albany NY) 11, 9025–9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gao L., Guo X., Feng C., Lian W., and Deng K., et al. (2019). Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging (Albany NY) 11, 12246–12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Ding Z., Qian J., Zhang J., Xu J., and Dong X., et al. (2016). Casp3/7-instructed intracellular aggregation of Fe3O4 nanoparticles enhances T2 MR imaging of tumor apoptosis. Nano Lett 16, 2686–2691 [DOI] [PubMed] [Google Scholar]
- Zhou M., Liu X., Li Z., Huang Q., Li F., and Li C.Y. (2018). Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int J Cancer 143, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.