Abstract
A tremendous loss of financial and human resources from seven large-scale HIV vaccine efficacy trials suggest a need for a systematic approach to vaccine selection. We conducted a systematic analysis of three important envelope glycoprotein (Env) vaccine candidates: BG505 SOSIP.664, 1086.C gp140, and 1086.C gp120 to determine the most promising by comparing their structure and antigenicity. We found that the BG505 SOSIP trimer and 1086.C gp140 clearly outperformed the 1086.C gp120 monomer. BG505 SOSIP.664 bound the strongest to the most potent and broadest broadly neutralizing antibodies (bnAbs) PG9, PGT145, VRC01, and PGT121. Of interest, although BG505 SOSIP.664 did not bind to the CH58 mAb, 1086.C gp140 bound strongly to this mAb, which belongs to a class of non-neutralizing antibodies that may be protective based on correlates of protection studies of the RV144 HIV vaccine trial. The 1086.C gp120 monomer was the least antigenic of the three vaccine immunogens, binding the weakest to bnAbs and CH58 mAb. Taken together, the evidence provided here combined with previous preclinical immunogenicity and efficacy data strongly argue that the BG505 SOSIP.664 trimer and 1086.C gp140 are likely to be better vaccine immunogens than the monomeric 1086.C gp120, which was just recently tested and shown to be nonefficacious in a phase IIb/III trial. Thus, to best utilize our financial and valuable human resources, we propose a systematic approach by not only comparing structure and antigenicity, but also immunogenicity and efficacy of Env vaccine candidates in the preclinical phase to the selection of only the most promising vaccine candidates for clinical testing.
Keywords: HIV, vaccine, immunogen, trimer, preclinical trial
Introduction
HIV/AIDS still remains one of the most devastating diseases affecting humans and a serious global public health threat. HIV currently afflicts ∼36.9 million people and has cost 35.4 million lives since the start of the pandemic in the 1980s (UNAIDS 2018 Global HIV and AIDS Statistics). Despite the availability of effective treatment with antiretroviral therapy, a preventive vaccine is desperately needed to stop the spread of HIV infection. The development of vaccine strategies that can elicit either protective B cell or T cell responses or both are being pursued. However, accruing evidence from correlates of protection studies from the human Thai RV144 trial along with passive transfer studies in the nonhuman primate model has shift the focus of efforts more toward the development of HIV-1 envelope glycoprotein (Env) vaccines that will induce protective antibodies to prevent infection.1–3
Induction of broadly neutralizing antibodies (bnAbs) against HIV Envs is a viable vaccine strategy because passive administration of bnAbs can fully protect from infection in the nonhuman primate model of AIDS.4 Moreover, some HIV patients develop potent bnAbs that can cross-neutralize a majority of global HIV isolates in vitro.5,6 Thus far, the most promising Env vaccine for generating bnAbs is the soluble BG505 SOSIP trimer that resembles the native Env spike that expose many bnAb epitopes.7 Initially created in 2013, the BG505 SOSIP trimer has undergone several iterations and with optimized antigen delivery, the trimer now has improved stability as well as antigenicity and immunogenicity.8 Optimized BG505 SOSIP trimers have been shown to elicit neutralizing antibodies with increasing breadth inhibiting most Tier 1, autologous Tier 2, and some heterologous Tier 2 viruses.8 Of importance, BG505 SOSIP trimer vaccine has been shown to protect against autologous Tier 2 SHIV challenge.9 This finding is significant in that circulating Tier 2 viruses seem to be responsible for natural human infections and the epidemic.10 Because of these promising preclinical data, the BG505 SOSIP trimer is now being tested by the International AIDS Vaccine Initiative (IAVI) in a phase I clinical trial (IAVI W001).
Developing a vaccine that elicits protective anti-V1/V2 loop antibodies could also be an effective strategy. The Thailand RV144 was the sole vaccine efficacy trial to indicate that it is possible to protect against HIV by vaccination, with a modest 31.2% protection. Post hoc analysis of the RV144 trial revealed that anti-V1/V2 loop apex functional antibody responses that were non-neutralizing, such as antibody-dependent cellular cytotoxicity (ADCC) correlated with protection.3 The partial success of the RV144 trial has led to the exploration of the Pox vector prime/Env boost approach by a number of laboratories that includes Env from HIV strains such as transmitted/founder 1086.C Clade C virus. The 1086.C Env gp140 is a good candidate because it was highly immunogenic.11,12 The Clade C 1086.C gp120 Env was selected in 2009 as a component of a bivalent vaccine to build on the RV144 results to address the HIV epidemic in sub-Saharan Africa where the majority of the population is infected by the Clade C virus.13 The HIV-1 Clade C-based prime-boost vaccine regimen uses ALVAC-HIV (vCP2438) based on the ALVAC vector backbone (as in RV144) with Clade B (gp41, Gag, and Protease Lai strain) and Clade C (96ZM651 gp120) HIV-1 gene inserts and bivalent subtype C recombinant HIV Env gp120 (1086.C gp120 and TV1.C gp120). This vaccine regimen was just recently tested and found to be nonefficacious in a large phase IIb/III trial (HVTN 702) in Africa.
For over three decades since the discovery of the HIV virus in 1983, there has been an overwhelming effort to develop a vaccine that will halt the HIV pandemic. Seven major efficacy trials (phase IIb/III) have been completed but none of the experimental vaccines tested have demonstrated significant efficacy for preventive measure. To date, >32,000 human volunteers have participated in the six completed efficacy trials.14–19 An additional 8,000 volunteers were scheduled to enroll in the HVTN 702 (Uhambo) and HVTN 705 (Imbokodo) phase IIb/III efficacy trials. Unfortunately, the HVTN 702 efficacy trial just recently was stopped early because the vaccine failed to protect against HIV infection. In addition, many human volunteers have participated in the testing of vaccine candidates in numerous early phase I/IIa trials that were not advanced because of safety issues or lack of immunogenicity. The financial cost and human resources utilized to conduct these trials are tremendous. With many more vaccine immunogens being created and tested on a regular basis, it is imperative that we implement a systematic approach to vaccine candidate selection to better utilize limited financial and precious human resources.
In this study, we propose a systematic approach to candidate Env vaccine selection. To begin, we compared side-by-side the structure and antigenicity of the three aforementioned vaccine candidates being developed to induce protective antibody responses: BG505 SOSIP.664, 1086.C gp140, and 1086.C gp120. We found that overall the BG505 SOSIP.664 trimer outperformed both 1086.C Env immunogens with an ability to bind to the most potent bnAbs. However, interestingly, the 1086.C gp140 was strongly recognized by an anti-V1/V2 loop antibody that may possibly be protective based on correlates of HIV protection studies of the RV144 trial.3 With these selection criteria (structure and antigenicity) along with historical data on the preclinical immunogenicity of these Env vaccines, the BG505 SOSIP.664 trimer and 1086.C gp140 seem to be better vaccine candidates than the monomeric 1086.C gp120, which was recently evaluated to be noneffective in a human HIV vaccine efficacy trial. The profound structural and antigenicity differences among the three Env candidates that we observed in this study strongly argue for a more comprehensive and stringent criteria for Env immunogen selection for testing in human clinical trials.
Materials and Methods
Antibodies and Env proteins
The HIV-1 Env 1086.C D7gp120 and 1086.C gp140C Envs, CD4-IgG2, anti-HIV-1 envelope monoclonal antibodies 2G12, PGT121, 3869, CH58, PG09, PGT145, IgG1 b12, and VRCO1 were obtained from the NIH AIDS Reagents Program (Germantown, MD). The BG505 SOSIP.664 envelope trimer proteins were kindly provided by Dr. John P. Moore, Cornell University (Ithaca, NY).7
Polyacrylamide gel electrophoresis analysis of HIV-1 envelope protein
For reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis, HIV-1 envelope proteins were prepared in Laemli sample buffer containing 10% 2-mercaptoethanol and incubated at 95°C, 5 min. Samples were loaded onto 4%–20% polyacrylamide mini-protean TGX (Bio-Rad Laboratories, Hercules, CA) and electrophoresis was conducted at 90 V for 2 h using 1 × Tris/glycine/SDS buffer (Bio-Rad Laboratories). For western blot analysis, envelope proteins transferred on Invitrolon PVDF membranes (Novex, Carlsbad, CA) were incubated in blocking buffer (0.25% Tween-20, 1% nonfat dried milk, 5% fetal bovine serum in phosphate-buffered saline [PBS]) and detected with 1 μg/mL HIVIg (NIH AIDS Reagent Program) and a secondary antihuman IgG-HRP antibody (Millipore, Billerica, MA). Membranes were then stained using Super Signal West Pico (Thermo Scientific, Rockford, IL) and exposed on CL-XPosure film (Thermo Scientific).
For nondenaturing blue native-PAGE (BN-PAGE), the HIV envelope proteins were resuspended in native PAGE sample buffer and loaded into native PAGE 4%–16% Bis-Tris gel (Invitrogen, Carlsbad, CA). Electrophoresis was conducted at 120 V for 2 h in a light blue solution containing native PAGE running buffer and native PAGE cathode additive (Invitrogen). The HIV proteins were then stained with GelCode Blue Safe Protein Stain (Thermo Scientific) for 1–1.5 h and analyzed using the Innotech FluorchemQ Image III System. For reducing and nonreducing PAGE analysis, Precision Plus Protein ladder (Bio-Rad Laboratories) and NativeMark Protein Standard (Invitrogen) were used as a molecular weight marker, respectively.
Negative staining electron microscopy analysis
Negative staining electron microscopy (NS-EM) was performed at the Harvard Medical School Electron Microscopy Facility (Boston, MA). Carbon-coated meshes were glow discharged at 25 mA for 20 s. The meshes were floated over with 5 μL of sample for 20 s. After drying with filter paper, the meshes were washed for 30 s, dried, and stained with 7.5% m/v uranyl formate. Images were captured on a Tecnai G2 Spirit BioTWIN microscope with 68,000 × direct magnification at 80 kV using Homamatsu ORCA HR camera.
Biolayer interferometry analysis
Biolayer interferometry (BLI) was performed at the Harvard University Center for Macromolecular Interactions (Boston, MA). Antihuman Fc capture biosensors (Fortebio, Fremont, CA) were loaded with 10 μg/mL antibody solution for 2 min. Biosensors underwent a cycle of binding and dissociation as follows: basal signal for 2 min, binding step for 5 min, and dissociation for 10 min. For each antibody, protein dilutions in running buffer (0.1% bovine serum albumin in PBS pH7.4) were assessed ranging from 3.9 to 250 nM. Data were processed using Fortebio software suite applying a global fit model. Binding measurement was obtained using a Fortebio Octet Red 384 system. Binding on-rate constant, Kon was expressed as 1/M × s and off-rate constant, Koff as 1/s. The affinity constant, KD was calculated using the kinetic equation: KD = Koff/Kon and expressed in molarity.
Results
Expression and structural analysis of candidate Env vaccines
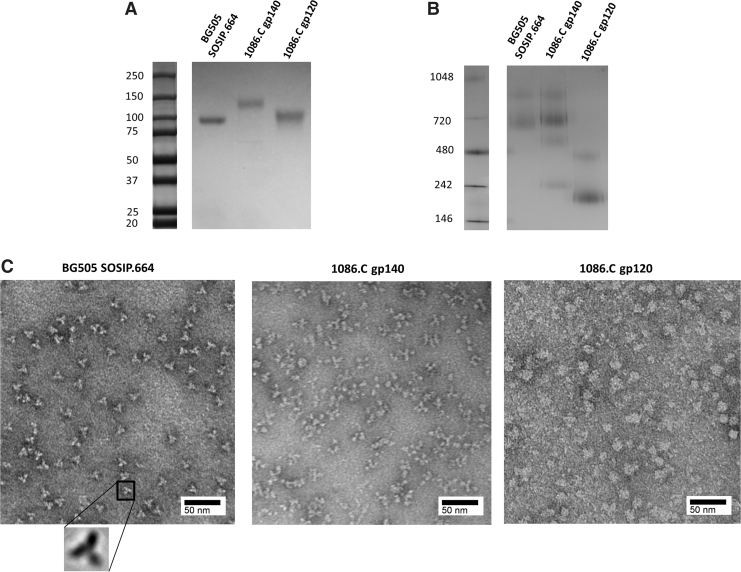
A number of studies have shown that soluble Env trimers are better than Env monomers at binding to bnAbs and are relatively more immunogenic.7,8,20 We analyzed both the expression and structure of BG505 SOSIP.664, 1086.C gp140, and 1086.C gp120. Western blot analysis under reducing condition showed that monomeric BG505 SOSIP.664 and 1086.C gp120 Env proteins have a molecular weight of ∼100 kDa and monomeric 1086.C Env gp140 has a molecular weight of ∼150 kDa (Fig. 1A). Under native condition using BN-PAGE, the BG505 SOSIP.664 and 1086.C gp140 are trimers with an approximate molecular weight of 700 and 720 kDa, respectively, similar to previous studies (Fig. 1B).12 The 1086.C gp120 migrated as a monomer, consistent with previous observation that this Env behaved as a stable monomer in solution (Fig. 1B).13 NS-EM confirms the BG505 SOSIP.664 native-like trimeric structure as previously described (Fig. 1C).7 Although the 1086.C gp140 showed a trimeric migration pattern on BN-PAGE gels, NS-EM analysis showed that 1086.C gp140 did not have the organized trimeric structure of the BG505 SOSIP.664 (Fig. 1C). The 1086.C gp140 formed complexes that were variable in shape and the 1086.C gp120 formed globular structures likely as a result of aggregation (Fig. 1C). As expected, these results demonstrate that only the BG505 SOSIP vaccine candidate and neither the 1086.C gp140 nor 1086.C gp120 was a native-like trimer.
FIG. 1.
NS-EM and PAGE analysis of HIV-1 Env vaccine candidates. (A) Western blot analysis of BG505 SOSIP.664, 1086.C gp140, and 1086.C gp120 Envs under reducing condition. (B) BN-PAGE analysis of BG505 SOSIP.664, 1086.C gp140, and gp120 (2 μg of each protein). The molecular weight of marker (M) proteins are indicated (B, C). (C) NS-EM analysis of BG505 SOSIP.664, 1086.C gp140, and 1086.C gp120 Envs stained with 7.5% uranyl formate. Electron microscopy was performed using Tecnai G2 Spirit BioTWIN at 80 kV. Scale bar, 50 nm. BN-PAGE, blue native–polyacrylamide gel electrophoresis; Env, envelope glycoprotein; NS-EM, negative staining electron microscopy.
Antigenicity and presence of bnAb epitopes on the candidate Env vaccines
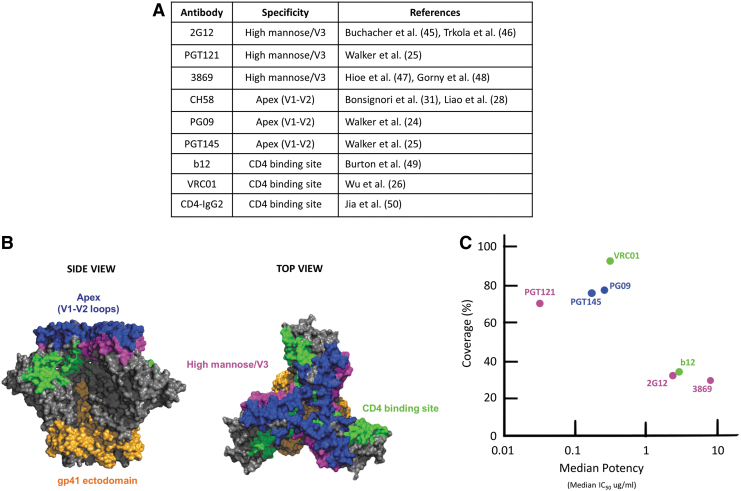
The antigenicity profile assessment of the HIV-1 Env vaccine is critical to demonstrate the presence of epitopes recognized by bnAbs against HIV-1. The presence of the bnAb epitopes on Env vaccines allows for exposure and recognition by B cells for eliciting bnAb responses in the vaccinated host. We analyzed Env antigenicity and the presence of important bnAb epitopes including the high mannose patch/V3, CD4 binding site (CD4bs), and V1-V2 loops (apex) by determining the binding affinity of the candidate Env vaccine immunogens to important non-neutralizing antibodies and bnAbs that recognize these vulnerable epitopes using BLI (Fig. 2A–C). The bnAbs selected have strong broadly neutralizing responses against HIV-1 (Fig. 2A–C).
FIG. 2.
Non-neutralizing antibodies and bnAbs to HIV-1 Env vaccine candidates. (A) Non-neutralizing and bnAbs used in this study with their specificities are listed in the table. (B) The crystal structure of BG505 SOSIP.664 trimer in a side view (left) and top view (right), and the location of bnAb epitopes: apex, V1-V2 loops (blue), CD4 binding site (green), high mannose patch/V3 (pink), and the gp41 ectodomain (orange). (C) Neutralization breadth as presented by neutralization coverage (%) (y-axis) of large panels of global isolates and potency (median IC50, in μg/mL) (x-axis) of bnAbs as determined previously.5,25 bnAbs, broadly neutralizing antibodies. Color images are available online.
Binding to high mannose patch/V3 bnAbs
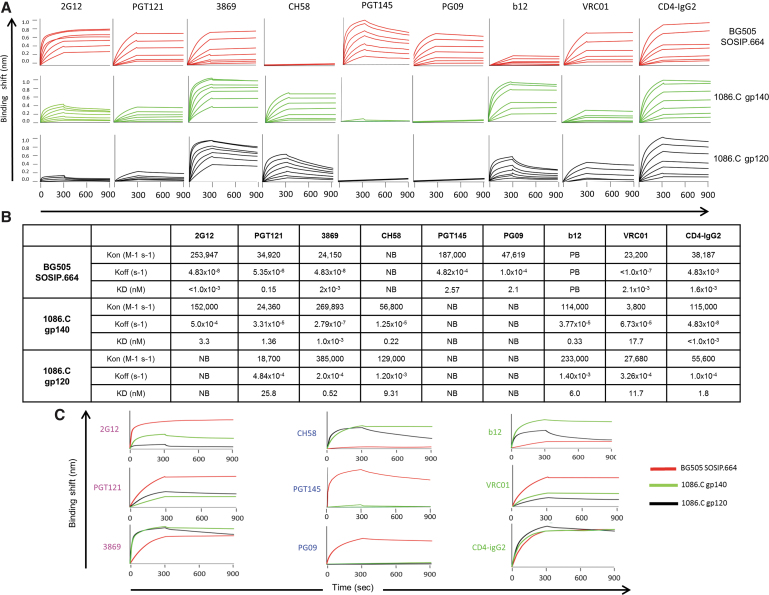
The 2G12, PGT121, and 3869 bnAbs recognize the high mannose patch/V3 (Fig. 2A–C).21–23 PGT121 was previously shown to neutralize roughly 70% of HIV-1 with high potency (Fig. 2C).24,25 BLI analysis revealed that 2G12 bound BG505 SOSIP.664 the strongest (<1 × 10−3 nM KD), followed by 1086.C gp140 (3.3 nM KD) (Fig. 3A–C). 2G12 did not bind the 1086.C gp120 immunogen (Fig. 3A–C). PGT121 bound BG505 SOSIP.664 with the highest affinity (0.153 nM KD), followed by 1086.C gp140 (1.36 nM KD) (Fig. 3A–C). Relatively weaker binding of PGT121 to the 1086.C gp120 (25.8 nM KD) (Fig. 3A–C) was observed. With the 3869 bnAb, binding to BG505 SOSIP.664 and 1086.C gp140 were strong (2 × 10−3 nM KD and 1.0 × 10−3 nM KD, respectively) (Fig. 3A–C), whereas binding affinity to 1086.C gp120 was significantly lower (0.52 nM KD) (Fig. 3A–C). These results show that the high mannose patch/V3 bnAbs bind much more strongly to the BG505 SOSIP trimer followed by the 1086.C gp140. The 1086.C gp120 was poorly antigenic to this class of bnAbs.
FIG. 3.
Comparison of non-neutralizing antibodies and bnAb binding to HIV-1 Env vaccine candidates. (A) BLI curves. Anti-HIV-1 Env bnAbs (2G12, PGT121, 3869, PGT145, PG09, IgG1 b12, and VRC01), the non-neutralizing antibody CH58 and CD4-IgG2 were immobilized on anti-hIgG Fc biosensors. In the association step, each sensor was incubated with Env proteins ranging from 3.9 to 250 nM. Binding curves of BG505 SOSIP.664 (red), 1086.C gp140 (green), and 1086.C gp120 (black) are shown. (B) The table indicates binding on-rate (Kon) and off-rate (Koff) constants and affinity (KD) of anti-HIV-1 Env antibodies to the HIV-1 Env glycoproteins. HIV Env with PB and NB are indicated. (C) BLI curves of BG505 SOSIP.664 (red), 1086.C gp140 (green), and 1086.C gp120 (black) binding to bnAbs (2G12, PGT121, 3869, PG09, PGT145, IgG1 b12, and VRC01), the non-neutralizing antibody CH58 and CD4-IgG2. BLI, biolayer interferometry; NB, no binding; PB, poor binding. Color images are available online.
Binding to CD4bs bnAbs and CD4-IgG2
The bnAbs IgG1 b12 (b12), VRCO1 and CD4-IgG2 recognize and bind to the CD4bs of the HIV-1 envelope trimer (Fig. 2A–C).26,27 BLI binding analysis showed that CD4-IgG2 bound 1086.C gp140 (<1 × 10−3 nM KD) the strongest, followed by BG505 SOSIP.664 (1.6 × 10−3 nM KD), and 1086.C gp120 the weakest (1.8 nM KD) (Fig. 3A–C). With regard to b12 binding, the 1086.C gp140 bound strongly (0.33 nM KD), 1086.C gp120 also bound but with less affinity (6.0 nM KD) (Fig. 3A–C). In contrast, BG505 showed no binding to b12. VRCO1 is one of the most potent bnAbs observed capable of neutralizing ∼91% of HIV-1 viruses (Fig. 2A–C).26 VRC01 bound BG505 with strong affinity (2.08 × 10−3 nM KD) and 1086.C gp140 and gp120 with markedly less affinity (17.7 nM KD and 11.7 nM KD, respectively) (Fig. 3A–C). Taken together, these results show that BG505 trimer contains a bnAb neutralization epitope, recognized strongly by one of the most potent bnAbs, VRC01. Of interest, the CD4bs on 1086.C gp140 is preferentially recognized by b12 and CD4-IgG2 over the BG505 SOSIP.664 (Fig. 3A–C). The 1086.C gp120 showed the weakest binding to the CD4bs bnAb class overall compared with the BG505 SOSIP trimer and 1086.C gp140 Env vaccines (Fig. 3A–C).
Binding to V1-V2 loops (apex)
Anti-apex bnAbs are found to be highly potent capable of neutralizing the majority of HIV-1 viruses. bnAbs to the trimer apex include PG09, which can neutralize roughly 84% of HIV-1 viruses (Fig. 2A–C).28,29 PG09 bound well to BG505 SOSIP.664 trimer (2.1 nM KD) but binding to 1086.C gp140 and gp120 immunogens were undetectable (Fig. 3A–C). The PGT145 binds to the quaternary structure of the V2 apex and is a highly potent bnAb, neutralizing ∼80% of HIV-1 isolates (Fig. 2A–C).25 PGT145 only binds specifically to the HIV-1 Env in trimeric configuration.30 PGT145 bound strongly to BG505 SOSIP.664 trimer (2.57 nM KD) but binding to 1086.C gp140 and gp120 immunogens were not observed (Fig. 3A–C).
Another class of anti-V2 apex antibodies that includes CH58 are nonbroadly neutralizing and are associated with vaccine-induced protection in the RV144 HIV vaccine trial.28,31 CH58 bound 1086.C gp140 with the highest affinity (0.22 nM KD) and 1086.C gp120 with lower affinity (9.31 nM KD) (Fig. 3A–C). Of interest, CH58 did not bind to the BG505 SOSIP.664 trimer (Fig. 3A–C). Taken together, these results show that the BG505 SOSIP.664 trimer is better than the nontrimer 1086.C gp140 and gp120 at binding to PG09 and trimer-specific PGT145 bnAb. However, the 1086.C gp140 clearly is a better immunogen at binding to the RV144 patient CH58 mAb.
Discussion
For almost four decades since the diagnosis of the first case of HIV/AIDS, seven preventive HIV-1 vaccine efficacy trials14–19 and numerous early-phase human trials have failed, amounting to a significant loss of financial and most importantly, human resources. A major lesson learnt from these trials is to establish a more systematic approach to selecting only the most promising vaccine strategies for testing in human clinical trials. To this end, we began to perform a comparative study on three HIV envelope proteins: 1086.C gp120, 1086.C gp140, and BG505 SOSIP that are considered promising HIV vaccine candidates.7,11–13,32 Each of these Env immunogens were selected for vaccine development based on one or more of the following criteria: structure, antigenicity, preclinical immunogenicity, and efficacy trials in small laboratory animals and nonhuman primates. The BG505 SOSIP trimer is currently being tested for immunogenicity and safety in a human phase I clinical trial (IAVI W001). The 1086.C gp120 is a component of a bivalent vaccine that was recently tested and found to be ineffective in preventing HIV infection in a large human vaccine efficacy trial (HVTN 702).33,34
Exterior HIV Envs are in a trimeric configuration on the virion surface. We confirmed in this study that the BG505 SOSIP.664 is a native-like trimer by NS-EM analysis and binding to the trimer-specific PGT145 bnAb. In contrast, the 1086.C gp120 and gp140 Envs were found to exist in monomeric and oligomeric or aggregate forms. HIV-1 gp120 and gp140 glycoproteins have the propensity to form aggregates because of the presence of noncanonical disulfide bonds not seen in BG505 SOSIP.664 trimers.35–37 These noncanonical disulfide bonds often result in aberrant intermolecular disulfide bonds that cause the crosslinking of Env glycoproteins into dimers and aggregates.35–37
Consistent with many studies demonstrating that HIV Env native-like trimer immunogens are better at binding and eliciting bnAbs than nontrimer and monomeric gp120 Env immunogens,20,29,38 we showed in this study that BG505 SOSIP.664 trimer for the most part outperformed both 1086.C gp140 and gp120 immunogens. The BG505 SOSIP trimer showed markedly the strongest affinity to PG09, VRC01, PGT145, and PGT121 that are considered among the most potent and broadest bnAbs.5,6 The BG505 SOSIP trimer strongly bound the bnAbs recognizing the high mannose patch/V3 region (2G12, PGT121, and 3869), the CD4bs bnAb VRC01, and the apex-specific PG09 and the trimer-specific PGT145 bnAbs. However, there was no detectible binding to IgG1 b12 bnAb. The binding of 2G12 to BG505 SOSIP trimer was stronger than previously observed by surface plasmon resonance (SPR) binding analysis probably because of differences between BLI and SPR technologies in dissociation constants measurements especially for some high-affinity antibodies.39 The 1086.C gp140 bound all the antibodies but with less affinity than the BG505 SOSIP trimer. The 1086.C gp140 did not bind to PG09 and PGT145, but it strongly bound to IgG1 b12 bnAb. The 1086.C gp120 monomer showed some affinity to the bnAbs PGT121, 3869, IgG1 b12, and VRCO1 and no binding to 2G12, PG09, and PGT145. However, the 1086.C gp120 monomer bound to these antibodies much weaker than either BG505 SOSIP.664 trimer or 1086.C gp140 immunogen.
Several studies have shown that the presence of bnAb epitopes on Env immunogens including trimers does not translate to immunogenicity in that these Env immunogens often do not elicit broadly neutralizing responses at a reasonable titer in vaccinated animals.40 However, despite the lack of direct correlation between HIV Env antigenicity and immunogenicity, the presence of bnAb epitopes on Env immunogens, especially native-like trimers such as BG505 SOSIP.664, is important for the recognition and induction of B cells capable of producing bnAbs. It must be noted that the structural and antigenicity studies conducted here are only the initial steps for selecting the most promising immunogens in the systematic evaluation of HIV Env vaccine candidates. Testing only Env immunogens that contain bnAb epitopes for further evaluation of immunogenicity in vaccinated animals is warranted.
The next critical steps for vaccine selection are immunogenicity and efficacy studies in laboratory animals. As these studies were not conducted here, we rely on historical immunogenicity and efficacy assessments of the three Env vaccine immunogens that also demonstrate that the BG505 SOSIP trimer is the most promising for possibly inducing bnAb responses.41 BG505 SOSIP.664 and a few other native-like trimer immunogens have been shown to elicit autologous and some heterologous Tier 2 virus neutralization.42 Furthermore, the BG505 SOSIP.664 trimer vaccine was shown to elicit protection against homologous Tier 2 SHIV infection.9 On the contrary, the monomeric envelope immunogens including 1086.C gp120 have only been found to elicit neutralization of Tier 1 viruses43 and protection against Tier 2 SHIV infection in vivo has not been observed.
Of interest, in a human correlate study, host IgG1 binding of the BG505 SOSIP trimer was the best predictor of HIV-1 neutralization breadth in the plasma of HIV-infected individuals,44 which suggests that in humans it might be possible to induce broadly neutralizing responses by immunization with the BG505 SOSIP trimer vaccine. Altogether, there is substantial evidence indicating that the BG505 SOSIP trimer is so far the most viable immunogen for eliciting bnAb responses. However, enthusiasm with the BG505 trimer vaccine is dampened by studies showing that the bnAb responses in vaccinated animals lacked significant potency and breadth against heterologous Tier 2 global virus isolates.40 It will be important to improve trimer immunogens and vaccination strategies to elicit more bnAb responses.
The RV144 trial was the only efficacy trial that showed some protection (31.2%) that was correlated with non-neutralizing antibodies binding to the V1-V2 apex and mediate ADCC.3 Antibodies that bind residue K169 in the V1-V2 region of the HIV-1 envelope correlated with reduced risk of infection. Here we tested Env vaccine binding to CH58 mAb that was one of RV144 vaccine-derived V1-V2 monoclonal antibodies isolated.28 The CH58 mAb recognizes residue K169, which underscores the importance of the CH58 epitope in vaccine-induced protection.28 We found that the 1086.C gp140 is better than the BG505 trimer in that it bound with high affinity to CH58, whereas BG505 trimer did not bind detectibly. Moreover, the 1086.C gp140 bound to CH58 markedly stronger than the 1086.C gp120. This suggests that the 1086.C gp140 would be a better choice than the 1086.C gp120 to improve the effectiveness of RV144 pox prime subunit Env vaccine regimen. It would be important to assess whether the 1086.C gp140 immunogen is capable of eliciting protective CH58-like ADCC responses.
The purpose of this study was to conduct a systematic approach to selecting which Env immunogens should be developed and advanced for clinical testing. Based on the results of our comparative structural and antigenicity studies combined with historical data on the preclinical immunogenicity and efficacy studies in nonhuman primates of the Env vaccines studied here, both the BG505 SOSIP.664 and 1086.C gp140 have outperformed the 1086.C gp120 vaccine immunogen. It is therefore not surprising that just recently, the HIV-1 Clade C gp120 bivalent vaccine regimen, which included the 1086.C gp120 immunogen, failed to protect against HIV infection in the large HVTN 702 vaccine efficacy trial. For financial but most importantly for ethical reasons, we propose in the future a collaborative effort among laboratories to conduct side-by-side comparative structural, antigenicity, immunogenicity, and efficacy of Env vaccine candidates in the preclinical phase to best select and prioritize only the most viable vaccine candidate(s) for human testing.
Acknowledgments
The authors thank the assistance of Kelly Arnett, Center for Macromolecular Interactions, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School for BLI analysis and Maria Ericsson, Electron Microscopy Facility, Harvard Medical School for NS-EM analysis. M.J.C is the founder of Pride Biologics, LLC (Boston, MA).
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Research reported in this publication was supported by NIH/NIDCR grant R01 DE027249 (to M.J.C). A.B and M.J.C. are American Association of Immunologist (AAI) Careers in Immunology fellowship recipients.
References
- 1. Moldt B, Rakasz EG, Schultz N, et al. : Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A 2012;109:18921–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hessell AJ, Rakasz EG, Poignard P, et al. : Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 2009;5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haynes BF, Gilbert PB, McElrath MJ, et al. . Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012;366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Gils M, Sanders R: In vivo protection by broadly neutralizing HIV antibodies. Trends Microbiol 2014;22:550–551 [DOI] [PubMed] [Google Scholar]
- 5. Sok D, Burton D: Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 2018;19:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burton D, Hangartner L: Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 2016;34:635–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanders R, Derking R, Cupo A, et al. : A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 2013;9:e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pauthner M, Havenar-Daughton C, Sok D, et al. : Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Cell 2017;46:1073–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pauthner MG, Nkolola JP, Havenar-Daughton C, et al. : Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 2019;50:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montefiori D, Roederer M, Lynn M, et al. : Neutralization tiers of HIV-1. Curr Opin HIV AIDS 2018;13:128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alexander J, Mendy J, Vang L, et al. : Pre-clinical development of a recombinant, replication-competent adenovirus serotype 4 vector vaccine expressing HIV-1 envelope 1086 clade C. PLoS One2 2013;8:e82380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao H, Tsao C, Alam S, et al. : Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol 2013;87:4185–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zambonelli C, Dey AK, Hilt S, et al. : Generation and characterization of a bivalent HIV-1 subtype C gp120 protein boost for proof-of-concept HIV vaccine efficacy trials in Southern Africa. PLoS One 2016;11:e0157391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flynn NM, Forthal DN, Harro CD, et al. : Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 2005;191:654–665 [DOI] [PubMed] [Google Scholar]
- 15. Pitisuttithum P, Gilbert P, Gurwith M, et al. : Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 2006;194:1661–1671 [DOI] [PubMed] [Google Scholar]
- 16. Buchbinder SP, Mehrotra DV, Duerr A, et al. : Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet (London, England) 2008;372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray GE, Allen M, Moodie Z, et al. : Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: A double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet 2011;11:507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. : Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009;361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 19. Hammer SM, Sobieszczyk ME, Janes H, et al. : Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 2013;369:2083–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sok D, van Gils MJ, Pauthner M, et al. : A recombinant HIV envelope trimer selects for quaternary dependent antibodies targeting the trimer apex. AIDS Res Hum Retrov 2014;30(S1):A7–A8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scanlan C, Pantophlet R, Wormald M, et al. : The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of 132 mannose residues on the outer face of gp120. J Virol 2002;76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorny M, Wang X, Williams C, et al. : Preferential use of the vh5-51 gene segment by the human immune response to code for antibodies against the v3 domain of HIV-1. Mol Immunol 2009;46:917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Julien J, Sok D, Khayat R, et al. : Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 2013;9:e1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walker LM, Phogat SK, Chan-Hui PY, et al. : Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science (80-) 2009;326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker L, Huber M, Doores K, et al. : Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011;477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu X, Yang Z, Li Y, et al. : Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (80-) 2010;329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roben P, Moore JP, Thali M, et al. : Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 1994;68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao HX, Bonsignori M, Alam SM, et al. : Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 2013;13AD;38:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Julien JP, Lee JH, Cupo A, et al. : Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 2013;110:4351–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yasmeen A, Ringe R, Derking R, et al. : Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology 2014;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonsignori M, Pollare J, Moody M: Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol 2012;86:11521–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pauthner M, Nkolola J, Havenar-Daughton C: Vaccine-induced protection from homologus tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 2018;50:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. NIAID: First new HIV vaccine efficacy study in seven years has begun. Available at https://www.niaid.nih.gov/news-events/first-new-hiv-vaccine-efficacy-study-seven-years-has-begun, accessed March24, 2020
- 34. Evaluating the safety and immunogenicity of ALVAC-HIV and MF 59®- or AS01B-adjuvanted bivalent subtype C gp120 in healthy, HIV-uninfected adult participants (HVTN 120). Available at https://clinicaltrials.gov/ct2/show/NCT03122223, accessed March24, 2020
- 35. Finzi A, Pacheco B, Zeng X, et al. : Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods 2010;168:155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Owens R, Compans R: The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 1990;179:827–833 [DOI] [PubMed] [Google Scholar]
- 37. Go E, Cupo A, Ringe R, et al. : Native conformation and canonical disulfide bond formation are interlinked properties of HIV-1 Env glycoproteins. J Virol 2016;90:2884–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bale S, Martiné A, Wilson R, et al. : Cleavage-independent HIV-1 trimers from CHO cell lines elicit robust autologous tier 2 neutralizing antibodies. Front Immunol 2018;9:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang D, Singh A, Wu H, et al. : Comparison of biosensor platforms in the evaluation of high affinity antibody-antigen binding kinetics. Anal Biochem 2016;508:78–96 [DOI] [PubMed] [Google Scholar]
- 40. Torrents de la Peña A, Sanders R: Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies. Retrovirology 2018;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanders R, Moore J: Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev 2017;275:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanders RW, Van Gils MJ, Derking R, et al. : HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science (80-) 2015;349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kovacs J, Nkolola J, Peng H, et al. : HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci U S A 2012;190:12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kadelka C, Liechti T, Ebner H, et al. : Distinct, IgG1-driven antibody response landscapes demarcate individuals with broadly HIV-1 neutralizing activity. J Exp Med 2018;215:1589–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buchacher A, Predl R, Strutzenberger K, et al. : Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retrov 1994;10:359–369 [DOI] [PubMed] [Google Scholar]
- 46. Trkola A, Purtscher M, Muster T, et al. : Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 1996;70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hioe CE, Wrin T, Seaman MS, et al. : Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One 2010;21:e10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gorny MK, Williams C, Volsky B, et al.: Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol 2006,80:6865–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burton DR, Pyiati J, Koduri R, et al. : Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 1994;266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 50. Jia M, Lu H, Markowitz M, et al. : Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J Virol 2016;90:4017–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]