Abstract
Altered gene expression is a characteristic feature of many disease states such as tumorigenesis, and in most cancers, it facilitates cancer cell survival and adaptation. Alterations in global gene expression is strongly impacted by post-transcriptional gene regulation. The RNA binding protein (RBP) HuR (ELAVL1) is an established regulator of post-transcriptional gene regulation and is overexpressed in most human cancers. In many cancerous settings, HuR is not only over-expressed, but it is “over-active” as denoted by increased subcellular localization within the cytoplasm. This dysregulation of HuR expression and cytoplasmic localization allows HuR to stabilize and increase the translation of various pro-survival mRNAs involved in the pathogenesis of numerous cancers and various diseases. Based on almost twenty years of work, HuR is now recognized as a therapeutic target. Herein, we will review the role HuR plays in the pathophysiology of different diseases and ongoing therapeutic strategies to target HuR. We will focus on three ongoing-targeted strategies: i) inhibiting HuR’s translocation from the nucleus to the cytoplasm; ii) inhibiting the ability of HuR to bind target RNA; and iii) silencing HuR expression levels. In an oncologic setting, HuR has been demonstrated to be critical for a cancer cell’s ability to survive a variety of cancer relevant stressors (including drugs and elements of the tumor microenvironment) and targeting this protein has been shown to sensitize cancer cells further to insult. We strongly believe that targeting HuR could be a powerful therapeutic target to treat different diseases, particularly cancer, in the near future.
Graphical Abstract
HuR, an RNA binding protein, is critical for many disease states and is particularly important for cancer survival and resistance to therapeutics. As shown, we highlight three current strategies being utilized to target HuR.
Introduction
RNA binding proteins (RBPs) are critical regulators of post-transcriptional gene regulation, in large part through effecting target mRNA localization, stability and translation (Pereira, Billaud, & Almeida, 2017). Individual RBPs can frequently act upon hundreds or thousands of target mRNAs, and thus, are important for driving tissue differentiation and maintaining homeostasis. Unsurprisingly, dysregulated RBPs can lead to various disease states (Lukong, Chang, Khandjian, & Richard, 2008). In particular, many RBPs have an impact on both cancer tumorigenesis (Pereira et al., 2017) and cancer cell survival under stressful tumor microenvironment settings. Of all the RBPs, one of the best characterized is Human Antigen R (HuR), the protein product of embryonic lethal and abnormal vision gene ELAVL1. HuR, among the entire ELAVL family members consisting of HuB (ELAVL2), HuC (ELAVL3), and HuD (ELAVL4), is the only one to be ubiquitously expressed in all human tissues, with the others being primarily expressed in neuronal cells (Good, 1995; Hinman & Lou, 2008). In part due to its ubiquitous expression, HuR has been demonstrated to be functionally important and associated with a number of disease states including cancer (Srikantan & Gorospe, 2012; J. Wang et al., 2013).
In general, HuR’s functional activity is regulated by dynamic subcellular localization (Grammatikakis, Abdelmohsen, & Gorospe, 2017; Hinman & Lou, 2008; H. H. Kim et al., 2008). Under normal cellular and physiologic conditions, HuR is primarily located in the nucleus, but upon exposure to intrinsic and/or extrinsic stress, HuR can translocate to the cytoplasm where it stabilizes and increases the translation of target mRNAs. Chronic activation of HuR leads to an inflammatory phenotype which underlies HuR’s contribution to many disease states, and is also thought to be a large part of HuR’s ability to promote tumorigenesis (Di Marco et al., 2005; Matsye et al., 2017; Nabors, Gillespie, Harkins, & King, 2001; W. Peng et al., 2018; Shin et al., 2016). Beyond this promotion of tumorigenesis, HuR has also been shown to be a lynchpin for driving resistance to a variety of stressful conditions that cancer cells face (Amreddy et al., 2018; Badawi, Hehlgans, Pfeilschifter, Rodel, & Eberhardt, 2017; Blanco, Jimbo, et al., 2016; Cai et al., 2019; Chand et al., 2017; Hostetter et al., 2008; G. L. Lin, Ting, Tseng, Juang, & Lo, 2017; Z. M. Liu, Tseng, Hong, & Huang, 2011; Mehta et al., 2016; Zarei et al., 2017; R. Zhang & Wang, 2018). For instance, intrinsic factors found within the tumor microenvironment such as hypoxia and hypoglycemia have been demonstrated to activate HuR (Levy, Chung, Furneaux, & Levy, 1998; Zarei et al., 2017), as have extrinsic factors such as radiation, chemotherapeutics and targeted agents which can all engage HuR’s RBP function as well (Blanco, Jimbo, et al., 2016; Chand et al., 2017; Filippova et al., 2011; Hostetter et al., 2008; Mazan-Mamczarz et al., 2003). HuR’s role in cancer cell therapeutic resistance has led to a new line of investigation, where targeting HuR can make cancer cells more sensitive to existing therapeutic strategies (e.g., chemotherapies)(Amreddy et al., 2018; Blanco, Jimbo, et al., 2016; Cai et al., 2019; Chand et al., 2017; Guo et al., 2016; Hostetter et al., 2008). While this strategy of targeting HuR to sensitize cancers to anti-cancer agents has been established in pre-clinical models, to date this strategy has been underutilized in the clinical setting. The paucity of clinical trials in this arena is primarily due to a lack of clinically available HuR inhibitors.
This review will detail the background of HuR in relation to the pathobiology of different diseases and focus on methods that have been utilized to target HuR. The strategies we will focus on include targeting: i) the translocation of HuR, ii) the ability of HuR to bind to target mRNA, and iii) the expression of HuR through nanoparticle delivery of siRNA oligonucleotides directed against HuR mRNA.
THE DISCOVERY OF ELAVL1, STRUCTURE AND FUNCTION
HuR, the protein product of the ELAVL1 gene, was first cloned and characterized in 1996 (Ma, Cheng, Campbell, Wright, & Furneaux, 1996) (Figure 1). While the other members of the ELAVL gene family (HuB, HuC and HuD) are almost exclusively expressed in neurons, HuR is ubiquitously expressed in human tissues (Good, 1995; Hinman & Lou, 2008). Functionally, HuR contains three RNA recognition motif (RRM) domains with which it binds target mRNAs (Figure 2) (Pabis et al., 2019; H. Wang et al., 2013). HuR primarily binds target mRNAs containing AU-rich elements (AREs) (Ripin et al., 2019) embedded primarily in the 3’UTR of target mRNAs. AREs predominantly function to destabilize mRNA through accelerated de-adenylation; however, when HuR is “active,” as an RBP, it is hypothesized that HuR competes for these ARE sites and confers increased stability and translation of bound mRNA targets (Blanco, Jimbo, et al., 2016; Lim, Lee, Joo, Song, & Choi, 2009; Winzen et al., 2004; Zarei et al., 2017).
Figure 1. Timeline of HuR discoveries, and discoveries specifically targeting HuR’s role in cancer:
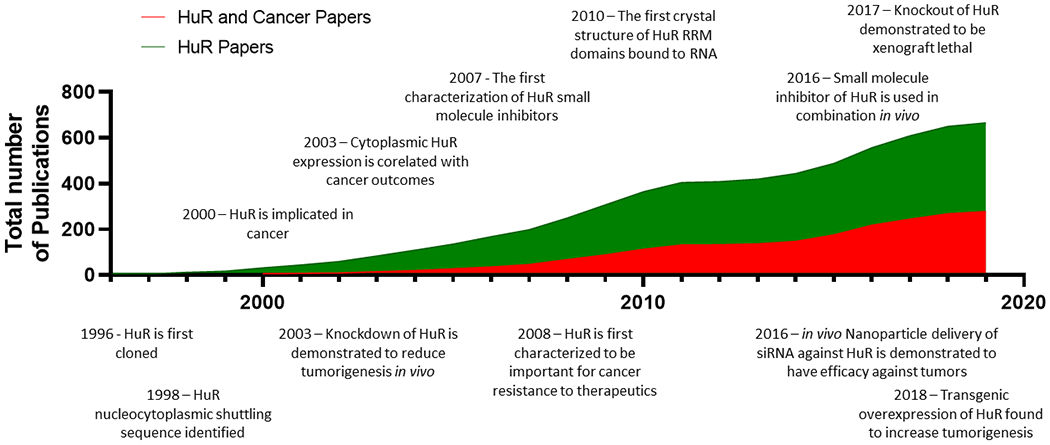
HuR was first cloned in 1996, however it has only been in the last decade that HuR has been more widely accepted as an important target in cancer. This chart details the total number of publications focused on HuR, and relevant discoveries relating to targeting HuR in cancer.
Figure 2. Structure and Function of HuR.
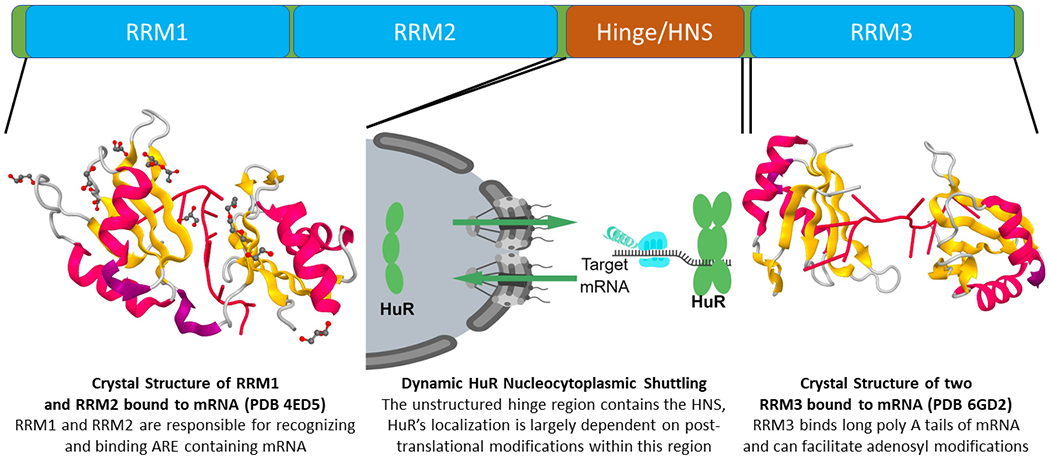
The crystal structure of RRMs 1 and 2 bound to RNA has been determined, as has that of two RRM3 (as could potentially be observed with an HuR dimer) moieties bound to RNA. While the crystal structure of the hinge region and thus the HNS has not been determined, studies have confirmed the overall importance of post-translational modifications in this region governing the localization of HuR between the cytoplasm and the nucleus.
“Active” HuR can be thought of as when HuR shuttles from the nucleus to the cytoplasm where HuR, as an RBP, stabilizes and increases the translation of target mRNAs (Blanco, Preet, et al., 2016; Guo et al., 2016; S. S. Peng, Chen, Xu, & Shyu, 1998). Studies have shown that HuR translocation is governed by the HuR Nucleocytoplasmic Shuttling sequence (HNS) located between the second and third RRM domains within the unstructured hinge region (Figure 2) (Fan & Steitz, 1998). Under certain cellular conditions, post-translational modifications in the HNS region can govern the dynamic subcellular localization of HuR by altering the ability of HuR to interact with co-factors such as importin α1 or transportin1, which are respectively required for import or export through the nuclear pore complex (Brennan, Gallouzi, & Steitz, 2000). As the altered expression of HuR and/or altered subcellular localization is a consistent feature observed in many disease states, this facet of HuR dysregulation underscores the significant role it plays in disease etiology.
HuR’s role in the pathobiology of disease
Cellular and tissue differentiation rely on coordinated large-scale regulation of target transcripts as part of the process of guiding signaling cascades that direct differentiation and homeostasis. HuR plays a key role in these pathways due to its ubiquitous expression and the ability to regulate thousands of targets. The importance of HuR in developmental biology is underscored by studies in mice demonstrating that loss of HuR leads to a midgestational lethal phenotype ((MGD), 2019). HuR has also been implicated later in development as shown by the key role it plays in muscle development (Beauchamp et al., 2010; Cuneo, Herrick, & Autieri, 2010; Figueroa et al., 2003; F. Li et al., 2010; van der Giessen, Di-Marco, Clair, & Gallouzi, 2003; van der Giessen & Gallouzi, 2007; von Roretz, Beauchamp, Di Marco, & Gallouzi, 2011), erythropoiesis (X. Li et al., 2014; X. Zhou et al., 2019) and immune cell differentiation (Diaz-Munoz et al., 2015). Beyond the developmental stages, HuR is still critical for organism survival, as postnatal HuR deletion is still lethal due to the loss of progenitor cells (Ghosh et al., 2009). As HuR is ubiquitously expressed, critical for developmental processes and differentiation, and critical for tissue homeostasis, it is not surprising that HuR dysregulation can contribute to disease phenotypes.
It is believed that chronic activation/cytoplasmic localization of HuR can facilitate the etiology of certain diseases by causing aberrant mRNA expression changes associated with inflammation(Srikantan & Gorospe, 2012). In normal cells, HuR principally resides in the nucleus where it can effect splicing (Akaike et al., 2014; Chang et al., 2014; Gauchotte et al., 2017; Izquierdo, 2008, 2010; Lebedeva et al., 2011; Lema et al., 2017; Srikantan & Gorospe, 2011) and alternative polyadenylation (Dutertre et al., 2014) of targets. Chronic activation and cytoplasmic localization of HuR; however, can lead to a strong pro-inflammatory response governed by HuR’s stabilization of pro-inflammatory cytokines such as IL-6, IL-8, TGF-β, TNF-α, IFN-γ, CCR6, pro-inflammatory enzymes COX-2 and iNOS (J. Chen et al., 2017; Di Marco et al., 2005; Gurgis et al., 2015; Matsye et al., 2017; Nabors et al., 2001; Shin et al., 2016; J. G. Wang et al., 2006; H. Zhou et al., 2007), and the inflammatory marker CRP (Y. Kim et al., 2015) among others. This inflammatory phenotype is in large part the driving force behind HuR’s implication in heart related diseases including vascular inflammation and atherosclerosis (Barton & Meyer, 2019; Fu, Zhai, & Yuan, 2018; F. Y. Lin et al., 2006), and inhibition of HuR has even been proven to be beneficial in animal models of cardiac hypertrophy (Green et al., 2019).
HuR dependent inflammation has also been linked to chronic diseases related to inflammation such as pancreatitis (W. Peng et al., 2018), rheumatoid arthritis (Sugihara et al., 2007; Suzuki et al., 2006), asthma (Atasoy et al., 2003; Esnault & Malter, 2003), and cachexia (Di Marco et al., 2005). Counterintuitively, HuR can drive an anti-inflammatory phenotype in myeloid cells leading to a phenotype where, when HuR is inhibited in myeloid cells, it can lead to an inflammatory phenotype (Christodoulou-Vafeiadou et al., 2018; Yiakouvaki et al., 2012). Still, in most studies and models, HuR drives the activation of a pro-inflammatory phenotype, and thus targeting HuR in most cases is expected to cause a decrease in inflammation, which is in most cases thought to inhibit disease progression.
All of the other ELAVL family members (HuB, HuC, HuD) have been shown to be critical to normal neuronal health, and thus, the dysregulation of these proteins has been linked to various neuronal disorders (Berto, Usui, Konopka, & Fogel, 2016; De Santis et al., 2019; Ogawa et al., 2018). Similarly, dysregulated HuR has been linked to several neuronal diseases (De Santis et al., 2019; Farooq et al., 2013; Farooq, Balabanian, Liu, Holcik, & MacKenzie, 2009; X. Li et al., 2009; Y. J. Liu, Lee, Lai, & Chern, 2015; Lu et al., 2014; Matsye et al., 2017; Perera et al., 2016; Wu et al., 2017). For instance, HuR has been shown to bind and potentially destabilize the neurofibromin transcript (Haeussler et al., 2000) contributing to the pathogenesis of Neurofibromatosis type 1 disorder. HuR has been characterized to affect the progression of Spinal Muscular Atrophy (SMA), which is caused by deficiency in survival motor neuron (SMN) protein encoded by SMN1 and to a lesser extent by SMN2 due to splicing defects inherent in SMN2. HuR forms aggregates with SMN protein (Perera et al., 2016), and can bind SMN RNA, stabilizing it and increasing SMN protein expression (Farooq et al., 2013; Farooq et al., 2009), which overall has a protective effect in this disease setting. To add to the complexity of HuR’s role in SMA, HuR has also been demonstrated to repress splicing of SMN2, and patients with SMN2 mutations causing a deficiency in HuR binding to SMN2 RNA have a less severe phenotype (Wu et al., 2017). Taken together, HuR may attenuate levels of SMN2 RNA through splicing in the nucleus, and thus, modulate SMN protein expression, but once in the cytoplasm, HuR may increase SMN mRNA stability and increase SMN protein expression. Future work may provide evidence that HuR is a target in a subset of these patients with Neurofibromatosis type I disorder.
HuR has also been demonstrated to have dual roles in the progression of amyotrophic lateral sclerosis (ALS), a disease characterized by loss of muscle motor neurons. Inhibiting HuR dependent regulation of VEGF, TDP-43 and FUS/TLS has been posited as a driver of ALS, and has been shown to cause neuronal toxicity (Lu et al., 2014). HuR has been shown to be inhibited in models of ALS either by mutated SOD1 (X. Li et al., 2009; Lu et al., 2007) or through the nuclear sequestration of HuR by over-activation of AMPK; both methods of HuR inhibition have been demonstrated as drivers of this disease (Y. J. Liu et al., 2015). Conversely, HuR is up-regulated in SOD1 mutant microglia (an ALS model), and inhibition of HuR (via siRNA or a HuR inhibitor) in these microglia inhibits their invasiveness and inflammatory properties (Matsye et al., 2017), leading to a less severe disease phenotype. These models, where HuR regulates both detrimental and protective pathways, demonstrates the complexity of studying a multifaceted molecule such as HuR in different models and cell types.
As HuR is ubiquitously expressed in humans, some parasitic organisms co-opt HuR biology to facilitate disease progression(Barnhart, Moon, Emch, Wilusz, & Wilusz, 2013). For example, viral infection normally causes the activation of HuR, and some viruses exploit active HuR function to preferentially stabilize viral RNAs (Jehung et al., 2018). HuR has been implicated to facilitate the infection of Hepatitis C virus (Korf, Jarczak, Beger, Manns, & Kruger, 2005; Shwetha et al., 2015), Sindbis virus (Sokoloski et al., 2010), Kaposi’s sarcoma herpes virus (Yoo et al., 2010), and HIV (Lemay et al., 2008), although HuR binding to HIV specific transcripts is debated (Ahn et al., 2010). In the case of HIV, HuR can also drive an inflammatory phenotype in response to HIV protease inhibitors commonly used to treat HIV, which can further exacerbate disease related complications (Zha et al., 2010; H. Zhou et al., 2007). These findings illustrate that viral infection is yet another example of HuR's involvement in the pathogenesis of different diseases, and thus may be a viable therapeutic target in some of these instances.
HuR as critical mediator of cancer progression
HuR has been studied as an important driver and facilitator of cancer for almost twenty years and has been identified to be a critical target in numerous models of cancer. HuR has been found to regulate and contribute to almost every hallmark of cancer (Hanahan & Weinberg, 2011): i) sustaining proliferative signaling (Holmes et al., 2018; W. Wang, Caldwell, Lin, Furneaux, & Gorospe, 2000; Yuan, Sanders, Ye, Wang, & Jiang, 2011; Z. Zhang, Huang, Zhang, & Zhou, 2017); ii) evading suppression of growth (Balkhi et al., 2013; Ghosh et al., 2009); iii) promoting invasion and metastasis (Z. Li, Wang, Hu, Xu, & Xu, 2018); iv) enabling replicative immortality (Tang et al., 2018); v) inducing angiogenesis (Goldberg-Cohen, Furneauxb, & Levy, 2002; Levy et al., 1998; Osera et al., 2015); vi) resisting cell death (Blanco, Jimbo, et al., 2016; Guo et al., 2016; G. L. Lin et al., 2017; H. Zhu et al., 2015), vii) deregulating cellular energetics (Cascajo et al., 2016; Diaz-Munoz et al., 2015; Zarei et al., 2017) viii) promoting tumor-associated inflammation (W. Peng et al., 2018; S. Sun et al., 2016), ix) and avoiding immune destruction (Brauss et al., 2017). Further underscoring the importance of HuR as a target in cancer, increased levels of HuR have been associated with tumor aggressiveness and poor outcomes in numerous tumor types (Miyata et al., 2013). For instance, in oral, ovarian, urothelial, esophageal, breast, lung, colorectal (CRC), and pancreatic (PDAC) cancers, high cytoplasmic HuR accumulation has been shown to correlate with worse histological grading, increased incidence of lymphatic spread and distant metastasis, and worse clinical outcomes (Danilin et al., 2010; Denkert et al., 2006; Grammatikakis et al., 2017; Lim et al., 2009; Miyata et al., 2013; Richards et al., 2010; Zucal et al., 2015).
Over the past almost twenty years there have been over a hundred papers demonstrating the importance of HuR in vitro in cancer cells; and genetic approaches used to inhibit HuR (e.g. siRNA or gene deletion) have recently been established as a potent means to inhibit tumor growth in vivo (Chand et al., 2017; Danilin et al., 2010; Filippova et al., 2011; Giammanco et al., 2014; P. Lal et al., 2017; Lopez de Silanes et al., 2003; H. Wang et al., 2018; Zarei et al., 2017). Interestingly, knockout of HuR utilizing clustered regularly interspersed palindromic sequence (CRISPR-Cas9) methodology in both PDAC and CRC cell lines demonstrated that lack of HuR severely attenuated in vivo tumor growth as compared to isogenic cells with proficient HuR (S. Lal et al., 2017). The engraftment potential of these knockout lines was rescued by re-expressing HuR or expressing HuR targets, which are frequently lost concomitantly with HuR. For example, IDH1 expression was shown to be lost in HuR knockout cells, and overexpression of IDH1 rescued the engraftment potential in HuR null lines (Zarei et al., 2017) in PDAC cells. While these pre-clinical models demonstrate the significant role HuR plays in tumorigenesis, targeting HuR alone may prove to be a challenge in the clinical setting and HuR inhibition is likely to be most effective in combination with other therapeutic strategies similar to the majority of cancer therapies.
TARGETING HUR TO SENSITIZE CANCER CELLS
As HuR is quickly activated by a number of anti-cancer therapies and can regulate numerous pro-survival pathways, targeting HuR can be viewed as a promising sensitizer for cancer therapies (Blanco, Jimbo, et al., 2016). For example, inhibition of HuR has been demonstrated to sensitize tumor cells to cancer associated stressors such as chemotherapeutics (Amreddy et al., 2018; Blanco, Jimbo, et al., 2016; Cai et al., 2019; Filippova et al., 2011; Hostetter et al., 2008; G. L. Lin et al., 2017; Z. M. Liu et al., 2011; Zarei et al., 2017), radiation (Badawi et al., 2017; Mehta et al., 2016; R. Zhang & Wang, 2018), targeted agents (Chand et al., 2017; Hostetter et al., 2008), hypoxia (Blanco, Jimbo, et al., 2016; Gauchotte et al., 2017), low glucose (Zarei et al., 2017; Zarei et al., 2019) and apoptosis activators (TRAIL, Caspases, apoptasome) (Badawi et al., 2017; Durie et al., 2011; Lal, Kawai, Yang, Mazan-Mamczarz, & Gorospe, 2005; Romeo et al., 2016) (Figure 3).
Figure 3. HuR as a sensitizer.
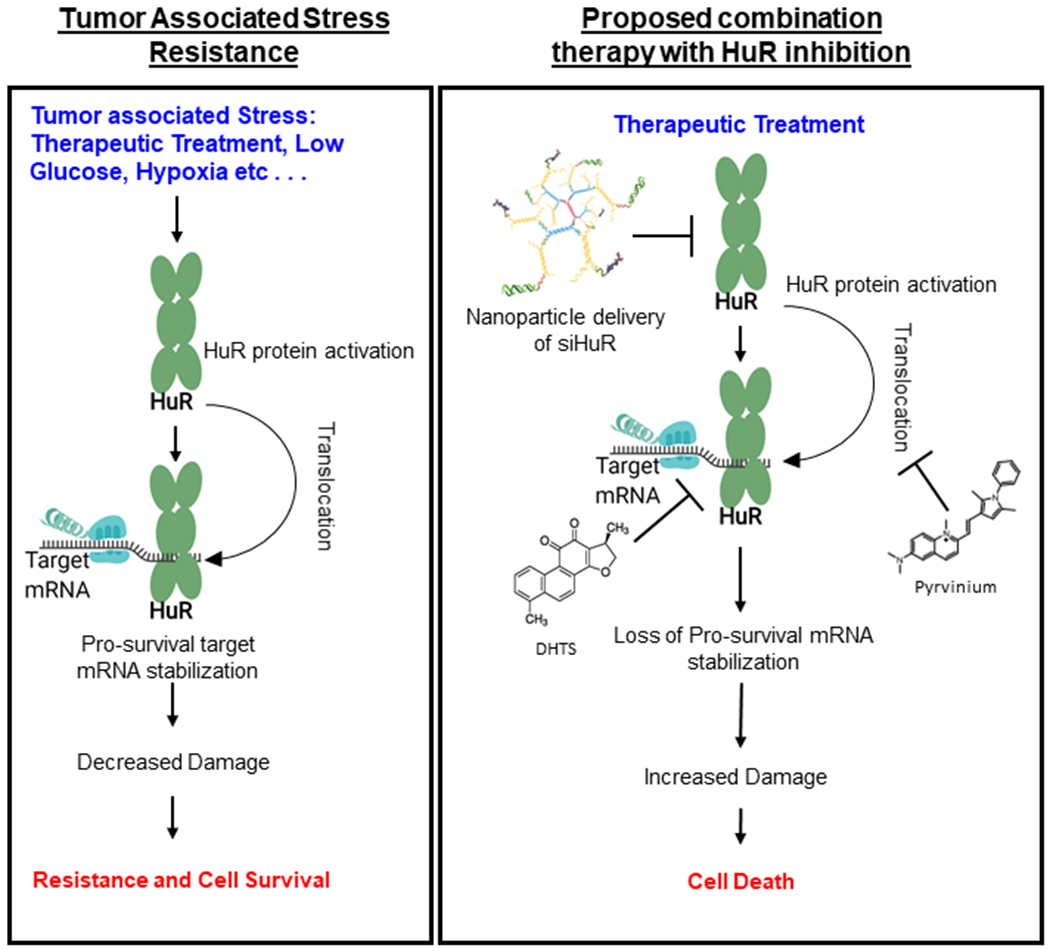
HuR inhibition has been used as a successful means to sensitize cancer calls. Standard of care chemotherapeutics cause damage which activates HuR causing a rapid response and stabilization of pro-survival mRNA. This leads to cell survival and chemotherapeutics resistance. Inhibition of HuR in response to DNA damaging agents can be accomplished through either inhibition of HuR expression (here shown with siRNA carrying nanoparticles), HuR translocation (here shown with pyrvinium), or target binding (here shown with DHTS), all of which would in this case lead to the loss of regulation of targets such as PIM1 or CDC6 and enhanced cell sensitivity.
Recently, HuR has been shown to be critical for resistance to the DNA damaging agent oxaliplatin in both normoxia through the regulation of CDC6 (Cai et al., 2019), and particularly in the setting of hypoxia through the regulation of the serine/threonine kinase involved in cell survival Pim1 (Blanco, Jimbo, et al., 2016). In hypoxia, the knockdown of either Pim1 or HuR causes a 3 to 10-fold increase in sensitivity to oxaliplatin (Blanco, Jimbo, et al., 2016). Furthermore, HuR has been shown to be critical for resistance to radiotherapy in breast and colorectal cancers (Badawi et al., 2017; Mehta et al., 2016). This was demonstrated through an approximately 2-fold increase in sensitivity upon HuR inhibition to low levels of radiation, with observed increases in γ-H2AX foci and ROS production (Mehta et al., 2016). This sensitization was demonstrated to be largely caused through inactivation of thioredoxin and an increase in Caspase 2 (Badawi et al., 2017; Mehta et al., 2016). The increase in Caspase 2 is intriguing, as HuR has been demonstrated to decrease translation of Caspase 2L through sequestration of the mRNA transcript, and loss of HuR causes an increase in protein expression (Winkler et al., 2014). This dual function of increasing the expression of pro-death proteins and decreasing the expression of pro-survival proteins makes targeting HuR applicable to sensitizing cancer cells to a wide variety of therapeutics. Thus, targeting HuR for sensitizing cancer cells to further insult may be key when ultimately evaluating the efficacy of potential HuR inhibitors for the clinic.
Tools for targeting HuR
As many laboratories have clearly implicated HuR’s role in tumorigenesis and pro-survival, various laboratories have initiated programs to develop inhibitors of HuR. Three main methods for inhibiting HuR that have been explored in the field (Figure 4). First, HuR has been targeted via inhibiting its cytoplasmic localization. Second, HuR has been targeted by inhibiting its ability to bind target mRNAs. Third, HuR has been targeted via decreasing its expression, which is normally accomplished via silencing with siRNA oligonucleotide delivery.
Figure 4. Methods of Targeting HuR:
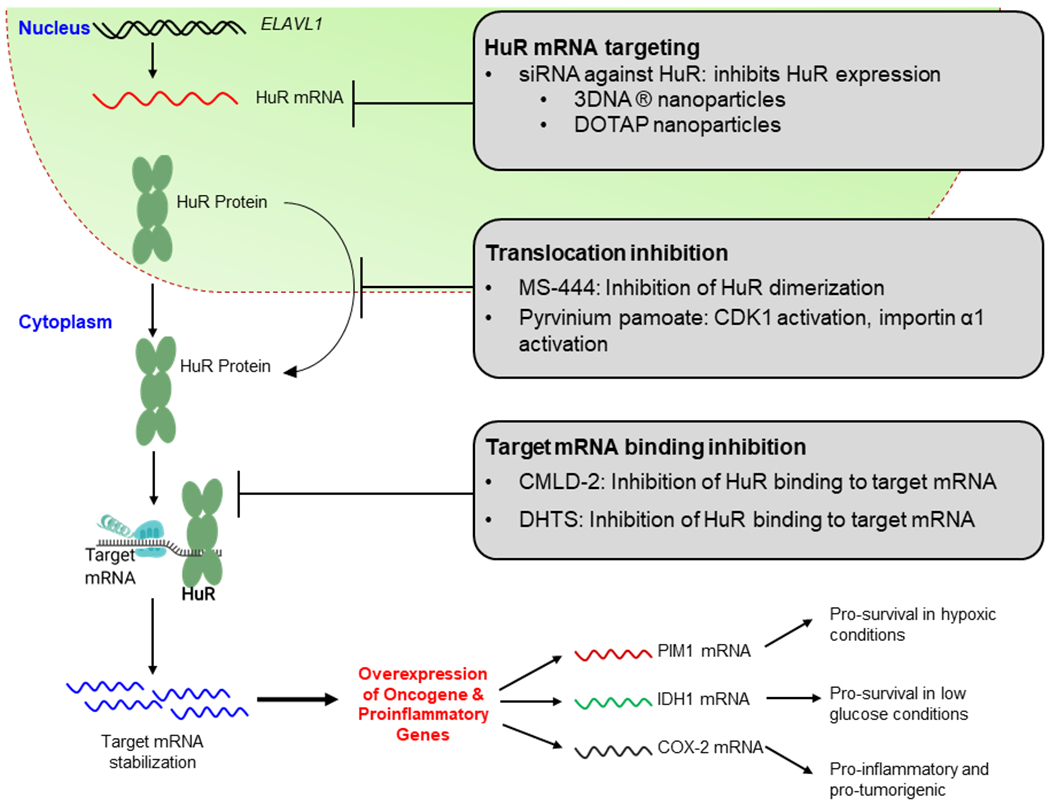
HuR is commonly targeted in one of three ways. 1) Targeting HuR’s expression levels, 2) translocation, and 3) target mRNA binding. There are various potential therapeutics that target HuR at each one of these 3 steps. Overall, the inhibition of HuR leads to an inhibition of the tumorigenic pathways that HuR promotes.
Inhibiting HuR Translocation
As stated previously, HuR in normal tissues is primarily localized to the nucleus where it can function in splicing (Akaike et al., 2014; Izquierdo, 2008; H. Zhu, Zhou, Hasman, & Lou, 2007) and alternative polyadenylation (Izquierdo, 2008; H. Zhu et al., 2007). However, HuR’s regulation of target stability in the cytoplasm is thought to be central to its pro-tumorigenic function (Fus, Pihowicz, Koperski, Marczewska, & Gornicka, 2018; Gallouzi et al., 2000; Gauchotte et al., 2017; Guo et al., 2016; Hostetter et al., 2008; Lim et al., 2009; Melling et al., 2016; Miyata et al., 2013; Nowotarski & Shantz, 2010; Tatarian et al., 2018; Toyota et al., 2018; D. Wang et al., 2014; Z. Zhu et al., 2013). As HuR cytoplasmic localization is commonly observed in cancer, inhibiting the translocation of HuR from the nucleus to the cytoplasm can be thought of as tantamount to inhibiting HuR’s ability to promote tumor progression. HuR translocates reversibly from the nucleus to the cytoplasm through the nuclear pore complex. This translocation is mediated by the association of HuR with cofactors such as transportin-1, transportin-2, importin-α1, and 14-3-3θ (H. H. Kim & Gorospe, 2008; Rebane, Aab, & Steitz, 2004; W. Wang et al., 2004), all of which when activated mainly lead to HuR’s nuclear localization. Conversely, the association of HuR with ANP32B, ANP32A, and XPO1 (Fries et al., 2007; Gallouzi, Brennan, & Steitz, 2003; Gravina et al., 2014; Williams et al., 2010) have been shown to be key mediators of HuR export. HuR localization may also be dependent upon RNA Polymerase II activity. This was demonstrated using a chimeric Pyruvate Kinase containing a portion of HuR’s hinge region containing the HNS (amino acids 205–237). This chimeric protein maintained nuclear localization, however treatment with the transcriptional inhibitor actinomycin D inhibited this nuclear retention (Fan & Steitz, 1998).
There is a complex network of proteins that interacts with HuR to determine HuR’s subcellular localization and HuR’s interaction with these different proteins is dependent upon numerous post-translation modifications that HuR undergoes including phosphorylation, neddylation, ubiquitination, methylation, acetylation and parylation (Chu, Chuang, Kulp, & Chen, 2012; Embade et al., 2012; Ke et al., 2017; H. H. Kim et al., 2008; H. Li et al., 2002). Most of these modifications that effect HuR’s localization take place in the HuR nucleocytoplasmic shuttling sequence (HNS) between HuR’s second and third RRM domain (Figure 2). This nucleocytoplasmic shuttling sequence and RRM3 are generally considered to be the most critical portions of the protein necessary for these interactions that dictate its localization (Doller, Schlepckow, Schwalbe, Pfeilschifter, & Eberhardt, 2010). For an extensive review on HuR post-translational modifications, please refer to: “Post-translational control of HuR function” (Grammatikakis et al., 2017).
As these post-translational modifications of HuR can impact cellular localization, targeting translocation becomes a complex process. This complexity causes inhibitors of HuR’s translocation to only be effective in certain situations and/or in specific cell lines. For instance, data from the Gorospe laboratory has shown in CRC cells that activation of AMPK causes the activation of importin-α1 leading to HuR’s nuclear re-uptake from the cytoplasm (W. Wang et al., 2002; W. Wang et al., 2004). Similar results were seen in C2Cl2 (mouse muscle cells) where AMPK activation with AICAR , an adenosine monophosphate analogue that is a strong activator of AMPK, caused a nuclear retention of HuR and inhibition of function (Di Marco et al., 2005). However, work in HepG2 cells (human liver carcinoma) demonstrated that treatment with AICAR can increase LDLR in an HuR-dependent manner. This HuR-dependent increase in LDLR implies that cytoplasmic localization of HuR was occurring through AICAR treatment (Yashiro, Nanmoku, Shimizu, Inoue, & Sato, 2013). It has also been demonstrated that in ARPE-19 (human retina) cells, AICAR treatment promoted HuR’s cytoplasmic translocation (Viiri et al., 2013). The distinct differences observed between these studies indicate that intrinsic differences in cell type and signaling may be a major factor when testing potential translocation inhibitors in multiple settings.
Small Molecule Inhibitors of HuR Translocation
Many different potential inhibitors of HuR translocation have been tested in vitro (Table 1). One of the first HuR inhibitors to be identified and characterized in vitro and in vivo is MS-444 (Blanco, Preet, et al., 2016). MS-444 was initially discovered as an inhibitor of myosin light chain kinase and was derived from the bacteria micromonospora (Nakanishi, Chiba, Yano, Kawamoto, & Matsuda, 1995). It was later found that MS-444, along with dehydromuctactin and okicenone, function as inhibitors through binding to HuR and impacting dimerization, which is necessary prior to subcellular trafficking (Meisner et al., 2007). MS-444 has been demonstrated to impact cancer growth in models of CRC, PDAC, and glioma, along with inhibiting HuR cytoplasmic translocation (Blanco, Preet, et al., 2016; Lang et al., 2017; Romeo et al., 2016; J. Wang, Hjelmeland, Nabors, & King, 2019). While MS-444 was not used in combination in vivo, it was found to significantly enhance sensitivity to PARP inhibitors, oxaliplatin, 5-Fluorouracil and sTRAIL in vitro (Blanco, Jimbo, et al., 2016; Chand et al., 2017; Romeo et al., 2016).
Table 1. Small Molecule HuR Inhibitors:
Small molecules that have been identified as inhibitors of HuR in vitro and in vivo. These drugs are categorized by their mechanism of action, and whether they have been utilized in vitro, in vivo or both.
| Published HuR inhibitors | Mechanism of action | IC50 (in vitro)* | Status | Reference |
|---|---|---|---|---|
| MS-444 | HuR translocation | 2.1-40.7 μM | In vitro/ in vivo | (Blanco, Jimbo, et al., 2016; Blanco, Preet, et al., 2016; Meisner et al., 2007) |
| Pyrvinium Pamoate | HuR translocation | 1.093 μM | In vitro/ in vivo | (Guo et al., 2016) |
| Cryptotanshinone | HuR translocation | 6.0 μM | In vitro/ in vivo | (Z. Zhu et al., 2016) |
| MPT0B098 | HuR translocation | 0.08 - 0.51 μM | In vitro/ in vivo | (Cheng et al., 2013) |
| Dehydromutactin | HuR translocation | 39.0 - 130.0 μM | In vitro only | (Meisner et al., 2007) |
| Okicenone | HuR translocation | 0.53 - 2.9 μg/ml | In vitro only | (Meisner et al., 2007) |
| SP600125 | HuR translocation | 0.12 - 16.0 μM | In vitro/ in vivo | (Hostetter et al., 2008) |
| 5-aza 2’ deoxycytidine / trichostatin A | HuR translocation | 0.1 uM - 0.5 uM | In vitro/ in vivo | (Hostetter et al., 2008) |
| N-Benzylcantharidinamide | HuR translocation | 50.0 - 100.0 μM | In vitro only | (Lee et al., 2014) |
| Triptolide | HuR translocation | 0.01 - 0.03 μM | In vitro/ in vivo | (L. Sun et al., 2011) |
| Leptomycin B | HuR translocation | 0.01 μM | In vitro/ in vivo | (Mutka et al., 2009) |
| Selinexor | HuR translocation | 0.12 - 0.22 μM | In vitro/ in vivo | (Hing et al., 2015) |
| Latrunculin A | HuR trafficking | 0.33 - 0.76 μM | In vitro/ in vivo | (Doller et al., 2015) |
| Blebbistatin | HuR trafficking | 0.43 - 22.8 μM | In vitro/ in vivo | (Doller et al., 2015) |
| CMLD2 | HuR target mRNA binding inhibition | 25.9 - 28.8 μM | In vitro only | (Muralidharan, Mehta, et al., 2017) |
| AZA-9 | HuR target mRNA binding inhibition | 51.9 - 67.6 μM | In vitro only | (Kaur et al., 2017) |
| 15,16-dihydrotanshinone-I (DHTS) | HuR target mRNA binding inhibition | 0.84 - 1.2 μM | In vitro/ in vivo | (D’Agostino et al., 2015; P. Lal et al., 2017) |
| Quercetin | HuR target mRNA binding inhibition | 5.78 - 31.04 μg/ml | In vitro/ in vivo | (Chae et al., 2009) |
| B-40 | HuR target mRNA binding inhibition | 0.38 μM | In vitro only | (Chae et al., 2009) |
| B-41 | HuR target mRNA binding inhibition | 6.21 μM | In vitro only | (Chae et al., 2009) |
| Tanshinone II | HuR target mRNA binding inhibition | 4.0 μM | In vitro only | (D’Agostino et al., 2015) |
| Cetylpyridinium choride | HuR target mRNA binding inhibition | 0.8-2.1 μg/ml | In vitro only | (D’Agostino, Adami, & Provenzani, 2013) |
|
Mitoxantrone |
HuR target mRNA binding inhibition | 0.01 - 1.13 μM | In vitro/ in vivo | (D’Agostino et al., 2013) |
| Suramin | HuR target mRNA binding inhibition | 732 μM | In vitro/in vivo | (Kakuguchi et al., 2018) |
| NSC# 5836 | HuR target mRNA binding inhibition | 14.7 μM | In vitro | (Z. Wang, Bhattacharya, & Ivanov, 2015) |
| NSC# 7572 | HuR target mRNA binding inhibition | 41.0 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 44750 | HuR target mRNA binding inhibition | 26.9 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 50648 | HuR target mRNA binding inhibition | 97.4 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 62685 | HuR target mRNA binding inhibition | 16.7 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 84126 | HuR target mRNA binding inhibition | 2.7 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 91438 | HuR target mRNA binding inhibition | 4.6 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 109292 | HuR target mRNA binding inhibition | 44.4 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 123418 | HuR target mRNA binding inhibition | 69.6 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 143491 | HuR target mRNA binding inhibition | 21.7 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 227186 | HuR target mRNA binding inhibition | 41.9 μM | In vitro | (Z. Wang et al., 2015) |
| NSC# 651084 | HuR target mRNA binding inhibition | 17.4 μM | In vitro | (Z. Wang et al., 2015) |
| KH-3 | Unknown | Unknown | In vitro/in vivo | (Green et al., 2019) |
IC50 values obtained in vitro through HuR binding inhibition and/or inhibition of cell proliferation/viability.
Another example of a HuR translocation inhibitor is the FDA-approved pyrvinium pamoate, which is currently approved to treat enterobiasis (pin worms). Pyrvinium has been shown to act as an HuR translocation inhibitor through indirect mechanisms instead of direct binding (Guo et al., 2016). Pyrvinium pamoate has been shown to work through both the activation of the AMPK pathway, and also through activating CDK1 leading to nuclear retention of HuR (Guo et al., 2016) through CDK1 dependent phosphorylation of HuR at Ser202. In vivo, pyrvinium pamoate was shown to inhibit the cytoplasmic accumulation of HuR and significantly increased the effectiveness of cisplatin in a bladder cancer xenograft model (Guo et al., 2016). These HuR inhibitor-based studies demonstrate the significance of cytoplasmic localization in HuR’s tumor-promoting activity and provide the framework for further validation to determine if translocation inhibitors may be effective as a single agent therapy and for sensitizing cells to further insults.
Inhibiting HuR Target Binding
HuR binds target mRNAs by recognizing adenine and uridine rich elements (AREs), generally in the 3’UTR of target mRNA. AREs are typically 50-150 nucleotide motifs and are classified into three major groups. Class I AREs have one to three of the ARE characteristic pentameric nucleotide sequence AUUUA scattered throughout this 50-150 bp region. Class II AREs contain several (generally 5 - 8) of these pentameric sequences with several overlapping AREs, while Class III AREs have long U rich stretches (C. Y. Chen & Shyu, 1995; C. Y. Chen, Xu, & Shyu, 2002). HuR can bind these different classes of AREs with a preference for U rich sequences, the top motif described as HUUUUHW (Bhandare, Goldberg, & Dowell, 2017). Inhibitors of HuR mRNA target binding have been identified using high-throughput approaches, utilizing purified HuR protein and a target mRNA that bears a fluorescent label. The binding of HuR protein and the fluorescently labeled mRNA can be screening for compounds that block mRNA target binding to HuR through changes in fluorescent polarization. While this is a useful screening approach, limitations exist in that these screens are performed outside of cells and typically against only one HuR-ARE target at a time. Subsequent validation of “hit” compounds require cell based assays in order to take into account the interactions of HuR with other mRNA stability factors, such as other RBPs, microRNAs, and long non-coding RNAs.
Identified Small Molecule Inhibitors of HuR (Target RNA binding inhibition)
As we described previously, target binding inhibitors can be used to inhibit HuR’s ability to bind target mRNAs. CMLD-2 is an example of an HuR target binding inhibitor that has been tested in vitro in colorectal, lung, and pancreatic cancer cells (Table 1). CMLD-2 was identified using a fluorescently labeled 16 nucleotide RNA sequence containing the binding site for the established HuR target Msi1 through library screening of 6000 compounds, and validated using cell-based assays (Wu et al., 2015). Other compounds found to inhibit HuR mRNA target binding also include AZA-9 (Kaur et al., 2017), quercetin (Chae et al., 2009), b-40 (Chae et al., 2009), and b-41 (Chae et al., 2009). It should be noted that some of these compounds have been utilized in combination with other drugs, suggesting that inhibition of HuR may be a contributing factor to the efficacy of these combinational studies.
One of the few HuR target binding inhibitors tested in vivo is 15,16-Dihydrotanshinone I (DHTS), which was shown to be effective at reducing tumor volume in a colorectal cancer xenograft model (P. Lal et al., 2017). Using DHTS-treated HeLa cells, transcript levels of HuR target mRNAs were reduced as expected, but some target transcript levels paradoxically increased. Accordingly, HuR ribonucleoprotein immunoprecipitation assays revealed that DHTS did not uniformly inhibit HuR binding to target mRNAs. DHTS was found to cause a shift from binding targets with lower affinities, to binding a smaller subset of mRNA that had higher affinities for HuR (P. Lal et al., 2017). While DHTS demonstrated HuR specificity, it may be RNA context-dependent and DHTS may cause an increase or decrease in HuR’s ability to bind specific target mRNAs. This may prove to be of importance if specific targets modulate stress responses (i.e. oxaliplatin resistance), and DHTS could be expected to either synergize with or antagonize oxaliplatin treatment.
Targeting HuR via silencing its expression
Genetic inhibition of HuR expression using knockdown (i.e. shRNA, siRNA) and knock out (i.e. CRISPR/Cas9) approaches have demonstrated feasibility for inhibiting cancer cell and tumor growth both in vitro and in vivo (Chand et al., 2017; Danilin et al., 2010; Filippova et al., 2011; Giammanco et al., 2014; P. Lal et al., 2017; H. Wang et al., 2018; Zarei et al., 2017), but a clinically viable method for inhibiting HuR expression has not been fully realized. One method for delivering HuR-specific siRNAs that has been demonstrated in vitro, takes advantage of extracellular vesicles which are naturally produced by cells. By conjugating siRNA with cholesterol as a means to efficiently load siRNAs into extracellular vesicles, silencing of HuR was observed in treated cell cultures (O’Loughlin et al., 2017). Another method for delivering siRNA-targeting HuR was developed with the goal to help treat diabetic retinopathy. Diabetic retinopathy progression is facilitated by HuR’s stabilization and increased translation of VEGF mRNA (Amadio et al., 2010; Yam & Kwok, 2007). Cationic liposomes (lipoplexes) complexed with siRNAs targeting HuR were able to decrease HuR protein expression after direct injection into the retina of rats with diabetic retinopathy. While downregulation of HuR expression was limited, a significant decrease in VEGF and retinal thickness was observed with lipoplex treatment (Amadio et al., 2016). Importantly, control naked siRNA injected into the retina produced similar results and were able to reduce of HuR and VEGF expression, undermining the potential efficacy of the lipoplexes at delivering siRNA, particularly if they are used in a systemic setting.
Another method utilized to systemically deliver siRNA oligonucleotides against HuR mRNA to cancer cells utilizes DNA dendrimer (i.e. 3DNA®) nanocarriers (Huang et al., 2016). These particles are made from building blocks of double stranded DNA containing 2 free non-complementary arms at each end of each chain. These monomers can be built upon each other in a stepwise fashion, with cross-linking after each round of addition. Targeting moieties can be attached to the free ends of the nanoparticle to achieve more selective targeting. As folic acid receptors are frequently up-regulated in cancer cells, 3DNA® nanoparticles can be coated with a folic acid moiety. Additionally, other drugs or deliverables such as siRNAs can be attached; the siRNA for these nanoparticles is not encapsulated, and is chemically modified for protection prior to delivery in vivo. Using this approach, in vivo treatment with 3DNA® nanoparticles containing a folic acid targeting moiety and modified siHuR oligos significantly reduced ascites development in an ovarian cancer model, along with extending the life of treated mice by 1.5 fold as compared to the control treated animals (Huang et al., 2016).
Liposome-based nanoparticles utilizing DOTAP:cholesterol as siRNA delivery agents have shown positive results in lung cancer cell models (Muralidharan et al., 2016). These nanoparticles are formed with a lipid bilayer of DOTAP (a cationic lipid) stabilized by small amounts of cholesterol. The positive charge of DOTAP allows these nanoparticles to stably interact with negatively charged siRNA oligonucleotides. A benefit of this approach is that the siRNA is encapsulated within the lipid bilayer and is protected from degradation. These nanoparticles also contained folic acid on the surface attached by short PEG linkers, which allowed for receptor-mediated endocytosis and subsequent silencing of HuR in folic acid receptor over-expressing cancer cells in vitro (Muralidharan et al., 2016). The folic acid-targeted DOTAP:cholesterol was not used in vivo, however modifications of the DOTAP:cholesterol particle to target HuR in transferrin receptor (CD71) expressing cells has been tested in a lung cancer model in vivo (Muralidharan, Babu, et al., 2017). Delivered using either intertumoral or intravenous injections, these CD71-targeted nanoparticles led to a decrease in lung cancer tumor size and an inhibition of metastatic growth (Muralidharan, Babu, et al., 2017).
As a means to target HuR in combination with chemotherapy, nanoparticles comprised of poly amidoamine (PAMAM) were used to deliver both siHuR and cisplatin (Amreddy et al., 2018). While the efficacy of this approach was demonstrated in in vitro models, its feasibility in vivo has not been established. With regard to systemic in vivo siRNA delivery using the 3DNA® and DOTAP:cholesterol nanoparticles, both were shown to be well tolerated and effective(Huang et al., 2016; Muralidharan, Babu, et al., 2017). Currently, there are no reports utilizing nanoparticles against HuR in combination with other therapeutics in vivo, and further development of these reagents in combination with conventional chemotherapeutic drugs will be needed for advancing this type of HuR-targeting approach.
Challenges and Potential Limitations for targeting HuR
There are many opportunities to target HuR through the array of factors that modulate its activity. Small molecules inhibiting HuR’s translocation and/or ability to bind target mRNAs have been the most well studied to date, with nanoparticle delivery of siRNA to silence of HuR expression only recently being developed (Figure 1). As each type of targeting strategy requires its own method of validation and testing, further studies are needed to confirm the mechanism of action and identify potential on- and off-target effects. With the field moving forward, validating these strategies in altering a disease phenotype will become a critical component. Aspects to consider when testing pre-clinical in vivo models can incorporate assays that examine: i) In vivo confirmation of nuclear HuR retention in tissues for HuR translocation inhibitors; ii) Tissue-specific confirmation of HuR knockdown using siHuR delivery approaches; iii) Conformation of disruption of HuR:RNA interaction using HuR target mRNA binding inhibitors; and iv) Evaluation of changes in HuR-target and global gene expression to demonstrate the downstream effects of targeting an RBP such as HuR.
While the targeting approaches described herein offer distinct advantages, each has its unique challenges and limitations. Targeting HuR translocation may be only impactful under specific scenarios given the number of cellular effectors that govern HuR’s localization, and that an incomplete inhibition could lead to a decrease in efficacy. Of note, targeting HuR molecular interactions (i.e. HuR dimerization), may prove to be beneficial as this mode of targeting could impact HuR translocation in all settings. Current high-throughput methods for identifying inhibitors that disrupt RNA binding have demonstrated success. However, this biochemical approach can yield false positive results through their interaction with target RNA and masking the HuR binding site or by stabilizing inhibitory RNA secondary structure (Meisner et al., 2004). As there are over a thousand RBPs (Gerstberger, Hafner, & Tuschl, 2014) with a high degree of homology between their conserved RRM domains, efforts to demonstrate HuR specificity are warranted. These RRM domains do however appear to have a large degree of variability both in target sequence, target structure, and protein structure (Maris, Dominguez, & Allain, 2005), and so this may not prove to be a significant hurdle. Nanoparticle delivery of siRNA offers a direct targeted approach, however tissue targeting and effective systemic delivery of siRNA-containing nanoparticles need further validation for clinical utility. Further advances in the nanoparticle delivery field, including exploring other potential methods for siRNA delivery such as using siRNA conjugated to ligands (i.e. small peptides) (Gandioso et al., 2017) and siRNA antibody conjugates (Tushir-Singh, 2017) are viable options to nanoparticle based delivery.
With any novel therapeutic approach lies the potential for off-target effects and toxicity. Based on HuR's ubiquitous expression pattern and necessity in early development, systemic inhibition of HuR could be detrimental. HuR has been shown to be critical for survival with knockout mice dying within ten days due to loss of progenitor cells, particularly those of myeloid and intestinal lineage (Ghosh et al., 2009). This, while concerning, is not uncommon as most chemotherapeutics that target rapidly dividing cells exhibit some levels of intestinal or myeloid toxicity (Boussios, Pentheroudakis, Katsanos, & Pavlidis, 2012; Kurtin, 2012). When examining potential HuR inhibitor toxicity, dosage may be an important parameter, since heterozygous loss of HuR displayed no harmful effects in mice, although HuR+/− mice did show increased sensitivity to whole body radiation induced toxicities with ~20 - 50% decrease in hematopoietic cell counts (Ghosh et al., 2009). In light of this, small molecule inhibitors of translocation may be less likely to cause toxicity, as they may not inhibit nuclear functions of HuR. Similarly, tissue-specific nanoparticle delivery of siRNAs targeting HuR may be able to selectively target tumor sites as opposed to normal tissue and circumvent the potential off-target toxicities of silencing HuR.
Concluding Remarks and Perspectives
Based on recent literature, HuR has been established as a novel post-transcriptional target in many cancer types and HuR inhibition may be more efficacious in combination with other agents. Targeting HuR to sensitize cancer cells to certain therapeutics is logical when one applies the basics of HuR biology. HuR activation can be thought of as a direct response to acute stressors, allowing for a quick and coordinated shift towards the cell having a pro-survival transcriptome. Inhibiting HuR thus allows other therapeutic agents to work virtually unopposed until other methods of transcriptional regulation (transcription factors etc.) can come in to play. Unsurprisingly, as a critical “first-responder” of sorts for a variety of therapeutics, inhibiting HuR has been shown to cause a dramatic sensitization to numerous different chemotherapeutics in many cancer types.
Bearing in mind that HuR inhibitors can be thought of as a critical sensitizing agent, the first clinical trials to determine clinical efficacy of HuR inhibitors most likely will include other agents. Realistically, nanoparticle-based methods of targeting HuR have more hurdles to go through prior to making it to the clinic, and so the first combination clinical trial will likely be utilizing a small molecule inhibitor of HuR in combination with another conventional therapeutic. Numerous studies have demonstrated the increased efficacy of platinum-based drugs (i.e. oxaliplatin and cisplatin) upon HuR inhibition (Amreddy et al., 2018; Blanco, Jimbo, et al., 2016; Guo et al., 2016). As platinum-based regimens are commonly used in many cancer types, a combination trial with a platinum-based therapy as the backbone may be the most appropriate, efficacious and fastest accruing trial to initiate.
HuR is likely to be the first RBP to be targeted clinically, either with innovative small molecules, or as one of many new targets made accessible with therapeutic siRNA delivery. The clinical inhibition of HuR is poised to be a potential breakthrough not only for cancer treatment, but also to help the biomedical field realize that RBPs as a gene regulatory class of proteins are not only therapeutically important, but also targetable. With notable investigations of targeting HuR ongoing, the clinical realization of a HuR inhibitor now seems not only likely but also inevitable, and has the potential to change the face of oncology.
Acknowledgments
Funding Information This work was supported by the National Institutes of Health [R01 CA212600 (JRB), R01 CA243445 (DAD)], American Cancer Society MRSG-14-019-01-CDD (JRB), NIH/NCI Cancer Center Support Grants P30 CA056036 and P30 CA168524, Fund A Cure and the Michele Barnett Rudnick Fund (JRB), and Pancreatic Cancer Action Network American Association for Cancer Research Acceleration Network Grant (15-90-25-BROD).
References
- (MGD), M. G. D. (2019). Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine http://www.informatics.jax.org/marker/MGI:1100851, January 2019.
- Ahn J, Byeon IJ, Dharmasena S, Huber K, Concel J, Gronenborn AM, & Sluis-Cremer N (2010). The RNA binding protein HuR does not interact directly with HIV-1 reverse transcriptase and does not affect reverse transcription in vitro. Retrovirology, 7, 40. doi: 10.1186/1742-4690-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike Y, Masuda K, Kuwano Y, Nishida K, Kajita K, Kurokawa K, … Rokutan K (2014). HuR regulates alternative splicing of the TRA2beta gene in human colon cancer cells under oxidative stress. Mol Cell Biol, 34(15), 2857–2873. doi: 10.1128/MCB.00333-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadio M, Bucolo C, Leggio GM, Drago F, Govoni S, & Pascale A (2010). The PKCbeta/HuR/VEGF pathway in diabetic retinopathy. Biochem Pharmacol, 80(8), 1230–1237. doi: 10.1016/j.bcp.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Amadio M, Pascale A, Cupri S, Pignatello R, Osera C, V, D. A, … Bucolo C (2016). Nanosystems based on siRNA silencing HuR expression counteract diabetic retinopathy in rat. Pharmacol Res, 111, 713–720. doi: 10.1016/j.phrs.2016.07.042 [DOI] [PubMed] [Google Scholar]
- Amreddy N, Babu A, Panneerselvam J, Srivastava A, Muralidharan R, Chen A, … Ramesh R (2018). Chemo-biologic combinatorial drug delivery using folate receptor-targeted dendrimer nanoparticles for lung cancer treatment. Nanomedicine, 14(2), 373–384. doi: 10.1016/j.nano.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U, Curry SL, Lopez de Silanes I, Shyu AB, Casolaro V, Gorospe M, & Stellato C (2003). Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J Immunol, 171(8), 4369–4378. doi: 10.4049/jimmunol.171.8.4369 [DOI] [PubMed] [Google Scholar]
- Badawi A, Hehlgans S, Pfeilschifter J, Rodel F, & Eberhardt W (2017). Silencing of the mRNA-binding protein HuR increases the sensitivity of colorectal cancer cells to ionizing radiation through upregulation of caspase-2. Cancer Lett, 393, 103–112. doi: 10.1016/j.canlet.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, … Guttridge DC (2013). miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal, 6(286), ra63. doi: 10.1126/scisignal.2004177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MD, Moon SL, Emch AW, Wilusz CJ, & Wilusz J (2013). Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep, 5(4), 909–917. doi: 10.1016/j.celrep.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M, & Meyer MR (2019). HuR-ry Up: How Hydrogen Sulfide Protects Against Atherosclerosis. Circulation, 139(1), 115–118. doi: 10.1161/CIRCULATIONAHA.118.036854 [DOI] [PubMed] [Google Scholar]
- Beauchamp P, Nassif C, Hillock S, van der Giessen K, von Roretz C, Jasmin BJ, & Gallouzi IE (2010). The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation. Cell Death Differ, 17(10), 1588–1599. doi: 10.1038/cdd.2010.34 [DOI] [PubMed] [Google Scholar]
- Berto S, Usui N, Konopka G, & Fogel BL (2016). ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum Mol Genet, 25(12), 2451–2464. doi: 10.1093/hmg/ddw110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandare S, Goldberg DS, & Dowell R (2017). Discriminating between HuR and TTP binding sites using the k-spectrum kernel method. PLoS One, 12(3), e0174052. doi: 10.1371/journal.pone.0174052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, … Brody JR (2016). The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene, 35(19), 2529–2541. doi: 10.1038/onc.2015.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, … Dixon DA (2016). Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget, 7(45), 74043–74058. doi: 10.18632/oncotarget.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussios S, Pentheroudakis G, Katsanos K, & Pavlidis N (2012). Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol, 25(2), 106–118. [PMC free article] [PubMed] [Google Scholar]
- Brauss TF, Winslow S, Lampe S, Scholz A, Weigert A, Dehne N, … Brune B (2017). The RNA-binding protein HuR inhibits expression of CCL5 and limits recruitment of macrophages into tumors. Mol Carcinog, 56(12), 2620–2629. doi: 10.1002/mc.22706 [DOI] [PubMed] [Google Scholar]
- Brennan CM, Gallouzi IE, & Steitz JA (2000). Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol, 151(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Wang H, Jiao X, Huang R, Qin Q, Zhang J, … Wang H (2019). The RNA-binding protein HuR confers oxaliplatin resistance of colorectal cancer by upregulating CDC6. Mol Cancer Ther. doi: 10.1158/1535-7163.MCT-18-0945 [DOI] [PubMed] [Google Scholar]
- Cascajo MV, Abdelmohsen K, Noh JH, Fernandez-Ayala DJ, Willers IM, Brea G, … Navas P (2016). RNA-binding proteins regulate cell respiration and coenzyme Q biosynthesis by post-transcriptional regulation of COQ7. RNA Biol, 13(7), 622–634. doi: 10.1080/15476286.2015.1119366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, … Park WY (2009). Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA. Exp Mol Med, 41(11), 824–831. doi: 10.3858/emm.2009.41.11.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S, … Brody JR (2017). Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors. Cancer Res, 77(18), 5011–5025. doi: 10.1158/0008-5472.CAN-16-2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Elemento O, Zhang J, Zhuang ZW, Simons M, & Hla T (2014). ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc Natl Acad Sci U S A, 111(51), 18309–18314. doi: 10.1073/pnas.1412172111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, & Shyu AB (1995). AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci, 20(11), 465–470. [DOI] [PubMed] [Google Scholar]
- Chen CY, Xu N, & Shyu AB (2002). Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol, 22(20), 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Martindale JL, Cramer C, Gorospe M, Atasoy U, Drew PD, & Yu S (2017). The RNA-binding protein HuR contributes to neuroinflammation by promoting C-C chemokine receptor 6 (CCR6) expression on Th17 cells. J Biol Chem, 292(35), 14532–14543. doi: 10.1074/jbc.M117.782771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Liou JP, Kuo CC, Lai WY, Shih KH, Chang CY, … Chang JY (2013). MPT0B098, a novel microtubule inhibitor that destabilizes the hypoxia-inducible factor-1alpha mRNA through decreasing nuclear-cytoplasmic translocation of RNA-binding protein HuR. Mol Cancer Ther, 12(7), 1202–1212. doi: 10.1158/1535-7163.MCT-12-0778 [DOI] [PubMed] [Google Scholar]
- Christodoulou-Vafeiadou E, Ioakeimidis F, Andreadou M, Giagkas G, Stamatakis G, Reczko M, … Kontoyiannis DL (2018). Divergent Innate and Epithelial Functions of the RNA-Binding Protein HuR in Intestinal Inflammation. Front Immunol, 9, 2732. doi: 10.3389/fimmu.2018.02732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PC, Chuang HC, Kulp SK, & Chen CS (2012). The mRNA-stabilizing factor HuR protein is targeted by beta-TrCP protein for degradation in response to glycolysis inhibition. J Biol Chem, 287(52), 43639–43650. doi: 10.1074/jbc.M112.393678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo AA, Herrick D, & Autieri MV (2010). Il-19 reduces VSMC activation by regulation of mRNA regulatory factor HuR and reduction of mRNA stability. J Mol Cell Cardiol, 49(4), 647–654. doi: 10.1016/j.yjmcc.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino VG, Adami V, & Provenzani A (2013). A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation. PLoS One, 8(8), e72426. doi: 10.1371/journal.pone.0072426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, … Provenzani A (2015). Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function. Sci Rep, 5, 16478. doi: 10.1038/srep16478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilin S, Sourbier C, Thomas L, Lindner V, Rothhut S, Dormoy V, … Massfelder T (2010). Role of the RNA-binding protein HuR in human renal cell carcinoma. Carcinogenesis, 31(6), 1018–1026. doi: 10.1093/carcin/bgq052 [DOI] [PubMed] [Google Scholar]
- De Santis R, Alfano V, de Turris V, Colantoni A, Santini L, Garone MG, … Rosa A (2019). Mutant FUS and ELAVL4 (HuD) Aberrant Crosstalk in Amyotrophic Lateral Sclerosis. Cell Rep, 27(13), 3818–3831 e3815. doi: 10.1016/j.celrep.2019.05.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, & Weichert W (2006). Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol, 19(9), 1261–1269. doi: 10.1038/modpathol.3800645 [DOI] [PubMed] [Google Scholar]
- Di Marco S, Mazroui R, Dallaire P, Chittur S, Tenenbaum SA, Radzioch D, … Gallouzi IE (2005). NF-kappa B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol, 25(15), 6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Munoz MD, Bell SE, Fairfax K, Monzon-Casanova E, Cunningham AF, Gonzalez-Porta M, … Turner M (2015). The RNA-binding protein HuR is essential for the B cell antibody response. Nat Immunol, 16(4), 415–425. doi: 10.1038/ni.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Badawi A, Schmid T, Brauss T, Pleli T, zu Heringdorf DM, … Eberhardt W (2015). The cytoskeletal inhibitors latrunculin A and blebbistatin exert antitumorigenic properties in human hepatocellular carcinoma cells by interfering with intracellular HuR trafficking. Exp Cell Res, 330(1), 66–80. doi: 10.1016/j.yexcr.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, & Eberhardt W (2010). Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol, 30(6), 1397–1410. doi: 10.1128/MCB.01373-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, & Holcik M (2011). RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene, 30(12), 1460–1469. doi: 10.1038/onc.2010.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Chakrama FZ, Combe E, Desmet FO, Mortada H, Polay Espinoza M, … Auboeuf D (2014). A recently evolved class of alternative 3’-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat Commun, 5, 3395. doi: 10.1038/ncomms4395 [DOI] [PubMed] [Google Scholar]
- Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V, … Martinez-Chantar ML (2012). Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology, 55(4), 1237–1248. doi: 10.1002/hep.24795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault S, & Malter JS (2003). Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J Immunol, 171(12), 6780–6787. [DOI] [PubMed] [Google Scholar]
- Fan XC, & Steitz JA (1998). HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci U S A, 95(26), 15293–15298. doi: 10.1073/pnas.95.26.15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq F, Abadia-Molina F, MacKenzie D, Hadwen J, Shamim F, O’Reilly S, … MacKenzie A (2013). Celecoxib increases SMN and survival in a severe spinal muscular atrophy mouse model via p38 pathway activation. Hum Mol Genet, 22(17), 3415–3424. doi: 10.1093/hmg/ddt191 [DOI] [PubMed] [Google Scholar]
- Farooq F, Balabanian S, Liu X, Holcik M, & MacKenzie A (2009). p38 Mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum Mol Genet, 18(21), 4035–4045. doi: 10.1093/hmg/ddp352 [DOI] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, … Munoz A (2003). Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol, 23(14), 4991–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, … Nabors LB (2011). The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res, 9(5), 648–659. doi: 10.1158/1541-7786.MCR-10-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries B, Heukeshoven J, Hauber I, Gruttner C, Stocking C, Kehlenbach RH, … Chemnitz J (2007). Analysis of nucleocytoplasmic trafficking of the HuR ligand APRIL and its influence on CD83 expression. J Biol Chem, 282(7), 4504–4515. doi: 10.1074/jbc.M608849200 [DOI] [PubMed] [Google Scholar]
- Fu X, Zhai S, & Yuan J (2018). Endothelial HuR deletion reduces the expression of proatherogenic molecules and attenuates atherosclerosis. Int Immunopharmacol, 65, 248–255. doi: 10.1016/j.intimp.2018.09.023 [DOI] [PubMed] [Google Scholar]
- Fus LP, Pihowicz P, Koperski L, Marczewska JM, & Gornicka B (2018). High cytoplasmic HuR expression is associated with advanced pT stage, high grade and increased microvessel density in urothelial bladder carcinoma. Ann Diagn Pathol, 33, 40–44. doi: 10.1016/j.anndiagpath.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, & Steitz JA (2003). Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA, 9(11), 1410. [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, & Steitz JA (2000). HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A, 97(7), 3073–3078. doi: 10.1073/pnas.97.7.3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandioso A, Massaguer A, Villegas N, Salvans C, Sanchez D, Brun-Heath I, … Terrazas M (2017). Efficient siRNA-peptide conjugation for specific targeted delivery into tumor cells. Chem Commun (Camb), 53(19), 2870–2873. doi: 10.1039/c6cc10287e [DOI] [PubMed] [Google Scholar]
- Gauchotte G, Hergalant S, Vigouroux C, Casse JM, Houlgatte R, Kaoma T, … Battaglia-Hsu SF (2017). Cytoplasmic overexpression of RNA-binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia. J Pathol, 242(4), 421–434. doi: 10.1002/path.4916 [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, & Tuschl T (2014). A census of human RNA-binding proteins. Nat Rev Genet, 15(12), 829–845. doi: 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, … Hla T (2009). Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest, 119(12), 3530–3543. doi: 10.1172/JCI38263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammanco A, Blanc V, Montenegro G, Klos C, Xie Y, Kennedy S, … Davidson NO (2014). Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res, 74(18), 5322–5335. doi: 10.1158/0008-5472.CAN-14-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg-Cohen I, Furneauxb H, & Levy AP (2002). A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem, 277(16), 13635–13640. doi: 10.1074/jbc.M108703200 [DOI] [PubMed] [Google Scholar]
- Good PJ (1995). A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci U S A, 92(10), 4557–4561. doi: 10.1073/pnas.92.10.4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I, Abdelmohsen K, & Gorospe M (2017). Posttranslational control of HuR function. Wiley Interdiscip Rev RNA, 8(1). doi: 10.1002/wrna.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina GL, Tortoreto M, Mancini A, Addis A, Di Cesare E, Lenzi A, … Festuccia C (2014). XPO1/CRM1-selective inhibitors of nuclear export (SINE) reduce tumor spreading and improve overall survival in preclinical models of prostate cancer (PCa). J Hematol Oncol, 7, 46. doi: 10.1186/1756-8722-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LC, Anthony SR, Slone S, Lanzillotta L, Nieman ML, Wu X, … Tranter M (2019). Human antigen R as a therapeutic target in pathological cardiac hypertrophy. JCI Insight. doi: 10.1172/jci.insight.121541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Lv J, Chang S, Chen Z, Lu W, Xu C, … Pang X (2016). Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder. Oncotarget, 7(29), 45249–45262. doi: 10.18632/oncotarget.9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgis FM, Yeung YT, Tang MX, Heng B, Buckland M, Ammit AJ, … Munoz L (2015). The p38-MK2-HuR pathway potentiates EGFRvIII-IL-1beta-driven IL-6 secretion in glioblastoma cells. Oncogene, 34(22), 2934–2942. doi: 10.1038/onc.2014.225 [DOI] [PubMed] [Google Scholar]
- Haeussler J, Haeusler J, Striebel AM, Assum G, Vogel W, Furneaux H, & Krone W (2000). Tumor antigen HuR binds specifically to one of five protein-binding segments in the 3’-untranslated region of the neurofibromin messenger RNA. Biochem Biophys Res Commun, 267(3), 726–732. doi: 10.1006/bbrc.1999.2019 [DOI] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hing ZA, Mantel R, Beckwith KA, Guinn D, Williams E, Smith LL, … Lapalombella R (2015). Selinexor is effective in acquired resistance to ibrutinib and synergizes with ibrutinib in chronic lymphocytic leukemia. Blood, 125(20), 3128–3132. doi: 10.1182/blood-2015-01-621391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, & Lou H (2008). Diverse molecular functions of Hu proteins. Cell Mol Life Sci, 65(20), 3168–3181. doi: 10.1007/s00018-008-8252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes B, Benavides-Serrato A, Freeman RS, Landon KA, Bashir T, Nishimura RN, & Gera J (2018). mTORC2/AKT/HSF1/HuR constitute a feed-forward loop regulating Rictor expression and tumor growth in glioblastoma. Oncogene, 37(6), 732–743. doi: 10.1038/onc.2017.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetter C, Licata LA, Witkiewicz A, Costantino CL, Yeo CJ, Brody JR, & Keen JC (2008). Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells. Cancer Biol Ther, 7(9), 1496–1506. [DOI] [PubMed] [Google Scholar]
- Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, … Sawicki JA (2016). Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res, 76(6), 1549–1559. doi: 10.1158/0008-5472.CAN-15-2073 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM (2008). Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem, 283(27), 19077–19084. doi: 10.1074/jbc.M800017200 [DOI] [PubMed] [Google Scholar]
- Izquierdo JM (2010). Heterogeneous ribonucleoprotein C displays a repressor activity mediated by T-cell intracellular antigen-1-related/like protein to modulate Fas exon 6 splicing through a mechanism involving Hu antigen R. Nucleic Acids Res, 38(22), 8001–8014. doi: 10.1093/nar/gkq698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehung JP, Kitamura T, Yanagawa-Matsuda A, Kuroshima T, Towfik A, Yasuda M, … Higashino F (2018). Adenovirus infection induces HuR relocalization to facilitate virus replication. Biochem Biophys Res Commun, 495(2), 1795–1800. doi: 10.1016/j.bbrc.2017.12.036 [DOI] [PubMed] [Google Scholar]
- Kakuguchi W, Nomura T, Kitamura T, Otsuguro S, Matsushita K, Sakaitani M, … Tei K (2018). Suramin, screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells. Cancer Med, 7(12), 6269–6280. doi: 10.1002/cam4.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur K, Wu X, Fields JK, Johnson DK, Lan L, Pratt M, … De Guzman RN (2017). The fungal natural product azaphilone-9 binds to HuR and inhibits HuR-RNA interaction in vitro. PLoS One, 12(4), e0175471. doi: 10.1371/journal.pone.0175471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, … Zeng X (2017). PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nat Commun, 8, 14632. doi: 10.1038/ncomms14632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr., Yang X, Galban S, … Gorospe M (2008). Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev, 22(13), 1804–1815. doi: 10.1101/gad.1645808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, & Gorospe M (2008). Phosphorylated HuR shuttles in cycles. Cell Cycle, 7(20), 3124–3126. doi: 10.4161/cc.7.20.6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Noren Hooten N, Dluzen DF, Martindale JL, Gorospe M, & Evans MK (2015). Posttranscriptional Regulation of the Inflammatory Marker C-Reactive Protein by the RNA-Binding Protein HuR and MicroRNA 637. Mol Cell Biol, 35(24), 4212–4221. doi: 10.1128/MCB.00645-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf M, Jarczak D, Beger C, Manns MP, & Kruger M (2005). Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol, 43(2), 225–234. doi: 10.1016/j.jhep.2005.02.046 [DOI] [PubMed] [Google Scholar]
- Kurtin S (2012). Myeloid toxicity of cancer treatment. J Adv Pract Oncol, 3(4), 209–224. [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kawai T, Yang X, Mazan-Mamczarz K, & Gorospe M (2005). Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J, 24(10), 1852–1862. doi: 10.1038/sj.emboj.7600661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal P, Cerofolini L, D’Agostino VG, Zucal C, Fuccio C, Bonomo I, … Provenzani A (2017). Regulation of HuR structure and function by dihydrotanshinone-I. Nucleic Acids Res, 45(16), 9514–9527. doi: 10.1093/nar/gkx623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, … Brody JR (2017). CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype. Mol Cancer Res, 15(6), 696–707. doi: 10.1158/1541-7786.MCR-16-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, … Gasche C (2017). HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer Res, 77(9), 2424–2438. doi: 10.1158/0008-5472.CAN-15-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, & Rajewsky N (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell, 43(3), 340–352. doi: 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Lee JY, Chung TW, Choi HJ, Lee CH, Eun JS, Han YT, … Ha KT (2014). A novel cantharidin analog N-benzylcantharidinamide reduces the expression of MMP-9 and invasive potentials of Hep3B via inhibiting cytosolic translocation of HuR. Biochem Biophys Res Commun, 447(2), 371–377. doi: 10.1016/j.bbrc.2014.04.035 [DOI] [PubMed] [Google Scholar]
- Lema I, Amazit L, Lamribet K, Fagart J, Blanchard A, Lombes M, … Viengchareun S (2017). HuR-Dependent Editing of a New Mineralocorticoid Receptor Splice Variant Reveals an Osmoregulatory Loop for Sodium Homeostasis. Sci Rep, 7(1), 4835. doi: 10.1038/s41598-017-04838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain JC, Benarous R, & Liu LX (2008). HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology, 5, 47. doi: 10.1186/1742-4690-5-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy NS, Chung S, Furneaux H, & Levy AP (1998). Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem, 273(11), 6417–6423. [DOI] [PubMed] [Google Scholar]
- Li F, Hu DY, Liu S, Mahavadi S, Yen W, Murthy KS, … Hu W (2010). RNA-binding protein HuR regulates RGS4 mRNA stability in rabbit colonic smooth muscle cells. Am J Physiol Cell Physiol, 299(6), C1418–1429. doi: 10.1152/ajpcell.00093.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, … Laird-Offringa IA (2002). Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem, 277(47), 44623–44630. doi: 10.1074/jbc.M206187200 [DOI] [PubMed] [Google Scholar]
- Li X, Lu L, Bush DJ, Zhang X, Zheng L, Suswam EA, & King PH (2009). Mutant copper-zinc superoxide dismutase associated with amyotrophic lateral sclerosis binds to adenine/uridine-rich stability elements in the vascular endothelial growth factor 3’-untranslated region. J Neurochem, 108(4), 1032–1044. doi: 10.1111/j.1471-4159.2008.05856.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lu YC, Dai K, Torregroza I, Hla T, & Evans T (2014). Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood, 123(9), 1384–1392. doi: 10.1182/blood-2013-09-526962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang Y, Hu R, Xu R, & Xu W (2018). LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Prolif, 51(6), e12504. doi: 10.1111/cpr.12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SJ, Lee SH, Joo SH, Song JY, & Choi SI (2009). Cytoplasmic expression of HuR is related to cyclooxygenase-2 expression in colon cancer. Cancer Res Treat, 41(2), 87–92. doi: 10.4143/crt.2009.41.2.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FY, Chen YH, Lin YW, Tsai JS, Chen JW, Wang HJ, … Lin SJ (2006). The role of human antigen R, an RNA-binding protein, in mediating the stabilization of toll-like receptor 4 mRNA induced by endotoxin: a novel mechanism involved in vascular inflammation. Arterioscler Thromb Vasc Biol, 26(12), 2622–2629. doi: 10.1161/01.ATV.0000246779.78003.cf [DOI] [PubMed] [Google Scholar]
- Lin GL, Ting HJ, Tseng TC, Juang V, & Lo YL (2017). Modulation of the mRNA-binding protein HuR as a novel reversal mechanism of epirubicin-triggered multidrug resistance in colorectal cancer cells. PLoS One, 12(10), e0185625. doi: 10.1371/journal.pone.0185625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Lee LM, Lai HL, & Chern Y (2015). Aberrant activation of AMP-activated protein kinase contributes to the abnormal distribution of HuR in amyotrophic lateral sclerosis. FEBS Lett, 589(4), 432–439. doi: 10.1016/j.febslet.2014.12.029 [DOI] [PubMed] [Google Scholar]
- Liu ZM, Tseng JT, Hong DY, & Huang HS (2011). Suppression of TG-interacting factor sensitizes arsenic trioxide-induced apoptosis in human hepatocellular carcinoma cells. Biochem J, 438(2), 349–358. doi: 10.1042/BJ20101653 [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, & Gorospe M (2003). Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene, 22(46), 7146–7154. doi: 10.1038/sj.onc.1206862 [DOI] [PubMed] [Google Scholar]
- Lu L, Zheng L, Si Y, Luo W, Dujardin G, Kwan T, … King PH (2014). Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis. J Biol Chem, 289(46), 31792–31804. doi: 10.1074/jbc.M114.573246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zheng L, Viera L, Suswam E, Li Y, Li X, … King PH (2007). Mutant Cu/Zn-superoxide dismutase associated with amyotrophic lateral sclerosis destabilizes vascular endothelial growth factor mRNA and downregulates its expression. J Neurosci, 27(30), 7929–7938. doi: 10.1523/JNEUROSCI.1877-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, & Richard S (2008). RNA-binding proteins in human genetic disease. Trends Genet, 24(8), 416–425. doi: 10.1016/j.tig.2008.05.004 [DOI] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, & Furneaux H (1996). Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem, 271(14), 8144–8151. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, & Allain FH (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J, 272(9), 2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Matsye P, Zheng L, Si Y, Kim S, Luo W, Crossman DK, … King PH (2017). HuR promotes the molecular signature and phenotype of activated microglia: Implications for amyotrophic lateral sclerosis and other neurodegenerative diseases. Glia, 65(6), 945–963. doi: 10.1002/glia.23137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, & Gorospe M (2003). RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A, 100(14), 8354–8359. doi: 10.1073/pnas.1432104100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M, Basalingappa K, Griffith JN, Andrade D, Babu A, Amreddy N, … Munshi A (2016). HuR silencing elicits oxidative stress and DNA damage and sensitizes human triple-negative breast cancer cells to radiotherapy. Oncotarget, 7(40), 64820–64835. doi: 10.18632/oncotarget.11706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, & Auer M (2004). mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem, 5(10), 1432–1447. doi: 10.1002/cbic.200400219 [DOI] [PubMed] [Google Scholar]
- Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, … Auer M (2007). Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol, 3(8), 508–515. doi: 10.1038/nchembio.2007.14 [DOI] [PubMed] [Google Scholar]
- Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, … Krech T (2016). Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer. Prostate, 76(3), 259–272. doi: 10.1002/pros.23120 [DOI] [PubMed] [Google Scholar]
- Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, & Sakai H (2013). High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One, 8(3), e59095. doi: 10.1371/journal.pone.0059095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, … Ramesh R (2016). Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration. J Nanobiotechnology, 14(1), 47. doi: 10.1186/s12951-016-0201-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan R, Babu A, Amreddy N, Srivastava A, Chen A, Zhao YD, … Ramesh R (2017). Tumor-targeted Nanoparticle Delivery of HuR siRNA Inhibits Lung Tumor Growth In Vitro and In Vivo By Disrupting the Oncogenic Activity of the RNA-binding Protein HuR. Mol Cancer Ther, 16(8), 1470–1486. doi: 10.1158/1535-7163.MCT-17-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, … Munshi A (2017). HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci Rep, 7(1), 9694. doi: 10.1038/s41598-017-07787-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB, & Murli S (2009). Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res, 69(2), 510–517. doi: 10.1158/0008-5472.CAN-08-0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabors LB, Gillespie GY, Harkins L, & King PH (2001). HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res, 61(5), 2154–2161. [PubMed] [Google Scholar]
- Nakanishi S, Chiba S, Yano H, Kawamoto I, & Matsuda Y (1995). MS-444, a new inhibitor of myosin light chain kinase from Micromonospora sp. KY7123. J Antibiot (Tokyo), 48(9), 948–951. [DOI] [PubMed] [Google Scholar]
- Nowotarski SL, & Shantz LM (2010). Cytoplasmic accumulation of the RNA-binding protein HuR stabilizes the ornithine decarboxylase transcript in a murine nonmelanoma skin cancer model. J Biol Chem, 285(41), 31885–31894. doi: 10.1074/jbc.M110.148767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin AJ, Mager I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, … Vader P (2017). Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol Ther, 25(7), 1580–1587. doi: 10.1016/j.ymthe.2017.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Kakumoto K, Yoshida T, Kuwako KI, Miyazaki T, Yamaguchi J, … Okano HJ (2018). Elavl3 is essential for the maintenance of Purkinje neuron axons. Sci Rep, 8(1), 2722. doi: 10.1038/s41598-018-21130-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osera C, Martindale JL, Amadio M, Kim J, Yang X, Moad CA, … Pascale A (2015). Induction of VEGFA mRNA translation by CoCl2 mediated by HuR. RNA Biol, 12(10), 1121–1130. doi: 10.1080/15476286.2015.1085276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, … Sattler M (2019). HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res, 47(2), 1011–1029. doi: 10.1093/nar/gky1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, & Shyu AB (1998). RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J, 17(12), 3461–3470. doi: 10.1093/emboj/17.12.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Furuuchi N, Aslanukova L, Huang YH, Brown SZ, Jiang W, … Sawicki JA (2018). Elevated HuR in Pancreas Promotes a Pancreatitis-Like Inflammatory Microenvironment That Facilitates Tumor Development. Mol Cell Biol, 38(3). doi: 10.1128/MCB.00427-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Billaud M, & Almeida R (2017). RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer, 3(7), 506–528. doi: 10.1016/j.trecan.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Perera ND, Sheean RK, Crouch PJ, White AR, Horne MK, & Turner BJ (2016). Enhancing survival motor neuron expression extends lifespan and attenuates neurodegeneration in mutant TDP-43 mice. Hum Mol Genet, 25(18), 4080–4093. doi: 10.1093/hmg/ddw247 [DOI] [PubMed] [Google Scholar]
- Rebane A, Aab A, & Steitz JA (2004). Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA, 10(4), 590–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, … Witkiewicz AK (2010). HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg, 252(3), 499–505; discussion 505-496. doi: 10.1097/SLA.0b013e3181f1fd44 [DOI] [PubMed] [Google Scholar]
- Ripin N, Boudet J, Duszczyk MM, Hinniger A, Faller M, Krepl M, … Allain FH (2019). Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc Natl Acad Sci U S A, 116(8), 2935–2944. doi: 10.1073/pnas.1808696116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, … Brody JR (2016). HuR Contributes to TRAIL Resistance by Restricting Death Receptor 4 Expression in Pancreatic Cancer Cells. Mol Cancer Res, 14(7), 599–611. doi: 10.1158/1541-7786.MCR-15-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JS, Choi HE, Seo S, Choi JH, Baek NI, & Lee KT (2016). Berberine Decreased Inducible Nitric Oxide Synthase mRNA Stability through Negative Regulation of Human Antigen R in Lipopolysaccharide-Induced Macrophages. J Pharmacol Exp Ther, 358(1), 3–13. doi: 10.1124/jpet.115.231043 [DOI] [PubMed] [Google Scholar]
- Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, & Das S (2015). HuR Displaces Polypyrimidine Tract Binding Protein To Facilitate La Binding to the 3’ Untranslated Region and Enhances Hepatitis C Virus Replication. J Virol, 89(22), 11356–11371. doi: 10.1128/JVI.01714-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, & Wilusz J (2010). Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe, 8(2), 196–207. doi: 10.1016/j.chom.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, & Gorospe M (2011). UneCLIPsing HuR nuclear function. Mol Cell, 43(3), 319–321. doi: 10.1016/j.molcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, & Gorospe M (2012). HuR function in disease. Front Biosci (Landmark Ed), 17, 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara M, Tsutsumi A, Suzuki E, Wakamatsu E, Suzuki T, Ogishima H, … Sumida T (2007). Effects of infliximab therapy on gene expression levels of tumor necrosis factor alpha, tristetraprolin, T cell intracellular antigen 1, and Hu antigen R in patients with rheumatoid arthritis. Arthritis Rheum, 56(7), 2160–2169. doi: 10.1002/art.22724 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang S, Jiang Z, Huang X, Wang T, Huang X, … Zhang L (2011). Triptolide inhibits COX-2 expression by regulating mRNA stability in TNF-alpha-treated A549 cells. Biochem Biophys Res Commun, 416(1-2), 99–105. doi: 10.1016/j.bbrc.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang X, Lyu L, Li X, Yao S, & Zhang J (2016). Autotaxin Expression Is Regulated at the Post-transcriptional Level by the RNA-binding Proteins HuR and AUF1. J Biol Chem, 291(50), 25823–25836. doi: 10.1074/jbc.M116.756908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E, Tsutsumi A, Sugihara M, Mamura M, Goto D, Matsumoto I, … Sumida T (2006). Expression of TNF-alpha, tristetraprolin, T-cell intracellular antigen-1 and Hu antigen R genes in synovium of patients with rheumatoid arthritis. Int J Mol Med, 18(2), 273–278. [PubMed] [Google Scholar]
- Tang H, Wang H, Cheng X, Fan X, Yang F, Zhang M, … Wang W (2018). HuR regulates telomerase activity through TERC methylation. Nat Commun, 9(1), 2213. doi: 10.1038/s41467-018-04617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarian T, Jiang W, Leiby BE, Grigoli A, Jimbo M, Dabbish N, … Brody JR (2018). Cytoplasmic HuR Status Predicts Disease-free Survival in Resected Pancreatic Cancer: A Post-hoc Analysis From the International Phase III ESPAC-3 Clinical Trial. Ann Surg, 267(2), 364–369. doi: 10.1097/SLA.0000000000002088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K, Murakami Y, Kondo N, Uemura K, Nakagawa N, Takahashi S, & Sueda T (2018). Cytoplasmic Hu-Antigen R (HuR) Expression is Associated with Poor Survival in Patients with Surgically Resected Cholangiocarcinoma Treated with Adjuvant Gemcitabine-Based Chemotherapy. Ann Surg Oncol, 25(5), 1202–1210. doi: 10.1245/s10434-018-6392-y [DOI] [PubMed] [Google Scholar]
- Tushir-Singh J (2017). Antibody-siRNA conjugates: drugging the undruggable for anti-leukemic therapy. Expert Opin Biol Ther, 17(3), 325–338. doi: 10.1080/14712598.2017.1273344 [DOI] [PubMed] [Google Scholar]
- van der Giessen K, Di-Marco S, Clair E, & Gallouzi IE (2003). RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem, 278(47), 47119–47128. doi: 10.1074/jbc.M308889200 [DOI] [PubMed] [Google Scholar]
- van der Giessen K, & Gallouzi IE (2007). Involvement of transportin 2-mediated HuR import in muscle cell differentiation. Mol Biol Cell, 18(7), 2619–2629. doi: 10.1091/mbc.e07-02-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, … Kaarniranta K (2013). Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One, 8(7), e69563. doi: 10.1371/journal.pone.0069563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Roretz C, Beauchamp P, Di Marco S, & Gallouzi IE (2011). HuR and myogenesis: being in the right place at the right time. Biochim Biophys Acta, 1813(9), 1663–1667. doi: 10.1016/j.bbamcr.2011.01.036 [DOI] [PubMed] [Google Scholar]
- Wang D, Wang M, Hu C, Shuang T, Zhou Y, & Yan X (2014). Expression of the ELAV-like protein HuR in the cytoplasm is associated with endometrial carcinoma progression. Tumour Biol, 35(12), 11939–11947. doi: 10.1007/s13277-014-2485-9 [DOI] [PubMed] [Google Scholar]
- Wang H, Chen Y, Guo J, Shan T, Deng K, Chen J, … Xia J (2018). Dysregulation of tristetraprolin and human antigen R promotes gastric cancer progressions partly by upregulation of the high-mobility group box 1. Sci Rep, 8(1), 7080. doi: 10.1038/s41598-018-25443-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zeng F, Liu Q, Liu H, Liu Z, Niu L, … Li X (2013). The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr D Biol Crystallogr, 69(Pt 3), 373–380. doi: 10.1107/S0907444912047828 [DOI] [PubMed] [Google Scholar]
- Wang J, Guo Y, Chu H, Guan Y, Bi J, & Wang B (2013). Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci, 14(5), 10015–10041. doi: 10.3390/ijms140510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hjelmeland AB, Nabors LB, & King PH (2019). Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther, 20(7), 979–988. doi: 10.1080/15384047.2019.1591673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, & Bender JR (2006). LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J Immunol, 176(4), 2105–2113. [DOI] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, & Gorospe M (2000). HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J, 19(10), 2340–2350. doi: 10.1093/emboj/19.10.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fan J, Yang X, Furer-Galban S, Lopez de Silanes I, von Kobbe C, … Gorospe M (2002). AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol, 22(10), 3425–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Kawai T, Lopez de Silanes I, Mazan-Mamczarz K, Chen P, … Gorospe M (2004). AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J Biol Chem, 279(46), 48376–48388. doi: 10.1074/jbc.M409014200 [DOI] [PubMed] [Google Scholar]
- Wang Z, Bhattacharya A, & Ivanov DN (2015). Identification of Small-Molecule Inhibitors of the HuR/RNA Interaction Using a Fluorescence Polarization Screening Assay Followed by NMR Validation. PLoS One, 10(9), e0138780. doi: 10.1371/journal.pone.0138780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TK, Costantino CL, Bildzukewicz NA, Richards NG, Rittenhouse DW, Einstein L, … Brody JR (2010). pp32 (ANP32A) expression inhibits pancreatic cancer cell growth and induces gemcitabine resistance by disrupting HuR binding to mRNAs. PLoS One, 5(11), e15455. doi: 10.1371/journal.pone.0015455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Doller A, Imre G, Badawi A, Schmid T, Schulz S, … Eberhardt W (2014). Attenuation of the ELAV1-like protein HuR sensitizes adenocarcinoma cells to the intrinsic apoptotic pathway by increasing the translation of caspase-2L. Cell Death Dis, 5, e1321. doi: 10.1038/cddis.2014.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen R, Gowrishankar G, Bollig F, Redich N, Resch K, & Holtmann H (2004). Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol, 24(11), 4835–4847. doi: 10.1128/MCB.24.11.4835-4847.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, … Xu L (2015). Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem Biol, 10(6), 1476–1484. doi: 10.1021/cb500851u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang SH, Sun J, Krainer AR, Hua Y, & Prior TW (2017). A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum Mol Genet, 26(14), 2768–2780. doi: 10.1093/hmg/ddx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam JC, & Kwok AK (2007). Update on the treatment of diabetic retinopathy. Hong Kong Med J, 13(1), 46–60. [PubMed] [Google Scholar]
- Yashiro T, Nanmoku M, Shimizu M, Inoue J, & Sato R (2013). 5-Aminoimidazole-4-carboxamide ribonucleoside stabilizes low density lipoprotein receptor mRNA in hepatocytes via ERK-dependent HuR binding to an AU-rich element. Atherosclerosis, 226(1), 95–101. doi: 10.1016/j.atherosclerosis.2012.09.033 [DOI] [PubMed] [Google Scholar]
- Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, & Kontoyiannis DL (2012). Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J Clin Invest, 122(1), 48–61. doi: 10.1172/JCI45021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Kang J, Lee HN, Aguilar B, Kafka D, Lee S, … Hong YK (2010). Kaposin-B enhances the PROX1 mRNA stability during lymphatic reprogramming of vascular endothelial cells by Kaposi's sarcoma herpes virus. PLoS Pathog, 6(8), e1001046. doi: 10.1371/journal.ppat.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Sanders AJ, Ye L, Wang Y, & Jiang WG (2011). Knockdown of human antigen R reduces the growth and invasion of breast cancer cells in vitro and affects expression of cyclin D1 and MMP-9. Oncol Rep, 26(1), 237–245. doi: 10.3892/or.2011.1271 [DOI] [PubMed] [Google Scholar]
- Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K, … Winter JM (2017). Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells. Cancer Res, 77(16), 4460–4471. doi: 10.1158/0008-5472.CAN-17-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei M, Lal S, Vaziri-Gohar A, O’Hayer K, Gunda V, Singh PK, … Winter JM (2019). RNA-Binding Protein HuR Regulates Both Mutant and Wild-Type IDH1 in IDH1-Mutated Cancer. Mol Cancer Res, 17(2), 508–520. doi: 10.1158/1541-7786.MCR-18-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha W, Liang G, Xiao J, Studer EJ, Hylemon PB, Pandak WM Jr., … Zhou H (2010). Berberine inhibits HIV protease inhibitor-induced inflammatory response by modulating ER stress signaling pathways in murine macrophages. PLoS One, 5(2), e9069. doi: 10.1371/journal.pone.0009069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, & Wang J (2018). HuR stabilizes TFAM mRNA in an ATM/p38-dependent manner in ionizing irradiated cancer cells. Cancer Sci, 109(8), 2446–2457. doi: 10.1111/cas.13657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huang A, Zhang A, & Zhou C (2017). HuR promotes breast cancer cell proliferation and survival via binding to CDK3 mRNA. Biomed Pharmacother, 91, 788–795. doi: 10.1016/j.biopha.2017.04.063 [DOI] [PubMed] [Google Scholar]
- Zhou H, Jarujaron S, Gurley EC, Chen L, Ding H, Studer E, … Hylemon PB (2007). HIV protease inhibitors increase TNF-alpha and IL-6 expression in macrophages: involvement of the RNA-binding protein HuR. Atherosclerosis, 195(1), e134–143. doi: 10.1016/j.atherosclerosis.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang S, Zheng M, Kuver A, Wan X, Dai K, & Li X (2019). Phosphorylation of ELAVL1 (Ser219/Ser316) mediated by PKC is required for erythropoiesis. Biochim Biophys Acta Mol Cell Res, 1866(2), 214–224. doi: 10.1016/j.bbamcr.2018.10.021 [DOI] [PubMed] [Google Scholar]
- Zhu H, Berkova Z, Mathur R, Sehgal L, Khashab T, Tao RH, … Samaniego F (2015). HuR Suppresses Fas Expression and Correlates with Patient Outcome in Liver Cancer. Mol Cancer Res, 13(5), 809–818. doi: 10.1158/1541-7786.MCR-14-0241 [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhou HL, Hasman RA, & Lou H (2007). Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem, 282(4), 2203–2210. doi: 10.1074/jbc.M609349200 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Wang B, Bi J, Zhang C, Guo Y, Chu H, … Wang J (2013). Cytoplasmic HuR expression correlates with P-gp, HER-2 positivity, and poor outcome in breast cancer. Tumour Biol, 34(4), 2299–2308. doi: 10.1007/s13277-013-0774-3 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhao Y, Li J, Tao L, Shi P, Wei Z, … Lu Y (2016). Cryptotanshinone, a novel tumor angiogenesis inhibitor, destabilizes tumor necrosis factor-alpha mRNA via decreasing nuclear-cytoplasmic translocation of RNA-binding protein HuR. Mol Carcinog, 55(10), 1399–1410. doi: 10.1002/mc.22383 [DOI] [PubMed] [Google Scholar]
- Zucal C, D'Agostino V, Loffredo R, Mantelli B, NatthakanThongon, Lal P, … Provenzani A (2015). Targeting the multifaceted HuR protein, benefits and caveats. Curr Drug Targets, 16(5), 499–515. [DOI] [PubMed] [Google Scholar]


