Abstract
The aim of the study was to investigate the effect of chemo-radiation on the genetic and immunological status of rectal cancer patients who were treated with preoperative chemoradiotherapy (CRT). The expression of immune response-associated genes was compared between rectal cancer patients treated (n = 9) and not-treated (n = 10) with preoperative CRT using volcano plot analysis. Apoptosis and epithelial-to-mesenchymal transition (EMT) marker genes were analysed by quantitative PCR (qPCR). Other markers associated with the tumor microenvironment (TME), such as tumor-infiltrating lymphocytes (TIL) and immune checkpoint molecules, were investigated using immunohistochemistry (IHC). The clinical responses of preoperative CRT for 9 rectal cancer patients were all rated as stable disease, while the pathological tumor regression score (TRG) revealed 6 cases of grade2 and 3 cases of grade1. According to the genetic signature of colon cancers, treated tumors belonged to consensus molecular subtype (CMS)4, while not-treated tumors had signatures of CMS2 or 3. CRT-treated tumors showed significant upregulation of EMT-associated genes, such as CDH2, TGF-beta and FGF, and cancer stem cell-associated genes. Additionally, qPCR and IHC demonstrated a suppressive immunological status derived from the upregulation of inflammatory cytokines (IL-6, IL-10 and TGF-beta) and immune checkpoint genes (B7-H3 and B7-H5) and from M2-type macrophage accumulation in the tumor. The induction of EMT and immune-suppressive status in the tumor after strong CRT treatment urges the development of a novel combined therapy that restores immune-suppression and inhibits EMT, ultimately leading to distant metastasis control.
Keywords: chemoradiotherapy (CRT), consensus molecular subtype (CMS), tumor microenvironment (TME), epithelial-to-mesenchymal transition (EMT), tumor-infiltrating lymphocytes (TIL)
INTRODUCTION
Currently, preoperative fluoropyrimidine-based chemoradiotherapy (CRT) followed by radical surgery is one of the standard treatments for locally advanced rectal cancer, and it has been successful in local control with a recurrence rate of ~5% [1–3]. However, the distant recurrence rate after preoperative CRT and surgery remains ~30%, and it is still a crucial clinical problem hindering the improvement of survival rates [4, 5].
Recently, a novel immunotherapy has been developed using specific monoclonal antibodies targeting immune checkpoint molecules, such as cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) and programmed death-1 (PD-1)/PD-ligand 1 (PD-L1). This type of therapy has been applied in many clinical trials [6–9] and shows promising clinical benefits for even advanced-stage or metastatic cancer patients. Use of an immune checkpoint antibody is not recommended in colorectal cancers in terms of its overall response rate; however, metastatic microsatellite instability-high (MSI-H) cases, which account for ~15% of all colorectal cancers [10], were approved for anti-PD-1 antibody and pembrolizumab-based treatment. In the future, new preoperative neoadjuvant regimens combined with pembrolizumab will be developed.
Two main parameters might be involved in the overall survival (OS) in rectal cancer patients with regard to the prognosis of locally advanced rectal cancers in terms of distant recurrence control. The first is molecular subtype classification based on genetic expression data; four consensus molecular subtypes (CMS), CMS1–4, were demonstrated to be associated with survival time, and the CMS4 group has the worst prognosis due to features of epithelial-to-mesenchymal transition (EMT) [11, 12]. The second parameter is memory T cell infiltration inside the tumor, which is used as a tumor microenvironment (TME) marker and is reported to be a good prognostic factor for OS and progression-free survival (PFS) [13–15].
In the present study, aimed to dissect the mechanism of distant recurrence developed after neoadjuvant regimen treatment in rectal cancer patients, we compared genetic and immunohistochemistry (IHC) profiling between preoperative CRT-treated and non-treated rectal cancer patients using whole exome sequencing (WES) and gene expression profiling (GEP) analyses conducted under the HOPE project in the Shizuoka Cancer Center (SCC) [16].
MATERIALS AND METHODS
Patient registration
In 2014, the SCC launched Project HOPE, which is based on multi-omics analyses including WES and GEP. Informed consent was obtained from all patients participating in Project HOPE, and the study was approved by the Institutional Review Board of the Shizuoka Cancer Center, Japan. All experiments using clinical samples were carried out in accordance with the Ethical Guidelines for Human Genome and Genetic Analysis Research. Nine rectal cancer patients given preoperative CRT and 10 rectal cancer patients without preoperative CRT participated, and both groups received surgery. The group without preoperative CRT was given CRT after surgery. At our institution, CRT was applied for patients with locally advanced rectal cancers for the purpose of obtaining clear resected margins after surgery and maximizing the chance of preserving anal sphincters and preventing urinary diversion [17]. The characteristics of both rectal cancer patient groups are shown in Table 1. The tumor tissue and surrounding normal tissue were collected by surgeons after surgery.
Table 1.
List of rectal cancer patients with or without preoperative chemoradiation
| Case | Age, years | Sex | Clinical stage | Preoperative therapy (RT dose) | Pathological effect (TRG) | Relapse |
|---|---|---|---|---|---|---|
| REC-001 | 60 | Male | IIIC | CRT (66 Gy) | 2 | - |
| REC-002 | 71 | Male | IIIC | CRT (50.4 Gy) | 2 | - |
| REC-003 | 63 | Male | IIIC | CRT (50.4 Gy) | 1a | - |
| REC-004 | 44 | Male | IIIC | CRT (50.4 Gy) | 2 | - |
| REC-005 | 47 | Male | IIIC | CRT (50.4 Gy) | 2 | - |
| REC-006 | 65 | Male | IIIB | CRT (50.4 Gy) | 2 | + (Liver) |
| REC-007 | 51 | Male | IIC | CRT (50.4 Gy) | 2 | + (Bone) |
| REC-008 | 60 | Female | IIIC | CRT (50.4 Gy) | 1b | - |
| REC-009 | 47 | Male | IIIC | CRT (50.4 Gy) | 1a | - |
| REC-010 | 40 | Male | IIIB | - | - | - |
| REC-011 | 59 | Male | IIA | - | - | - |
| REC-012 | 65 | Female | IIIC | - | - | - |
| REC-013 | 71 | Male | IIIB | - | - | - |
| REC-014 | 60 | Male | IIIC | - | - | - |
| REC-015 | 57 | Male | IIIC | - | - | - |
| REC-016 | 43 | Female | IIIC | - | - | - |
| REC-017 | 70 | Male | IIIC | - | - | - |
| REC-018 | 63 | Male | IVA | - | - | + (Liver) |
| REC-019 | 61 | Male | IIA | - | - | - |
DNA microarray-based GEP and WES using next-generation sequencing
The method used to perform GEP and WES analyses were described previously [16, 18]. Briefly, for GFP the ratio of the expression intensity between the tumor tissue (T) and the surrounding normal tissue (N) was calculated from the normalized values. The expression values for all probes were log (base 2)-transformed before performing the statistical analysis. In WES, all the variants called by the variant caller were filtered to discard sequences with a quality <30, variant allele frequency <10% or depth of coverage <20. Those mutations that were identified in tumor samples and not observed in matched normal samples were extracted as somatic mutations. Single-nucleotide variants (SNVs) of the total exonic mutations for each sequenced tumor included nonsynonymous, synonymous and indels/frameshift mutations.
Determination of tumor mutation burden and copy number variation
Tumor mutation burden (TMB), representing the number of somatic mutations per megabase, was defined as follows:
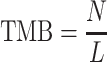 |
where N is the number of somatic mutations identified in RefSeq exons targeted by the WES amplicon, and L is the length of the target region with a depth of coverage ≥20 in both the tumor and normal samples. N and L are calculated for each sample. On average, L is 33.16 Mb in our cohort; therefore, a TMB value of 20 corresponds to 660 mutations in the exons of 18 835 protein-coding genes in the human genome.
A joint segmentation-based approach that accounts for the relative ratio of the read depth in tumor and matched normal samples and variant allele frequency was applied for copy number variation (CNV) analysis. SAAS-CNV (a joint segmentation approach on aggregated and allele specific signals for the identification of somatic copy number alterations with next-generation sequencing data) by Zhang and Hao [19] is the method we utilized in the current study. The following information was extracted from the output of Torrent Variant Caller (TVC) and used as input for SAAS-CNV: (i) chromosome, (ii) position, (iii) reference sequence, (iv) alternative sequence, (v) quality score, (vi) genotype in normal sample, (vii) reference read depth in normal sample, (viii) variant read depth in normal samples, (ix) genotype in tumor samples, (x) reference read depth in tumor samples and (xi) variant read depth in tumor samples. SNVs with a quality score ≥100 and depth of coverage ≥20 were used in the CNV analysis. Those regions were extracted as CNVs.
Immune response-associated genes and SCC820 gene panel expression profiling
An updated list of immune response-associated genes is shown in Supplementary Table 1, see online supplementary material. In total, 203 immune response-associated genes [97 antigen-presenting cell (APC)-associated and T-cell-associated genes, 34 cytokine- and metabolism-associated genes, 47 tumor necrosis factor (TNF) and TNF receptor superfamily genes and 25 regulatory T-cell-associated genes] were used for comparing gene expression levels between the CRT-treated and not-treated group. Additionally, the expression levels of genes of the SCC820 cancer-associated panel, which was reported previously [16], were also compared between the CRT-treated and not-treated groups.
IHC
For the tumor-infiltrating immune cells, antibodies against CD4 and CD8 (Thermo Fisher Scientific, Waltham, MA, USA), CD204 (Transgenic Inc., Kobe, Japan), IL-6 (Abcam, Cambridge, UK), TGF-beta1 (R and D Systems, Inc., Minneapolis, MN, USA), cytokeratin (AE1 and AE3, Nichirei Bio., Tokyo, Japan), FoxP3 (Abcam), Granzyme B (Dako, Glostrup, Denmark) and PD-L1 (Abcam) were purchased and used for the IHC and immuno-fluorescence analyses. For reference, the staining level of tumor-infiltrating immune cells was assessed by a semiquantitative estimation of the density of CD8+ T cells inside the tumor site as follows: score 0, no or sporadic CD8+ T cells; score 1, moderate number of CD8+ T cells; score 2, abundant CD8+ T cells; and score 3, highly abundant CD8+ T cells [20].
For immuno-fluorescence staining, the TGF-beta1, IL-6 and CD204 stains were conducted using an Opal 4-color IHC kit (PerkinElmer Inc., Waltham, MA, USA) and evaluated on a fluorescent Zeiss imager Z1 microscope (Carl Zeiss, Oberkochen, Germany).
Real-time PCR analysis
Real-time PCR analysis of the apoptosis genes (BAX, BCL2, BCL2L1 and TP53), cytokine genes (STAT3, VEGFA and IL-6), stem cell genes (CD44, OCT3/4 and SOX2) and EMT-associated genes (CDH1, CDH2, COL1A2, MMP2, CTNNB1, RSPO1, TCF4, TGFB1, TGFB3, SMAD2, SNAIL2, TWIST1, NOTCH1, WNT5A and ZEB1) was performed using a QuantStudio 12 K Flex Real Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) as described previously [21]. Total RNA was isolated from formalin-fixed and paraffin-embedded (FFPE) rectal cancer specimens using a High Pure RNA Paraffin Kit (Roche Diagnostics, Indianapolis, IN, USA).
Statistical analysis
The genes from the 203 immune response-associated gene panel that were upregulated in the CRT-treated group compared to the not-treated group were identified using the volcano plot method. The proportion of categorical variables that were different between the CRT-treated and not-treated group was calculated with Pearson’s chi-square test and the unpaired two-tailed t-test. Values with P < 0.05 were considered significant. Data analysis was performed using GeneSpring GX software version 13.1.1 (Agilent Technologies) and Microsoft Excel.
RESULTS
Characterization of rectal cancer patients
The number of locally advanced rectal cancer patients with and without preoperative CRT was 9 and 10, respectively (Table 1). We did not observe a significant difference in age, sex or clinical staging between the group with therapy and without therapy. The preoperative therapy regimen consisted of a combination of FOLFIRI (5-FU + l-LV + CPT-11) or Capecitabine and local radiation with a dose of 50.4 to ~66 Gy. The clinical responses of preoperative CRT for 9 rectal cancer patients were all rated as stable disease, while the pathological tumor regression score (TRG) revealed 6 cases of grade2 and 3 cases of grade1.
Characterization of genetic signatures from rectal cancer patients
There was a significant reduction in exon SNV and Vogelstein driver gene mutation numbers in the preoperative CRT-treated rectal cancer group (Table 2). Similarly, there was a significant reduction in tumor mutation burden (TMB) number and CNV size. The decrease in tumor mutation frequency might be a result of preoperative CRT; however, the difference in tumor content was not statistically significant.
Table 2.
Genetic characteristics of rectal cancer patients with or without preoperative chemoradiation
| Case | Preoperative therapy | Duration (days) | SNV (exon) | SNV (nonsynonymous) | Driver gene mutations | TMB (mutations/Mb) | CNV size (Mb) | Tumor content (%) |
|---|---|---|---|---|---|---|---|---|
| REC-001 | CRT | 86 | 9 | 6 | None | 0.122 | 2.27 | 18.5 |
| REC-002 | CRT | 42 | 3 | 1 | None | 0.091 | 175.9 | 93.5 |
| REC-003 | CRT | 41 | 4 | 1 | None | 0.06 | 66.4 | 23.4 |
| REC-004 | CRT | 50 | 7 | 5 | None | 0.091 | 14.6 | 18.4 |
| REC-005 | CRT | 56 | 9 | 8 | None | 0.12 | 131.5 | 19.2 |
| REC-006 | CRT | 42 | 7 | 5 | LMO1 | 0.06 | 833.2 | 20.4 |
| REC-007 | CRT | 47 | 11 | 8 | None | 0.325 | NA | NA |
| REC-008 | CRT | 43 | 19 | 13 | None | 0.565 | NA | NA |
| REC-009 | CRT | 56 | 4 | 3 | None | 0.117 | NA | NA |
| REC-010 | - | - | 65 | 38 | APC, NOTCH1, NOTCH2, TP53 | 1.982 | 2447 | 68.6 |
| REC-011 | - | - | 101 | 65 | APC, FBXW7, KRAS | 3.249 | 655.5 | 47.1 |
| REC-012 | - | - | 107 | 73 | BRAF, TP53 | 3.025 | 611.5 | 53.8 |
| REC-013 | - | - | 146 | 108 | APC, CREBBP, TP53 | 4.371 | 2545 | 28.5 |
| REC-014 | - | - | 155 | 107 | APC, BRCA2, KRAS | 4.702 | 835.0 | 36.1 |
| REC-015 | - | - | 144 | 89 | APC, FBXW7, KRAS, TP53 | 4.399 | 785.3 | 39.6 |
| REC-016 | - | - | 94 | 61 | APC, BRAF, PTEN | 2.845 | 1087 | 27.4 |
| REC-017 | - | - | 70 | 47 | TP53 | 2.136 | 1700 | 33.7 |
| REC-018 | - | - | 79 | 52 | AMER1, APC, KDM6A, KMT2C, TP53 | 2.238 | 1874 | 83.3 |
| REC-019 | - | - | 80 | 54 | APC, ATM, KRAS, PTEN | 2.41 | 1668 | 47.5 |
Expression profiling of immune response-associated and SCC820 gene panels in rectal cancers treated with CRT
Volcano plot analysis demonstrated that the expression of 13 and 5 immune response-associated genes was upregulated and downregulated, respectively, in preoperative CRT-treated rectal cancers (Fig. 1A, Supplementary Table S1). Additionally, the expression analysis of SCC820 cancer-associated genes revealed that 8 were upregulated and 7 were downregulated (Fig. 1B, Supplementary Table S2, see online supplementary material). Among the genes with altered expression, 7 upregulated genes (FGF7, HGF, IL-6, PTGS2, TGFB3, CDH2 and NGFR) and 2 downregulated genes (CDH1 and CDH3) were closely associated with EMT occurrence.
Fig. 1.
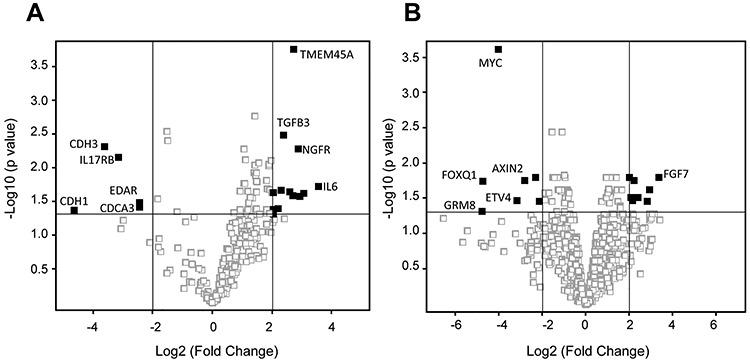
Immune response-associated gene and SCC820 cancer-associated gene expression profiling in CRT-treated rectal cancers. Upregulated or downregulated immune response-associated genes (A) and SCC820 cancer-associated genes (B) with changes >4-fold identified using a volcano plot with Benjamini–Hochberg correction. The horizontal grey line represents a P value of 0.05. The vertical gray lines show 4- and 0.25-fold changes in gene expression. The closed squares represent the upregulated and downregulated genes with changes >4-fold, respectively.
Interestingly, TMEM45A, which belongs to a large family of genes encoding predicted transmembrane (TMEM) proteins, was obviously upregulated in preoperative CRT-treated rectal cancers.
Apoptosis and EMT marker gene expression measured by quantitative PCR
Quantitative PCR revealed that the expression of apoptosis-associated genes (BCL2) and the EMT-associated genes (STAT3, SOX2, CDH2, COL1A2, MMP2, CTNNB1, RSPO1, TCF4, SNAIL2 and TWIST1) was upregulated in preoperative CRT-treated rectal cancers (Fig. 2). Meanwhile, the expression of TP53, c-Myc, CDH1 and NOTCH1 was downregulated. In Fig. 3, a scheme of gene groups that were upregulated in CRT-treated rectal cancers and promoted EMT mediated by SNAIL and TWIST signaling, is provided.
Fig. 2.
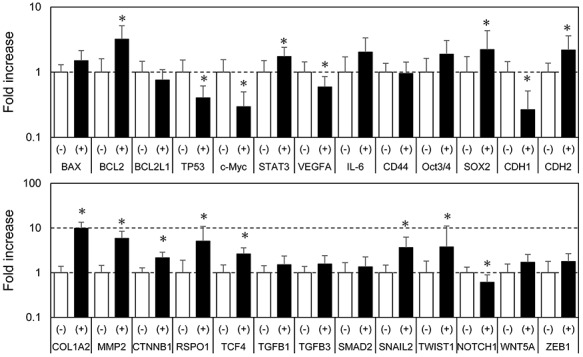
Apoptosis and EMT-associated gene expression profiling using real-time PCR. Total RNA was isolated from the formalin-fixed paraffin-embedded (FFPE) rectal cancer specimens and real-time PCR was performed using a QuantStudio 12 K Flex Real Time PCR System. Open columns represent the control group, and shaded columns the CRT-treated group. Each column represents the mean value of triplicate experiments. *P < 0.05, statistically significant.
Fig. 3.
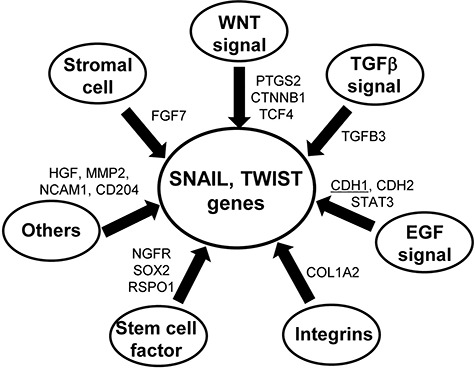
Scheme of differentially expressed genes in CRT-treated rectal cancers leading to EMT induction. The upregulated genes (FGF7, PTFS2, CTNNB1, TCF4, TGFB3, CDH2, STAT3, COL1A2, NGFR, SOX2, RSPO1, HGF, NCAM1 and CD204) and downregulated genes (underlined, CDH1) were associated with EMT events, as determined from the gene profiling of immune response-associated and SCC820 panel genes.
B7 Family and cytokine gene expression profiling in rectal cancers treated with chemoradiation
The heatmap analysis of the B7 family and cytokine gene profiling indicated upregulation of immune-suppressive cytokines (IL-6, IL-10), upregulation of immune checkpoint genes (B7-H3, B7-H5), upregulation of the M2-type macrophage-associated gene (CD204) and downregulation of the immune-activating B7 family genes (B7-H6) (Fig. 4).
Fig. 4.
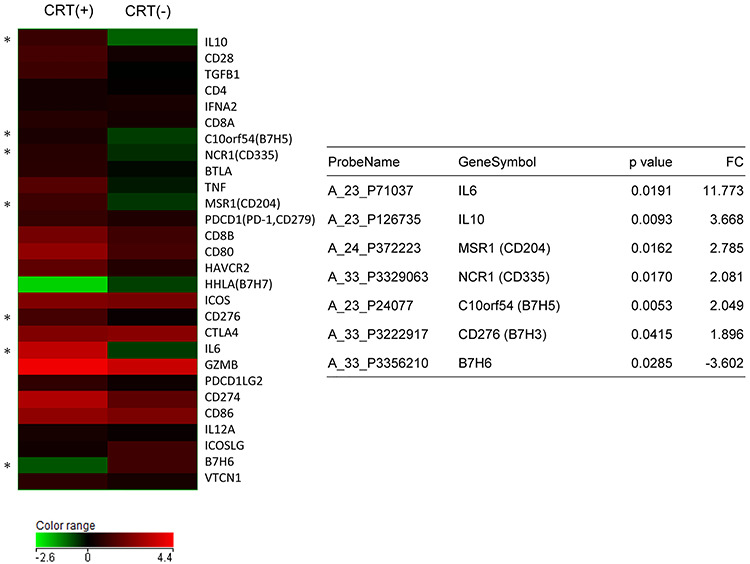
B7 Family and cytokine gene expression profiling in CRT-treated rectal cancers. The relative expression ratio of each tumor tissue versus surrounding normal tissue was calculated from normalized values. The color log scale indicates the average of each group. Genes with significantly altered expression between CRT-treated (6 cases) and not-treated (8 cases) rectal cancers were selected. *P < 0.05, statistically significant. A summary of the statistical analysis is shown in the table.
Tumor-infiltrating immune cell characterization in the TME from rectal cancer patients using IHC
The number of CD4+ T-cells tended to decrease and the number of FoxP3+ regulatory T-cells tended to increase inside the tumor nest between CRT-treated and not-treated rectal cancers. CD8+ T-cell infiltration was neither remarkable nor different between groups with and without therapy. However, CD204+ macrophage infiltration was higher in CRT-treated rectal cancers (Fig. 5A, Supplementary Table S3, see online supplementary material). Meanwhile, immuno-fluorescence staining showed that TGF-beta was derived from tumor cells and IL-6 might be produced from macrophages (Fig. 5B).
Fig. 5.
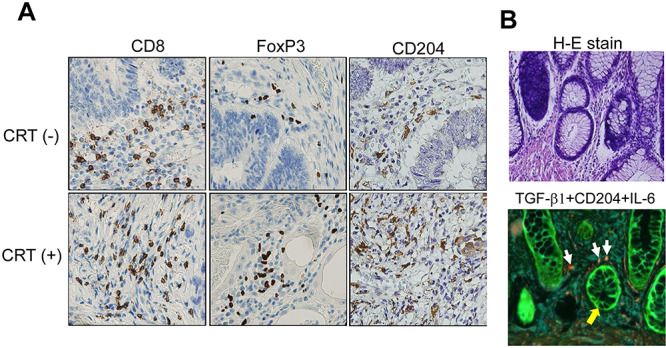
Tumor-infiltrating immune cell characterization using IHC and immuno-fluorescence staining. (A) IHC staining against CD8, Foxp3 and CD204. CRT(−) and CRT(+) specimens were derived from casees REC011 and REC007, respectively. (B) H-E staining and immuno-fluorescent staining for TGF-beta1 (green), IL-6 (blue) and CD204 (red) using the Opal 4-color kit. The white arrows show CD204+ macrophages producing IL-6. The yellow arrows show TGF-beta1+ rectal cancer cells. The specimen was derived from case REC006 with CRT(+) which showed the upregulation of IL-6 gene in a quantitative PCR. Magnification x400.
DISCUSSION
From the results of past clinical trials that targeted locally advanced rectal cancers, preoperative CRT has been reported to show better control of local recurrence; however, it showed no significant difference in cumulative incidence of distant metastasis compared with postoperative CRT [2]. With the aim of improving overall survival after chemoradiation therapy, it is therefore important to evaluate the patient status after treatment in terms of genetic and immunological observations. Few studies have investigated the direct effect of radiation or chemoradiation on rectal cancers using NGS-based genetic analysis [22–24].
Molecular subtype classification based on genetic expression data has been reported by Guinney et al. to facilitate large-scale data sharing and analysis [25]. They found that CMS 1–4 were associated with survival time.
In the present study, most rectal cancers without preoperative CRT are considered CMS2 or 3, and rectal cancers with preoperative CRT show CMS4 typing with obvious EMT. Specifically, many EMT marker genes were recognized as upregulated (STAT3, SOX2, CDH2, COL1A2, MMP2, CTNNB1, RSPO1, TCF4, SNAIL2 and TWIST1) and downregulated (CDH1) (Fig. 2). Such an EMT-promoting genetic signature induced by CRT might lead to immunological escape, resulting in the occurrence of distant metastasis. In particular, TGF-beta is known to perform a central role in constructing the immune-suppressive TME by inducing regulatory T cells and MDSCs in the tumor site and attracting M2-type macrophages into the tumors [24]. Additionally, elevated TGF-beta production can promote IL-11 and IL-6 secretion from cancer-associated macrophages and fibroblasts, and this secretion is associated with distant metastasis of colorectal cancers by activating STAT3 signaling [26]. On the other hand, changes in FGF signaling-associated gene, such as FGF7, was identified, and this is an important observation because CAFs are associated with poor prognosis and resistance to radiotherapy in untreated colorectal cancers [27]. One more upregulated gene, TMEM45A, was verified in CRT-treated rectal cancers. Functionally, little is known about the TEMM45A gene; however, several studies with siRNA-based inhibition indicated similar results of strong inhibition of cancer cell proliferation and invasion, which partially suppressed in vivo tumors [28–30]. These results suggest that TMEM45A might be a novel target for anti-EMT therapeutics development.
Interestingly, another genetic observation was noted in CRT-treated rectal cancer patients. The numbers of SNV, TMB and Vogelstein driver gene mutations were remarkably decreased after CRT, as shown in Table 2. The reduction in c-MYC and ETV4 gene expression might be further evidence of the decrease of cancer cells with driver gene mutations. The tumor content reduction after CRT was partially responsible for this effect because tumor content with CRT (32.2%) tended to decrease compared to that without CRT (46.5%).
The IHC study showed similar results from genetic expression profiling of rectal cancer tissues. Upregulation of immune-suppressive cytokines (TGF-beta and IL-6) and immune checkpoint proteins (B7-H3 and B7-H5) and higher infiltration of M2-type macrophages were observed. The frequency of other tumor-infiltrating lymphocytes (TILs) (CD4+, CD8+ and FoxP3+) was not significantly different between the rectal cancer groups with and without CRT (Supplementary Table S3). Matsutani et al. investigated CD8+ TIL numbers before and after neoadjuvant CRT in the same rectal cancer patients and demonstrated that the density of CD8+ TILs in post-treatment resected tumors was significantly increased compared with that in pretreatment biopsy samples [31]. Additionally, Shinto et al. used multivariate analyses to demonstrate that high levels of stromal CD8+ T cells post-CRT and a high pretreatment intraepithelial CD8/FoxP3 ratio were associated with better prognosis in CRT-treated rectal cancers [32]. Our study demonstrated that CD4+ T-cells tended to decrease and FoxP3+ regulatory T-cells tended to increase inside the tumor nest between the CRT-treated and not-treated rectal cancers, which could reflect the immunological effect on TME of CRT; observations of patient survival time are necessary in the near future. These observations suggest that the timing of posttreatment resection and the dose of radiation might be important factors for evaluating TIL status.
Considering the immunological observations in the current study, some EMT markers may be therapeutic targets for the development of an efficient combination regimen with preoperative CRT. The first potential treatments for combination are the immune checkpoint blockade molecules like anti-PD-1/PD-L1 antibodies. Recently, based on the evidence that radiotherapy might upregulate the PD-L1 expression level in the tumor, the combination therapy of radiotherapy with anti-PD-L1 antibody has been performed for recurrent non-small-cell lung cancer and other cancers [33]. Considering that our study demonstrated that the expression of immune checkpoint genes like B7-H3 and B7-H5 was upregulated after CRT, the combination of RT and anti-immune checkpoint blockade could be a possible regimen.
The second target is the TGF-beta signaling pathway. Anti-TGF-beta monoclonal antibody (fresolimumab) [34] and the small molecule compounds regorafenib [35] and galunisertib are under development. Fresolimumab was used in a phase I clinical study against advanced malignant melanoma or renal cell carcinoma [34]. Regorafenib prevents TGF-beta1-induced EMT by activating SH2-domain-containing phosphatase 1 (SHP-1)-dependent p-STAT3 Tyr705 suppression [35]. Galunisertib is an oral small molecule inhibitor of TGF-beta receptor I kinase and has recently been studied in glioblastoma and pancreatic cancer patients.
The third target is tumor-associated macrophages producing inflammatory cytokines like IL-6 and 1L-10, which can contribute to rectal cancer metastasis through cytokine-mediated COX-2/β-catenin signal-induced EMT [36]. However, an efficient therapeutic device has not been available for clinical trial to date.
There were several limitations to this study. It was a retrospective study conducted in a single institution with a small number of patients. In Japan, lateral lymph node dissection is the standard treatment for patients with locally advanced low rectal cancer [37] and therefore a limited number of patients received neoadjuvant CRT at our institution. This made it difficult to enroll more patients and compare these patients with those of well-matched patients who underwent surgery alone. Also, the chemotherapy regimens were not unified. In addition, patients with a clinically complete response after CRT were not enrolled in this study. This might affect assessments of the microenvironment after CRT.
CONCLUSION
The genetic and immunological features were compared between preoperative CRT-treated and non-treated rectal cancer patients. CRT-treated tumors showed significant upregulation of EMT-associated genes and cancer stem cell-associated genes, and a suppressive immunological status, which was derived from the upregulation of inflammatory cytokines and immune checkpoint genes and from M2-type macrophage accumulation in the tumor. The induction of EMT and immune-suppressive status in the tumor after strong CRT might be one of the mechanisms leading to the development of distant recurrence.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the staff at the Shizuoka Cancer Center Hospital for clinical support and sample preparation. This work was supported by the Shizuoka Prefectural Government, Japan.
CONFLICT OF INTEREST
None declared.
REFERENCES
- 1. Valentini V, Stiphout RG, Lammering G et al. . Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol 2011;29:3163–72. [DOI] [PubMed] [Google Scholar]
- 2. Sauer R, Liersch T, Merkel S et al. . Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol 2012;30:1926–33. [DOI] [PubMed] [Google Scholar]
- 3. Hofheinz RD, Wenz F, Post S et al. . Chemoradiotherapy with capecitabine versus fluorouracil for locally advanced rectal cancer: A randomised, multicentre, non-inferiority, phase 3 trial. Lancet Oncol 2012;13:579–88. [DOI] [PubMed] [Google Scholar]
- 4. Rödel C, Graeven U, Fietkau R et al. . German rectal cancer group. Oxaliplatin added to fluorouracil-based preoperative chemoradiotherapy and postoperative chemotherapy of locally advanced rectal cancer (the German CAO/ARO/AIO-04 study): Final results of the multicenter, open-label, randomized, phase 3 trial. Lancet Oncol 2015;16:979–89. [DOI] [PubMed] [Google Scholar]
- 5. Deng Y, Chi P, Lan P et al. . Modified FOLFOX6 with or without radiation versus fluorouracil and leucovorin with radiation in neoadjuvant treatment of locally advanced rectal cancer: Initial results of the Chinese FOWARC multicenter, open-label, randomized three-arm phase III trial. J Clin Oncol 2016;34:3300–7. [DOI] [PubMed] [Google Scholar]
- 6. Weber JS, O’Day S, Urba W et al. . Phase I/II study of ipilimumab for patients with metastatic melanoma. J Clin Oncol 2008;26:5950–6. [DOI] [PubMed] [Google Scholar]
- 7. Topalian SL, Hodi FS, Brahmer JR et al. . Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brahmer JR, Tykodi SS, Chow LQ et al. . Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolchok JD, Kluger H, Callahan MK et al. . Nivolomab plus öipilimumab in advanced melanoma. N Engl J Med 2013;369:122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tato-Costa J, Casimiro S, Pacheco T et al. . Therapy-induced cellular senescence induces epitherial-to-mesebchymal transition and increases invasiveness in rectal cancer. Clin Colorectal Cancer 2016;15:170–8. [DOI] [PubMed] [Google Scholar]
- 12. Bhangu A, Wood G, Mirnezami A et al. . Epithelial mesenchymal transition in colorectal cancer: Seminal role in promoting disease progression and resistance to neoadjuvant therapy. Surg Oncol 2012;21:316–23. [DOI] [PubMed] [Google Scholar]
- 13. Pagès F, Berger A, Camus M et al. . Effector momory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66. [DOI] [PubMed] [Google Scholar]
- 14. Kim Y, Bae JM, Li G et al. . Image analyzer-based assessment of tumor-infiltrating T cell subsets and their prognostic values in colorectal carcinomas. PLoS One 2015;10:e0122183. doi: 10.1371/journal.pone.0122183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galon J, Costes A, Sanchez-Cabo F et al. . Type, density, and location of immune cells within human corelectal tumors predict clinical outcome. Science 2006;313:1960–4. [DOI] [PubMed] [Google Scholar]
- 16. Ohshima K, Hatakeyama K, Nagashima T et al. . Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci Rep 2017;7:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hino H, Yamaguchi T, Kinugasa Y et al. . Robotic-assisted multivisceral resection for rectal cancer: Short-term outcomes at a single center. Tech Coloproctol 2017;21:879–86. [DOI] [PubMed] [Google Scholar]
- 18. Hatakeyama K, Ohshima K, Nagashima T et al. . Molecular profiling and sequential somatic mutation shift in hypermutator tumors harbouring POLE mutations. Sci Rep 2018;8:8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Z, Hao K. SAAS-CNV: A joint segmentation approach on aggregated and allele specific signals for the identification of somatic copy number alterations with next-generation sequencing data. PLoS Comput Biol 2015;11:e1004618. doi: 10.1371/journal.pcbi.1004618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dahlin AM, Henriksson ML, Van Guelpen B et al. . Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol 2011;24:671–82. [DOI] [PubMed] [Google Scholar]
- 21. Ashizawa T, Iizuka A, Nonomura C et al. . Antitumor effect of programmed death-1 (PD-1) blockade in humanized the NOG-MHC double knockout mouse. Clin Cancer Res 2017;23:149–58. [DOI] [PubMed] [Google Scholar]
- 22. Findlay VJ, Wang C, Watson DK et al. . Epithelial-to-mesenchymal transition and the cancer stem cell phenotype: Insights from cancer biology with therapeutic implications for colorectal cancer. Cancer Gene Ther 2014;21:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawamoto A, Yokoe T, Tanaka K et al. . Radiation induces epitherial-mesenchymal transition in colorectal cancer cells. Oncol Rep 2012;27:51–7. [DOI] [PubMed] [Google Scholar]
- 24. Dienstmann R, Vermeulen L, Guinney J et al. . Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79–92. [DOI] [PubMed] [Google Scholar]
- 25. Guinney J, Dienstmann R, Wang X et al. . The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calon A, Espinet E, Palomo-Ponce S et al. . Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastatic initiation. Cancer Cell 2012;22:571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Isella C, Terrasi A, Bellomo SE et al. . Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015;47:312–9. [DOI] [PubMed] [Google Scholar]
- 28. Guo J, Chen L, Luo N et al. . Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep 2015;33:3124–30. [DOI] [PubMed] [Google Scholar]
- 29. Sun W, Qiu G, Zou Y et al. . Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int J Clin Exp Pathol 2015;8:12657–67. [PMC free article] [PubMed] [Google Scholar]
- 30. Manawapat-Klopfer A, Thomsen LT, Martus P et al. . TMEM45A, SERPINB5 and p16NK4A transcript levels are predictive for development of high-grade cervical lesions. Am J Cancer Res 2016;6:1524–36. [PMC free article] [PubMed] [Google Scholar]
- 31. Matsutani S, Shibutani M, Maeda K et al. . Significance of tumor-infiltrating lymphocytes before and after neoadjuvant therapy for rectal cancer. Cancer Sci 2018;109:966–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shinto E, Hase K, Hashiguchi Y et al. . CD8+ and FOXP3+ tumor-infiltrating T cells before and after chemoradiotherapy for rectal cancer. Ann Surg Oncol 2014;Suppl 3:S414–21. [DOI] [PubMed] [Google Scholar]
- 33. Gong X, Li X, Jiang T et al. . Combined radiotherapy and anti-PD-L1 antibody synergistically enhances antitumor effect in non-small cell lung cancer. J Thorac Oncol 2017;12:1085–97. [DOI] [PubMed] [Google Scholar]
- 34. Morris JC, Tan AR, Olencki TE et al. . Phase I study of GC1008 (fresolimumab) : A human anti-transforming growth factor-beta (TGFb) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PLoS One 2014;9:e90353. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan LC, Teng HW, Shiau CW et al. . Regorafenib (Stivarga) pharmacologically targets epithelial-mesenchymal transition in colorectal cancer. Oncotarget 2016;7:64136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Che D, Zhang S, Jing Z et al. . Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE2/β-catenin signalling pathway. Mol Immunol 2017;90:197–210. [DOI] [PubMed] [Google Scholar]
- 37. Hashiguchi Y, Muro K, Saito Y et al. . Japanese Society for Cancer of the colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol 2020;25:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


