Abstract
Cell-based therapies are gaining prominence in treating a wide variety of diseases and using synthetic polymers to manipulate these cells provides an opportunity to impart function that could not be achieved using solely genetic means. Herein, we describe the utility of functional block copolymers synthesized by ring-opening metathesis polymerization (ROMP) that can insert directly into the cell membrane via the incorporation of long alkyl chains into a short polymer block leading to non-covalent, hydrophobic interactions with the lipid bilayer. Furthermore, we demonstrate that these polymers can be imbued with advanced functionalities. A photosensitizer was incorporated into these polymers to enable spatially controlled cell death by the localized generation of 1O2 at the cell surface in response to red-light irradiation. In a broader context, we believe our polymer insertion strategy could be used as a general methodology to impart functionality onto cell-surfaces.
Keywords: block copolymers, cell-surface engineering, hydrophobic insertion, photosensitizer, ring-opening metathesis polymerization
Graphical Abstract

Polymer-Cell Conjugates: By incorporating a short polymer block (2-5 repeat units) containing long hydrophobic chains, block copolymers can be synthesized that quickly label cell surfaces and can incorporate advanced functionalities.
Cell-based therapies have recently emerged as next-generation treatments for a variety of disease states. These therapies have been engineered to be efficacious for hematopoietic malignancies, tissue engineering scaffolds and therapeutic hormone factories.[1] The success of cell-based therapies is largely contingent on the interactions between biomacromolecules on cell surfaces, providing impetus to find mew methods to engineer these interfaces. Genetic modifications are a powerful strategy to tailor the cell surface however, the process of engineering cells on the genetic level can be technically difficult and laborious with unpredictable outcomes.[1] An alternative to genetic expression of biomacromolecules on the cell surface is the modification of the cell-surface with synthetic macromolecules to impart unique functionalities.[2-4]
Researchers have modified the surface of therapeutically relevant cell lines with synthetic polymers for a variety of applications such as camouflaging transplanted cells from a hostile immune response,[5,6] promoting neural differentiation of embryonic stem cells[7,8] as well as directing migration of mesenchymal stem cells to damaged heart tissues.[9] These cell-polymer conjugates can be constructed either by: 1) covalent polymer attachment to membrane proteins, 2) electrostatic layer-by-layer (LBL) deposition or 3) non-covalent hydrophobic insertion into the cell membrane.[10] While polymer incorporation by covalent chemistry and LBL both generate stable cell-polymer conjugates, these modifications can disrupt the function of surface-exposed proteins, compromising the efficacy of cell-based therapies.[9] Non-covalent membrane insertion affords cell-polymer constructs in which the functional integrity of surface-exposed proteins is maintained. Furthermore, insertion into the cell membrane occurs rapidly, usually in minutes.[11,12]
The most common method of membrane insertion involves synthetic manipulation of the lipid constituents of the cell membrane to anchor synthetic polymers, most often long linear polyethylene glycol (PEG) chains, to the cell surface.[13] However, the utility of PEG is constrained by its capacity to incorporate multiple functionalities within a single polymer. Additionally, a growing proportion of the population have developed anti-PEG antibodies which would have detrimental effects on any PEG-based approach.[14] The development of novel synthetic polymers that can effectively interact at the cell interface will be required to overcome these challenges.
Living polymerization techniques allow the incorporation of multiple orthogonal functionalities within a single polymer chain as well as control over the distribution of those functionalities along the polymer backbone, significantly expanding the scope of materials that can be attached to a cell surface. For example, phospholipid-terminated synthetic glycopolymers synthesized by reversible addition-fragmentation chain-transfer (RAFT) have been used to mimic native glycoproteins and promote embryonic stem cell differentiation.[7,8,15]
To our knowledge, polymers synthesized by ring-opening metathesis polymerization (ROMP) have not been employed for cell-engineering applications. ROMP has distinct advantages over other controlled polymerization techniques, including rapid and mild reaction conditions, broad functional group tolerance and high fidelity of the terminal ruthenium carbene for block copolymer formation. We envisioned a diblock copolymer, where one block consists of hydrophobic alkyl chains to facilitate membrane insertion, followed by a second block to impart water solubility, immune shielding and other functionalities for downstream biomedical applications (Scheme 1). Herein, we describe poly(oxanorbornene) diblock copolymers synthesized by ROMP in which key parameters such as the number of membrane insertion moieties, polymer molecular weight and monomer compositions are evaluated for their effect on polymer insertion into cell membranes.
Scheme 1.

Depiction of poly(oxanorbornene) polymers decorating cell membrane surface via hydrophobic insertion into the phospholipid bilayer.
To synthesize membrane-inserting block copolymers, we chose a ROMP graft-through approach to first install a discrete hydrophobic block to anchor the polymer to the cell surface followed by a copolymerization of a water-soluble monomer and fluorescent dye (Figure 1). The hydrophobic block consisted of a polyoxyethylene fatty ether derived from cetyl alcohol (Brij C20), containing a C16 alkyl chain. This hydrocarbon length has been demonstrated in both phospholipid and nonionic detergents to enable rapid insertion as well as enhanced retention on the membrane surface.[13,16] Coupling of Brij C20 to exo-5-norbornene-2-carboxylic acid provided NB-Brij monomer in good overall yield. Monomers oNB-PEG and oNB-Zwit were synthesized according to literature procedures to explore the effects and scope of water-soluble monomers.[17,18] A fluorescent monomer (oNB-FITC) was also synthesized (See SI) and incorporated into the hydrophilic block to image the resulting polymers on the cell surface by confocal laser scanning microscopy (CLSM) and evaluate insertion via flow cytometry.
Figure 1.

Polymerization strategy of Brij-containing poly(oxanorbornene) block copolymers.
Grubbs 3rd generation catalyst was used to make the diblock copolymer by first polymerizing the NB-Brij block followed by addition of the hydrophilic monomer (either oNB-PEG or oNB-Zwit) and oNB-FITC (Figure 1). Polymers consisting of 25 and 50 repeat units of oNB-PEG and oNB-Zwit were targeted to explore effects of molecular weight. Additionally, polymers containing 0, 2 and 5 equivalents of NB-Brij were synthesized to investigate the effect of increasing number of Brij moieties on cell membrane insertion and retention. Polymers containing Brij units greater than 5 were not appreciably soluble in PBS and precluded their use in this study. It is important to note that the targeted block lengths represent a statistical average within the polymer backbone. However, the ability of ROMP to make well-defined polymer blocks gives us confidence that the distribution of block lengths is narrow and contributes toward the validity of comparisons. Unfortunately, sulfobetaine polymers containing 5 equivalents of NB-Brij were minimally soluble and precluded their use from this study. The molecular weights of the resulting polymers were obtained by gel permeation chromatography (GPC) (Table 1, Figure S1). While experimental molecular weights deviated slightly from the theoretical molecular weights, we attribute this observation to polymer associations with the column and the use of dissimilar polymer standards. Furthermore, 1H NMR analysis shows nearly full monomer consumption (Figure S18).
Table 1.
ROMP block copolymers synthesized for this study.
| Polymer[a] | Theoretical MW (kDa) |
Actual Mn (kDa)[b] |
Ɖ | Brij : PEG[c] |
Brij : FITC[c] |
|---|---|---|---|---|---|
| Brij0PEG25 | 14.2 | 14.8 | 1.62 | N/A | N/A |
| Brij2PEG25 | 16.7 | 14.3 | 1.25 | 2 : 23 | 2 :1 |
| Brij5PEG25 | 20.5 | 17.5 | 1.25 | 5 : 20 | 5 : 1.5 |
| Brij2PEG50 | 30.9 | 20.7 | 1.35 | 2 : 49 | 2 : 1 |
| Brij5PEG50 | 34.7 | 24.4 | 1.32 | 5 : 46 | 5 : 1.5 |
| Brij0Zwit25 | 10.7 | 7.5 | 1.24 | N/A | N/A |
| Brij2Zwit25 | 12.5 | 10.6 | 1.53 | N/A | N/A |
| Brij2Zwit50 | 22.2 | 18.2 | 1.15 | N/A | N/A |
All polymers were synthesized with 3 equivalents of oNB-FITC.
The molecular weights of polymers containing oNB-PEG were determined by SEC-GPC in THF using polystyrene standards. The molecular weights of polymers containing oNB-Zwit were determined by SEC-GPC in TFE using PMMA standards.
Ratios were determined by 1H – NMR in CDCl3. Polymers containing oNB-Zwit were insoluble in typical deuterated solvents.
To test the ability of polymers to insert into the cell membrane, adherent 3T3 fibroblasts were chosen as a robust cell line. All polymers exhibited no significant cytotoxicity after 24 hours over a range of concentrations (Figure S2). For cell labelling experiments, 3T3 cells were incubated with 5 μM polymer at 37 °C for 20 min in serum-free media. This concentration lies below the experimentally determined critical micelle concentration (Figure S8), which we hypothesized should facilitate insertion of the polymer unimers into the cell membrane.[16] Flow cytometry was used to determine the effect of the Brij moiety on cell uptake. Gratifyingly, only polymers containing a Brij block exhibited significant membrane-insertion and a large shift in mean fluorescence intensity (MFI), whereas Brij0PEG25 and Brij0Zwit25 showed MFI values comparable to untreated cells (Figure 2A). Subsequent CLSM clearly shows Brij-containing polymers are primarily labelled around the periphery of the cell membrane with minimal internalization (Figure 2B).
Figure 2.
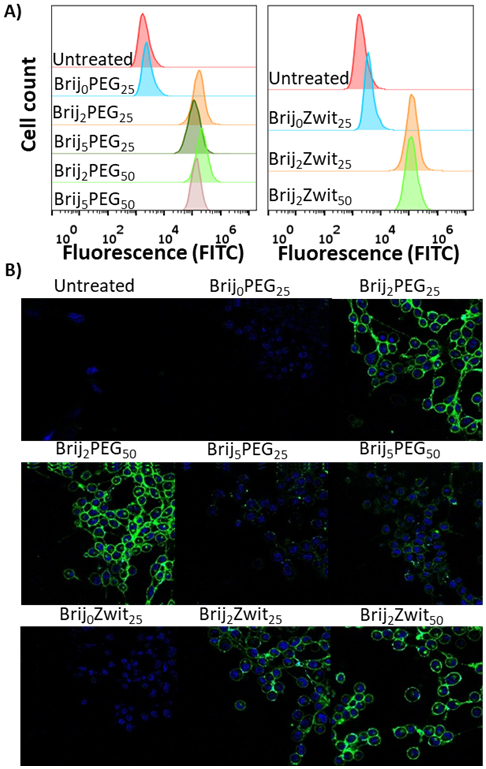
A) Flow cytometry data of 3T3 cell line labeled with polymers. B) CLSM images of 3T3 cells with nucleus stain (blue; Hoechst dye) and polymer (green; FITC).
With the recent FDA approval of CAR-T cell therapy for the treatment of certain hematological malignancies, we were interested in expanding our methodology to a relevant model cell line to demonstrate the potential of our surface engineering strategy. For this reason, Jurkat T-cells were chosen as a mimic for CAR-T therapeutic cells. Jurkat cells maintained high viability after cell labeling over a range of concentrations up to 72 hours (Figure 3). Additionally, a CFSE cell proliferation assay was conducted to asses any inhibition on cell division due to the presence of our polymers inserted into the cell membrane. No differences in cell division were observed between untreated cells and those treated with either Brij2 of Brij5 containing polymers (Figure S3).
Figure 3.

MTT assay of Jurkat cells at varying concentrations of polymer over 24 and 72 hours.
As with the 3T3 cell line, only Brij containing polymers inserted into the Jurkat cell membrane and displayed on the cell surface (Figure 4A). Subsequent membrane staining further illustrates localization of the polymers on the cell membranes (Figure 4B).
Figure 4.

A) Flow cytometry data of Jurkat cell line labeled with polymers. B) Confocal microscopy images of Jurkat cells with nucleus stain (blue; Hoechst dye), membrane stain (red; WGA-647) and polymer (green; FITC). C) Monitoring fraction of polymer retained on the cell surface for Brij2PEG25 and Brij5PEG25. Fraction of polymer retention determined by comparing mean fluorescence intensity at a given time point relative to intensity immediately after polymer labeling.
By flow cytometry, no apparent dependence on hydrophilic monomer composition for polymer insertion into Jurkats was observed when comparing Brij2PEG25 and Brij2Zwit25 as well as their corresponding high molecular weight counterparts (Figure 4A; Figure S4C). A slight decrease in the MFI was observed as the molecular weight of the polymer increased. This is likely due to the larger hydrodynamic radius of the higher molecular weight polymer already attached at the cell surface hindering the approach of additional polymer chains.
A notable difference in polymer incorporation is observed when comparing polymers with a longer Brij polymer block. There is an approximate 2-fold increase in the MFI for polymers with two equivalents of NB-Brij compared to polymers with 5 equivalents of NB-Brij. We attribute this to self-association of the Brij block within unimers, making the alkyl chains inaccessible for insertion.
A valuable component of this block copolymer strategy is the potential to modulate polymer retention on the cell surface by tuning the Brij block length. To test this hypothesis, Jurkat cells modified with Brij2PEG25 and Brij5PEG25 were incubated in complete media at 37 °C and the fraction of remaining polymers on the cell surface was determined by monitoring the decrease of MFI over time relative to the initial MFI values immediately after polymer labeling (Figure 4C). Both polymers exhibited significant desorption within 24 hours, which is consistent with previous reports on lipid and cholesterol modification of cell surfaces.[12,13,15] However as expected, Brij5PEG25 polymers were retained on the cell surface more effectively than Brij2PEG25 polymers. Even after 24 hours, polymers were still observed primarily on the cell surface, albeit with significantly less intensity (Figure S7). These results demonstrate that this block copolymer strategy is a viable method to tailor polymer retention on the cell surface.
As a further demonstration of the potential for this cell-labelling methodology, we explored more advanced functionalities that could be incorporated into the polymer and thus imparted on the polymer-labeled cells. As proof of concept, we explored the use of a photosensitizer that could be used to generate a highly localized concentration of 1O2 near the cell surface, triggering cell death in a spatiotemporal manner without harming healthy unlabeled cells.[19] This ability would be particularly useful in mitigating the off-target effects associated with some cell-based therapies, such as in the case of CAR-T cells.[20] While genetic strategies incorporating bispecific T-cell engagers (BiTEs)[21,22] to further gain “on-target” tumor specificity and suicide genes[23,24] to eradicate “on-target, off-tumor” T cell activity have been exploited to address these concerns, the cellular engineering required to incorporate such modalities are not straightforward.
Pheophorbide A (Pheo) is a photosensitizer with a high 1O2 quantum yield that has shown promise in various polymeric formulations in vitro and in vivo.[25-28] To that end, Pheo was conjugated to amine terminated oxanorbornene, giving oNB-Pheo and subsequently incorporated into a Brij containing polymer, giving Brij2PEG25Pheo (Figure 5A). Jurkat cells were incubated with Brij2PEG25Pheo as well as Brij2PEG25 as a control. CLSM confirmed the surface localization of Brij2PEG25Pheo (Figure 5B). Cell-polymer conjugates were irradiated with 660 nm red LED light at 10.4 mW/cm2 for 15 minutes. After 24 h, cell viability was assessed by MTT. Figure 5C shows that triggered cell death can be achieved for cells labeled with Pheo-containing polymers in the presence of 660 nm light.
Figure 5.

A) Structure of Brij2PEG25Pheo. B) CLSM of Jurkat cells modified with Brij2PEG25Pheo, (red, Pheo) and nucleus stain (blue, Hoechst) dye. C) Cell viability of Jurkat cells modified with either Brij2PEG25 or Brij2PEG25Pheo in the dark or irradiated with 660 nm light (10.4 mW/cm2, 15 minutes).
In conclusion, we have demonstrated that poly(oxanorbornene) block copolymers containing a short Brij block can be used to insert into cell membranes. Polymer retention on the cell surface can be tuned by changing the length of the Brij block. Furthermore, advanced functionalities can be incorporated into the polymer construct. As proof of concept, we have incorporated a photosensitizer into the polymer backbone to trigger spatially controlled on-demand cell killing. We envision that the functionality these materials can impart at the cell surface can be utilized to improve upon cell-based therapies.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the NIH (R21 EB024874) and NSF (1808031) for funding. CLSM images was made possible by the UCSD Microscopy Core (NINDS NS047101). We would also like to thank Prof. Todd Emrick and Le Zhou at UMass Amherst for their assistance in characterizing the zwitterionic polymers by GPC.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Institute and/or researcher Twitter usernames: @pokorski_lab
References
- [1].Woodsworth DJ, Holt RA, Trends in Molecular Medicine 2017, 23, 104–115. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Fu L, Chen X, Journal of Controlled Release 2015, 219, 560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Csizmar CM, Petersburg JR, Wagner CR, Cell Chemical Biology 2018, 25, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Abbina S, Siren EMJ, Moon H, Kizhakkedathu JN, ACS Biomaterials Science & Engineering 2018, 4, 3658–3677. [DOI] [PubMed] [Google Scholar]
- [5].Bradley AJ, Test ST, Murad KL, Mitsuyoshi J, Scott MD, Transfusion 2001, 41, 1225–1233. [DOI] [PubMed] [Google Scholar]
- [6].Totani T, Teramura Y, Iwata H, Biomaterials 2008, 29, 2878–2883. [DOI] [PubMed] [Google Scholar]
- [7].Liu Q, Lyu Z, Yu Y, Zhao Z-A, Hu S, Yuan L, Chen G, Chen H, ACS Applied Materials & Interfaces 2017, 9, 11518–11527. [DOI] [PubMed] [Google Scholar]
- [8].Huang ML, Smith RAA, Trieger GW, Godula K, Journal of the American Chemical Society 2014, 136, 10565–10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee DY, Cha B-H, Jung M, Kim AS, Bull DA, Won Y-W, Journal of Biological Engineering 2018, 12, DOI 10.1186/s13036-018-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Teramura Y, Iwata H, Soft Matter 2010, 6, 1081. [Google Scholar]
- [11].Kato K, Itoh C, Yasukouchi T, Nagamune T, Biotechnology Progress 2004, 20, 897–904. [DOI] [PubMed] [Google Scholar]
- [12].Teramura Y, Kaneda Y, Totani T, Iwata H, Biomaterials 2008, 29, 1345–1355. [DOI] [PubMed] [Google Scholar]
- [13].Itagaki T, Arima Y, Kuwabara R, Kitamura N, Iwata H, Colloids and Surfaces B: Biointerfaces 2015, 135, 765–773. [DOI] [PubMed] [Google Scholar]
- [14].Schellekens H, Hennink WE, Brinks V, Pharmaceutical Research 2013, 30, 1729–1734. [DOI] [PubMed] [Google Scholar]
- [15].Woods EC, Yee NA, Shen J, Bertozzi CR, Angewandte Chemie International Edition 2015, 54, 15782–15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Casadei BR, Domingues CC, Clop EM, Couto VM, Perillo MA, de Paula E, Colloids and Surfaces B: Biointerfaces 2018, 166, 152–160. [DOI] [PubMed] [Google Scholar]
- [17].Rankin DA, Lowe AB, Macromolecules 2008, 41, 614–622. [Google Scholar]
- [18].Adams M, Richmond V, Smith D, Wang Y, Fan F, Sokolov AP, Waldow DA, Polymer 2017, 116, 218–225. [Google Scholar]
- [19].Zhang A, Jung K, Li A, Liu J, Boyer C, Progress in Polymer Science 2019, 99, 101164. [Google Scholar]
- [20].Srivastava S, Riddell SR, The Journal of Immunology 2018, 200, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Choi BD, Cai M, Bigner DD, Mehta AI, Kuan C-T, Sampson JH, Expert Opinion on Biological Therapy 2011, 11, 843–853. [DOI] [PubMed] [Google Scholar]
- [22].Choi BD, Yu X, Castano AP, Bouffard AA, Schmidts A, Larson RC, Bailey SR, Boroughs AC, Frigault MJ, Leick MB, et al. , Nature Biotechnology 2019, DOI 10.1038/s41587-019-0192-1. [DOI] [PubMed] [Google Scholar]
- [23].Casucci M, Hawkins RE, Dotti G, Bondanza A, Cancer Immunology, Immunotherapy 2015, 64, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Diaconu I, Ballard B, Zhang M, Chen Y, West J, Dotti G, Savoldo B, Molecular Therapy 2017, 25, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rapozzi V, Zorzet S, Zacchigna M, Drioli S, Xodo LE, Investigational New Drugs 2013, 31, 192–199. [DOI] [PubMed] [Google Scholar]
- [26].Kim WL, Cho H, Li L, Kang HC, Huh KM, Biomacromolecules 2014, 15, 2224–2234. [DOI] [PubMed] [Google Scholar]
- [27].Zagami R, Rapozzi V, Piperno A, Scala A, Triolo C, Trapani M, Xodo LE, Monsù Scolaro L, Mazzaglia A, Biomacromolecules 2019, 20, 2530–2544. [DOI] [PubMed] [Google Scholar]
- [28].Kato T, Jin CS, Ujiie H, Lee D, Fujino K, Wada H, Hu H, Weersink RA, Chen J, Kaji M, et al. , Lung Cancer 2017, 113, 59–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


