Abstract
The mammalian preimplantation embryo is a highly tractable, self-organizing developmental system in which three cell types are consistently specified without the need for maternal factors or external signals. Studies in the mouse over the past decades have greatly improved our understanding of the cues that trigger symmetry breaking in the embryo, the transcription factors that control lineage specification and commitment, and the mechanical forces that drive morphogenesis and inform cell fate decisions. These studies have also uncovered how these multiple inputs are integrated to allocate the right number of cells to each lineage despite inherent biological noise, and as a response to perturbations. In this review, we summarize our current understanding of how these processes are coordinated to ensure a robust and precise developmental outcome during early mouse development.
This article is part of a discussion meeting issue ‘Contemporary morphogenesis'.
Keywords: mouse blastocyst, patterning, morphogenesis, self-organization, population size control, robustness
1. Introduction
Tissue morphogenesis, patterning and growth control are fundamental processes during the development of multicellular organisms. Both local and systemic mechanisms ensure coordination between these processes to generate reproducible outcomes during embryonic development and to maintain homeostasis in the adult [1–3]. Understanding this coordination of events in space and over time can be challenging, given their dynamic nature, the complex interactions between cells and the multitude of signalling pathways involved. The mouse preimplantation embryo represents a simple and experimentally tractable system to understand how self-organizing developmental systems solve some of these challenges. In this review, we discuss our current understanding of how patterning, morphogenesis and growth (whereby we refer to changes in cell numbers) are coordinated to guarantee precision and robustness during the earliest stages of mammalian development.
2. Overview of preimplantation development
Mammalian preimplantation development encompasses the period between fertilization and implantation of the embryo into the maternal uterine wall. Cleavage of the zygote (one cell) generates a morula of eight cells (referred to as blastomeres) which undergo patterning and morphogenetic events to generate the blastocyst, a universal structure across mammals, which is capable of attaching to the uterus and eventually developing into a fetus (figure 1a). Beside the morphological changes in the embryo, during this period, the embryonic genome is activated and undergoes dramatic modifications—including epigenetic reprogramming and X chromosome inactivation in female embryos—which are fundamental for embryonic development. These processes are reviewed in detail elsewhere [4–6].
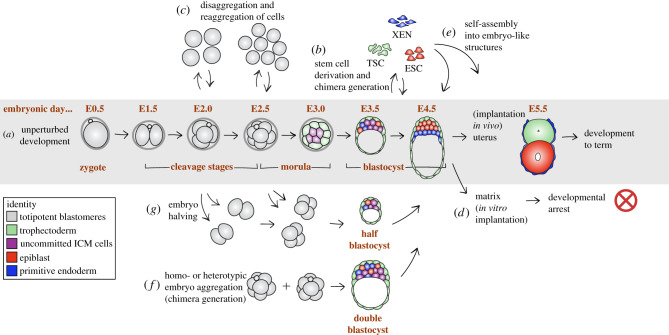
Figure 1.
Natural and experimental paths for preimplantation development. (a) Normal developmental progression of the preimplantation mouse embryo, from fertilization, at embryonic day (E)0, until implantation (around E4.5) and the immediate post-implantation stage (E5.5). (b) Stem cell lines can be derived and maintained from each of the three blastocyst populations: embryonic stem cells (ESCs) from the epiblast, trophectoderm stem cells (TSCs) from the trophectoderm and extraembryonic endoderm (XEN) stem cells from the primitive endoderm. ESCs and XEN cells can be reintroduced into the embryo to generate chimeras. (c) Cleavage-stage embryos (from 2- to 8-cell stage) can be disaggregated into constituent cells and these can be re-aggregated with their sister cells or with cells from another embryo to generate viable embryos. (d) Blastocysts can implant into the uterus to develop into a fetus, but also initiate implantation into certain synthetic matrices. (e) Embryo-derived stem cells can be combined using different protocols to generate blastocyst-like or egg cylinder-like structures reminiscent of embryos and capable of attaching to the uterine wall. (f) Whole cleavage-stage embryos can be aggregated to give rise to double or triple blastocysts, which have two or three times as many cells as an intact blastocyst of the same age, but the same anatomy. (g) Cleavage-stage embryos can be split into clusters of cells and give rise to half- (or even quarter-)blastocysts, which display a cyst structure and half (or quarter) the number of cells of an age-matched intact blastocyst.
The blastocyst comprises three tissues: the embryonic epiblast and two mainly extraembryonic epithelia, the trophectoderm (TE) and the primitive endoderm (PrE, or hypoblast in non-rodents). The epiblast is the pluripotent tissue that gives rise to most of the soma and the germ cells [7,8]. The TE develops into the embryonic part of the placenta, while the PrE gives rise to the parietal and visceral endoderm (VE) of the yolk sac, as well as to parts of the embryonic gut endoderm [8–12] (reviewed in [13]). During implantation, the PrE develops into the VE, evolving from a cuboidal to an eventually squamous epithelium that is essential both for the growth of the epiblast—as it undergoes rapid proliferation prior to gastrulation—and for its patterning—to establish the future antero-posterior axis of the embryo proper (reviewed in [14–16]). In the mouse, these three cell types can give rise to stable stem cell lines when embryos are cultured under the appropriate in vitro conditions: embryonic stem cells (ESCs) arise from and represent the epiblast [17–20], trophoblast stem (TS) cells the TE [21] and extraembryonic endoderm (XEN) stem cells the PrE [22] (figure 1b). For detailed reviews on these stem cells, see [23,24].
3. The mammalian preimplantation embryo is a self-organizing system
The preimplantation embryo displays the three features associated with biological self-organizing systems: (i) self-assembly, (ii) self-patterning and (iii) self-morphogenesis [25]. A mammalian zygote can autonomously develop into a blastocyst on a culture dish, in minimal medium, without the need for exogenous cytokines. Self-patterning and self-morphogenesis are evident in this process. Moreover, dissociated blastomeres readily aggregate when placed into close proximity and give rise to a normal blastocyst [26–32] (figure 1c). These self-organizing abilities are not restricted to the preimplantation period: blastocysts can attach to a synthetic matrix and grow into correctly patterned egg cylinders without maternal input [33–36] (figure 1d), while aggregates of embryo-derived stem cells can assemble into structures that resemble blastocysts and early post-implantation embryos [37–41] (figure 1e).
The integration of patterning, morphogenesis and tissue growth in these embryos is perhaps best manifest in two notable phenomena: the formation of chimeras and the ability of embryos to scale. Just as dissociated blastomeres spontaneously reaggregate and resume development (figure 1c), intact embryos can incorporate foreign cells, to generate chimeras that develop into normal mice [26,27,42–44] (figure 1b). In extreme cases, the grafted cells can even displace host cells to produce the entire adult body [45–48]. Chimeras can be also produced by aggregating two or more embryos [27,49–52] (figure 1f), which nonetheless give rise to normal-sized mice [27,49,53–55]. On the other hand, cleavage-stage embryos can be separated into halves, which develop into smaller blastocysts that can give rise to viable adults of normal size [56–58] (figure 1g). In both double and half embryos, the number of cells of each type in the blastocyst scales up or down, respectively, so as to maintain consistent lineage proportions [59,60]. These findings highlight that pattering and morphogenesis in the mouse embryo are very robust and independent of embryo size.
A multitude of strategies exist to scale patterns with tissue size across developmental systems [1,61–64]. Often, these patterns are dictated by morphogen gradients that can adapt to changes in tissue size. As we will discuss in the next sections, patterning and morphogenesis in the mouse blastocyst occur autonomously, without instructive signals from neighbouring tissues or organizers. Instead, location within the embryo, mechanical cues and short-range signalling are integrated to establish cell fates. A fundamental emerging property of this system is robustness, which acts to dampen the effect of biological noise in such a small cell population.
4. Position precedes trophectoderm or inner cell mass identity
The first cell fate decision that cells make during mouse development is whether to become TE or inner cell mass (ICM) (figure 1a). The TE is an epithelium that mediates attachment to and invasion of the uterine wall and that eventually contributes to the placenta, the mother–embryo exchange interface [9,65]. Its cells are bound by extensive intercellular junctions [66–69], display apicobasal polarity [70–72] and secrete a basement membrane [73,74]. Conversely, apolar ICM cells remain bound only by adherens and GAP junctions [66] and present a mesenchymal character.
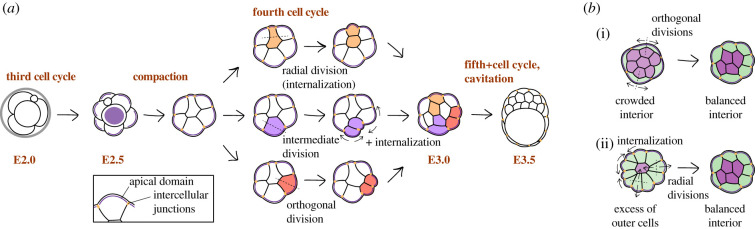
This epithelialization process begins at the 8-cell stage, as the embryo undergoes a process of compaction, whereby blastomeres become tightly attached and sequentially assemble adherens and tight junctions [66–69] (figure 2a). Intercellular junctions consequently isolate the distinct apical domain of 8-cell stage blastomeres from basolateral membrane domains [70,71,75–78]. The establishment of apical polarity is in turn necessary for the development of tight junctions [71,79,80]. Subsequent cell divisions, during the fourth and fifth cell cycle, generate internal cells (figure 2a). Inner cells are generated through the asymmetric division of polarized blastomeres, cell internalization, as well as through the symmetric division of existing inner cells [81–83]. The generation of inner and outer cells represents the first symmetry breaking event in the embryo—the differential position of these populations results in exposure to different stimuli, which in turn results in the acquisition of different fates.
Figure 2.
Allocation of inner and outer cells in morula-stage embryos. (a) After compaction, mitoses during the fourth and fifth cell cycles can take place along angles that range from perfectly radial to orthogonal to the embryo radius (tangential to its surface). Different angles of division lead to differential inheritance of the apical domain. Ultimately, the cell position is affected by mechanical properties and interaction with neighbouring cells. (b) Excess of inner (i) or outer (ii) cells can be compensated for through changes in the orientation of cell divisions and apical constriction.
Despite this differential position, all cells in the morula (approx. E2.5–E3.25) co-express TE and ICM-specific transcription factors, such as CDX2 (TE), OCT4 and NANOG (ICM) [84], and remain totipotent [29,30,32,85]. Although not essential for preimplantation development, the activity of the Notch pathway contributes to this early expression of CDX2 [86,87]. This symmetry in gene expression is broken with the segregation of inner and outer cells and the differential activation of the intracellular Hippo signalling pathway [88,89]. Hippo and Notch synergistically restrict gene expression and trigger cell fate specification among inner and outer cells [86,87,89]. Apical polarization of outer cells is fundamental for lineage specification, as it prevents activation of the Hippo signalling cascade through the restriction of angiomotin (AMOT) to the apical domain [72,80,90]. Consequently, the transcriptional coactivator YAP binds TEAD4 to maintain expression of the transcription factor CDX2, which activates the TE genetic programme [72,89–93], and to inhibit expression of the ICM transcription factor SOX2 [94–96]. In addition, Notch activity promotes an outer position and TE identity via upregulation of CDX2 [86,87]. Conversely, in apolar (inner) cells, AMOT and NF2 activate the LATS1/2 kinase, which phosphorylates YAP, triggering its degradation and thus promoting SOX2 and inhibiting CDX2 expression [89,94–97]. Detailed reviews of these molecular interactions can be found in [98–100].
5. Finding a balance through geometry
Given the small number of cells that compose the embryo at these stages (8–32 cells), a small difference in cell numbers can have a large effect in the relative size of the TE and ICM, and so, control over cell-type proportions is necessary. This is a fundamental decision, and errors in cell allocation can result in developmental failure. The TE : ICM ratio has been found to be highly consistent, with most embryos having ratios between 17 : 15 and 22 : 10 (TE : ICM) cells at the 32-cell stage [83,101]. The numbers of inner and outer cells generated during the fourth and fifth cell cycles were found to be inversely correlated, so as to ultimately yield a balanced ratio [83]. Accordingly, grafting supernumerary inner cells onto an 8-cell embryo (before the fourth cell cycle) shifted the relative contribution of the host cells toward the TE to accommodate for this perturbation and maintain the overall TE : ICM ratio [101].
The Hippo pathway provides an elegant link between cell position, polarity and cell fate, but it does not explain this balanced generation of cell fates. How is this ratio consistently achieved? Recent studies suggest geometry and energy minimization as the main inputs. During compaction, both embryo surface energy and area are reduced, while cell positions become constrained through the stabilization of adherens junctions between blastomeres, which are primarily composed of E-cadherin at this stage [66,67]. Accordingly, embryos lacking E-cadherin (CDH1) form a disorganized mass of loosely attached cells and fail to generate a blastocyst [102,103]. Despite the need for E-cadherin for compaction and for maintaining embryo integrity, it has been shown that, in contrast to previous suggestions [104], it is not the main regulator of embryo compaction. Instead, compaction is driven by contractile pulses generated by the actomyosin cortex [105]. The intercellular junctions between blastomeres anchor the cytoskeleton and restrict these contractile forces to the apical surfaces [105], thus causing apical constriction, in a process reminiscent of those taking place in the epidermis of the Drosophila embryo and during Caenorhabditis elegans gastrulation [106,107].
The presence of intercellular junctions and this overall increase in contractility reduce the surface area of the morula and impose a geometric constraint on a cell's position and cell division. A division along the radial axis of the embryo can readily generate an inner cell and an outer cell, while a division orthogonal to it generates two outer cells (figure 2a). However, the angle of division can vary between these two extremes, thus yielding a range of intermediate positions [108–111]. The final position of most cells is thus determined not by the angle of division itself, but by mechanical interactions with their neighbours so as to reduce the overall surface energy (figure 2a) [108–110]. Let us consider two extreme examples: (i) 12 inner cells versus 4 outer cells and (ii) 3 inner cells versus 13 outer cells (figure 2b). A crowded inner population figure 2b(i) will push and flatten outer cells, thus stretching the apical domain and facilitating divisions parallel to the surface, which will counter the excess of inner cells (figure 2b). Broad apical domains are associated with lower contractility, an outside position and ultimately a TE fate [71,108,112,113], both in mouse and in human embryos [113]. On the other hand, an excess of outer cells will create competition for an outside position (figure 2b(ii)) to try and minimize surface energy. This situation facilitates both a radial orientation of the division spindle and the internalization of cells with higher contractility (and a narrow apical domain) [112]. Throughout the fourth and fifth cell cycles, these local interactions will orient division angles and internalize (or fail to) cells in intermediate positions, effectively sorting them out to produce a stable configuration.
Therefore, geometrical constraints and energy minimization can account for the balanced physical allocation of cells within the morula/early blastocyst [83], which in turn determines cell fate [88]. Low contractility facilitates an outer position and a broad apical domain, which inhibits the Hippo pathway and promotes TE identity. Accordingly, mechanically forcing more cells into an outer position expands their apical surface and leads to an increase in YAP nuclear localization and CDX2 expression [113]. Conversely, cells with a narrow or no apical domain adopt an internal position and eventually an ICM fate [72,90,94–96,111–115]. The stabilization of a TE genetic programme causes commitment of most outer cells to a TE fate by the 32-cell stage [29,30,32,84]. By contrast, ICM cells have been shown to remain plastic for a wider developmental window [30,32,116–118]. This may, in turn, reflect an unstable state, one that rapidly transitions towards epiblast or PrE [32].
6. PrE or epiblast identity precedes position
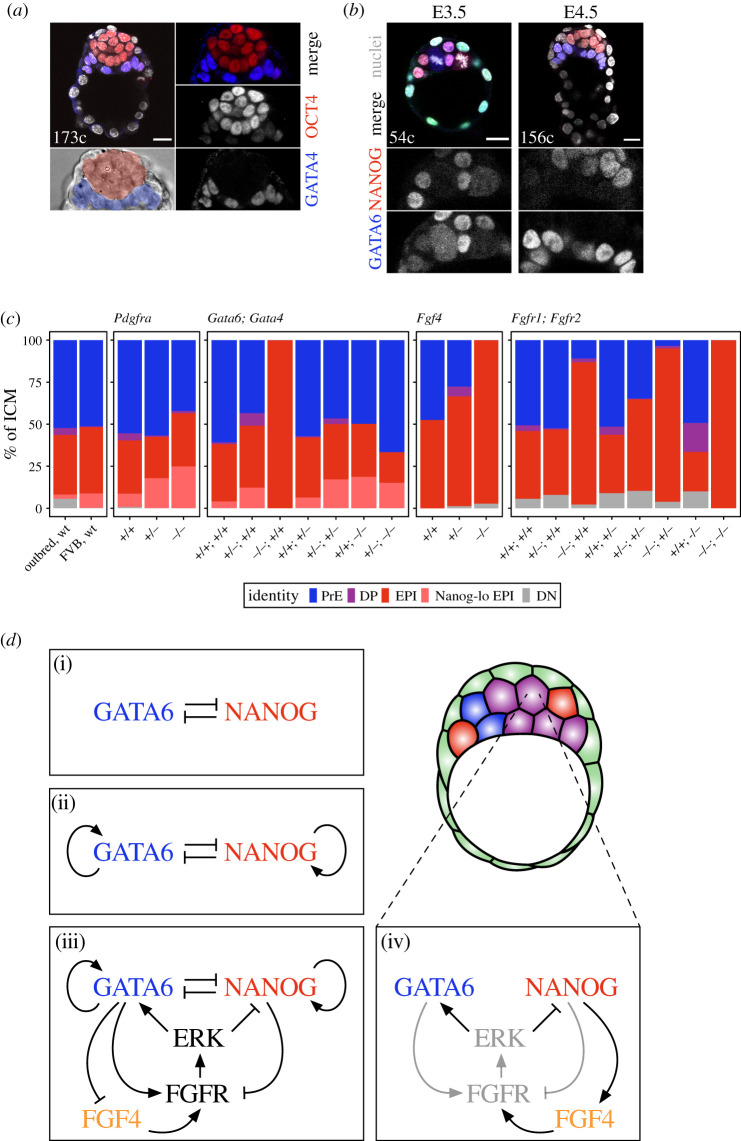
ICM cells are generated during the fourth and fifth cell cycles of mouse development (as the embryo goes from comprising 8 to comprising 32 cells). During the following two cycles (32 to 128 cells), ICM cells further segregate into two populations: the epiblast and the PrE. Cells in the ICM of the early blastocyst (approx. 32 cells) show no overt differences, whereas at the time of implantation (embryonic day (E)4.5, approx. 128–256 cells), PrE cells form a morphologically distinct epithelial layer on the surface of the ICM, covering the epiblast (figure 3a). Since there is no influx of outer cells to the ICM after the fifth cell cycle [30,32], ICM cells in the early blastocyst give rise to both cell types. All ICM cells may therefore be equivalent in potency and capable of becoming either epiblast or PrE. This bipotency is supported by the co-expression of markers specific to both lineages, such as NANOG, SOX2, OCT4, KLF5 and GATA6 [12,84,120–128] (figure 3b). The transcription factors such as NANOG, SOX2, OCT4 and KLF5 are essential for epiblast specification [128–135]. Expression of NANOG, SOX2 and OCT4 is restricted to the epiblast in the peri-implantation blastocyst (E4.5) [121,136–138], coinciding with epiblast fate restriction [8,137]. Conversely, the transcription factor GATA6 is required for PrE specification [138,139] and identifies PrE cells at E4.5, alongside KLF5 and later PrE markers, such as PDGFRα, SOX17, GATA4 and SOX7 [12,120,121,125,128,140–143] (figure 3a,b).
Figure 3.
Ways to achieve a balanced lineage composition in the ICM. (a) Immunofluorescence images of an E4.5 blastocyst with sorted epiblast (marked by OCT4, red) and PrE (marked by GATA4, blue). At this stage, the PrE forms a coherent epithelial layer between the epiblast and the blastocyst cavity. (b) Immunofluorescence images of an E3.5 and an E4.5 embryo stained for NANOG (epiblast marker, red) and GATA6 (PrE marker, blue). At E3.5, all or most ICM cells co-express both NANOG and GATA6. At E4.5, the two markers label the sorted populations, as shown in (a). (c) Stacked bar plots showing ICM composition in late blastocysts (greater than 100 cells) of the genotypes indicated. Composition for embryos from an outbred (CD1) and an inbred (FVB) line is shown in the first box. (d) Different theoretical models to explain lineage specification within the ICM: direct mutual inhibition between GATA6 and NANOG (i), direct mutual inhibition with auto-activation (ii), direct mutual inhibition with auto-activation and modulation by FGF–MAPK signalling (iii) and indirect mutual inhibition via FGF–MAPK signalling (iv). Black arrows in (iv) represent interactions explicitly modelled in [119], whereas grey arrows represent interactions that are implicit in that model. Note how the net effect of these interactions is equivalent to the model shown in (ii) (e.g. GATA6 inhibits NANOG via the activation of FGFR, not directly). See the text for details. Colour coding is indicated. EPI, epiblast; Nanog-lo EPI, NANOG-low epiblast; PrE, primitive endoderm; DP, double-positive (for NANOG and GATA6); DN, double negative (for NANOG and GATA6). Scale bar, 20 µm.
Given the final arrangement of epiblast and PrE cells at the time of implantation (E4.5), it was originally proposed that, as for the TE, the position played a key role in symmetry breaking within the ICM [144]. However, it was later discovered that these two cell types emerge scattered throughout the ICM, with a salt-and-pepper distribution [145,146]. These cells express either NANOG or GATA6, indicating that they have adopted epiblast or PrE identity, respectively [146]. Further analyses have shown that cells specified as either epiblast or PrE coexist with uncommitted ICM cells, which still co-express NANOG and GATA6 [60,121] (figure 3b). These NANOG, GATA6 double-positive (DP) progenitor cells adopt epiblast or PrE identity asynchronously as the blastocyst develops [60,137,147]. This lack of spatial pattern is progressively resolved, concomitantly with cell fate acquisition, through the segregation of epiblast and PrE cells into distinct layers [121]. ICM cells are motile and undergo extensive neighbour exchange over the course of blastocyst development [148]. As nascent PrE cells reach the ICM surface, they stabilize their position and their fate, resulting in the gradual coalescence of PrE cells and the eventual separation of both compartments [121]. PrE cells initiate an epithelial programme when they become specified and develop apicobasal polarity as they come into contact with the blastocyst cavity [73,149,150]. This ability to polarize is critical for PrE survival and positioning [150] and consequently for correct cell sorting and epithelialization in the embryo.
If not differential position, what triggers symmetry breaking within the ICM? And why do epiblast and PrE cells emerge with no evident spatial pattern? In recent years, it has become clear that the answer to these questions is intercellular signalling. Beside the cell-autonomous need for GATA6 [138,139], PrE specification in the mouse embryo requires activation of the receptor tyrosine kinase (RTK)–mitogen-activated protein kinase (MAPK) pathway by a non-cell-autonomous signal: fibroblast growth factor (FGF)4 [125,136,146,151–157]. Fgf4 is expressed primarily in emerging epiblast cells, as well as in unspecified ICM cells [12,120,125,133,158] downstream of NANOG, SOX2, OCT4 and KLF5 [94,128,132–135,159,160]. Activation of the RTK–MAPK pathway by FGF4 is required to maintain GATA6 expression in ICM cells, and without it the PrE fails to form [136,146,152,153,157]. However, while PrE specification requires the presence of FGF4 produced by epiblast cells, the maturation of the epiblast lineage also requires FGF4 and the presence of PrE cells [125]. Although uncommitted ICM cells default to a functional, naive epiblast state when the RTK–MAPK pathway is inactive [136], FGF4 is necessary directly to downregulate NANOG levels as cells transition from a naive to a primed epiblast state [60,152,154], and indirectly, since extracellular matrix components produced by PrE cells are required for epiblast maturation (and potentially for the commitment of cells to an epiblast fate) [20,141]. Therefore, the formation of these two cell types is intimately linked via FGF4.
7. Finding a balance through communication
Activation of the RTK–MAPK pathway by FGF4 is necessary for the specification of both PrE and epiblast in the mouse and in the rat [161] blastocyst, although this does not seem to be the case in all mammals [162,163]. Despite the evidence available for the mouse, one may ask why is this pathway necessary in rodents? The answer may lie in the need for a robust outcome in a small population of cells. The ICM of the mouse blastocyst has a very conserved composition [60]. Genotypes in which the ICM contains 40–60% of either lineage are viable past implantation and into fertile adults (figure 3c) [60,138,143,152,154,164]. However, a lack of PrE invariably results in peri-implantation lethality [138,139,146,152–155,165,166], presumably due to the underdevelopment of the epiblast. It is therefore likely that comparable amounts of epiblast and PrE cells are necessary for successful developmental progression and that this outcome needs to be precisely controlled—although the underlying reasons are unknown and could be manifold: too few PrE cells could impose a mechanical constraint on epiblast growth or fail to synthesize sufficient extracellular membrane components, whereas an epiblast that is too small may not be able to reach the appropriate size to acquire an anterior–posterior pattern, consequently initiate gastrulation and eventually produce a viable fetus [167].
To test whether there is control over the ratio of epiblast to PrE, we experimentally altered ICM composition by either (i) introducing ESCs into embryos to increase the number of epiblast cells or (ii) eliminating epiblast or PrE cells using laser ablation [119]. Both perturbations triggered a compensatory differentiation pattern in uncommitted ICM progenitors to restore the normal lineage composition: increased PrE differentiation when the epiblast was experimentally expanded and differentiation towards the reduced lineage in ablation experiments [119]. Predictably, the ability of the system to fully recover from these perturbations depended on both the available number of uncommitted ICM cells and the magnitude of the perturbation.
But how might this population-level control of cell-type proportions be achieved? Gene regulatory networks that involve only cell-autonomous interactions between transcription factors generate two stable fates (figure 3d(i,ii)) [168,169] and can yield reproducible cell-type proportions irrespective of population size. Such a network, centred around direct mutual inhibition between NANOG and GATA6, has been proposed to underpin multilineage priming in ESCs [170–172], although no evidence currently exists for this direct interaction taking place in vivo in the embryo. More importantly, such a cell-autonomous system cannot account for the regulatory behaviour observed in the ICM of the mouse embryo [119]. This behaviour and the need for non-cell-autonomous activation of the RTK–MAPK cascade in this process suggest that the role of FGF4 may be to ensure robustness in this cell fate decision.
Several models of cell fate specification in the ICM have combined (i) direct mutual inhibition between NANOG and GATA6, (ii) auto-activation of each transcription factor and (iii) feedback through the FGF4 signalling (figure 3d(iii)) [139,173–175]. These models (reviewed in [176,177]) elegantly recapitulate many experimental observations and have certain predictive power. However, their complexity precludes dissection of the contribution of each regulatory module to the behaviour of the system or testing for sufficiency of any of its individual elements. To better understand the regulative nature of this system, we recently built a model in which the mutual repression between NANOG and GATA6 is mediated indirectly by a growth factor, instead of directly through mutual inhibition (figure 3d(iv)) [119]. Like previous models, our growth factor-mediated lineage specification model recapitulates the progressive emergence of lineages observed in embryos, as well as the ability of lineage size (as defined by the number of cells) to scale with absolute embryo size [119]. However, in this simpler model, the contribution and sufficiency of the single variable (growth factor feedback) is evident (figure 3d(iv)). Moreover, this minimal model predicts that the uncommitted progenitor state is unstable and it rapidly evolves towards one of two stable states—epiblast or PrE—in line with the available experimental evidence (i.e. a stable, DP-equivalent stem cell state has not yet been readily captured in vitro).
These results reveal an intrinsic mechanism for active control of cell-type composition in the ICM mediated non-cell autonomously by FGF4. This control relies on the presence of uncommitted ICM progenitors [119], the cells whose fate depends on activity of the FGF–MAPK pathway [60,136]. Accordingly, when FGF4 is provided to Fgf4-null ICM cells by grafting FGF4-producing ESCs onto mutant embryos, PrE fate is induced among mutant uncommitted host cells in a manner that recapitulates cell fate specification in wild-type embryos [119].
8. An autonomous, scalable way to robustly generate cell-type diversity
The proposed growth factor-mediated switch [119] makes two important predictions: (i) the system is robust to changes in the relative proportions of each lineage, as demonstrated experimentally [119], and (ii) the lateral inhibition of fates leads to the scattered emergence of cell types first described by Chazaud, Rossant and co-workers [145,146]. Multiple explanations have been put forward for the salt-and-pepper distribution of epiblast and PrE cells. These include endogenous differences in RTK–MAPK activity [137,151–153,158], lineage history [142,178] and reaction–diffusion mechanisms [174]. In all of these scenarios, however, FGF4 maintains Gata6 expression [152,153] and is primarily produced by cells with high NANOG levels [12,125,158]. While acknowledging the existence of two receptors for FGF4 (FGFR1 and FGFR2) and multiple feedback regulatory interactions taking place between receptor activation and transcription of their target genes [12,125,154–157], if the pathway is distilled to its net output, as proposed (figure 3d(iv)), it results in a lateral inhibition mechanism, in which cells producing FGF4 generate a local high concentration of ligand that induces PrE fate among their neighbours (and consequently inhibits epiblast fate). In this scenario, FGF4 is assumed to be bound to the extracellular matrix, rather than to diffuse throughout the ICM [179,180]. This model does not explain how (and if) FGF4 producers are less sensitive to the ligand than their neighbours, so as to ensure alternative fates. Perhaps, a higher number of receptor molecules, or FGFR1–R2 heterodimers, are necessary to fully transduce the signal; perhaps, FGFR1 becomes quickly desensitized, or putative epiblast cells become locked in their fate early on [137]. At this point, we can only speculate. Nevertheless, this behaviour is compatible with the existing experimental evidence and agnostic to both the origin of the differences in NANOG/FGF4 levels and the nature of the signalling pathway.
Growth factor-mediated control of lineage size is a common strategy employed in developmental systems. Neuron-produced GDF11, a TGF-β superfamily ligand, inhibits neuronal differentiation in the mouse olfactory epithelium [181], while epidermal growth factor receptor (EGFR) stimulation by Spitz is necessary to coordinate the size of anterior and posterior compartments of Drosophila epidermal segments [182]. However, in the case of the blastocyst, the lateral inhibition of fates is remarkably similar to that observed in Dictyostelium discoideum [183]. In Dictyostelium, a facultative social amoeba, aggregation of individual cells upon starvation is followed by the specification of two broad fates—prespore and prestalk—through a differential response to the factor DIF-1, secreted by prespore cells, which activates a GATA-family transcription factor [184–186]. These two cell types emerge interspersed and eventually sort out and contribute to different regions of the fruiting body (the parallels between spore versus stalk and embryonic versus extraembryonic fates are immediate) [183,187]. In the mouse blastocyst, the differential response to FGF4 spontaneously generates a salt-and-pepper distribution of cell identities that are resolved (and stabilized) through cell sorting [121]. A system of this type inevitably yields a consistent ratio of cell types and is independent of both population size (scaling behaviour) and an external source of growth factor. Therefore, it is suited for patterning autonomous, self-organizing systems, such as the preimplantation embryo or the social amoeba.
9. What triggers the decision?
Despite our current understanding of the transcription factors and signals mediating cell fate specification in the ICM of the mouse blastocyst, perhaps the biggest open question concerns how is this process initiated? Many (but not all) ICM cells in the early blastocyst express Fgf4 [12,125,154]. These include emerging epiblast cells, but certainly also many (if not all) uncommitted progenitors. All these cells express high levels of NANOG, which has been shown to, directly or indirectly, promote Fgf4 expression [133,139,173]. Different explanations have been proposed for the differential production of FGF4. It could be the result of noisy gene expression: bursts in NANOG levels or some other coactivator of Fgf4 could result in stochastic foci of high ligand concentration that would trigger symmetry breaking. It has also been postulated that certain ICM cells are biased towards epiblast or PrE as a result of their lineage history and/or differential inheritance of FGFR2 [142,167]. FGFR2 has since been shown to be dispensable for PrE specification [154,155], and correlations always call for cautious interpretation. Nevertheless, differences due to metabolic status or a number of other internal checkpoints can arise within a population of otherwise equipotent cells and bias their behaviour. For instance, nutritional status has been shown to affect DIF-1 sensitivity and cell fate choice in Dictyostelium (reviewed in [188]), providing another potential parallel for the mammalian ICM. Moreover, epiblast cells undergo an active selection process through cell competition in the early post-implantation embryo, prior to gastrulation [189,190], which might be the result of metabolic differences [191,192]. Accordingly, the Hippo pathway component YAP and TEAD1 have been proposed to mediate the elimination of unspecified epiblast cells through cell competition in the preimplantation embryo [193]. It is therefore easy to envision how either differential production or sensitivity to FGF4 could arise from intrinsic differences between ICM cells. These differences need not determine ultimate cell fate but can destabilize the progenitor state to initiate lineage divergence.
Another key element to consider is the cell cycle. Cell divisions during preimplantation development become progressively asynchronous as embryos grow, providing yet another source of heterogeneity among blastomeres. For instance, all ICM cells could conceivably have the same potential to produce FGF4 but do so only during G1. In this scenario, faster-cycling cells will always initiate symmetry breaking and might end up in the epiblast as a result, in agreement with previous observations [142]. Another possibility is that the sensitivity of ICM cells to differentiation cues changes in different phases of the cell cycle, as shown in ESCs [194]. This could be due to varying expression levels of signal transducers along the cell cycle. In this case, asynchrony in the cell cycle phase would create a buffer against sudden changes in FGF4 levels, by ensuring only a subset of cells are competent to respond to the growth factor signal at any given time [119]. This has been shown to be the case in Dictyostelium [195], where asynchrony ensures correct cell-type proportioning in the multicellular slug. Therefore, cell cycle asynchrony in the mouse blastocyst may underlie both the asynchrony in cell fate specification within the ICM and the robustness observed in this process [60], while also providing an explanation for seemingly disparate observations.
10. Conclusion
The preimplantation mouse embryo is a highly tractable system and an example of in vivo self-organization. Classic studies during the second half of the twentieth century established the mouse blastocyst as a truly fascinating biological system. Over the past two decades, the combination of those classic experimental techniques with mathematical modelling and novel genomic and microscopy tools has shed new light on how signalling, mechanical forces and positional information drive patterning and morphogenesis in this system. Perhaps not surprisingly, we have learned that it is the integration of these multiple cues that ensures a precise and reproducible developmental outcome. The application of these tools to study the preimplantation stages of other mammalian species has begun to reveal differences between the early development of rodents and that of larger mammals, including ourselves. The continued use of novel technologies on a diversity of biological models will paint a clearer picture of the strategies used by mammalian embryos to overcome challenges and ensure developmental success. These principles not only will improve our understanding of human development but will help us engineer better self-organizing developmental systems and models.
Acknowledgements
We thank Dr Berenika Plusa for insightful comments on the manuscript.
Data accessibility
Data presented in figure 3 are publicly available with the data from our article ‘Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development’ [119].
Authors' contributions
N.S. conceived the manuscript. N.S. and A.-K.H. wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
Work in the Hadjantonakis Lab is supported by the National Institutes of Health (grant nos R01-HD094868, R01-DK084391 and P30-CA008748) and by the MSKCC Functional Genomics Initiative.
References
- 1.Kicheva A, Bollenbach T, Ribeiro A, Valle HP, Lovell-Badge R, Episkopou V, Briscoe J. 2014. Coordination of progenitor specification and growth in mouse and chick spinal cord. Science 345, 1254927 ( 10.1126/science.1254927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C-C, et al. 2015. Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 161, 277–290. ( 10.1016/j.cell.2015.02.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roselló-Díez A, Madisen L, Bastide S, Zeng H, Joyner AL. 2018. Cell-nonautonomous local and systemic responses to cell arrest enable long-bone catch-up growth in developing mice. PLoS Biol. 16, e2005086 ( 10.1371/journal.pbio.2005086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seah MKY, Messerschmidt DM. 2018. From germline to soma: epigenetic dynamics in the mouse preimplantation embryo. Curr. Top. Dev. Biol. 128, 203–235. ( 10.1016/bs.ctdb.2017.10.011) [DOI] [PubMed] [Google Scholar]
- 5.Brockdorff N, Turner BM. 2015. Dosage compensation in mammals. Cold Spring Harb. Perspect. Biol. 7, a019406 ( 10.1101/cshperspect.a019406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galupa R, Heard E. 2018. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. ( 10.1146/annurev-genet-120116-024611) [DOI] [PubMed] [Google Scholar]
- 7.Gardner RL, Papaioannou VE. 1975. Differentiation in the trophectoderm and inner cell mass. In The early development of mammals: 2nd Symp. Br. Soc. Dev. Biol., 9–12 September 1974, Norwich, UK (eds Balls M, Wild AE), pp. 107–132. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Gardner RL, Rossant J. 1979. Investigation of the fate of 4–5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 52, 141–152. [PubMed] [Google Scholar]
- 9.Gardner RL. 1983. Origin and differentiation of extraembryonic tissues in the mouse. Int. Rev. Exp. Pathol. 24, 63–133. [PubMed] [Google Scholar]
- 10.Kwon GS, Viotti M, Hadjantonakis A-K. 2008. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509–520. ( 10.1016/j.devcel.2008.07.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viotti M, Nowotschin S, Hadjantonakis A-K. 2014. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 16, 1146–1156. ( 10.1038/ncb3070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowotschin S, et al. 2019. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367. ( 10.1038/s41586-019-1127-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowotschin S, Hadjantonakis A-K, Campbell K. 2019. The endoderm: a divergent cell lineage with many commonalities. Development 146, dev150920 ( 10.1242/dev.150920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephenson RO, Rossant J, Tam PPL. 2012. Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb. Perspect. Biol. 4, a008235 ( 10.1101/cshperspect.a008235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takaoka K, Hamada H. 2012. Cell fate decisions and axis determination in the early mouse embryo. Development 139, 3–14. ( 10.1242/dev.060095) [DOI] [PubMed] [Google Scholar]
- 16.Stower MJ, Srinivas S. 2018. The head's tale: anterior-posterior axis formation in the mouse embryo. Curr. Top. Dev. Biol. 128, 365–390. ( 10.1016/bs.ctdb.2017.11.003) [DOI] [PubMed] [Google Scholar]
- 17.Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638. ( 10.1073/pnas.78.12.7634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. ( 10.1038/292154a0) [DOI] [PubMed] [Google Scholar]
- 19.Brook FA, Gardner RL. 1997. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl Acad. Sci. USA 94, 5709–5712. ( 10.1073/pnas.94.11.5709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boroviak T, Loos R, Bertone P, Smith A, Nichols J. 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516–528. ( 10.1038/ncb2965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka S, Kunath T, Hadjantonakis A-K, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075. ( 10.1126/science.282.5396.2072) [DOI] [PubMed] [Google Scholar]
- 22.Kunath T, et al. 2005. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661. ( 10.1242/dev.01715) [DOI] [PubMed] [Google Scholar]
- 23.Garg V, Morgani S, Hadjantonakis A-K. 2016. Capturing identity and fate ex vivo: stem cells from the mouse blastocyst. Curr. Top. Dev. Biol. 120, 361–400. ( 10.1016/bs.ctdb.2016.04.007) [DOI] [PubMed] [Google Scholar]
- 24.Morgani S, Nichols J, Hadjantonakis A-K. 2017. The many faces of pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev. Biol. 17, 7 ( 10.1186/s12861-017-0150-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasai Y. 2013. Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–326. ( 10.1038/nature11859) [DOI] [PubMed] [Google Scholar]
- 26.Kelly SJ. 1977. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J. Exp. Zool. 200, 365–376. ( 10.1002/jez.1402000307) [DOI] [PubMed] [Google Scholar]
- 27.Mintz B. 1964. Formation of genetically mosaic mouse embryos, and early development of ‘lethal (t12/t12)-normal’ mosaics. J. Exp. Zool. 157, 273–292. ( 10.1002/jez.1401570210) [DOI] [PubMed] [Google Scholar]
- 28.Hillman N, Sherman MI, Graham C. 1972. The effect of spatial arrangement on cell determination during mouse development. J. Embryol. Exp. Morphol. 28, 263–278. [PubMed] [Google Scholar]
- 29.Tarkowski AK, Suwińska A, Czołowska R, Ozdzeński W. 2010. Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev. Biol. 348, 190–198. ( 10.1016/j.ydbio.2010.09.022) [DOI] [PubMed] [Google Scholar]
- 30.Suwińska A, Czołowska R, Ozdzeński W, Tarkowski AK. 2008. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev. Biol. 322, 133–144. ( 10.1016/j.ydbio.2008.07.019) [DOI] [PubMed] [Google Scholar]
- 31.Krupa M, Mazur E, Szczepanska K, Filimonow K, Maleszewski M, Suwińska A. 2014. Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8→16- and 16→32-cells). Dev. Biol. 385, 136–148. ( 10.1016/j.ydbio.2013.09.008) [DOI] [PubMed] [Google Scholar]
- 32.Posfai E, Petropoulos S, de Barros FR, Schell JP, Jurisica I, Sandberg R, Lanner F, Rossant J.. 2017. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. eLife 6, e22906 ( 10.7554/eLife.22906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris SA, Grewal S, Barrios F, Patankar SN, Strauss B, Buttery L, Alexander M, Shakesheff KM, Zernicka-Goetz M. 2012. Dynamics of anterior–posterior axis formation in the developing mouse embryo. Nat. Commun. 3, 673–710. ( 10.1038/ncomms1671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedzhov I, Zernicka-Goetz M. 2014. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156, 1032–1044. ( 10.1016/j.cell.2014.01.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. 2016. Self-organization of the in vitro attached human embryo. Nature 533, 251–254. ( 10.1038/nature17948) [DOI] [PubMed] [Google Scholar]
- 36.Shahbazi MN, et al. 2016. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18, 700–708. ( 10.1038/ncb3347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M. 2017. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810 ( 10.1126/science.aal1810) [DOI] [PubMed] [Google Scholar]
- 38.Sozen B., et al. 2018. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20, 979–989. ( 10.1038/s41556-018-0147-7) [DOI] [PubMed] [Google Scholar]
- 39.Rivron NC, et al. 2018. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. ( 10.1038/s41586-018-0051-0) [DOI] [PubMed] [Google Scholar]
- 40.Li R, et al. 2019. Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell 179, 687–702. ( 10.1016/j.cell.2019.09.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahbazi MN, Siggia ED, Zernicka-Goetz M. 2019. Self-organization of stem cells into embryos: a window on early mammalian development. Science 364, 948–951. ( 10.1126/science.aax0164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mintz B, Illmensee K. 1975. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc. Natl Acad. Sci. USA 72, 3585–3589. ( 10.1073/pnas.72.9.3585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardner RL. 1968. Mouse chimeras obtained by the injection of cells into the blastocyst. Nature 220, 596–597. ( 10.1038/220596a0) [DOI] [PubMed] [Google Scholar]
- 44.Bradley A, Evans M, Kaufman MH, Robertson E. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256. ( 10.1038/309255a0) [DOI] [PubMed] [Google Scholar]
- 45.Tokunaga T, Tsunoda Y. 1992. Efficacious production of viable germ-line chimeras between embryonic stem (ES) cells and 8-cell stage embryos. Dev. Growth Differ. 34, 561–566. ( 10.1111/j.1440-169X.1992.00561.x) [DOI] [PubMed] [Google Scholar]
- 46.Nagy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, Markkula M, Rossant J. 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110, 815–821. [DOI] [PubMed] [Google Scholar]
- 47.Lallemand Y, Brûlet P. 1990. An in situ assessment of the routes and extents of colonisation of the mouse embryo by embryonic stem cells and their descendants. Development 110, 1241–1248. [DOI] [PubMed] [Google Scholar]
- 48.Poueymirou WT, et al. 2006. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat. Biotechnol. 25, 91–99. ( 10.1038/nbt1263) [DOI] [PubMed] [Google Scholar]
- 49.Tarkowski AK. 1961. Mouse chimaeras developed from fused eggs. Nature 190, 857–860. ( 10.1038/190857a0) [DOI] [PubMed] [Google Scholar]
- 50.Markert CL, Petters RM. 1978. Manufactured hexaparental mice show that adults are derived from three embyronic cells. Science 202, 56–58. ( 10.1126/science.694518) [DOI] [PubMed] [Google Scholar]
- 51.Petters RM, Markert CL. 1980. Production and reproductive performance of hexaparental and octaparental mice. J. Hered. 71, 70–74. ( 10.1093/oxfordjournals.jhered.a109334) [DOI] [PubMed] [Google Scholar]
- 52.Bowman P, McLaren A. 1970. Viability and growth of mouse embryos after in vitro culture and fusion. J. Embryol. Exp. Morphol. 23, 693–704. [PubMed] [Google Scholar]
- 53.Mintz B. 1967. Gene control of mammalian pigmentary differentiation. I. Clonal origin of melanocytes. Proc. Natl Acad. Sci. USA 58, 344–351. ( 10.1073/pnas.58.1.344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis NE, Rossant J. 1982. Mechanism of size regulation in mouse embryo aggregates. J. Embryol. Exp. Morphol. 72, 169–181. [PubMed] [Google Scholar]
- 55.Buehr M, McLaren A. 1974. Size regulation in chimaeric mouse embryos. J. Embryol. Exp. Morphol. 31, 229–234. [PubMed] [Google Scholar]
- 56.Tarkowski AK. 1959. Experiments on the development of isolated blastomers of mouse eggs. Nature 184, 1286–1287. ( 10.1038/1841286a0) [DOI] [PubMed] [Google Scholar]
- 57.Tsunoda Y, McLaren A. 1983. Effect of various procedures on the viability of mouse embryos containing half the normal number of blastomeres. J. Reprod. Fertil. 69, 315–322. ( 10.1530/jrf.0.0690315) [DOI] [PubMed] [Google Scholar]
- 58.Papaioannou VE, Mkandawire J, Biggers JD. 1989. Development and phenotypic variability of genetically identical half mouse embryos. Development 106, 817–827. [DOI] [PubMed] [Google Scholar]
- 59.Papaioannou VE, Ebert KM. 1995. Mouse half embryos: viability and allocation of cells in the blastocyst. Dev. Dyn. 203, 393–398. ( 10.1002/aja.1002030402) [DOI] [PubMed] [Google Scholar]
- 60.Saiz N, Williams KM, Seshan VE, Hadjantonakis A-K. 2016. Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst. Nat. Commun. 7, 13463 ( 10.1038/ncomms13463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-Zvi D, Shilo B-Z, Fainsod A, Barkai N. 2008. Scaling of the BMP activation gradient in Xenopus embryos. Nature 453, 1205–1211. ( 10.1038/nature07059) [DOI] [PubMed] [Google Scholar]
- 62.Reversade B, De Robertis EM.. 2005. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell 123, 1147–1160. ( 10.1016/j.cell.2005.08.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uygur A, Young J, Huycke TR, Koska M, Briscoe J, Tabin CJ. 2016. Scaling pattern to variations in size during development of the vertebrate neural tube. Dev. Cell 37, 127–135. ( 10.1016/j.devcel.2016.03.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishimatsu K, Hiscock TW, Collins ZM, Sari DWK, Lischer K, Richmond DL, Bessho Y, Matsui T, Megason SG. 2018. Size-reduced embryos reveal a gradient scaling-based mechanism for zebrafish somite formation. Development 145, dev161257 ( 10.1242/dev.161257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gardner RL, Johnson MH. 1972. An investigation of inner cell mass and trophoblast tissues following their isolation from the mouse blastocyst. J. Embryol. Exp. Morphol. 28, 279–312. [PubMed] [Google Scholar]
- 66.Ducibella T, Albertini DF, Anderson E, Biggers JD. 1975. The preimplantation mammalian embryo: characterization of intercellular junctions and their appearance during development. Dev. Biol. 45, 231–250. ( 10.1016/0012-1606(75)90063-9) [DOI] [PubMed] [Google Scholar]
- 67.Vestweber D, Gossler A, Boller K, Kemler R. 1987. Expression and distribution of cell adhesion molecule uvomorulin in mouse preimplantation embryos. Dev. Biol. 124, 451–456. ( 10.1016/0012-1606(87)90498-2) [DOI] [PubMed] [Google Scholar]
- 68.Fleming TP, Garrod DR, Elsmore AJ. 1991. Desmosome biogenesis in the mouse preimplantation embryo. Development 112, 527–539. [DOI] [PubMed] [Google Scholar]
- 69.Fleming TP, McConnell J, Johnson MH, Stevenson BR. 1989. Development of tight junctions de novo in the mouse early embryo: control of assembly of the tight junction-specific protein, ZO-1. J. Cell Biol. 108, 1407–1418. ( 10.1083/jcb.108.4.1407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Louvet S, Aghion J, Santa-Maria A, Mangeat P, Maro B. 1996. Ezrin becomes restricted to outer cells following asymmetrical division in the preimplantation mouse embryo. Dev. Biol. 177, 568–579. ( 10.1006/dbio.1996.0186) [DOI] [PubMed] [Google Scholar]
- 71.Plusa B, Frankenberg S, Chalmers A, Hadjantonakis A-K, Moore C, Papalopulu N, Papaioannou VE, Glover DM, Zernicka-Goetz M. 2005. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 118, 505–515. ( 10.1242/jcs.01666) [DOI] [PubMed] [Google Scholar]
- 72.Hirate Y, et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23, 8–14. ( 10.1016/j.cub.2013.05.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, Edgar D. 1999. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J. Cell Biol. 144, 151–160. ( 10.1083/jcb.144.1.151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dziadek M, Timpl R. 1985. Expression of nidogen and laminin in basement membranes during mouse embryogenesis and in teratocarcinoma cells. Dev. Biol. 111, 372–382. ( 10.1016/0012-1606(85)90491-9) [DOI] [PubMed] [Google Scholar]
- 75.Ziomek CA, Johnson MH. 1980. Cell surface interaction induces polarization of mouse 8-cell blastomeres at compaction. Cell 21, 935–942. ( 10.1016/0092-8674(80)90457-2) [DOI] [PubMed] [Google Scholar]
- 76.Reeve WJ, Ziomek CA. 1981. Distribution of microvilli on dissociated blastomeres from mouse embryos: evidence for surface polarization at compaction. J. Embryol. Exp. Morphol. 62, 339–350. [PubMed] [Google Scholar]
- 77.Handyside AH. 1980. Distribution of antibody- and lectin-binding sites on dissociated blastomeres from mouse morulae: evidence for polarization at compaction. J. Embryol. Exp. Morphol. 60, 99–116. [PubMed] [Google Scholar]
- 78.Fleming TP, Pickering SJ, Qasim F, Maro B. 1986. The generation of cell surface polarity in mouse 8-cell blastomeres: the role of cortical microfilaments analysed using cytochalasin D. J. Embryol. Exp. Morphol. 95, 169–191. [PubMed] [Google Scholar]
- 79.Eckert JJ, McCallum A, Mears A, Rumsby MG, Cameron IT, Fleming TP. 2004. PKC signalling regulates tight junction membrane assembly in the pre-implantation mouse embryo. Reproduction 127, 653–667. ( 10.1530/rep.1.00150) [DOI] [PubMed] [Google Scholar]
- 80.Alarcon VB. 2010. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347–358. ( 10.1095/biolreprod.110.084400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bałakier H, Pedersen RA. 1982. Allocation of cells to inner cell mass and trophectoderm lineages in preimplantation mouse embryos. Dev. Biol. 90, 352–362. ( 10.1016/0012-1606(82)90384-0) [DOI] [PubMed] [Google Scholar]
- 82.Pedersen RA, Wu K, Bałakier H. 1986. Origin of the inner cell mass in mouse embryos: cell lineage analysis by microinjection. Dev. Biol. 117, 581–595. ( 10.1016/0012-1606(86)90327-1) [DOI] [PubMed] [Google Scholar]
- 83.Fleming TP. 1987. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev. Biol. 119, 520–531. ( 10.1016/0012-1606(87)90055-8) [DOI] [PubMed] [Google Scholar]
- 84.Dietrich JE, Hiiragi T. 2007. Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219–4231. ( 10.1242/dev.003798) [DOI] [PubMed] [Google Scholar]
- 85.Rossant J, Vijh KM. 1980. Ability of outside cells from pre-implantation mouse embryos to form inner cell mass derivatives. Dev. Biol. 76, 475–482. ( 10.1016/0012-1606(80)90395-4) [DOI] [PubMed] [Google Scholar]
- 86.Rayon T, et al. 2014. Notch and Hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell 30, 410–422. ( 10.1016/j.devcel.2014.06.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Menchero S, et al. 2019. Transitions in cell potency during early mouse development are driven by Notch. eLife 8, 2813 ( 10.7554/eLife.42930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tarkowski AK, Wróblewska J. 1967. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155–180. [PubMed] [Google Scholar]
- 89.Nishioka N, et al. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410. ( 10.1016/j.devcel.2009.02.003) [DOI] [PubMed] [Google Scholar]
- 90.Leung CY, Zernicka-Goetz M. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 4, 2251 ( 10.1038/ncomms3251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strumpf D, Mao C, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. ( 10.1242/dev.01801) [DOI] [PubMed] [Google Scholar]
- 92.Ralston A, Rossant J. 2008. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 313, 614–629. ( 10.1016/j.ydbio.2007.10.054) [DOI] [PubMed] [Google Scholar]
- 93.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. 2008. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283. ( 10.1016/j.mod.2007.11.002) [DOI] [PubMed] [Google Scholar]
- 94.Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A. 2014. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet. 10, e1004618 ( 10.1371/journal.pgen.1004618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frum T, Murphy TM, Ralston A. 2018. HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. eLife 7, 347 ( 10.7554/eLife.42298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frum T, Watts JL, Ralston A. 2019. TEAD4, YAP1 and WWTR1 prevent the premature onset of pluripotency prior to the 16-cell stage. Development 146, dev179861 ( 10.1242/dev.179861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cockburn K, Biechele S, Garner J, Rossant J. 2013. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195–1201. ( 10.1016/j.cub.2013.05.044) [DOI] [PubMed] [Google Scholar]
- 98.Frum T, Ralston A. 2015. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 31, 402–410. ( 10.1016/j.tig.2015.04.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sasaki H. 2017. Roles and regulations of Hippo signaling during preimplantation mouse development. Dev. Growth Differ. 59, 12–20. ( 10.1111/dgd.12335) [DOI] [PubMed] [Google Scholar]
- 100.Menchero S, Sainz de Aja J, Manzanares M. 2018. Our first choice: cellular and genetic underpinnings of trophectoderm identity and differentiation in the mammalian embryo. Curr. Top. Dev. Biol. 128, 59–80. ( 10.1016/bs.ctdb.2017.10.009) [DOI] [PubMed] [Google Scholar]
- 101.Humięcka M, Krupa M, Guzewska MM, Maleszewski M, Suwińska A. 2016. ESCs injected into the 8-cell stage mouse embryo modify pattern of cleavage and cell lineage specification. Mech. Dev. 141, 40–50. ( 10.1016/j.mod.2016.06.002) [DOI] [PubMed] [Google Scholar]
- 102.Larue L, Ohsugi M, Hirchenhain J, Kemler R. 1994. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl Acad. Sci. USA 91, 8263–8267. ( 10.1073/pnas.91.17.8263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stephenson RO, Yamanaka Y, Rossant J. 2010. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 137, 3383–3391. ( 10.1242/dev.050195) [DOI] [PubMed] [Google Scholar]
- 104.Fierro-González JC, White MD, Silva JC, Plachta N. 2013. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat. Cell Biol. 15, 1424–1433. ( 10.1038/ncb2875) [DOI] [PubMed] [Google Scholar]
- 105.Maître J-L, Niwayama R, Turlier H, Nédélec F, Hiiragi T. 2015. Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol. 17, 849–855. ( 10.1038/ncb3185) [DOI] [PubMed] [Google Scholar]
- 106.Martin AC, Kaschube M, Wieschaus EF. 2008. Pulsed contractions of an actin–myosin network drive apical constriction. Nature 457, 495–499. ( 10.1038/nature07522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Solon J, Kaya-Çopur A, Colombelli J, Brunner D. 2009. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell 137, 1331–1342. ( 10.1016/j.cell.2009.03.050) [DOI] [PubMed] [Google Scholar]
- 108.McDole K, Xiong Y, Iglesias PA, Zheng Y. 2011. Lineage mapping the pre-implantation mouse embryo by two-photon microscopy, new insights into the segregation of cell fates. Dev. Biol. 355, 239–249. ( 10.1016/j.ydbio.2011.04.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dard N, Louvet-Vallée S, Maro B. 2009. Orientation of mitotic spindles during the 8- to 16-cell stage transition in mouse embryos. PLoS ONE 4, e8171 ( 10.1371/journal.pone.0008171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Watanabe T, Biggins JS, Tannan NB, Srinivas S. 2014. Limited predictive value of blastomere angle of division in trophectoderm and inner cell mass specification. Development 141, 2279–2288. ( 10.1242/dev.103267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, Hiiragi T. 2017. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev. Cell 40, 235–247. ( 10.1016/j.devcel.2017.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maître J.-L., Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F, Hiiragi T. 2016. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 536, 344–348. ( 10.1038/nature18958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Royer C, et al. 2019. Position-sensing established during compaction dictates cell fate in the mammalian embryo. bioRxiv, 856161. (doi:10.1101/856161)
- 114.Hirate Y, Hirahara S, Inoue K-I, Kiyonari H, Niwa H, Sasaki H. 2015. Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev. Growth Differ. 57, 544–556. ( 10.1111/dgd.12235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y. 2014. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 141, 2813–2824. ( 10.1242/dev.107276) [DOI] [PubMed] [Google Scholar]
- 116.Handyside AH. 1978. Time of commitment of inside cells isolated from preimplantation mouse embryos. J. Embryol. Exp. Morphol. 45, 37–53. [PubMed] [Google Scholar]
- 117.Hogan B, Tilly R. 1978. In vitro development of inner cell masses isolated immunosurgically from mouse blastocysts. II. Inner cell masses from 3.5- to 4.0-day p.c. blastocysts. J. Embryol. Exp. Morphol. 45, 107–121. [PubMed] [Google Scholar]
- 118.Rossant J, Lis WT. 1979. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev. Biol. 70, 255–261. ( 10.1016/0012-1606(79)90022-8) [DOI] [PubMed] [Google Scholar]
- 119.Saiz N, Mora-Bitria L, Rahman S, George H, Herder JP, Garcia-Ojalvo J, Hadjantonakis A-K. In press. Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development. eLife. See https://elifesciences.org/articles/56079 . [DOI] [PMC free article] [PubMed]
- 120.Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 34, e42 ( 10.1093/nar/gkl050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis A-K. 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081–3091. ( 10.1242/dev.021519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Palmieri SL, Peter W, Hess H, Schöler HR. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166, 259–267. ( 10.1006/dbio.1994.1312) [DOI] [PubMed] [Google Scholar]
- 123.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929. ( 10.1016/j.cell.2005.08.040) [DOI] [PubMed] [Google Scholar]
- 124.Lin S-CJ, Wani MA, Whitsett JA, Wells JM. 2010. Klf5 regulates lineage formation in the pre-implantation mouse embryo. Development 137, 3953–3963. ( 10.1242/dev.054775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ohnishi Y, et al. 2014. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 16, 27–37. ( 10.1038/ncb2881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A, Bertone P. 2015. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell 35, 366–382. ( 10.1016/j.devcel.2015.10.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mohammed H, et al. 2017. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228. ( 10.1016/j.celrep.2017.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Azami T, et al. 2017. Klf5 maintains the balance of primitive endoderm versus epiblast specification during mouse embryonic development by suppression of Fgf4. Development 144, 3706–3718. ( 10.1242/dev.150755) [DOI] [PubMed] [Google Scholar]
- 129.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. ( 10.1016/S0092-8674(03)00393-3) [DOI] [PubMed] [Google Scholar]
- 130.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. ( 10.1016/S0092-8674(03)00392-1) [DOI] [PubMed] [Google Scholar]
- 131.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140. ( 10.1101/gad.224503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391. ( 10.1016/S0092-8674(00)81769-9) [DOI] [PubMed] [Google Scholar]
- 133.Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. 2011. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 21, 1005–1013. ( 10.1016/j.devcel.2011.10.019) [DOI] [PubMed] [Google Scholar]
- 134.Frum T, Halbisen MA, Wang C, Amiri H, Robson P, Ralston A. 2013. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell 25, 610–622. ( 10.1016/j.devcel.2013.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Le Bin GC, et al. 2014. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141, 1001–1010. ( 10.1242/dev.096875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nichols J, Silva J, Roode M, Smith A. 2009. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222. ( 10.1242/dev.038893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grabarek JB, Zyzyńska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis A-K, Plusa B. 2012. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development 139, 129–139. ( 10.1242/dev.067702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schrode N, Saiz N, Di Talia S, Hadjantonakis A-K.. 2014. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell 29, 454–467. ( 10.1016/j.devcel.2014.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, Chazaud C.. 2014. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development 141, 3637–3648. ( 10.1242/dev.109678) [DOI] [PubMed] [Google Scholar]
- 140.Cai KQ, Capo-Chichi CD, Rula ME, Yang D-H, Xu X-X. 2008. Dynamic GATA6 expression in primitive endoderm formation and maturation in early mouse embryogenesis. Dev. Dyn. 237, 2820–2829. ( 10.1002/dvdy.21703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Niakan KK, et al. 2010. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 24, 312–326. ( 10.1101/gad.1833510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morris SA, Teo RTY, Li H, Robson P, Glover DM, Zernicka-Goetz M. 2010. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl Acad. Sci. USA 107, 6364–6369. ( 10.1073/pnas.0915063107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Artus J, Piliszek A, Hadjantonakis A-K. 2011. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev. Biol. 350, 393–404. ( 10.1016/j.ydbio.2010.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rossant J. 1975. Investigation of the determinative state of the mouse inner cell mass. II. The fate of isolated inner cell masses transferred to the oviduct. J. Embryol. Exp. Morphol. 33, 991–1001. [PubMed] [Google Scholar]
- 145.Rossant J, Chazaud C, Yamanaka Y. 2003. Lineage allocation and asymmetries in the early mouse embryo. Phil. Trans. R. Soc. Lond. B 358, 1341–1349. ( 10.1098/rstb.2003.1329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chazaud C, Yamanaka Y, Pawson T, Rossant J. 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624. ( 10.1016/j.devcel.2006.02.020) [DOI] [PubMed] [Google Scholar]
- 147.Bessonnard S, Coqueran S, Vandormael-Pournin S, Dufour A, Artus J, Cohen-Tannoudji M. 2017. ICM conversion to epiblast by FGF/ERK inhibition is limited in time and requires transcription and protein degradation. Scient. Rep. 7, 12285 ( 10.1038/s41598-017-12120-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec J-F, Zernicka-Goetz M. 2009. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev. Biol. 331, 210–221. ( 10.1016/j.ydbio.2009.04.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gerbe F, Cox B, Rossant J, Chazaud C. 2008. Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Dev. Biol. 313, 594–602. ( 10.1016/j.ydbio.2007.10.048) [DOI] [PubMed] [Google Scholar]
- 150.Saiz N, Grabarek JB, Sabherwal N, Papalopulu N, Plusa B. 2013. Atypical protein kinase C couples cell sorting with primitive endoderm maturation in the mouse blastocyst. Development 140, 4311–4322. ( 10.1242/dev.093922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamanaka Y, Lanner F, Rossant J. 2010. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724. ( 10.1242/dev.043471) [DOI] [PubMed] [Google Scholar]
- 152.Kang M, Piliszek A, Artus J, Hadjantonakis A-K. 2013. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140, 267–279. ( 10.1242/dev.084996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krawchuk D, Honma-Yamanaka N, Anani S, Yamanaka Y. 2013. FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Dev. Biol. 384, 65–71. ( 10.1016/j.ydbio.2013.09.023) [DOI] [PubMed] [Google Scholar]
- 154.Kang M, Garg V, Hadjantonakis A-K. 2017. Lineage establishment and progression within the inner cell mass of the mouse blastocyst requires FGFR1 and FGFR2. Dev. Cell 41, 496–510. ( 10.1016/j.devcel.2017.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Molotkov A, Mazot P, Brewer JR, Cinalli RM, Soriano P. 2017. Distinct requirements for FGFR1 and FGFR2 in primitive endoderm development and exit from pluripotency. Dev. Cell 41, 511–526. ( 10.1016/j.devcel.2017.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brewer JR, Molotkov A, Mazot P, Hoch RV, Soriano P. 2015. Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes Dev. 29, 1863–1874. ( 10.1101/gad.264994.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Azami T, Bassalert C, Allègre N, Valverde-Estrella L, Pouchin P, Ema M, Chazaud C. 2019. Regulation of the ERK signalling pathway in the developing mouse blastocyst. Development 146, dev177139 ( 10.1242/dev.177139) [DOI] [PubMed] [Google Scholar]
- 158.Guo G, Huss M, Tong GQ, Wang C, Sun LL, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685. ( 10.1016/j.devcel.2010.02.012) [DOI] [PubMed] [Google Scholar]
- 159.Yuan H, Corbi N, Basilico C, Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9, 2635 ( 10.1101/gad.9.21.2635) [DOI] [PubMed] [Google Scholar]
- 160.Ambrosetti DC, Basilico C, Dailey L. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17, 6321–6329. ( 10.1128/MCB.17.11.6321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roode M, Blair K, Snell P, Elder K, Marchant S, Smith A, Nichols J. 2012. Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 361, 358–363. ( 10.1016/j.ydbio.2011.10.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Soszyńska A, Klimczewska K, Suwińska A. 2019. FGF/ERK signaling pathway: how it operates in mammalian preimplantation embryos and embryo-derived stem cells. Int. J. Dev. Biol. 63, 171–186. ( 10.1387/ijdb.180408as) [DOI] [PubMed] [Google Scholar]
- 163.Piliszek A, Grabarek JB, Frankenberg SR, Plusa B. 2016. Cell fate in animal and human blastocysts and the determination of viability. Mol. Hum. Reprod. 22, 681–690. ( 10.1093/molehr/gaw002) [DOI] [PubMed] [Google Scholar]
- 164.Artus J, Panthier J-J, Hadjantonakis A-K. 2010. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development 137, 3361–3372. ( 10.1242/dev.050864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. 1998. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl Acad. Sci. USA 95, 5082–5087. ( 10.1073/pnas.95.9.5082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang DH, Smith ER, Roland IH, Sheng Z, Martin WD, Hamilton TC, Lambeth JD, Xu X-X. 2002. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev. Biol. 251, 27–44. ( 10.1006/dbio.2002.0810) [DOI] [PubMed] [Google Scholar]
- 167.Morris SA, Guo Y, Zernicka-Goetz M. 2012. Developmental plasticity is bound by pluripotency and the Fgf and Wnt signaling pathways. Cell Rep. 2, 756–765. ( 10.1016/j.celrep.2012.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Huang S, Guo Y-P, May G, Enver T. 2007. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713. ( 10.1016/j.ydbio.2007.02.036) [DOI] [PubMed] [Google Scholar]
- 169.Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342. ( 10.1038/35002131) [DOI] [PubMed] [Google Scholar]
- 170.Chickarmane V, Peterson C. 2008. A computational model for understanding stem cell, trophectoderm and endoderm lineage determination. PLoS ONE 3, e3478 ( 10.1371/journal.pone.0003478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schröter C, Rué P, Mackenzie JP, Martinez Arias A. 2015. FGF/MAPK signaling sets the switching threshold of a bistable circuit controlling cell fate decisions in embryonic stem cells. Development 142, 4205–4216. ( 10.1242/dev.127530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Singh AM, Hamazaki T, Hankowski KE, Terada N. 2007. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25, 2534–2542. ( 10.1634/stemcells.2007-0126) [DOI] [PubMed] [Google Scholar]
- 173.De Mot L, Gonze D, Bessonnard S, Chazaud C, Goldbeter A, Dupont G. 2016. Cell fate specification based on tristability in the inner cell mass of mouse blastocysts. Biophys. J. 110, 710–722. ( 10.1016/j.bpj.2015.12.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nissen SB, Perera M, Gonzalez JM, Morgani SM, Jensen MH, Sneppen K, Brickman JM, Trusina A. 2017. Four simple rules that are sufficient to generate the mammalian blastocyst. PLoS Biol. 15, e2000737 ( 10.1371/journal.pbio.2000737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tosenberger A, Gonze D, Bessonnard S, Cohen-Tannoudji M, Chazaud C, Dupont G. 2017. A multiscale model of early cell lineage specification including cell division. npj. Syst. Biol. Appl. 3, 32 ( 10.1038/s41540-017-0017-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Simon CS, Hadjantonakis A-K, Schröter C. 2018. Making lineage decisions with biological noise: lessons from the early mouse embryo. Wiley Interdiscip. Rev. Dev. Biol. 61, e319 ( 10.1002/wdev.319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tosenberger A, Gonze D, Chazaud C, Dupont G. 2019. Computational models for the dynamics of early mouse embryogenesis. Int. J. Dev. Biol. 63, 131–142. ( 10.1387/ijdb.180418gd) [DOI] [PubMed] [Google Scholar]
- 178.Morris SA, Graham SJL, Jedrusik A, Zernicka-Goetz M. 2013. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open Biol. 3, 130104 ( 10.1098/rsob.130104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lanner F, Lee KL, Sohl M, Holmborn K, Yang H, Wilbertz J, Poellinger L, Rossant J, Farnebo F. 2010. Heparan sulfation–dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells 28, 191–200. ( 10.1002/stem.265) [DOI] [PubMed] [Google Scholar]
- 180.Matsuo I, Kimura-Yoshida C. 2014. Extracellular distribution of diffusible growth factors controlled by heparan sulfate proteoglycans during mammalian embryogenesis. Phil. Trans. R. Soc. B 369, 20130545 ( 10.1098/rstb.2013.0545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu H-H, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. 2003. Autoregulation of neurogenesis by GDF11. Neuron 37, 197–207. ( 10.1016/S0896-6273(02)01172-8) [DOI] [PubMed] [Google Scholar]
- 182.Parker J. 2006. Control of compartment size by an EGF ligand from neighboring cells. Curr. Biol. 16, 2058–2065. ( 10.1016/j.cub.2006.08.092) [DOI] [PubMed] [Google Scholar]
- 183.Kay RR, Thompson CRL. 2009. Forming patterns in development without morphogen gradients: scattered differentiation and sorting out. Cold Spring Harb. Perspect. Biol. 1, a001503 ( 10.1101/cshperspect.a001503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kay RR, Thompson CR. 2001. Cross-induction of cell types in Dictyostelium: evidence that DIF-1 is made by prespore cells. Development 128, 4959–4966. [DOI] [PubMed] [Google Scholar]
- 185.Keller T, Thompson CRL. 2008. Cell type specificity of a diffusible inducer is determined by a GATA family transcription factor. Development 135, 1635–1645. ( 10.1242/dev.020883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chattwood A, Nagayama K, Bolourani P, Harkin L, Kamjoo M, Weeks G, Thompson CR. 2013. Developmental lineage priming in Dictyostelium by heterogeneous Ras activation. eLife 2, e01067 ( 10.7554/eLife.01067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thompson CRL, Reichelt S, Kay RR. 2004. A demonstration of pattern formation without positional information in Dictyostelium. Dev. Growth Differ. 46, 363–369. ( 10.1111/j.1440-169x.2004.00753.x) [DOI] [PubMed] [Google Scholar]
- 188.Chattwood A, Thompson CRL. 2011. Non-genetic heterogeneity and cell fate choice in Dictyostelium discoideum. Dev. Growth Differ. 53, 558–566. ( 10.1111/j.1440-169X.2011.01270.x) [DOI] [PubMed] [Google Scholar]
- 189.Sancho M, Di-Gregorio A, George N, Pozzi S, Sánchez JM, Pernaute B, Rodriguez TA. 2013. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev. Cell 26, 19–30. ( 10.1016/j.devcel.2013.06.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Clavería C, Giovinazzo G, Sierra R, Torres M. 2013. Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500, 39–44. ( 10.1038/nature12389) [DOI] [PubMed] [Google Scholar]
- 191.Bowling S, et al. 2018. P53 and mTOR signalling determine fitness selection through cell competition during early mouse embryonic development. Nat. Commun. 9, 1763 ( 10.1038/s41467-018-04167-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lima A, et al. 2020. Differences in mitochondrial activity trigger cell competition during early mouse development. bioRxiv, 2020.01.15.900613. (doi:10.1101/2020.01.15.900613)
- 193.Hashimoto M, Sasaki H. 2019. Epiblast formation by TEAD-YAP-dependent expression of pluripotency factors and competitive elimination of unspecified cells. Dev. Cell 50, 139–154. ( 10.1016/j.devcel.2019.05.024) [DOI] [PubMed] [Google Scholar]
- 194.Pauklin S, Vallier L. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147. ( 10.1016/j.cell.2013.08.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gruenheit N, et al. 2018. Cell cycle heterogeneity can generate robust cell type proportioning. Dev. Cell 47, 494–508. ( 10.1016/j.devcel.2018.09.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data presented in figure 3 are publicly available with the data from our article ‘Growth factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development’ [119].