Abstract
Correct cell shape is indispensable for tissue architecture, with cell shape being determined by cortical actin and surface adhesion. The role of adhesion in remodelling tissue is to counteract the deformation of cells by force, resulting from actomyosin contractility, and to maintain tissue integrity. The dynamics of this adhesion are critical to the processes of cell shape formation and maintenance. Here, we show that the trafficking molecule Arf6 has a direct impact on cell elongation, by acting to stabilize E-cadherin-based adhesion complexes at the cell surface, in addition to its canonical role in endocytosis. We demonstrate that these functions of Arf6 are dependent on the molecule Flotillin1, which recruits Arf6 to the plasma membrane. Our data suggest that Arf6 and Flotillin1 operate in a pathway distinct from clathrin-mediated endocytosis. Altogether, we demonstrate that Arf6- and Flotillin1-dependent regulation of the dynamics of cell adhesion contribute to moulding tissue in vivo.
This article is part of the discussion meeting issue ‘Contemporary morphogenesis’.
Keywords: Arf6, cell shape, epithelium, adhesion, actin, endocytosis
1. Introduction
Tissue architecture is determined by the shapes of the individual cells of which it is composed, with cell shape in the plane of cell–cell adhesion being crucial for the architecture of epithelial monolayers [1]. In these epithelia, cell shape in the apical adhesion plane is the product of cortical actomyosin and intercellular adhesion [2]. This intercellular adhesion is mediated by proteins on the cell surface that bind to molecules on adjacent cells and form stable attachments. The primary adhesion molecule in epithelial cells is E-cadherin (E-cad), which is instrumental in both the formation and maintenance of tissues [3,4]. In morphogenesis, E-cad stabilizes new contacts during cell rearrangements [5] and expands contact length between neighbouring cells [6], regulating final tissue architecture. However, these adhesion sites are not static but highly dynamic, with alterations in these dynamics resulting in abnormal cell shapes and rearrangements [7–10]. Therefore, both the dynamics of adhesion and levels are critical to tissue architecture.
E-cad molecules at the cell surface are constantly removed from the plasma membrane (PM) by the process of endocytosis [9]. These molecules are then either recycled or degraded and replaced by newly synthesized molecules [11,12]. Multiple pathways of endocytosis exist, with the clathrin-mediated pathway being the best characterized [13]. We have recently discovered that clathrin-mediated endocytosis of E-cad from the PM regulates shape of epidermal cells in the late Drosophila embryo [7]. At the same time, several clathrin-independent pathways have also been identified [14], and clathrin-independent endocytosis has been implicated in Wingless and Notch signalling in Drosophila cells [15,16]. What all of these pathways share is the necessity for specific protein mediators to target and traffic surface proteins between the surface and cytoplasm. One of these mediator groups is the evolutionarily conserved small GTPase Arf proteins, a family that participates in all major protein trafficking routes known in cells [17,18]. One of its members—Arf6, the sole member of the class III Arfs [17,19,20]—acts at the PM to sort cargo, particularly adhesion molecules, in mammalian cells [18,21] and assists the internalization of these cargoes including E-cad [22,23].
The dynamics of E-cad at the PM are determined by both endocytosis and the motion of the molecule itself within the lipid bilayer [12,24]. Techniques such as Fluorescence Recovery After Photobleaching (FRAP) have been instrumental in dissecting the mechanisms of E-cad dynamics at junctions [12,25–29]. This has revealed the existence of at least several subpopulations of E-cad at the cell surface, which are distinguished by their dynamic properties and might correspond to E-cad clusters of different sizes and functions [24,29–31]. Protein–membrane domains within the PM serve as one of the general mechanisms to promote such protein clustering [32]. These domains have distinct protein and lipid compositions, and their assembly is facilitated by membrane-associated proteins, such as Flotillins [33]. Flotillins are a family of proteins embedded within the PM lipid bilayer, where they form protrusions into the cytoplasm enabling interactions that can modify the structure of the surroundings [33]. Flotillins participate in a range of fundamental cellular processes across species: endocytosis, signal transduction and cortical actin dynamics [33]. In mammalian cells, Flotillins have been reported to recruit E-cad and stabilize it at the PM [34,35], but also could be potentially involved in E-cad endocytosis and recycling [35,36].
Here, we demonstrate that Arf6 function contributes to normal tissue architecture in an intact tissue in vivo using the Drosophila embryonic epidermis as a model. Arf6 acts both by regulating the levels of E-cad at the PM and by enabling the formation of stable adhesion. We further show that Arf6 recruitment to the PM is dependent on Flotillin1, and downregulation of Flotillin1 has the same effect on E-cad dynamics and cell shape as that of Arf6. Taken together, our data demonstrate that Arf6 recruitment to the PM by Flotillin1 has the effect of stabilizing E-cad intercellular adhesion and thus regulating junctional stability and tissue architecture.
2. Results and discussion
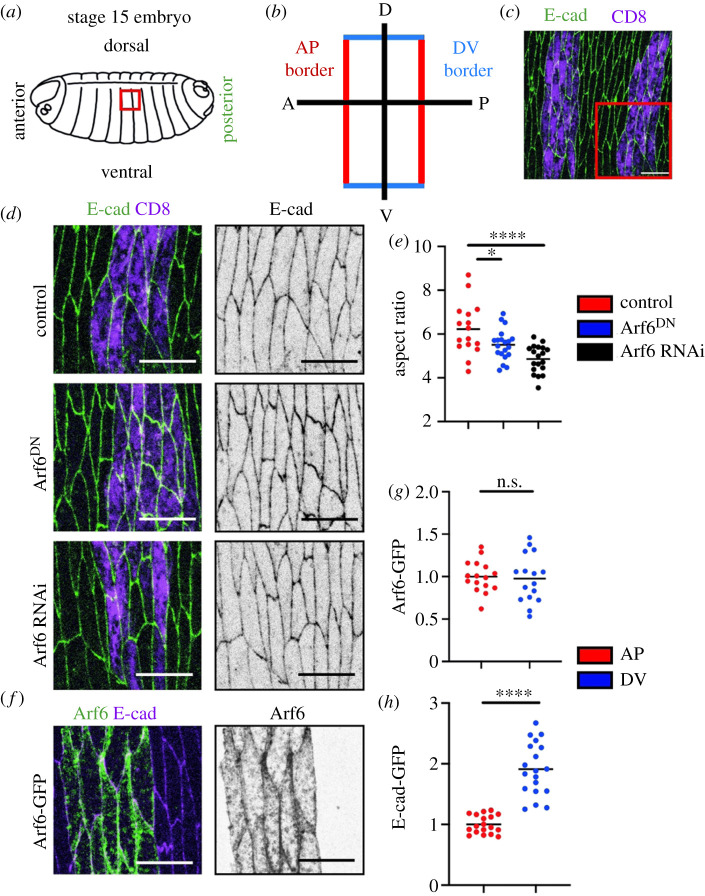
To assess the function of Arf6 in general tissue architecture, we used the stage 15 Drosophila embryonic epidermis, in which cells are highly elongated in the apical plane and rectangular in appearance (figure 1a–d). This elongation produces two distinct cell borders: a short border which is orthogonal to the dorsal–ventral axis (DV), and a long border which faces the anterior–posterior axis (AP) of the embryo (figure 1a–c). We downregulated Arf6 function with a dominant-negative variant (Arf6DN) using the engrailed promoter driving GAL4 in the posterior halves of each embryonic segment (en::Gal4, figure 1c,d). We co-expressed Arf6DN with UAS::CD8-Cherry to label the cells. The cells expressing this construct were visibly abnormal (figure 1d). Using the aspect ratio of the cells as a measure of correct elongation (the ratio between the long and short cell axis, see Material and methods), we found that cells expressing Arf6DN were less elongated than in the control expressing UAS::CD8-Cherry alone (p = 0.04, figure 1d,e). A comparable decrease in the aspect ratio was detected in cells upon downregulation of Arf6 levels with RNAi (p < 0.0001, figure 1d,e). To confirm the efficacy of this knock-down, we used Arf6 transgenically tagged with GFP (UAS::Arf6-GFP, hereafter Arf6-GFP) expressed using the en::Gal4 (figure 1f). In the presence of the RNAi, we observed a reduction in Arf6-GFP signal relative to control which co-expressed Arf6-GFP with UAS::CD8-Cherry (p = 0.002, electronic supplementary material, figure S1A).
Figure 1.
Arf6 contributes to correct cell shape and is localized to the adherens junction. (a–c) Overview of the stage 15 Drosophila embryonic epidermis. (a) Cartoon sketch of the embryo; area indicated by red box is the dorsolateral epidermis imaged in this study. (b) Schematic of the cell morphology in the tissue and the two cell borders present in these cells: the long anterior–posterior (AP, red) and short dorsal–ventral (DV, blue). (c) Apical view of the epidermis highlighted in the red box in (a). Cells are outlined by E-cad-GFP (green) and the engrailed compartments which express all transgenic construct used in this study are highlighted by CD8-Cherry (magenta). (d,e) Examples (d) and aspect ratio (e) of cells expressing UAS::CD8-Cherry alone (magenta, control in c and top in d), and co-expressing it with the Arf6DN (magenta, middle in d) and Arf6 RNAi (magenta, bottom in d). E-cad-GFP marks cell morphology (green, left and grey, right images in d). The area of images in (d) is indicated by the box in (c). (f,g) Localization of Arf6-GFP in the apical region of the epidermal cells corresponding with the plane of adherens junctions, with a representative image (f) and quantification (g). Arf6-GFP was expressed in the engrailed compartments (green, left; and grey, right in f). (h) Levels of E-cad-GFP at the two cell borders of the epidermal cell. Statistical analysis was performed using a two-tailed Students t-test with Welch's correction. Scale bars, 10 µm. n.s., not significant; *p < 0.05; ****p < 0.0001. Each dot represents an individual embryo; n (number) was 10–20 embryos per genotype with a minimum of 28 cells imaged per embryo.
To determine where Arf6 acts within the cell to promote cell elongation, we examined its localization in our system using Arf6-GFP. In the plane of cell–cell adhesion, Arf6-GFP localized predominantly at the PM, overlapping with E-cad (figure 1f). In these epidermal cells, E-cad-GFP localizes in a thin belt of approximately 1 µm depth at the cell–cell junctions, below the apical surface, in an approximate 1 : 2 ratio between the AP and DV cell borders (figure 1d,h), consistent with previous observations [7,12,37,38]. Here, we used E-cad tagged with GFP and expressed from a ubiquitous Ubi-p63E promoter (E-cad-GFP) [39]. Arf6-GFP signal was distributed uniformly around the cell periphery, in contrast to E-cad (p = 0.77 and p < 0.0001, respectively, figure 1g,h). These observations were consistent with previous reports [40] and suggested that Arf6 acts at intercellular junctions. This result raised two related questions. First, once at the PM, what does Arf6 do to promote cell elongation? Second, what determines the specific enrichment of Arf6 at the PM? We next sought to answer these questions.
Two known functions of Arf6 could explain its role in cell elongation: vesicle trafficking and regulation of the actin cytoskeleton [22]. Indeed, one of the potential Arf6 regulators—the ArfGEF Steppke—is important for cell morphology and tissue architecture in the Drosophila embryonic epidermis [41]. First, we tested whether the interaction with cortical actomyosin was important for the role of Arf6 in cell elongation. We used a YFP-tagged variant of non-muscle Myosin-II under the spaghetti-squash promoter (hereafter, MyoII-YFP) to visualize the actomyosin cortex (figure 2a,b). In control cells at this stage of embryonic development, MyoII-YFP was enriched at the AP cell borders, consistent with previous observations [7,12,38]. Expression of Arf6DN led to a significant increase in the levels of MyoII-YFP at both cell borders (p = 0.0001, p = 0.04, figure 2b), which is consistent with Arf6 preventing the formation of actin stress fibres by downregulating RhoA [41,42]. To explore this further, we decided to measure the activity of RhoA directly. To this end, we used a biosensor of RhoA activity, the Rho-binding domain of Anillin (RBD-GFP) [43]. This biosensor localized in a similar fashion to MyoII-YFP, with enrichment at the AP cell borders but strong cytoplasmic signal (electronic supplementary material, figure S1B-B’). The expression of Arf6DN had no effect on the localization and levels of RBD-GFP (p = 0.17, electronic supplementary material, figure S1B-B’). This indicated either that this biosensor is not sensitive enough to reflect the changes in RhoA activity, or that Arf6 influences MyoII independently of RhoA activity in this system. Overall, the impairment of Arf6 function leads to an increase in MyoII-YFP at the cortex, potentially through a RhoA-independent mechanism, the existence of which has been reported [44,45].
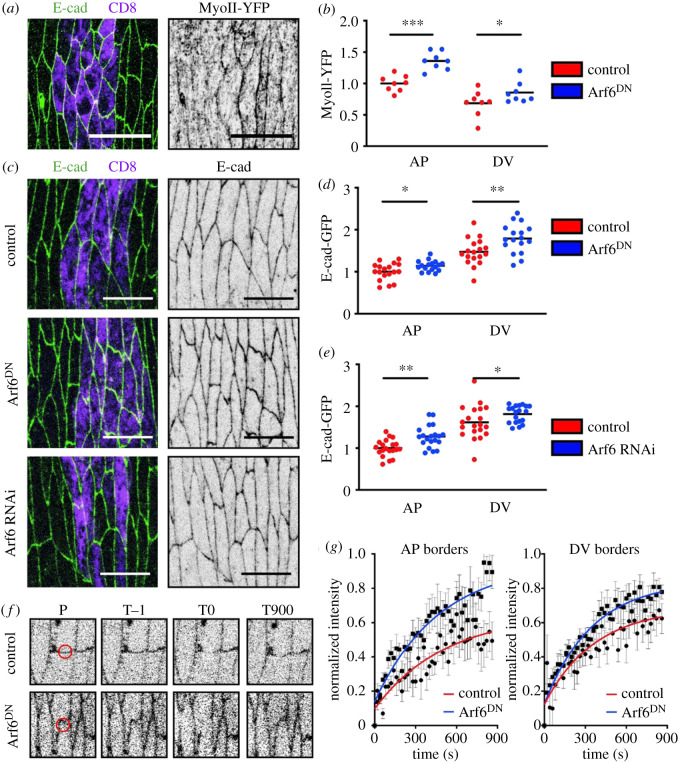
Figure 2.
Arf6 function alters both myosin-II localization and the levels and dynamics of E-cad. (a,b) Representative images (a) and levels (b) of MyoII-YFP in the epidermis of internal control and Arf6DN-expressing cells. Cell borders were visualized with antibody against E-cad (green in a) and cells expressing Arf6DN transgene marked by UAS::CD8-Cherry (magenta). (c–g) Levels (c–e) and dynamics (f,g) of E-cad following downregulation of Arf6. (c–e) Representative images (c) and levels (d,e) of E-cad in cells expressing UAS::CD8-Cherry alone (control, top), Arf6DN (middle) or an RNAi against Arf6 (bottom). (f–g) Representative examples (f) and quantification (g) of E-cad-GFP FRAP in control and Arf6DN-expressing cells. Panels in (f) show the region bleached (position P, red circle) at the prebleach (time T−1), bleach (time T0) and end (time T900) time points. Time is in seconds. Average recovery curves (mean ± s.e.m.) and the best-fit curves (solid lines) are shown in (g). All best-fit and membrane intensity data are in electronic supplementary material, table S1. Scale bars, 10 µm. Statistical analysis of the two cell borders between the genotypes was performed using a two-way ANOVA. Individual p-values are shown for each border between the genotypes. *p < 0.05; **p < 0.01; ***p < 0.001. Each dot represents an individual embryo; n (number) was 10–20 embryos per genotype, with a minimum of 29 cells imaged per embryo. For FRAP 8–10 embryos were used, with two AP and DV cell borders measured per embryo and averaged to give the final embryo value.
MyoII contractility reduces contact length between cells and contributes to cell shape [2]. However, the increase in MyoII at the cortex following Arf6 downregulation was uniform—about 20% at both AP and DV borders (23 ± 7 and 15 ± 9%, respectively). At the same time, no change in the apical cell area was observed (p = 0.47 and p = 0.83 for Arf6 RNAi and Arf6DN, respectively, electronic supplementary material, figure S1C), which would have been expected in the case of a uniform contact length reduction. This suggested that the anisotropic change in the cell shape following Arf6 downregulation, namely the reduction in cell elongation, is likely to be through a different mechanism rather than only cortical actomyosin activity. We next sought to determine the contribution of the trafficking function of Arf6 to the cell shape phenotype.
Our recent studies have demonstrated key roles for membrane trafficking in the regulation of cell shape and specifically in the elongation of cells in the embryonic epidermis [7]. Arf6 regulates vesicle trafficking between the PM and endosomal system [22,46,47]. Indeed, Arf6 is the primary member of the Arf family that is enriched at the PM, making it an obvious candidate to be involved in endocytic trafficking of adhesion proteins. Therefore, we explored the effects of Arf6 downregulation on intercellular adhesion by measuring the distribution and dynamics of E-cad. In cells expressing Arf6DN, E-cad-GFP was elevated at both the AP and DV cell borders in comparison with control (p = 0.02 and p = 0.007, figure 2d), indicating a uniform effect consistent with the uniform distribution of Arf6. We confirmed the role of Arf6 in E-cad levels at intercellular junctions using an independent approach to inhibit Arf6 function: the expression of an RNAi targeted to Arf6 led to a similar increase in E-cad-GFP at both the AP and DV cell borders (p = 0.0023, p = 0.031, figure 2c,e).
As we have previously found that both the levels of E-cad and its dynamics contribute to cell shape [7], we decided to explore the effect of Arf6DN on the dynamics of E-cad at the cell surface using FRAP. In our previous work we showed E-cad-GFP recovered to 70 and 50% of prebleach signal at the DV and AP cell borders, respectively [12,37]. In cells expressing Arf6DN we observed an increased recovery of the E-cad-GFP signal at both cell borders (figure 2f,g; electronic supplementary material, table S1). Therefore, impairing Arf6 function results in the reduction of the immobile E-cad fraction, suggesting that it is involved in stabilizing E-cad within the cell–cell junctions. Overall, these data support a model whereby Arf6 function has a dual effect on cell adhesion: to modulate E-cad levels and facilitate the formation of stable adhesion complexes.
Neither the elevation of E-cad at the PM nor the increase in the mobile fraction could be explained purely by the effect on cortical actomyosin. Myosin-II promotes E-cad endocytosis in both Drosophila and mammalian cells, and its reduction rather than elevation increases E-cad mobility and vice versa [7,8,12,48]. The observed elevation of E-cad at the PM by the impairment of Arf6 function was however consistent with defects in E-cad endocytosis and the known Arf6 function in vesicle trafficking. Such elevation is also observed when E-cad endocytosis is perturbed by other means: for example, by hyperactivating RhoA signalling or overexpressing p120-catenin (p120ctn), which stabilize E-cad [7,49]. However, if the effect of Arf6 was exclusively through an impairment of endocytosis it would be predicted to produce an increased amount of immobile E-cad, as reported previously when E-cad endocytosis was prevented by inhibiting the function of dynamin [12]. This further suggested that Arf6 has an additional function and contributes to the formation of the stable fraction of E-cad in addition to its known role in endocytosis.
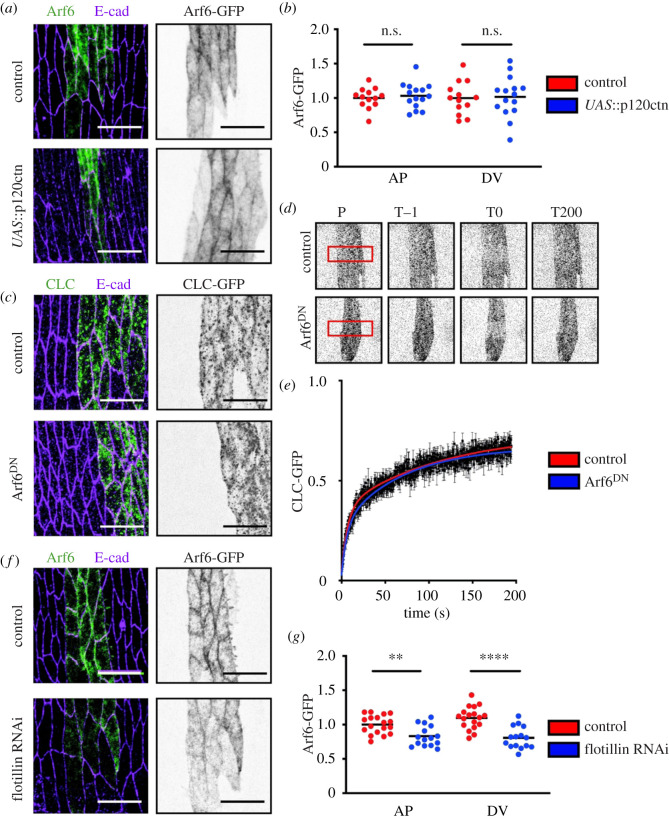
Therefore, we next examined the role of Arf6 in E-cad stability. A key regulator of E-cad stability at the cell surface is the molecule p120-catenin (p120ctn) [7,37,50,51]. The overexpression of p120ctn stabilizes E-cad within junctions in both mammalian and Drosophila cells [7,49,52]. If Arf6 were stabilizing E-cad downstream of p120ctn, one might expect its increased localization to the PM upon p120ctn overexpression. However, we found that the overexpression of p120ctn, which also elevates Myosin-II [7], did not alter the localization of Arf6 (figure 3a,b), from which we concluded that Arf6 is likely to act independently of p120ctn.
Figure 3.
Arf6 acts independently of clathrin and p120ctn but requires Flotillin1 for recruitment to the cell surface. (a,b). Representative images (a) and levels (b) of Arf6-GFP (grey, right and green, left in a) in control (top in a) and the overexpression of p120ctn (bottom in a). (c–e). Localization (c) and FRAP (d,e) of CLC-GFP (grey, d and right in c; magenta, left in c) in the plane of cell–cell adhesion, expressed in the engrailed compartment in control cells (top in c,d) and Arf6DN-expressing cells (bottom in c,d). Panels in (d) show the region bleached (position P, red rectangle) at the prebleach (time T−1), bleach (time T0) and the end (time T200) time points. Time in seconds. Average recovery curves (mean ± s.e.m.) and the best-fit curves (solid lines) are shown in (e). All best-fit and membrane intensity data are in electronic supplementary material, table S1. (f,g) Representative images (f) and levels (g) of Arf6-GFP (grey, right and green, left in f) in control (top in f) and Flotillin RNAi-expressing cells (bottom in f). Scale bars, 10 µm. Cell borders were visualized with antibody against E-cad (magenta, left in a, c and f). Statistical analysis was performed using a two-way ANOVA. p-values are shown for each border type between the genotypes. n.s., not significant; **p < 0.01; ****p < 0.0001. Each dot represents an individual embryo; n (number) was 10–20 embryos per genotype with a minimum of 35 cells imaged per embryo. For FRAP, 8–10 embryos were used, with a single rectangular region of interest (ROI) encompassing an engrailed stripe used per embryo.
We then asked whether Arf6 was involved in clathrin-mediated endocytosis. We used a transgenically tagged variant of the clathrin light chain (CLC-GFP), and analysed the effect of Arf6DN on clathrin dynamics by measuring the exchange of CLC-GFP in the plane of cell–cell adhesion using FRAP (figure 3c–e). This approach has previously been used to identify defects in clathrin-mediated endocytosis in both mammalian and Drosophila cells [7,53]. We observed no difference in the CLC-GFP recovery between the control and Arf6DN-expressing cells (p = 0.28, figure 3e). Thus, clathrin dynamics were insensitive to the downregulation of Arf6 function, suggesting that Arf6 operates in a clathrin-independent endocytic pathway in this system.
Therefore, we explored Arf6 involvement in clathrin-independent endocytic pathways. As Drosophila has no orthologues of the mammalian caveolae, we turned to Flotillins [54,55]. To explore the connection between Flotillin and Arf6, we measured the effect of knocking-down Flotillin1 on Arf6-GFP (figure 3f,g) [33,56]. We downregulated Flotillin1 using two independent RNAi lines (results were identical for both). A significant decrease of Arf6-GFP at the PM was detected at both cell borders in cells expressing the Flotillin1 RNAi (p = 0.002, p < 0.0001, figure 3f,g). This decrease was accompanied by an increase in Arf6-GFP signal in the cytoplasm, suggesting a defect in Arf6 recruitment rather than total protein levels (p = 0.018, electronic supplementary material, figure S1D). Therefore, Arf6 is recruited to the PM downstream of Flotillin1, suggesting that Arf6 functions in a Flotillin-mediated pathway.
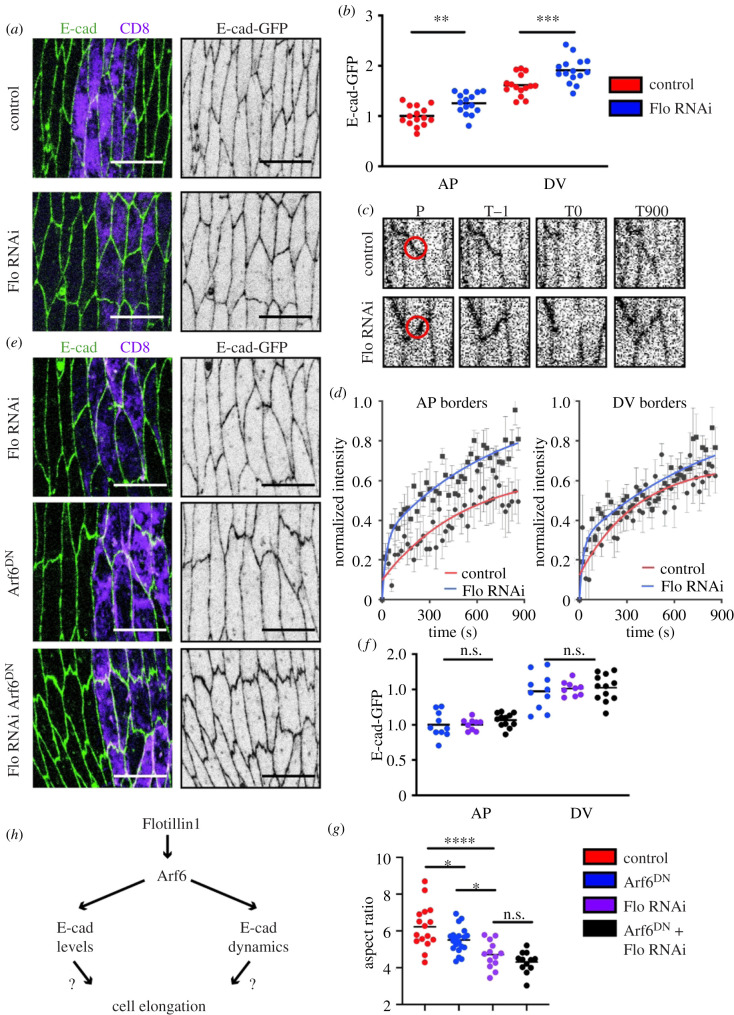
Therefore, we wondered if Flotillin1 could be upstream of the stabilizing effect that Arf6 had on E-cad, and if the cell shape defect we observed when perturbing Arf6 was due to this pathway. We found that the levels of E-cad-GFP were elevated at both the AP and DV cell borders upon Flotillin1 downregulation with RNAi (p = 0.0025, p = 0.0005, figure 4a,b), similar to what we observed for Arf6 (figure 2c–e). To determine if Flotillin1 was also altering the turnover of cell adhesion, we measured the dynamics of E-cad-GFP in cells expressing Flotillin1 RNAi using FRAP (figure 4c,d; electronic supplementary material, table S1). In these cells, the recovery of E-cad-GFP was elevated at both the AP and DV cell borders, again reminiscent of the effect of Arf6DN (figure 2f,g). Altogether, these results suggested that Arf6 and Flotillin1 act in the same pathway to stabilize E-cad, with Flotillin1 being required for Arf6 localization to the PM. Indeed, downregulation of Flotillin1 by RNAi simultaneous with the expression of Arf6DN did not increase E-cad levels beyond the increase caused by either of them alone (p = 0.96, p = 0.97, figure 4f), which further confirms that Flotillin1 and Arf6 act in a single pathway. To explore any effect of Flotillin1 on actomyosin, we examined the distribution of MyoII-YFP in cells expressing Flotillin1 RNAi. We found no significant difference between control cells and those expressing Flotillin1 RNAi at both the AP and DV cell borders (p = 0.33, p = 0.83, respectively, electronic supplementary material, figure S1E-E’). This finding contrasts with Arf6DN, in which an elevation of MyoII-YFP was observed (figure 2a,b), potentially due to the magnitude of the Arf6 reduction in cells expressing Flotillin1 RNAi being insufficient to exert an effect on MyoII. The similarities and differences in the effects of Flotillin1 RNAi and Arf6 impairment additionally suggest that Arf6 regulates E-cad and MyoII through different and independent mechanisms.
Figure 4.
Flotillin determines cell shape and E-cad dynamics upstream of Arf6. (a,b). Representative images (a) and levels (b) of E-cad-GFP (green, left and grey, right in a) in control (top in a) and Flotillin1 RNAi-expressing cells (bottom in a). (c,d) Representative examples (c) and quantification (d) of E-cad-GFP FRAP in control and Flotillin1 RNAi-expressing cells. Panels in (c) show the region bleached (position P, red circle) at the prebleach (time T−1), bleach (time T0), and the end (time T900) time points. Time is in seconds. Average recovery curves (mean ± s.e.m.) and the best-fit curves (solid lines) are shown in (d). (e,f) Representative images (e) and levels (f) of E-cad-GFP (green, left: grey, right in e) in cells expressing either: Flotillin1 RNAi alone (top in e), Arf6DN alone (middle in e), or both Flotillin1 RNAi and Arf6DN (bottom in e). (f) E-cad-GFP levels at cell borders in cells expressing: Arf6DN alone (blue), Flotillin1 RNAi alone (purple), or both Arf6DN and Flotillin1 RNAi (black). (g) Aspect ratio of the cells expressing Flotillin1 RNAi alone, Arf6DN alone, or both Flotillin1 RNAi and Arf6DN. Scale bars: 10 µm. RNAi-expressing cells marked by UAS::CD8-Cherry (magenta, left in a and e). Statistical analysis was performed using a two-way ANOVA. p-values are shown for each border type between the genotypes. For analysis of aspect ratios, a one-way ANOVA with Tukeys multiple comparison test was used. n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Each dot represents an individual embryo; n (number) was; 10–20 embryos per genotype with a minimum of 29 cells imaged per embryo. For FRAP, 8–10 embryos were used, with two AP and DV cell borders measured per embryo and averaged to give the final embryo value.
Finally, we explored the role of Flotillins in tissue architecture and compared it with that of Arf6 (figure 4e,g). Similar to the inhibition of Arf6, Flotillin1 RNAi reduced cell elongation, measured by aspect ratio, by comparison with control cells (p < 0.0001 and p = 0.0001 for two RNAi lines, figure 4e,g, electronic supplementary material, table S1). As this reduction of cell elongation occurred without any effect on MyoII (see electronic supplementary material, figure S1E), we concluded that it is the dynamics of adhesion that are the primary contributory mechanism in this phenotype. Furthermore, when Flotillin1 RNAi was expressed simultaneously with Arf6DN there was no additive phenotype: cell elongation was affected, but it was reduced to the same degree as Flotillin1 RNAi alone (p < 0.0001 in comparison with control and p = 0.59 in comparison with Flotillin1 RNAi, figure 4e,g). Therefore, the stabilization of E-cad by Flotillin-Arf6 is required for correct cell elongation.
We recently found that hyperstabilization of E-cad, by overexpressing p120ctn or inhibiting endocytosis by expressing the dominant-negative variant of dynamin Shibire, has a similar phenotype: reduced cell elongation [7]. The current study alongside our previous work suggests that the correct dynamics of intercellular adhesion are crucial for cell elongation and tissue architecture: if E-cad is too stable or too dynamic, cells fail to expand their long borders to a normal extent. This further highlights the fact that all E-cad subcomplexes contribute to cell shape regulation. However, it is tempting to speculate that they do so through different mechanisms: while endocytosis of mobile E-cad enables membrane and thus junctional remodelling required for elongation, immobilization of stable E-cad is rather more likely to stabilize the junctions following remodelling. Therefore, it is the nature of adhesion rather than simply levels that is crucial for correct tissue architecture.
One interesting question that we were unable to address is what accounts for this dual function of Arf6 and Flotillin: they are involved in both stabilization and the regulation of E-cad levels at the PM, with the latter suggesting an endocytic component. There is evidence that ‘immobile’ E-cad despite not exchanging at the cell junctions on the time-scale of a typical FRAP experiment is still turned-over on the scale of approximately 2 h [26]. It is possible that Arf6-Flotllin targets this slowly turning-over population of E-cad: following E-cad recruitment to Flotillin domains in the PM it is internalized owing to unknown cues.
In summary, we have examined the role of stable cell adhesion in tissue architecture using the trafficking protein Arf6. We discovered that Arf6 function contributes to tissue architecture by regulating cell shape. While Arf6 counteracts cortical actomyosin in what appears to be a RhoA-independent manner, this counteraction is not sufficient to explain the Arf6 function in tissue architecture. Rather, this function is due to the dual role of Arf6 on cell adhesion: both stabilizing and modulating the levels of E-cad. We then uncovered the mechanism of this dual role by identifying that Arf6 operates in a clathrin-independent pathway in Drosophila and that the Arf6-mediated stabilization of E-cad requires Flotillin1. Finally, we showed that Flotillins support the formation of stable adhesion complexes and that they function upstream of Arf6 by recruiting it to the PM, which enables the formation and regulation of stable cell adhesion and thus correct tissue architecture.
3. Material and methods
(a). Fly stocks and genetics
Flies were raised on standard medium. The GAL4/UAS system [57] was used for expression of transgenic and RNAi constructs. The GAL4 expressional driver used for all experiments was engrailed::GAL4 (en::GAL4, Bloomington number 30564). The following fly stocks were used in this study (Bloomington, or Kyoto numbers included where applicable): E-cad-GFP (Ubi::E-cadherin-GFP) (109007), E-cad-mCherry (59014), UAS::CD8-mCherry (27393), UAS::CLC-GFP (7109), UAS::Arf6-GFP (65867), UAS::Arf6-T27N (DN) [58], UAS::Arf6 RNAi (27261), Myosin-II-YFP (Kyoto Stock Center, 115082), Anillin-GFP (RBD-GFP) [59], UAS::p120ctn [7] and Flotillin RNAi (36649 and 36700; note, the latter stock is currently not available at Bloomington).
(b). Embryo collection and fixation
Embryos were collected at 25°C at 3 h time intervals and allowed to develop at 18°C for 21 h to reach the desired developmental stage. Then embryos were dechorionated using 50% sodium hypochlorite (bleach, Invitrogen) in water for 4 min, and extensively washed with deionized water prior to fixation. Fixation was performed with a 1 : 1 solution of 4% formaldehyde (Sigma) in phosphate-buffered saline (PBS) and heptane (Sigma) for 20 min on an orbital shaker at room temperature. Embryos were then devitellinized in 1 : 1 solution of methanol and heptane for 20 s with vigorous agitation. Following subsequent methanol washes the fixed embryo specimens were stored at −20°C in methanol until required.
(c). Embryo live imaging
Embryos were collected and dechorionated as described above. Once washed with deionized water, embryos were transferred to apple juice agar segments upon microscope slides. Correct genotypes were selected under a fluorescent microscope (Leica) using a needle. Embryos were positioned and orientated in a row consisting of 6–10 embryos per genotype. Following this, embryos were transferred to pre-prepared microscope slides with Scotch tape and embedded in Halocarbon oil 27 (Sigma). Embryos were left to aerate for 10 min prior to covering with a coverslip and imaging.
(d). Immunostaining
The only embryos that were immunostained for E-cad are those expressing Arf6-GFP and CLC-GFP (figures 1f, 2a and 3a,c,d). In all other cases, the native fluorescence of GFP and mCherry was imaged. The embryos were washed three times in 1 ml of PBST (PBS with 0.05% Triton) with gentle rocking. Blocking of the embryos prior to staining was done in 300 µl of 1% normal goat serum (NGS) in PBST for 1 h at room temperature with gentle rocking. For staining, the blocking solution was removed, 300 µl of the primary antibody 1 : 100 rat anti-E-cad (DCAD2, DSHB) in fresh blocking solution was added and the embryos were incubated overnight at 4°C with orbital rotation. Then, embryos were washed three times with 1 ml of PBST. A 300 µl 1 : 300 dilution of the secondary antibody (goat Cy3-conjugated IgG, Invitrogen) was added, and the embryos incubated either overnight at 4°C with orbital rotation or for 2 h at room temperature. Then embryos were washed three times with PBST, following which they were incubated with 50–70 µl of Vectashield (Vector Laboratories) and allowed to equilibrate for a period of 2 h before being mounted on microscope slides (Thermo).
(e). Microscopy, data acquisition and FRAP
All experiments were performed using an upright Olympus FV1000 confocal microscope with a 60×/1.40 NA oil immersion objective. All measurements were made on dorsolateral epidermal cells of embryos, which were near or just after completion of dorsal closure, corresponding to the end of stage 15 of embryogenesis. An area encompassing two adjacent engrailed domains was imaged to ensure a minimum of 25 cells would be analysed for each embryo. For fixed samples, 16-bit images were taken at a magnification of 0.051 µm pixel−1 (1024 × 1024 pixel XY-image) with a pixel dwell of 4 µs pixel−1. For each embryo, a Z-axis sectional stack through the plane of adherens junctions was taken, which consisted of six sections with a 0.38 µm intersectional spacing. The images were saved in the Olympus binary image format for further processing.
For E-cad FRAP (adapted from [12]) 16-bit images were taken at a magnification of 0.093 µm pixel−1 (320 × 320 pixel XY-image). In each embryo, several circular regions of 1 µm radius were photobleached at either DV or AP junctions, resulting in one bleach event per cell. Photobleaching was performed with 8 scans at 2 µs pixel−1 at 50–70% 488 nm laser power, resulting in the reduction of E-cad-GFP signal by 60–80%. A stack of six Z-sections spaced by 0.38 µm was imaged just before photobleaching, and immediately after photobleaching, and then at 20 s intervals, for a total of 15 min.
For CLC-GFP FRAP, 16-bit images were taken at a magnification of 0.051 µm pixel−1 (256 × 256 pixel XY-image). In each embryo, a single plane was selected in the centre of the cell–cell adhesion band using E-cad-mCherry for positioning. An area encompassing a transverse region orthogonal to the axis of the engrailed-expressing cells was selected (140 × 60 pixels) and photobleached with 1 scan at 2 µm pixel−1 using 100% 488 nm laser power resulting in reduction of CLC-GFP signal by 70–80%. Images were taken using continuous acquisition at a frame rate of 2 s−1. Prior to bleaching a sequence of 10 images was taken, and a total of 400 frames corresponding to 3.5 min were taken.
(f). Data processing and statistical analysis
(i). Membrane intensity and cell shape
Images were processed in Fiji (https://fiji.sc) by generating average intensity projections of the channel required for quantification. Masks were created by processing background-subtracted maximum intensity projections using the Tissue Analyzer plugin in Fiji [60]. Quantification of the membrane intensity at the AP and DV borders and cell elongation (aspect ratio) was done as described previously using a custom-built Matlab script [37], which can be found at https://github.com/nbul/Intensity. In short, cells were identified as individual objects using the created masks, and their eccentricities were calculated. The aspect ratio was calculated from the eccentricity as , where e is eccentricity. At the same time, the individual borders were identified as objects by subtracting a dilated mask of vertices from a dilated mask of cell outlines. The mean intensity and orientation of each border were calculated. The average border intensities (0–10° for the AP and 40–90° for the DV borders relative to cell mean orientation) were calculated for each embryo and used as individual data points to compare datasets. The average cytoplasmic intensity was used for the background subtraction. Statistical analysis was performed in GraphPad Prism (https://www.graphpad.com/scientific-software/prism/). First, the data were cleaned using ROUT detection of outliers in Prism, followed by testing for normal distribution (D'Agostino - Pearson normality test). Then, the significance for parametric data was tested by either a two-way ANOVA or a two-tailed t-test with Welch's correction.
(ii). E-cad FRAP
Images were processed by using the Grouped ZProjector plugin in Fiji to generate average intensity projections for each time point. Following this the bleached region of interest (ROI), control ROI and background intensity were manually measured for each time point. These data were processed in Microsoft Excel. First, the intensity of the bleached ROI at each time point was background-subtracted and normalized as follows: where: Fn is intensity of the bleached ROI at the time point n; FCn, intensity of the control unbleached ROI of the same size at the plasma membrane at the time point n; and BGn, background intensity, measured with the same size ROI in cytoplasm at the time point n. Then the relative recovery at each time point was calculated using the following formula: , where In, I1 and I0 are normalized intensities of bleached ROI at time point n, immediately after photobleaching, and before photobleaching, respectively. These values were input to Prism and nonlinear regression analysis was performed to test for best-fit model and if recoveries were significantly different between cell borders or genotypes. The recovery was fitted to either a single exponential model of the form , or a bi-exponential model of the form , where Fim is the size of the immobile fraction, Tfast and Tslow are the half-times, and A1 and A2 are amplitudes of the fast and slow components of the recovery, respectively. An F-test was used to choose the model and compare datasets.
(iii). CLC-GFP FRAP
Measurements of all intensities, i.e. the bleached ROI, control ROI and the background, and normalization were done by means of a custom-build Matlab script (http://github.com/nbul/FRAP) using the same algorithm as described for E-cad FRAP. Curve fitting and statistical analysis were performed in GraphPad Prism using a nonlinear regression analysis as described for E-cad FRAP.
Supplementary Material
Supplementary Material
Acknowledgements
The authors wish to acknowledge the help of Professor David Strutt and Dr Kyra Campbell, both of the University of Sheffield. The authors thank the technical staff of the Wolfson Light Microscopy Facility (LMF) and the Fly Facility, the University of Sheffield, without whom this work would not have been possible.
Data accessibility
This article has no additional data.
Authors' contributions
J.G. and N.A.B. designed and performed experiments and wrote the manuscript.
Competing interest
The authors declare and confirm that there is no conflict of interest for the work presented in this paper.
Funding
This work was supported by grant no. BB/P007503/1 from the UK Biotechnology and Biological Sciences Research Council.
References
- 1.Farhadifar R, Röper J-C, Aigouy B, Eaton S, Jülicher F. 2007. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 17, 2095–2104. ( 10.1016/j.cub.2007.11.049) [DOI] [PubMed] [Google Scholar]
- 2.Lecuit T, Lenne P-F. 2007. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644. ( 10.1038/nrm2222) [DOI] [PubMed] [Google Scholar]
- 3.Harris TJC, Tepass U. 2010. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514. ( 10.1038/nrm2927) [DOI] [PubMed] [Google Scholar]
- 4.Maître J-L, Heisenberg C-P. 2013. Three functions of cadherins in cell adhesion. Curr. Biol. 23, R626–R633. ( 10.1016/j.cub.2013.06.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collinet C, Rauzi M, Lenne P-F, Lecuit T. 2015. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat. Cell Biol. 17, 1247–1258. ( 10.1038/ncb3226) [DOI] [PubMed] [Google Scholar]
- 6.de Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. 2004. Maternal β-catenin and E-cadherin in mouse development. Development 131, 4435–4445. ( 10.1242/dev.01316) [DOI] [PubMed] [Google Scholar]
- 7.Greig J, Bulgakova NA.2019. Interplay between cortical actin and E-cadherin dynamics regulates cell shape in the Drosophila embryonic epidermis. bioRxiv , 801456. ( ) [DOI]
- 8.Levayer R, Pelissier-Monier A, Lecuit T. 2011. Spatial regulation of Dia and myosin-II by RhoGEF2 controls initiation of E—cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 13, 529–540. ( 10.1038/ncb2224) [DOI] [PubMed] [Google Scholar]
- 9.Brüser L, Bogdan S. 2017. Adherens junctions on the move—membrane trafficking of E-cadherin. Cold Spring Harb. Perspect. Biol. 9, a029140 ( 10.1101/cshperspect.a029140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West JJ, Harris TJC. 2016. Cadherin trafficking for tissue morphogenesis: control and consequences. Traffic 17, 1233–1243. ( 10.1111/tra.12407) [DOI] [PubMed] [Google Scholar]
- 11.Le TL, Yap AS, Stow JL.. 1999. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219–232. ( 10.1083/jcb.146.1.219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulgakova NA, Grigoriev I, Yap AS, Akhmanova A, Brown NH. 2013. Dynamic microtubules produce an asymmetric E-cadherin–Bazooka complex to maintain segment boundaries. J. Cell Biol. 201, 887–901. ( 10.1083/jcb.201211159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilar RC, Wendland B. 2005. Endocytosis of membrane receptors: two pathways are better than one. Proc. Natl Acad. Sci. USA 102, 2679–2680. ( 10.1073/pnas.0500213102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemalatha A, Mayor S. 2019. Recent advances in clathrin-independent endocytosis. F1000Res 8, 138 ( 10.12688/f1000research.16549.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemalatha A, Prabhakara C, Mayor S. 2016. Endocytosis of Wingless via a dynamin-independent pathway is necessary for signaling in Drosophila wing discs. Proc. Natl Acad. Sci. USA 113, E6993–E7002. ( 10.1073/pnas.1610565113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu H, Woodcock SA, Wilkin MB, Trubenová B, Monk NAM, Baron M. 2014. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 157, 1160–1174. ( 10.1016/j.cell.2014.03.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza-Schorey C, Chavrier P. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, nrm1910 ( 10.1038/nrm1910) [DOI] [PubMed] [Google Scholar]
- 18.Palacios F. 2001. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 20, 4973–4986. ( 10.1093/emboj/20.17.4973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson JG, Jackson CL. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375. ( 10.1038/nrm3117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson CL, Bouvet S. 2014. Arfs at a glance. J. Cell Sci. 127, 4103–4109. ( 10.1242/jcs.144899) [DOI] [PubMed] [Google Scholar]
- 21.Sabe H. 2003. Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. J. Biochem. 134, 485–489. ( 10.1093/jb/mvg181) [DOI] [PubMed] [Google Scholar]
- 22.Donaldson JG. 2003. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41 573–41 576. ( 10.1074/jbc.R300026200) [DOI] [PubMed] [Google Scholar]
- 23.Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. 2011. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin. Cell Dev. Biol. 22, 39–47. ( 10.1016/j.semcdb.2010.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erami Z, Timpson P, Yao W, Zaidel-Bar R, Anderson KI. 2015. There are four dynamically and functionally distinct populations of E-cadherin in cell junctions. Biol. Open 4, 1481–1489. ( 10.1242/bio.014159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etournay R, et al. 2015. Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. eLife 4, e07090 ( 10.7554/eLife.07090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer KV, Piscitello-Gómez R, Paijmans J, Jülicher F, Eaton S. 2019. Epithelial viscoelasticity is regulated by mechanosensitive E-cadherin turnover. Curr. Biol. 29, 578–591.e5. ( 10.1016/j.cub.2019.01.021) [DOI] [PubMed] [Google Scholar]
- 27.Erami Z, et al. 2015. Intravital FRAP imaging using an E-cadherin-GFP mouse reveals disease- and drug-dependent dynamic regulation of cell-cell junctions in live tissue. Cell Rep. 14, 152–167. ( 10.1016/j.celrep.2015.12.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Huang L, Chen Y-J, Austin E, Devor CE, Roegiers F, Hong Y. 2011. Differential regulation of adherens junction dynamics during apical–basal polarization. J. Cell Sci. 124, 4001–4013. ( 10.1242/jcs.086694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavey M, Rauzi M, Lenne P-F, Lecuit T. 2008. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature 453, 751–756. ( 10.1038/nature06953) [DOI] [PubMed] [Google Scholar]
- 30.Strale P-O, et al. 2015. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell–cell contact fluidity. J. Cell Biol. 210, 333–346. ( 10.1083/jcb.201410111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Indra I, Choi J, Chen C-S, Troyanovsky RB, Shapiro L, Honig B, Troyanovsky SM. 2018. Spatial and temporal organization of cadherin in punctate adherens junctions. Proc. Natl Acad. Sci. USA 115, E4406–E4415. ( 10.1073/pnas.1720826115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong-Quang B-A, Lenne P-F. 2014. Membrane microdomains: from seeing to understanding. Front. Plant Sci. 5, 18 ( 10.3389/fpls.2014.00018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otto GP, Nichols BJ. 2011. The roles of flotillin microdomains – endocytosis and beyond. J. Cell Sci. 124, 3933–3940. ( 10.1242/jcs.092015) [DOI] [PubMed] [Google Scholar]
- 34.Chartier NT, Lainé MG, Ducarouge B, Oddou C, Bonaz B, Albiges-Rizo C, Jacquier-Sarlin MR. 2011. Enterocytic differentiation is modulated by lipid rafts-dependent assembly of adherens junctions. Exp. Cell Res. 317, 1422–1436. ( 10.1016/j.yexcr.2011.03.009) [DOI] [PubMed] [Google Scholar]
- 35.Guillaume E, Comunale F, Do Khoa N, Planchon D, Bodin S, Gauthier-Rouvière C. 2013. Flotillin microdomains stabilize cadherins at cell–cell junctions. J. Cell. Sci. 126, 5293–5304. ( 10.1242/jcs.133975) [DOI] [PubMed] [Google Scholar]
- 36.Solis GP, Hülsbusch N, Radon Y, Katanaev VL, Plattner H, Stuermer CAO. 2013. Reggies/flotillins interact with Rab11a and SNX4 at the tubulovesicular recycling compartment and function in transferrin receptor and E-cadherin trafficking. Mol. Biol. Cell 24, 2689–2702. ( 10.1091/mbc.e12-12-0854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulgakova NA, Brown NH. 2016. Drosophila p120-catenin is crucial for endocytosis of the dynamic E-cadherin-Bazooka complex. J. Cell Sci. 129, 477–482. ( 10.1242/jcs.177527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Matos Simões S, Blankenship JT, Weitz O, Farrell DL, Tamada M, Fernandez-Gonzalez R, Zallen JA.. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell 19, 377–388. ( 10.1016/j.devcel.2010.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oda H, Tsukita S. 2001. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J. Cell Sci. 114, 493–501. [DOI] [PubMed] [Google Scholar]
- 40.Lee DM, Rodrigues FF, Yu CG, Swan M, Harris TJC. 2015. PH domain-Arf G protein interactions localize the Arf-GEF Steppke for cleavage furrow regulation in Drosophila. PLoS ONE 10, e0142562 ( 10.1371/journal.pone.0142562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West JJ, Zulueta-Coarasa T, Maier JA, Lee DM, Bruce AEE, Fernandez-Gonzalez R, Harris TJC. 2017. An actomyosin-Arf-GEF negative feedback loop for tissue elongation under stress. Curr. Biol. 27, 2260–2270. ( 10.1016/j.cub.2017.06.038) [DOI] [PubMed] [Google Scholar]
- 42.Boshans RL, Szanto S, van Aelst L, D'Souza-Schorey C. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20, 3685–3694. ( 10.1128/MCB.20.10.3685-3694.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason FM, Xie S, Vasquez CG, Tworoger M, Martin AC. 2016. RhoA GTPase inhibition organizes contraction during epithelial morphogenesis. J. Cell Biol. 214, 603–617. ( 10.1083/jcb.201603077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uehara R, Goshima G, Mabuchi I, Vale RD, Spudich JA, Griffis ER. 2010. Determinants of myosin II cortical localization during cytokinesis. Curr. Biol. 20, 1080–1085. ( 10.1016/j.cub.2010.04.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borges RM, Lamers ML, Forti FL, Santos MFD, Yan CYI. 2011. Rho signaling pathway and apical constriction in the early lens placode. Genesis 49, 368–379. ( 10.1002/dvg.20723) [DOI] [PubMed] [Google Scholar]
- 46.D'Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ. 1998. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J. Cell Biol. 140, 603–616. ( 10.1083/jcb.140.3.603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Acker T, Tavernier J, Peelman F.. 2019. The small GTPase Arf6: an overview of its mechanisms of action and of its role in host–pathogen interactions and innate immunity. Int. J. Mol. Sci. 20, 2209. ( 10.3390/ijms20092209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratheesh A, et al. 2012. Centralspindlin and α-catenin regulate Rho signalling at the epithelial zonula adherens. Nat. Cell Biol. 14, 818–828. ( 10.1038/ncb2532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter MV, Lee DM, Harris TJC, Fernandez-Gonzalez R. 2015. Polarized E-cadherin endocytosis directs actomyosin remodeling during embryonic wound repair. J. Cell Biol. 210, 801–816. ( 10.1083/jcb.201501076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds AB. 2007. p120-catenin: past and present. Biochim. Biophys. Acta Mol. Cell Res. 1773, 2–7. ( 10.1016/j.bbamcr.2006.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ireton RC, et al. 2002. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476. ( 10.1083/jcb.200205115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadwell CM, Su W, Kowalczyk AP. 2016. Cadherin tales: regulation of cadherin function by endocytic membrane trafficking. Traffic 17, 1262–1271. ( 10.1111/tra.12448) [DOI] [PubMed] [Google Scholar]
- 53.Wu X, Zhao X, Baylor L, Kaushal S, Eisenberg E, Greene LE. 2001. Clathrin exchange during clathrin-mediated endocytosis. J. Cell Biol. 155, 291–300. ( 10.1083/jcb.200104085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bodin S, Planchon D, Rios Morris E, Comunale F, Gauthier-Rouvière C. 2014. Flotillins in intercellular adhesion – from cellular physiology to human diseases. J. Cell Sci. 127, 5139–5147. ( 10.1242/jcs.159764) [DOI] [PubMed] [Google Scholar]
- 55.Katanaev VL, Solis GP, Hausmann G, Buestorf S, Katanayeva N, Schrock Y, Stuermer CAO, Basler K. 2008. Reggie-1/flotillin-2 promotes secretion of the long-range signalling forms of Wingless and Hedgehog in Drosophila. EMBO J. 27, 509–521. ( 10.1038/sj.emboj.7601981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Babuke T, Ruonala M, Meister M, Amaddii M, Genzler C, Esposito A, Tikkanen R. 2009. Hetero-oligomerization of reggie-1/flotillin-2 and reggie-2/flotillin-1 is required for their endocytosis. Cell. Signal. 21, 1287–1297. ( 10.1016/j.cellsig.2009.03.012) [DOI] [PubMed] [Google Scholar]
- 57.Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 58.Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. 2003. Control of myoblast fusion by a guanine nucleotide exchange factor, Loner, and its effector ARF6. Cell 114, 751–762. ( 10.1016/S0092-8674(03)00720-7) [DOI] [PubMed] [Google Scholar]
- 59.Munjal A, Philippe J-M, Munro E, Lecuit T. 2015. A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature 524, 351–355. ( 10.1038/nature14603) [DOI] [PubMed] [Google Scholar]
- 60.Aigouy B, Umetsu D, Eaton S. 2016. Segmentation and quantitative analysis of epithelial tissues. Methods Mol. Biol. 1478, 227–239. ( 10.1007/978-1-4939-6371-3_13) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.