Abstract
To predict if a threatened species can adapt to changing selective pressures, it is crucial to understand the genetic basis of adaptive traits, especially in species historically affected by severe bottlenecks. We estimated the heritability of three hihi (Notiomystis cincta) morphological traits known to be under selection (nestling tarsus length, body mass and head–bill length) using 523 individuals and 39 699 single nucleotide polymorphisms (SNPs) from a 50 K Affymetrix SNP chip. We then examined the genetic architecture of the traits via chromosome partitioning analyses and genome-wide association scans (GWAS). Heritabilities estimated using pedigree relatedness or genomic relatedness were low. For tarsus length, the proportion of genetic variance explained by each chromosome was positively correlated with its size, and more than one chromosome explained significant variation for body mass and head–bill length. Finally, GWAS analyses suggested many loci of small effect contributing to trait variation for all three traits, although one locus (an SNP within an intron of the transcription factor HEY2) was tentatively associated with tarsus length. Our findings suggest a polygenic nature for the morphological traits, with many small effect size loci contributing to the majority of the variation, similar to results from many other wild populations. However, the small effective population size, polygenic architecture and already low heritabilities suggest that both the total response and rate of response to selection are likely to be limited in hihi.
Keywords: genomic relatedness matrix, chromosome partitioning, genome-wide association, polygenic traits, pedigree study, Notiomystis cincta
1. Introduction
In small populations, genetic diversity is eroded over time through genetic drift, including events such as population bottlenecks and founding events [1]. The continual loss of genetic diversity is of particular interest in species of conservation concern with already low levels of variation. The erosion of genetic diversity may result in the loss of putatively adaptive alleles, leaving the species with low adaptive potential when facing new selection pressures, such as a change in climate or habitat fragmentation [2–4]. However, the complex genomic architecture of traits in non-model organisms is often unknown and, consequently, the underlying evolutionary processes and possible ecological consequences for a population or a species subject to selection cannot be directly assessed [5]. Recently, with the advent of modern and inexpensive sequencing and genotyping technologies, the field of conservation genomics has entered a new era where it is possible to genotype a large number of markers for threatened species with small population sizes [6]. Now, the challenge at hand is to adequately analyse and interpret these genomic snapshots in a way that we can predict the adaptive potential for threatened species and begin to inform conservation management [7,8].
Genomic approaches have already proven to be a powerful tool to better understand the structure and evolution of adaptive traits [9,10]. For example, a closer look at the genetic basis of horn polymorphism in Soay sheep (Ovis aries) revealed that variation is maintained by heterozygote advantage at a single gene [11], which would be impossible to infer from pedigree-based methods alone. Hence, today, many population genomic studies aim to investigate the genetic basis of variation in life-history, fitness and morphological traits in their organism of interest [10]. To make assumptions about adaptive potential, it is crucial to understand the proportion of the phenotypic variation observed in a population (VP) that is explained by the additive genetic component (VA), a factor referred to as narrow-sense heritability (h2) [12]. Traditionally, h2 has been estimated using the expected relatedness between individuals captured by pedigree data. However, those pedigrees may contain errors, not capture all relationships between sampled individuals due to shallow and inconsistent pedigree depth, and only pick up expected, rather than realized, genome sharing between two related individuals [13]. Therefore, it has recently become common practice to instead use genomic data when estimating trait heritabilities [14], for example, via inferring the realized genome sharing between individuals described by a genomic relatedness matrix (GRM). Assuming that marker density is sufficient to accurately capture genomic relatedness, GRM-based heritabilities can be more precise, providing valuable insight into the evolutionary potential of the species [14–16].
In addition to characterizing the overall GRM-based heritability, the heritability can be partitioned across regions of the genome to give further insight into the genomic basis of trait variation [17]. If large chromosomes, containing more genes, explain a more substantial proportion of the genetic variation compared with smaller chromosomes, the trait is likely to be polygenic (i.e. there are many loci, scattered throughout the genome, contributing to trait differences [17,18]). More than one chromosome explaining significant variation also suggests that many loci contribute to trait variation [19].
Another way of investigating the genetic basis of adaptive traits using genomic data involves a genome-wide association scan (GWAS). This detects specific loci with small to large effect on the phenotype of the study species. The power to detect association is dependent, among other factors, on the effect size of the locus. If a trait with significant heritability has no significant GWAS peaks, it suggests that the trait is polygenic, with many loci of small effect contributing to variation [20]. A further hurdle to finding significant genome-wide associations is presented when individuals are closely related, which is commonly the case when sampling from natural populations, particularly those with small population size. The similarity in genetic background must first be adjusted for before testing for an association, which may further reduce the power to detect an association [21].
It appears that a polygenic architecture underlies many evolutionarily significant traits under investigation [16,22]. Variation in these traits is maintained by the balance between genetic drift, mutation and selection, with smaller heritabilities predicted in populations with small effective population size [2]. For these populations, theoretical and simulation results suggest that polygenic adaptation in response to selection is likely to proceed via allele frequency changes at a subset of loci with large effect and/or high allele frequency, rather than frequency shifts across all loci [23]. Variation at other loci is susceptible to be lost via genetic drift and will therefore constrain both the rate of response and the total response to selection [24]. In addition, the evolution of polygenic traits can be further constrained by linkage disequilibrium between deleterious and beneficial alleles, and trade-offs with other (potentially unmeasured) traits [25–27]. Therefore, understanding the genetic architecture of adaptive traits is likely to enable better predictions of the adaptive potential of small populations.
The hihi is an endemic passerine of Aotearoa|New Zealand and is becoming a model species for studying the genetics of endangered birds, and species with small population sizes more generally [4,28,29]. The hihi dataset on the island of Tiritiri Matangi (36°36′ S, 174°53′ E) contains a multi-generational pedigree and morphological and life-history data for every individual born since 1995 when the population was established through reintroduction. Recently, de Villemereuil et al. [4,29] predicted little adaptive potential of this species based on signatures of low genetic diversity, low additive genetic variance of fitness and small pedigree-based heritabilities for traits under selection. All but one life-history trait had very small heritabilities (less than 0.0001); however, there was good support for low but non-zero heritabilities for beak length, body mass and tarsus length [4]. While previous hihi studies have examined the effect of body size on reproduction (e.g. [30]) and survival (e.g. [31]), the large dataset (greater than 2000 birds) analysed by de Villemereuil et al. [4] revealed that all three morphological traits had positive linear selection and negative nonlinear selection gradients. These gradients suggest that there are optimal values for nestling tarsus length, head–bill length and mass that maximize fitness, but that all trait means are smaller than these optima, and hence are under directional selection to increase body size. Morphological traits are commonly recorded in avian studies, with body size directly associated with fitness in many species (reviewed in [32,33]), and are the focus of a growing number of genomic studies (reviewed in [34]).
Here, we used genomic data to reveal previously undiscovered genetic patterns underlying these three adaptive morphological phenotypes in the Tiritiri Matangi population. We employed state-of-the-art software and data for 523 individuals genotyped on a recently generated custom SNP array to generate heritability estimates from pedigree relatedness and genomic relatedness, testing whether a genomic snapshot provides comparable results to a pedigree over many generations for the additive genetic variance component of the traits. To explore the genomic architecture of each trait, we further partitioned the variance explained by the genotyped SNPs across chromosomes to test for a possible polygenic architecture. Finally, we performed a genome-wide association study in order to find loci of small, medium and large effect on any of the traits under investigation. For traits with low heritability, we wanted to discern whether a GWAS could detect loci of large effect, or whether there was evidence that traits are controlled by many loci of small effect, in agreement with studies on many other wild populations. From a conservation management perspective, a low heritability estimate and polygenic makeup of these morphological traits will complement our previous work demonstrating that hihi has little adaptive potential when facing future changes in selection pressures.
2. Methods
(a). Study population and phenotypic data
The hihi was once abundant on the North Island of Aotearoa, until habitat loss and predation by introduced mammals confined its range to only one naturally occurring population on Te Hauturu-o-Toi (Little Barrier Island), in the Hauraki Gulf (36°12′ S, 175°05′ E) [28]. In 1995, the hihi was successfully introduced to the nearby island of Tiritiri Matangi, with a population size of 210 birds prior to the 2019/2020 breeding season [35]. As a result of extensive monitoring since its translocation, we now have a wealth of environmental, life history, fitness, morphological and pedigree data for the population [4]. Hihi do not migrate between the populations and predominantly nest in nest-boxes on Tiritiri Matangi Island. During every breeding season (ranging from October through to February), nestlings are banded and measured at 21 days for tarsus length (mm) and head–bill length (mm), both of which remain unchanged from this stage [36], and also for body mass (g). Blood has been routinely taken (around 60 µl per individual via brachial vein puncture) from nestlings since 2004.
(b). Genotyping and pedigree data
Due to a very high rate of extra-pair copulations in this species, the paternity is determined via routine genotyping of a panel of 19 microsatellite markers, allowing the construction of a multi-generational pedigree for the population (for details, see [29,37]). For nestlings without blood samples available (i.e. those born before 2004), fathers are assumed to be unknown in the pedigree. In addition, in 2016, 1536 birds (from five different hihi populations) were genotyped using a custom 50 K Affymetrix single nucleotide polymorphism (SNP) array [38]. This array was developed based on the identification of SNPs from de novo assembly of restriction site-associated DNA sequencing (RAD-seq) of 31 individuals from the Te Hauturu-o-Toi and Tiritiri Matangi populations, and low-coverage whole-genome sequencing of 10 of these birds (three of which were from Tiritiri Matangi). The flanking sequences of identified variants were aligned to the Ensemble 86 zebra finch (Taeniopygia guttata) genome, which, given the very high synteny of avian genomes [39], is used in the following analyses as a proxy for the distribution of markers in the hihi genome (electronic supplementary material, table S1).
Of the 58 466 SNPs included on the array, 45 553 markers were designated as polymorphic high resolution according to the default quality control metrics in the Axiom Analysis Suite software. PLINK [40] filtering (--maf 0.01, --hwe 1e-20) resulted in the removal of 325 and 69 loci among the autosomes, respectively. Two thousand seven hundred and thirty-four SNPs appeared as duplicates (i.e. mapped to the same chromosome at the same position) and hence only one SNP from each position was retained. To avoid population structure as a confounding factor, we only included birds from Tiritiri Matangi Island in this study, which shows no structure (electronic supplementary material, figure S1). No individual was removed due to low genotyping rates (--mind), but some were excluded for having incomplete phenotypic information. The final dataset contained 39 699 autosomal genomic loci, with a mean call rate of 0.997398, and complete sex and phenotype information on all three traits for 523 birds.
(c). Trait heritabilities
We employed several different methods of retrieving heritability estimates for our three traits under investigation, using generalized linear mixed models (an ‘animal model’ [41]) and either the pedigree relatedness or the relatedness estimated from the genotyped markers (i.e. the GRM). First, pedigree- and GRM-based heritabilities (, ) were estimated in a Bayesian framework using the R [42] package MCMCglmm [43]. To estimate , we fitted the relatedness matrix calculated from the full pedigree. To estimate , the GRM was constructed using an approach that scales by the actual variance in relatedness (approach 3 [18]). Following de Villemereuil [4], the additive genetic variance (VA) for each trait was estimated by fitting sex and clutch size as fixed effects and accounting for the random effects of mother, social father, year and month when the nestling was born. Models were run for 503 000 iterations with a burn-in period of 3000, sampling every 10th output from the chain, and convergence checked graphically and using the Heidelberger and Welch convergence test. The heritability was then calculated as the ratio of VA to VP (total phenotypic variance), where VP and thus h2 accounts for the variance explained by fixed effects [44].
Furthermore, to be able to compare our results with the heritability of morphological traits reported in other avian studies (reviewed in [34]), we also used the ‘prefitModel’ function in the R package RepeatABEL [45]. While RepeatABEL is mainly known for allowing the inclusion of repeated measures, it was chosen as it allows the inclusion of multiple random effects into the model. The GRM-based heritability () was estimated by fitting the same fixed and random effects as above.
Finally, the package BayesR [46] was used to infer the phenotypic variation explained by SNPs in different effect size distributions and to estimate the SNP-based heritability (). The SNP-based heritability differs from the GRM-based heritability as it estimates the proportion of variance accounted for by linear regression on a set of genotyped SNPs [47]. As BayesR does not allow for the inclusion of any fixed or random effects, the raw phenotypes were corrected for those effects in MCMCglmm (same model as for the heritability analysis, but excluding VA) and the residuals then used as phenotypes in BayesR. We set the mean effect sizes of 0.01, 0.001, 0.0001 or 0 for the four mixture distributions and ran the programme for 50 000 iterations, with a burn-in of 20 000, recording every 10th sample in the chain after that (as suggested in the default settings).
(d). Chromosome partitioning of genetic variance
For each chromosome, we estimated the contribution to overall trait variance by fitting a mixed model in MCMCglmm with a GRM estimated from markers on the focal chromosome, a GRM calculated from all other markers in the genome, and accounting for sex, clutch size, mother, social father, year and month of birth. As above, the GRMs were calculated using an approach that scales by the actual variance in relatedness. We ran 203 000 iterations with a burn-in period of 3000, sampling every 10th output from the chain. Owing to convergence issues for chromosomes with fewer than 850 markers, we merged multiple chromosomes based on their size according to the zebra finch karyotypes (electronic supplementary material, table S1). The variance was estimated individually for chromosomes 1–14, 1A and 20 and each merged chromosome group.
We also used GCTA (V1.24) [48] to estimate the variance explained by each chromosome. GRMs were generated with the --make-grm option for each individual chromosome and all but the focal chromosome. Because the model failed to converge when fitting all autosomes simultaneously, we fitted the GRM pairs (e.g. GRM for Chr 1 and GRM for all but Chr 1, using the --mgrm option) in the default REML model one by one to estimate the additive genetic variance explained by each chromosome. Both pathways, however, should theoretically result in the same heritability estimate per autosomal chromosome [48]. Fixed effects included sex and clutch size, and we adjusted for population stratification using the --qcovar function on the top 10 principal components.
For both methods, we used the cor.test function in R to test the significance of a positive Pearson's correlation between chromosome size (based on the zebra finch genome) and variance explained. To account for p-value inflation as a result of heteroscedasticity and censoring, we used 100 000 permutations to calculate corrected p-values following the approach of Kemppainen & Husby [49].
(e). Genome-wide association scan
To test for association between trait and genotype, we used the RepeatABEL package. The ‘prefitModel’ function first fits a model without the SNP effects and computes the variance components for the trait, using the hglm package. SNPs are then tested for association by accounting for these random effects in the ‘rGLS’ function in RepeatABEL, using the same fixed and random effects as in the heritability analysis. p-values for each SNP are computed using the Wald statistics, together with the SNP effect for each marker. We used a Bonferroni correction to control for family-wise error rate from multiple testing (significance value of 0.05 divided by the number of markers, giving a critical p-value of 1.26 × 10−6). All genomic SNPs were tested for association, including those that mapped to random or the Unknown zebra finch chromosomes, or were not mapped to a zebra finch chromosome (which we combine as ‘chromosome 77’; electronic supplementary material, table S1). SNPs of interest were further investigated based on the zebra finch genome annotation in the Ensembl database (version 100.12: bTaeGut1_v1.p [50]). In addition, we calculated the genomic inflation factor (λ, computed using the lower 90% of the distribution), to determine whether population structure was likely to have led to inflation of the test statistics.
3. Results
(a). Trait heritability
The pedigree-based estimates for the heritability of nestling morphological traits tended to be higher than the GRM-based heritabilities, and MCMCglmm GRM-based estimates () were lower than corresponding RepeatABEL heritabilities (; table 1; electronic supplementary material, table S2, table S3 and figure S2). The SNP-based heritabilities from BayesR were 0.117, 0.079 and 0.076, respectively (electronic supplementary material, table S5). All methods agreed on tarsus length being the ‘most heritable’ of the three morphological traits.
Table 1.
Summary table of the outputs (electronic supplementary material, tables S2 and S3) for the trait mean, total phenotypic variance (VP), pedigree-based and GRM-based heritability (h2) estimates and additive genetic variance (VA) for tarsus length (mm), body mass (g) and head–bill length (mm). We report the mean h2 and VA for the MCMCglmm (ped, GRM1) and RepeatABEL (GRM2) estimates for 523 hihi (268 males and 255 females). Credible/confidence intervals (MCMCglmm) or standard error (RepeatABEL) are reported in square brackets.
| trait | mean | VP | h2ped | VA,ped | h2GRM1 | VA,GRM1 | h2GRM2 | VA,GRM2 |
|---|---|---|---|---|---|---|---|---|
| tarsus length | 27.576 | 1.440 | 0.133 [0.028, 0.253] |
0.190 [0.040, 0.362] |
0.077 [1.02 × 10−9, 0.154] |
0.111 [1.39 × 10−9, 0.219] |
0.081 [−0.020, 0.182] |
0.082 [0.054, 0.126] |
| body mass | 37.011 | 43.662 | 0.066 [1.02 × 10−10, 0.151] |
2.880 [4.18 × 10−9, 6.646] |
0.042 [2.44 × 10−10, 0.105] |
1.812 [1.14 × 10−8, 4.548] |
0.065 [−0.031, 0.161] |
2.050 [1.283, 3.276] |
| head–bill length | 38.793 | 2.845 | 0.120 [5.89 × 10−8, 0.252] |
0.343 [1.45 × 10−7, 0.727] |
0.032 [5.15 × 10−16, 0.094] |
0.091 [1.42 × 10−15, 0.270] |
0.070 [−0.028, 0.168] |
0.159 [0.101, 0.251] |
The correlation between off-diagonal elements of the pedigree-based and the GRM-based relatedness matrices was 0.749 (electronic supplementary material, figure S3). A correlation of less than one is expected due to factors such as Mendelian sampling and agrees with other studies (e.g. [15,51,52]).
(b). Chromosome partitioning of genetic variance
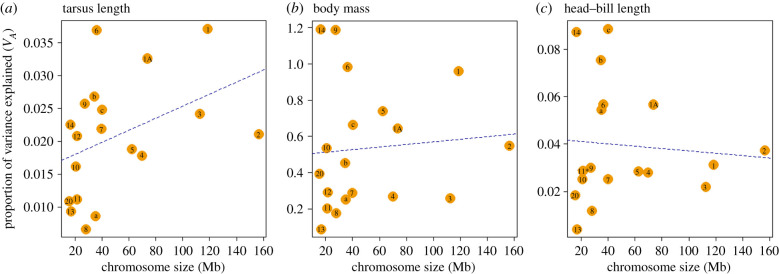
There was evidence that multiple loci contributed to the variance in all three morphological traits (figure 1; electronic supplementary material, figure S4). For tarsus length, we found a positive but non-significant correlation between the proportion of additive genetic variance explained by each chromosome and the chromosome size (HC corrected p-value = 0.113). Chromosomes 1, 1A and 6 explain the most variation, although the largest chromosome (2) explained only moderate variation.
Figure 1.
The variation explained by each chromosome or chromosome composite (in the circles, excluding ‘Chr 77’) for all three traits ((a) tarsus length, (b) body mass and (c) head–bill length) as calculated in MCMCglmm. Plotted is the additive genetic variance explained by each chromosome (VA) against the length of the chromosome (in Mb) based on the zebra finch reference genome. After HC correction, all p-values were non-significant (0.113, 0.422 and 0.574). a = chromosomes 15 and 4A, b = chromosomes 17–19, c = chromosomes 21–28 and 1B (electronic supplementary material, table S1). (Online version in colour.)
For body mass, we found a weak and non-significant positive correlation between chromosome size and phenotypic variance explained (HC corrected p-value = 0.422). Here, chromosomes 14 and 9 explain the most variation.
Finally, for head–bill length, we found a weak negative correlation between the variation and the size of the genomic region. The three largest chromosomes explain relatively little of the overall variation, while chromosome 14, and micro-chromosome composites ‘b’ and ‘c’ each explain the most variation. Chromosome partitioning in GCTA resulted in a comparable picture for all three traits (electronic supplementary material, figure S5).
(c). Genome-wide association scan
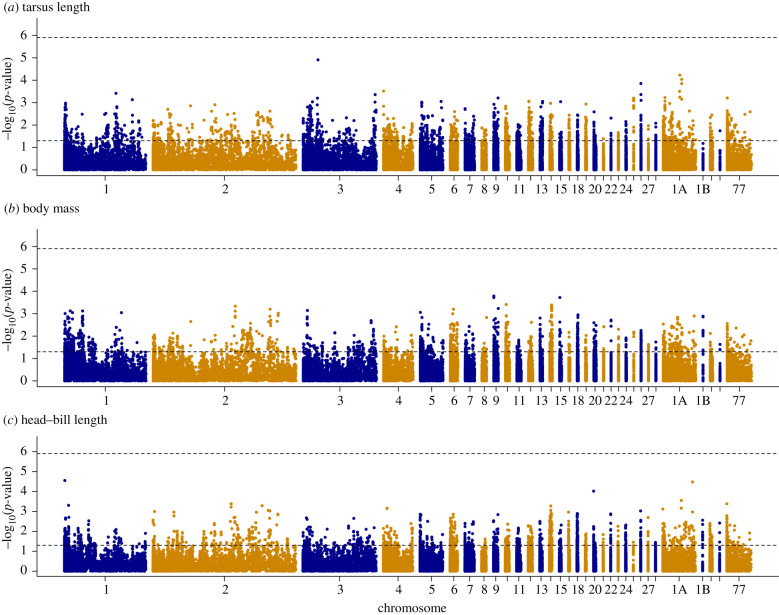
No SNP was significantly associated with the morphological traits after testing for association in RepeatABEL (figure 2). According to the well-curated zebra finch genome (which the hihi chromosome assignment is derived from), the SNP with the highest support for association with tarsus length is located in one of the intronic regions of the HEY2 transcription factor (electronic supplementary material, figure S6), which is involved in cardiac morphogenesis, neurogenesis and somitogenesis [53].
Figure 2.
Manhattan plots for the association between SNPs and (a) tarsus length, (b) body mass and (c) head–bill length in 523 hihi nestlings of Tiritiri Matangi. Top dashed line: Bonferroni significance threshold, bottom dashed line: nominal significance (p-value = 0.05). A description of the chromosome order and naming can be found in the electronic supplementary material, table S1. (Online version in colour.)
For head–bill length, there are two SNPs that, while not significant, show a stronger association with the trait than all others. One of them is placed in an intron of an uncharacterized protein-coding gene (ENSTGUG00000006844) on chromosome 1, the other in between two genes associated with cat eye syndrome (CECR1, CECR2) on chromosome 1A. The third most associated SNP for head–bill length was found on chromosome 20, a chromosome previously associated with bill length [54,55].
The top 10 SNPs with the lowest p-values across all traits can be found in electronic supplementary material, tables S4. We refrained from calculating the additive genetic variance explained by each of the most associated SNPs, as p-values were generally high. The genomic inflation factor values ranged from 0.96 to 1.05 for all three phenotypic traits under investigation (electronic supplementary material, figure S7), suggesting that population structure has been adequately accounted for.
The BayesR analyses confirmed that all traits under investigation appear polygenic, with less than 9% of the SNPs affecting trait variation, the majority of which contributed to the smallest effect size distribution (electronic supplementary material, table S5). A comparison of the SNPs with the lowest p-values from the GWAS showed reasonable agreement with those inferred to have a high probability of inclusion in the largest effect size distribution from BayesR (electronic supplementary material, table S4).
4. Discussion
Adaptive response requires that there is genetic variation in traits linked to fitness, and further, that response to selection is unconstrained by trade-offs with other traits and loci. Here, we examined the genetic basis of variation in nestling tarsus length, body mass and head–bill length in the hihi, a threatened yet well-monitored bird species of Aotearoa|New Zealand, in order to understand whether these traits can respond to current and future selection pressures. Our analyses confirmed low heritability estimates, and support a polygenic basis for all three traits, with no loci of large effect contributing to trait variation. Given the low effective population size and the polygenic architecture, such that adaptive variation lost via drift is unlikely to be replenished by mutation in the short term, we conclude that evolutionary potential in hihi is likely to be constrained even further than the already low heritabilities suggest.
We have three reasons to believe that a polygenic architecture underlies all three morphological traits under investigation: (i) for tarsus length, the variance explained by each chromosome scaled with chromosome size, and for all traits more than one chromosome contributed to variation; (ii) there were no significant SNP associations in the GWAS; and (iii) the majority of loci explaining variation are of small effect size. This study is in good agreement with studies in other passerines (e.g. great tits [56–60], collared flycatchers [22,55,61], house sparrows [55,62], great reed warblers [63], reviewed by Husby et al. [34] as well as studies from other wild animal populations (e.g. [64])), where many loci with small effect are responsible for variation in adaptive phenotypes, rather than a few large-effect loci.
Our pedigree- and GRM-based heritability estimates support a small but non-zero heritability for all traits and agree with previous pedigree heritabilities estimated from a much larger number (greater than 2000) of individuals [4]. Interestingly, the GRM-based heritabilities were lower than the pedigree-based estimates, although the credible intervals are overlapping in all cases. One reason for this may be that our SNP array is not dense enough to have captured true genome sharing more accurately than the pedigree has done. Given the number of SNPs genotyped, this seems unlikely [15], although we note that we have not successfully genotyped SNPs from a number of microchromosomes, including chromosome 16. We acknowledge that, while 523 individuals is a large dataset for a threatened species, our sample size is likely to be somewhat limiting and has certainly contributed to the large credible intervals and standard errors around both the pedigree- and GRM-based heritability estimates, which may also explain some of the differences between estimates. Interestingly, a study of Corsican blue tits (Cyanistes caeruleus [65]) suggests that a sample size of around 500 birds and data from a 50 K SNP chip can outperform heritability estimates from a seven-generation pedigree. The ability to estimate heritabilities without long-term and near-complete monitoring of a population offers exciting opportunity to infer adaptive potential in other wild populations and will be particularly valuable in assessing the vulnerability of endangered species [14]. The sample size, number of markers and low heritabilities is also likely to have limited power to detect association in the GWAS analyses. Linkage disequilibrium between neighbouring markers is moderate (mean of 0.518, calculated with all 523 individuals using PLINK --r2 and reporting all pairwise values with --ld_window_r2 0.0) and suggests that power to detect a locus explaining all of the trait heritability ranges from 0.306 to 0.516 [66] (electronic supplementary material, table S6). As expected, nestling mass, with the lowest heritability due to substantial maternal and month effects compared to the other traits [4], has the lowest power to detect association.
Morphological traits are often the subject of wild bird studies, as they can reveal evolutionary patterns over time and in turn shed light on the adaptive response of traits known to correlate with fitness [32,33]. The three morphological traits studied here are all under selection as previous analyses suggest that the population mean of all three traits is smaller than the fitness optimum [4]. Although there was no significant association detected in the GWAS, it is interesting to note the regions that were most highly associated with tarsus length and head–bill length. The tentative association of a locus on chromosome 3 with tarsus length and of variants on chromosomes 1 and 1A with head–bill length does not appear to be replicated in some studies of passerine body size [55,59], while Lawson & Petren [67] highlight a region on chromosome 1A in increased linkage with regard to beak shape in Darwin finches (Geospiza). Additionally, the third-highest association for head–bill length, an SNP on chromosome 20, stands within an intron of NFS1 cysteine desulfurase and is 7–12 Mb distant from QTLs previously associated with beak morphology [54,55]. The challenges in identifying significant loci in a genome-wide association analysis include low SNP density and small sample sizes [68], both of which are likely to apply to the hihi study system to some degree. All the above-mentioned loci are interesting candidates for further study if sample size and SNP density in these regions can be increased. Creating a larger sample size through combining all monitored populations could improve the precision of estimates, and shed more light on the effects of translocations on trait variation and responses to selection.
Despite the low heritabilities of the traits, our genomic analyses have provided important further insights for hihi, and threatened species more generally. A more nuanced understanding of the genomic architecture of a trait under selection helps assess the likely speed of the adaptive response in a species and the longer-term total response to selection [23,24]. We have demonstrated that many loci of small effect are likely to contribute the majority of variation to all three morphological traits. Given the small effective population size of all hihi populations, many adaptive variants are likely to be lost to drift at a rate that is faster than the input of new variation via mutation (reviewed in [69,70]). This is likely to slow the speed of adaptation and lead to further constraint on the adaptive potential of the species. Most importantly, other wild populations may be facing similar threats as the hihi, highlighting the need for continued collection and use of genomic resources to aid conservation measures. We therefore encourage further characterization of the heritability and genomic architecture for species that face anthropogenic threats including habitat decimation and climate change, in order to better predict their evolutionary potential to adapt to such challenges [71]. Finally, we acknowledge that future studies will need to go beyond describing quantitative parameters in order to successfully combine genomic insights with active species conservation, including informing management actions that can buffer species with small population sizes from the impacts of low adaptive potential.
Supplementary Material
Acknowledgements
We extend many thanks to the volunteers, past students and Department of Conservation staff who have contributed to monitoring the Tiritiri Matangi hihi population, and to the Hihi Recovery Group, Department of Conservation and Supporters of Tiritiri Matangi for maintaining such a long-term vision in monitoring and management of this population, with a special thank you to the current hihi conservation officer Mhairi McCready. We acknowledge Ngati Manuhiri as Mana Whenua and Kaitiaki of Te Hauturu-o-Toi and its taonga, including hihi. Many thanks to Rachel Tucker and Jon Slate at the NERC Biomolecular Analysis Facility, University of Sheffield, for completing DNA extractions for the RAD-seq data and to Johanna Nielsen and Kang-Wook Kim for help with microsatellite genotyping. We acknowledge the use of New Zealand eScience Infrastructure (NeSI) high-performance computing facilities and greatly appreciated help from Catarina Silva and Lars Rönnegård with RepeatABEL. Finally, we are very grateful to associate editor Nancy Chen, Arild Husby and an anonymous reviewer for their insightful and helpful comments on an earlier version of the manuscript.
Ethics
Permissions to conduct research and collect blood samples on Tiritiri Matangi were granted by the New Zealand Department of Conservation, permit numbers 36186-FAU, 15073-RES, 24128-FAU, 13939-RES and 44300-FAU.
Data accessibility
Supporting figures and tables are provided in the electronic supplementary material. Genotype and phenotype data, along with R code to run the heritability, chromosome partitioning and GWAS analyses, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.98sf7m0g1 [72].
Competing interests
We declare we have no competing interests.
Funding
P.d.V., A.W.S., K.D.L., P.B. and J.G.E. were supported by a Marsden Grant (grant no. UOA1408) awarded to A.W.S. from the New Zealand Royal Society Te Aparangi. A.W.S. was also supported by a New Zealand National Science Challenge Biological Heritage Project Grant, Project 1.4. P.B. and the pedigree genotyping were also supported by a ZSL Fellowship. L.D. is supported by a University of Auckland Doctoral Scholarship.
References
- 1.Bouzat JL. 2010. Conservation genetics of population bottlenecks: the role of chance, selection, and history. Conserv. Genet. 11, 463–478. ( 10.1007/s10592-010-0049-0) [DOI] [Google Scholar]
- 2.Hoffmann AA, Sgrò CM, Kristensen TN. 2017. Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 32, 506–517. ( 10.1016/j.tree.2017.03.012) [DOI] [PubMed] [Google Scholar]
- 3.Chen N, Cosgrove EJ, Bowman R, Fitzpatrick JW, Clark AG. 2016. Genomic consequences of population decline in the endangered Florida scrub-jay. Curr. Biol. 26, 2974–2979. ( 10.1016/j.cub.2016.08.062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Villemereuil P, Rutschmann A, Lee KD, Ewen JG, Brekke P, Santure AW. 2019. Little adaptive potential in a threatened passerine bird. Curr. Biol. 29, 889–894. ( 10.1016/j.cub.2019.01.072) [DOI] [PubMed] [Google Scholar]
- 5.Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820. ( 10.1038/nrg3522) [DOI] [PubMed] [Google Scholar]
- 6.Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. 2013. Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22, 2841–2847. ( 10.1111/mec.12350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P. 2014. Using genomics to characterize evolutionary potential for conservation of wild populations. Evol. Appl. 7, 1008–1025. ( 10.1111/eva.12149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafer AB, et al. 2015. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30, 78–87. ( 10.1016/j.tree.2014.11.009) [DOI] [PubMed] [Google Scholar]
- 9.Pardo-Diaz C, Salazar C, Jiggins CD. 2015. Towards the identification of the loci of adaptive evolution. Methods Ecol. Evol. 6, 445–464. ( 10.1111/2041-210X.12324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luikart G, Kardos M, Hand BK, Rajora OP, Aitken SN, Hohenlohe PA. 2018. Population genomics: advancing understanding of nature. In Population genomics (ed. Rajora OP.), pp. 3–79. Cham, Switzerland: Springer. [Google Scholar]
- 11.Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM, Slate J. 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502, 93–95. ( 10.1038/nature12489) [DOI] [PubMed] [Google Scholar]
- 12.Falconer DS, Mackay TF. 1996. Introduction to quantitative genetics. Harlow, UK: Longmans Green. [Google Scholar]
- 13.Visscher PM, Medland SE, Ferreira MA, Morley KI, Zhu G, Cornes BK, Montgomery GW, Martin NG. 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2, e41 ( 10.1371/journal.pgen.0020041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gienapp P, Fior S, Guillaume F, Lasky JR, Sork VL, Csilléry K. 2017. Genomic quantitative genetics to study evolution in the wild. Trends Ecol. Evol. 32, 897–908. ( 10.1016/j.tree.2017.09.004) [DOI] [PubMed] [Google Scholar]
- 15.Bérénos C, Ellis PA, Pilkington JG, Pemberton JM. 2014. Estimating quantitative genetic parameters in wild populations: a comparison of pedigree and genomic approaches. Mol. Ecol. 23, 3434–3451. ( 10.1111/mec.12827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen H, Szulkin M, Slate J. 2014. Molecular quantitative genetics. In Quantitative genetics in the wild (eds Charmantier A, Garant D, Kruuk LEB), pp. 209–227. Oxford, UK: Oxford University Press. [Google Scholar]
- 17.Yang J, et al. 2011. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519 ( 10.1038/ng.823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MR, Santure AW, DeCauwer I, Sheldon BC, Slate J. 2013. Partitioning of genetic variation across the genome using multimarker methods in a wild bird population. Mol. Ecol. 22, 3963–3980. ( 10.1111/mec.12375) [DOI] [PubMed] [Google Scholar]
- 19.Kemppainen P, Husby A. 2018. Inference of genetic architecture from chromosome partitioning analyses is sensitive to genome variation, sample size, heritability and effect size distribution. Mol. Ecol. Resour. 18, 767–777. ( 10.1111/1755-0998.12774) [DOI] [PubMed] [Google Scholar]
- 20.Manolio TA, et al. 2009. Finding the missing heritability of complex diseases. Nature 461, 747–753. ( 10.1038/nature08494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. 2014. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46, 100 ( 10.1038/ng.2876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kardos M, Husby A, McFarlane SE, Qvarnström A, Ellegren H. 2016. Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol. Ecol. Resour. 16, 727–741. ( 10.1111/1755-0998.12498) [DOI] [PubMed] [Google Scholar]
- 23.Höllinger I, Pennings PS, Hermisson J. 2019. Polygenic adaptation: from sweeps to subtle frequency shifts. PLoS Genet. 15, e1008035 ( 10.1371/journal.pgen.1008035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John S, Stephan W. 2020. Important role of genetic drift in rapid polygenic adaptation. Ecol. Evol. 10, 1278–1287. ( 10.1002/ece3.5981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294. ( 10.1017/S0016672300010156) [DOI] [PubMed] [Google Scholar]
- 26.Hospital F, Chevalet C. 1996. Interactions of selection, linkage and drift in the dynamics of polygenic characters. Genet. Res. 67, 77–87. ( 10.1017/S0016672300033498) [DOI] [PubMed] [Google Scholar]
- 27.Lande R. 1982. A quantitative genetic theory of life history evolution. Ecology 63, 607–615. ( 10.2307/1936778) [DOI] [Google Scholar]
- 28.Brekke P, Bennett PM, Santure AW, Ewen JG. 2011. High genetic diversity in the remnant island population of hihi and the genetic consequences of re-introduction. Mol. Ecol. 20, 29–45. ( 10.1111/j.1365-294X.2010.04923.x) [DOI] [PubMed] [Google Scholar]
- 29.de Villemereuil P, Rutschmann A, Ewen JG, Santure AW, Brekke P. 2019. Can threatened species adapt in a restored habitat? No expected evolutionary response in lay date for the New Zealand hihi. Evol. Appl. 12, 482–497. ( 10.1111/eva.12727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brekke P, Ewen JG, Clucas G, Santure AW. 2015. Determinants of male floating behaviour and floater reproduction in a threatened population of the hihi (Notiomystis cincta). Evol. Appl. 8, 796–806. ( 10.1111/eva.12287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low M, Pärt T. 2009. Patterns of mortality for each life-history stage in a population of the endangered New Zealand stitchbird. J. Anim. Ecol. 78, 761–771. ( 10.1111/j.1365-2656.2009.01543.x) [DOI] [PubMed] [Google Scholar]
- 32.Merilä J, Sheldon BC. 2001. Avian quantitative genetics. In Current ornithology (eds Nolan V, Ketterson E), pp. 179–255. Boston, MA: Springer. [Google Scholar]
- 33.Gardner JL, Peters A, Kearney MR, Joseph L, Heinsohn R. 2011. Declining body size: a third universal response to warming? Trends Ecol. Evol. 26, 285–291. ( 10.1016/j.tree.2011.03.005) [DOI] [PubMed] [Google Scholar]
- 34.Husby A, McFarlane SE, Qvarnström A. 2019. Avian population studies in the Genomic Era. In Avian genomics in ecology and evolution (ed. Kraus R.), pp. 267–293. Cham, Switzerland: Springer. [Google Scholar]
- 35.Parlato EH, Ewen JG, McCready M, Parker KA, Armstrong DP. In preparataion. A modelling framework for integrating reproduction, survival and count data when projecting the fates of threatened populations. [DOI] [PubMed] [Google Scholar]
- 36.Low M. 2006. Sex, age and season influence morphometrics in the New Zealand Stitchbird (or Hihi; Notiomystis cincta). Emu 106, 297–304. ( 10.1071/MU06003) [DOI] [Google Scholar]
- 37.Brekke P, Wang J, Bennett PM, Cassey P, Dawson DA, Horsburgh GJ, Ewen JG. 2012. Postcopulatory mechanisms of inbreeding avoidance in the island endemic hihi (Notiomystis cincta). Behav. Ecol. 23, 278–284. ( 10.1093/beheco/arr183) [DOI] [Google Scholar]
- 38.Lee KD, Millar CD, Brekke P, Ewen JG, Santure AW. doi: 10.1111/1755-0998.13480. In preparation. The design and application of a 50K SNP microarray for a threatened Aotearoa | New Zealand passerine, the hihi. [DOI] [PubMed] [Google Scholar]
- 39.Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320. ( 10.1126/science.1251385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson AJ, Reale D, Clements MN, Morrissey MM, Postma E, Walling CA, Kruuk LE, Nussey DH. 2010. An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26. ( 10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- 42.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 43.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 44.de Villemereuil P, Morrissey MB, Nakagawa S, Schielzeth H. 2018. Fixed-effect variance and the estimation of repeatabilities and heritabilities: issues and solutions. J. Evol. Biol. 31, 621–632. ( 10.1111/jeb.13232) [DOI] [PubMed] [Google Scholar]
- 45.Rönnegård L, McFarlane SE, Husby A, Kawakami T, Ellegren H, Qvarnström A. 2016. Increasing the power of genome wide association studies in natural populations using repeated measures—evaluation and implementation. Methods Ecol. Evol. 7, 792–799. ( 10.1111/2041-210X.12535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moser G, Lee SH, Hayes BJ, Goddard ME, Wray NR, Visscher PM. 2015. Simultaneous discovery, estimation and prediction analysis of complex traits using a Bayesian mixture model. PLoS Genet. 11, e1004969 ( 10.1371/journal.pgen.1004969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de los Campos G, Sorensen D, Gianola D. 2015. Genomic heritability: what is it? PLoS Genet. 11, e1005048 ( 10.1371/journal.pgen.1005048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Lee SH, Goddard ME, Visscher PM. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. ( 10.1016/j.ajhg.2010.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kemppainen P, Husby A. 2018. Accounting for heteroscedasticity and censoring in chromosome partitioning analyses. Evol. Lett. 2, 599–609. ( 10.1002/evl3.88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham F, et al. 2019. Ensembl 2019. Nucleic Acids Res. 47, D745–D751. ( 10.1093/nar/gky1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pryce JE, Hayes BJ, Goddard ME. 2012. Novel strategies to minimize progeny inbreeding while maximizing genetic gain using genomic information. J. Dairy Sci. 95, 377–388. ( 10.3168/jds.2011-4254) [DOI] [PubMed] [Google Scholar]
- 52.Galla SJ, et al. 2020. A comparison of pedigree, genetic, and genomic estimates of relatedness for informing pairing decisions in two critically endangered birds: implications for conservation breeding programmes worldwide. Evol. Appl. 13, 991–1008. ( 10.1111/eva.12916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihara D, et al. 2020. Expression of Hey2 transcription factor in the early embryonic ventricles is controlled through a distal enhancer by Tbx20 and Gata transcription factors. Dev. Biol. 461, 124–131. ( 10.1016/j.ydbio.2020.02.001) [DOI] [PubMed] [Google Scholar]
- 54.Knief U, Schielzeth H, Kempenaers B, Ellegren H, Forstmeier W. 2012. QTL and quantitative genetic analysis of beak morphology reveals patterns of standing genetic variation in an Estrildid finch. Mol. Ecol. 21, 3704–3717. ( 10.1111/j.1365-294X.2012.05661.x) [DOI] [PubMed] [Google Scholar]
- 55.Silva CN, et al. 2017. Insights into the genetic architecture of morphological traits in two passerine bird species. Heredity 119, 197–205. ( 10.1038/hdy.2017.29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santure AW, De Cauwer I, Robinson MR, Poissant J, Sheldon BC, Slate J. 2013. Genomic dissection of variation in clutch size and egg mass in a wild great tit (Parus major) population. Mol. Ecol. 22, 3949–3962. ( 10.1111/mec.12376) [DOI] [PubMed] [Google Scholar]
- 57.Santure AW, et al. 2015. Replicated analysis of the genetic architecture of quantitative traits in two wild great tit populations. Mol. Ecol. 24, 6148–6162. ( 10.1111/mec.13452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gienapp P, Laine VN, Mateman AC, van Oers K, Visser ME. 2017. Environment-dependent genotype-phenotype associations in avian breeding time. Front. Genet. 8, 102 ( 10.3389/fgene.2017.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosse M, et al. 2017. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science 358, 365–368. [DOI] [PubMed] [Google Scholar]
- 60.Kim JM, et al. 2018. A high-density SNP chip for genotyping great tit (Parus major) populations and its application to studying the genetic architecture of exploration behaviour. Mol. Ecol. Resour. 18, 877–891. ( 10.1111/1755-0998.12778) [DOI] [PubMed] [Google Scholar]
- 61.Husby A, Kawakami T, Rönnegård L, Smeds L, Ellegren H, Qvarnström A. 2015. Genome-wide association mapping in a wild avian population identifies a link between genetic and phenotypic variation in a life-history trait. Proc. R. Soc. B 282, 20150156 ( 10.1098/rspb.2015.0156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lundregan SL, et al. 2018. Inferences of genetic architecture of bill morphology in house sparrow using a high-density SNP array point to a polygenic basis. Mol. Ecol. 27, 3498–3514. ( 10.1111/mec.14811) [DOI] [PubMed] [Google Scholar]
- 63.Hansson B, Sigeman H, Stervander M, Tarka M, Ponnikas S, Strandh M, Westerdahl H, Hasselquist D. 2018. Contrasting results from GWAS and QTL mapping on wing length in great reed warblers. Mol. Ecol. Resour. 18, 867–876. ( 10.1111/1755-0998.12785) [DOI] [PubMed] [Google Scholar]
- 64.Bérénos C, Ellis PA, Pilkington JG, Lee SH, Gratten J, Pemberton JM. 2015. Heterogeneity of genetic architecture of body size traits in a free-living population. Mol. Ecol. 24, 1810–1830. ( 10.1111/mec.13146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrier C, Delahaie B, Charmantier A. 2018. Heritability estimates from genomewide relatedness matrices in wild populations: application to a passerine, using a small sample size. Mol. Ecol. Resour. 18, 838–853. ( 10.1111/1755-0998.12886) [DOI] [PubMed] [Google Scholar]
- 66.Wang M, Xu S. 2019. Statistical power in genome-wide association studies and quantitative trait locus mapping. Heredity 123, 287–306. ( 10.1038/s41437-019-0205-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lawson LP, Petren K. 2017. The adaptive genomic landscape of beak morphology in Darwin's finches. Mol. Ecol. 26, 4978–4989. ( 10.1111/mec.14166) [DOI] [PubMed] [Google Scholar]
- 68.Santure AW, Garant D. 2018. Wild GWAS—association mapping in natural populations. Mol. Ecol. Resour. 18, 729–738. ( 10.1111/1755-0998.12901) [DOI] [PubMed] [Google Scholar]
- 69.Johnson T, Barton N. 2005. Theoretical models of selection and mutation on quantitative traits. Phil. Trans. R. Soc. B 360, 1411–1425. ( 10.1098/rstb.2005.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willi Y, Van Buskirk J, Hoffmann AA. 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458. ( 10.1146/annurev.ecolsys.37.091305.110145) [DOI] [Google Scholar]
- 71.Gienapp P, Brommer JE. 2014. Evolutionary dynamics in response to climate change. In Quantitative genetics in the wild (eds Charmantier A, Garant D, Kruuk LEB), pp. 254–273. Oxford, UK: Oxford University Press. [Google Scholar]
- 72.Duntsch L, Tomotani BM, de Villemereuil P, Brekke P, Lee KD, Ewen JG, Santure AW. 2020. Data from: Polygenic basis for adaptive morphological variation in a threatened Aotearoa | New Zealand bird, the hihi (Notiomystis cincta) Dryad Digital Repository. ( 10.5061/dryad.98sf7m0g1) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Duntsch L, Tomotani BM, de Villemereuil P, Brekke P, Lee KD, Ewen JG, Santure AW. 2020. Data from: Polygenic basis for adaptive morphological variation in a threatened Aotearoa | New Zealand bird, the hihi (Notiomystis cincta) Dryad Digital Repository. ( 10.5061/dryad.98sf7m0g1) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Supporting figures and tables are provided in the electronic supplementary material. Genotype and phenotype data, along with R code to run the heritability, chromosome partitioning and GWAS analyses, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.98sf7m0g1 [72].