Abstract
The gut microbiome plays a critical role in the health of many animals. Honeybees are no exception, as they host a core microbiome that affects their nutrition and immune function. However, the relationship between the honeybee immune system and its gut symbionts is poorly understood. Here, we explore how the beneficial symbiont Snodgrassella alvi affects honeybee immune gene expression. We show that both live and heat-killed S. alvi protect honeybees from the opportunistic pathogen Serratia marcescens and lead to the expression of host antimicrobial peptides. Honeybee immune genes respond differently to live S. alvi compared to heat-killed S. alvi, the latter causing a more extensive immune expression response. We show a preference for Toll pathway upregulation over the Imd pathway in the presence of both live and heat-killed S. alvi. Finally, we find that live S. alvi aids in clearance of S. marcescens from the honeybee gut, supporting a potential role for the symbiont in colonization resistance. Our results show that colonization by the beneficial symbiont S. alvi triggers a replicable honeybee immune response. These responses may benefit the host and the symbiont, by helping to regulate gut microbial members and preventing overgrowth or invasion by opportunists.
Keywords: honeybees, microbiome, innate immunity, symbiosis, colonization resistance, immune priming
1. Introduction
Honeybees (Apis mellifera) harbour a distinctive gut microbiome that is key to their health [1]. Having co-evolved with social bees for over 80 million years, 95% of gut-dwelling organisms fall within nine species clusters of host-specific bacteria spatially organized within bee hindgut compartments [2–4]. The honeybee gut provides an enticing model system for studying host–microbe interactions and understanding the mechanisms by which gut bacteria influence their hosts [5].
Snodgrassella alvi colonizes the honeybee ileum and grows in contact with the gut epithelia. Cells of another core gut species, Gilliamella apicola, grow on top of this S. alvi layer to form a dense biofilm [2,6]. Honeybees with perturbed gut communities die at higher rates when challenged by the opportunistic pathogen Serratia marcescens [7]. Thus, the bee gut community protects the host from infection. This protection, or ‘colonization resistance', is one of the most widespread benefits provided by symbiotic communities to hosts, including mammalian hosts [8,9]. However, the mechanism of this protection in bees is unclear. Microbiome-derived protection may be caused by active symbiont colonization that physically blocks or antagonizes pathogens.
The dense biofilm formed by S. alvi could block pathogen access to host epithelial cells and sequester nutrients [6]. S. alvi may directly antagonize invaders with its type VI secretion system and diverse array of effectors [10]. Additionally, metabolism by bee gut microbial members lowers gut lumen pH and oxygen levels [11], and produces short-chain fatty acids (SCFAs) which can inhibit pathogen virulence and growth in mice [12]. Finally, immune priming may also be responsible, a process by which bacteria activate the host innate immune system, making the host more resistant to subsequent bacterial encounters [1,12].
Insect antibacterial immunity relies heavily on the Toll and Imd pathways of the innate immune system [13,14]. These pathways are best-studied in Drosophila melanogaster, in which Toll receptors and peptidoglycan recognition proteins (PGRPs) react to bacterial motifs [13]. For Toll, binding of the endogenous ligand Spaetzle with the Toll receptor triggers the degradation of Cactus, an inhibitor of the NF-kB transcription factor Dorsal [15]. The Imd pathway is triggered by direct binding of peptidoglycan with PGRPs, signalling cleavage of a self-inhibitory region of the NF-kB-like transcription factor Relish by the protein Dredd [16]. Dorsal and Relish then translocate into the host nucleus and upregulate immune effectors, including antimicrobial peptides (AMPs)—short peptides that perforate bacterial membranes and inhibit protein folding [14,17]. Honeybees encode orthologues for all core proteins of these pathways as well as a unique ensemble of AMPs: abaecin, apidaecin, defensin and hymenoptaecin [18–22].
Honeybees upregulate expression of apidaecin in response to S. alvi colonization [23,24]. Combined with previous data supporting symbiont-based pathogen protection, these results support a role for S. alvi in honeybee immune priming. However, neither study measured transcriptional responses of fat bodies—the centre of the insect immune response [14]. Additionally, previous studies detailing this protective effect used only live bacterial inoculations, so they were unable to separate the impacts of colonization and immune priming.
In this study, we investigated the role S. alvi plays in activating the honeybee immune system and protecting hosts from pathogens. Using both live and heat-killed bacteria, we show that colonization is not required for pathogen protection, but aids in pathogen clearance, and that heat-killed S. alvi causes a more extensive immune expression response than the live symbiont. These results suggest that S. alvi is not only priming the honeybee immune system but also potentially modulating it.
2. Methods
(a). Bacterial strains
Escherichia coli strain MG1655 was grown at 37°C on LB agar. S. alvi strain wkB2 and S. marcescens strain N10 were grown as previously described, on Columbia agar supplemented with sterile sheep blood at 35°C and 5% CO2 [2]. S. alvi and S. marcescens isolates were previously derived from honeybee guts.
(b). Preparation of heat-killed cells
Overnight growths of E. coli and S. alvi on agar plates were pooled individually and suspended in 500 μl PBS. The OD600 of bacterial cultures was measured, and the amount of S. alvi and E. coli used for heat-killing was diluted to an OD representative of 5 × 108 CFUs. Heat-killed cells were prepared by then suspending S. alvi or E. coli in 1 ml of PBS and heating to 80°C. Cells were left at 80°C for 30 min for S. alvi and 10 min for E. coli. Direct plating of heat-killed bacterial suspensions confirmed that no live cells were present.
(c). Honeybee collection and containment
All Apis mellifera samples were obtained from hives at the University of Texas at Austin. To obtain microbiota-free bees, dark-eyed pupae were removed from capped brood cells using sterilized forceps and placed in sterile plastic cages as previously described [25]. Bees obtained this way are ‘microbiota-free' and lack the usual gut bacterial species [26]. Newly emerged bees were fed sterile pollen and 1 : 1 filter-sterilized sucrose water until fully matured 3 days later. Bees were randomly assigned to groups of approximately 20 adult bees and then chilled at 4°C and placed in 50 ml centrifuge tubes corresponding to their treatment group. Bee groups were inoculated by feeding with 1 ml filter-sterilized sucrose water (in a 1 : 1 mix of sucrose and water) or 800 μl filter-sterilized sucrose water and 200 μl of their bacterial or heat-killed bacterial treatment. Bees were then transferred to plastic cup cages and fed sterile pollen. Each cage was given 10 ml of 1 : 1 filter-sterilized sucrose water mixed with their respective treatment.
(d). Gene expression analysis
Bees were sampled on their respective days post bacterial inoculation and moved into 15 ml centrifuge tubes to be frozen at −80°C. Abdomens were removed from each bee and placed into 600 μl of RNA lysis buffer and pestle homogenized. RNA was then isolated using a Zymo Research Quick-RNA Tissue/Insect Microprep Kit (catalogue no. R2030). RNA was eluted in 50 μl of RNase-free water and stored at −80°C. Concentrations were quantified using a Thermo Fisher Scientific NanoDrop Lite Spectrophotometer. RNA was reverse-transcribed using a QuantaBio qScript cDNA Synthesis Kit (catalogue no. 95047-500) with all reactions normalized to 500 ng μl−1 of input RNA. Relative gene expression was determined using quantitative PCR. The source of primers used in qPCR assays can be found in electronic supplementary material, table S1. Absolute quantification of the housekeeping gene RPS18 was accomplished by cloning RPS18 into the Promega pGEM-T vector (catalogue no. A1360). Primer efficiency was determined using 10-fold serial dilutions of primer standards. Standards for target genes not cloned into a pGEM-T vector were created by PCR amplification of the control group cDNA with target gene qPCR primers. Reactions were run in an Eppendorf MasterCycler Realplex machine using technical replicates for each sample and Bio-Rad iTaq Universal SYBR Green Supermix (catalogue no. 1725121) for fluorescence. Relative gene expression for the genes, abaecin, apidaecin, hymenoptaecin, cactus-1, cactus-2, dorsal, relish, dredd, pgrp-lc, pirk and toll, were determined using a 2-ΔΔCt method and log2 transformed [27].
(e). Survival and clearance assays
Microbiota-free bees were fed their respective bacterial, heat-killed bacterial or PBS treatment. Five days post-inoculation bees were chilled and placed in 0.5 ml microcentrifuge tubes, each with a hole cut in the bottom. Each bee was then hand-fed 5 μl of a mixture of 20% sucrose and 80% PBS or this same mixture plus a 24 h growth of S. marcescens diluted to an OD600 of 1 for survival assays and 0.5 for clearance assays. In survival assays, bees were then returned to their respective cup cages and the number of dead bees was recorded daily for 10 days. Survival data were pooled from three independent trials. A Cox proportional hazard model was used to test any effect independent trials might have on bee survivorship (electronic supplementary material, figure S1). In the clearance assay, whole guts were extracted from bees 1 day and 3 days post-inoculation with S. marcescens. These guts were immediately homogenized into 200 μl PBS. We prepared serial dilutions and spot-plated 10 μl of each dilution onto LB agar plates. S. marcescens colonies were counted after 1 day at 37°C in aerobic conditions and total CFUs per gut calculated. Less than 10 discernible CFUs were found in bees given no bacterial exposure.
(f). Statistical analysis
Survival curves were created using the survminer and survival packages in RStudio (https://rstudio.com/) [28,29]. Curves are based on a Kaplan–Meier fit [30]. The Forrest plot was based on a Cox proportional hazard model [31]. Statistical analysis was accomplished using a pairwise log-rank test with a Benjamini–Hochberg correction for multiple testing [32,33]. Gene expression graphs were created using the ggplot2 library in RStudio [34]. Statistical analysis for gene expression assays and S. marcescens clearance assays was accomplished using the Tukey honest significant difference method [35].
3. Results
(a). Heat-killed Snodgrassella alvi protects against the pathogen Serratia marcescens
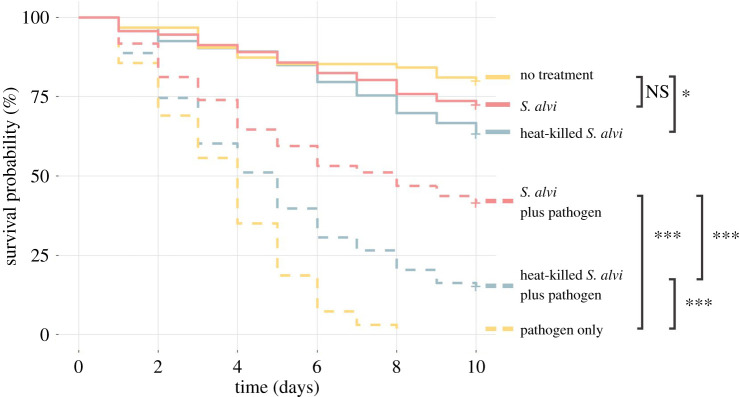
Honeybees containing core microbiome members have higher survival following challenge with the opportunistic pathogen S. marcescens [7]. This protection may happen by physically blocking pathogen colonization, actively killing or suppressing pathogens, or priming the host immune system. Therefore, to determine if symbiont colonization is required for protection, microbiota-free honeybees were treated with live S. alvi, heat-killed S. alvi or sterile sugar syrup. Five days post-treatment, each bee was hand-fed S. marcescens or sterile PBS. Across treatments, bees receiving no pathogen had higher survival than bees challenged with the pathogen (figure 1). Bees inoculated with live S. alvi showed higher survival compared to those lacking S. alvi when challenged with S. marcescens (figure 1). Despite lacking the ability to colonize the gut, heat-killed S. alvi still provided significant protection compared to bees not exposed to any form of S. alvi. But this protection was less than that provided by live S. alvi (figure 1). Additionally, bees inoculated with heat-killed S. alvi experienced lower survival, suggesting a possible adverse effect to this treatment (figure 1). These trends are consistent across individual experiments (electronic supplementary material, figure S1). Thus, colonization is not required for improved survival after pathogen challenge.
Figure 1.
Inoculation with live and heat-killed S. alvi provides pathogen protection. Survival of bees from different treatment groups after inoculation with PBS or 5 μl of the pathogen S. marcescens at OD600 = 1. When challenged with S. marcescens, bees previously treated with live S. alvi or heat-killed S. alvi survived better than bees given no treatment prior to pathogen exposure. p-values obtained using a log-rank test with Benjamini–Hochberg correction. Total N = 570 bees across three replicate experiments. *** = p < 0.001, ** = p < 0.01, * = p < 0.05, NS = not significant. (Online version in colour.)
(b). Both live and heat-killed Snodgrassella alvi trigger upregulation of antimicrobial peptides
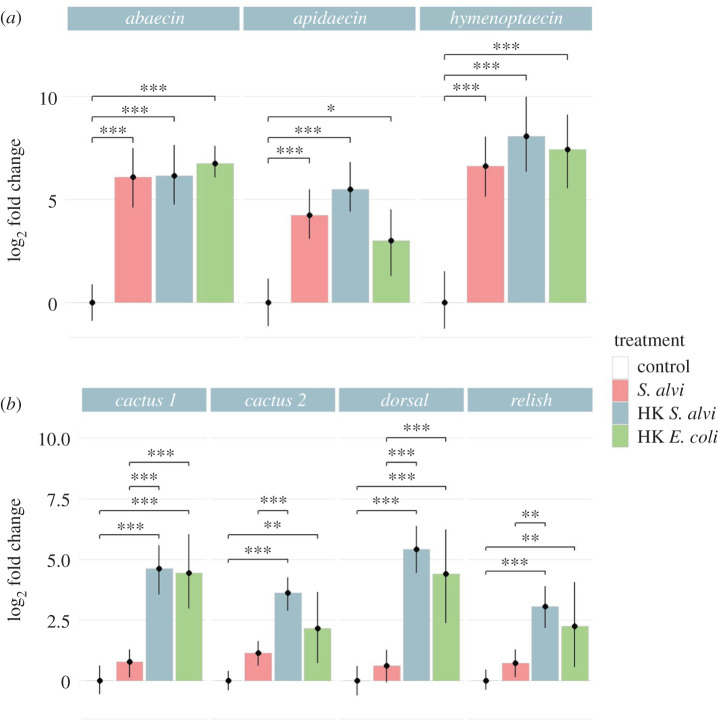
Protection against S. marcescens by heat-killed S. alvi supports immune priming as a factor in pathogen defence. To determine if live and dead S. alvi trigger a host immune response, we treated microbiota-free bees with live S. alvi, heat-killed S. alvi and heat-killed E. coli and assessed expression of immune genes 5 days later. While S. alvi is a co-evolved symbiont of honeybees, E. coli is not a typical bee gut microbiome member. However, both bacteria are Gram-negative. Therefore, by including heat-killed E. coli we could assess whether gene expression responses were general to Gram-negative bacterial components or if they were symbiont-specific. In all treatments, we found significant upregulation of the AMPs abaecin, apidaecin and hymenoptaecin relative to uninoculated microbiota-free bees (figure 2a). In a replicated experiment, sampling bees 1, 2 and 5 days post-inoculation, we saw similar upregulation, suggesting that this response spans sampling dates (electronic supplementary material, figure S2A). These results show that both live and dead S. alvi trigger an immune response and that this response is not symbiont-specific, as heat-killed E. coli treatments show similar upregulation of AMPs.
Figure 2.
Live and heat-killed S. alvi trigger differential host immune gene expression. Bee gene expression relative to the housekeeping gene RPS18 measured from bee whole abdomens 5 days post-treatment. HK = heat-killed. (a) Bees inoculated with live S. alvi, heat-killed S. alvi, and heat-killed E. coli trigger upregulation of AMPs compared to the uninoculated control group. (b) Bees inoculated with heat-killed S. alvi or E. coli had significantly higher expression of immune regulatory genes than the control or live S. alvi groups. Total N = 73 bees from one hive. *** = p < 0.001, ** = p < 0.01, * = p < 0.05. p-values found using Tukey honest significant difference method. (Online version in colour.)
(c). Heat-killed Snodgrassella alvi trigger a more extensive immune expression response than live Snodgrassella alvi
In Drosophila, Dorsal and Relish play critical roles as transcription factors in the expression of AMPs [14,17]. To further explore the upregulation of immune effectors following exposure to live or dead S. alvi, we measured relative gene expression via qPCR of dorsal, relish and cactus after inoculation with live S. alvi, heat-killed S. alvi or heat-killed E. coli. Surprisingly, bees inoculated with live S. alvi did not have as high upregulation compared to both heat-killed treatments (Figure 2b). In a replicated experiment, sampling bees 1, 2 and 5 days post-inoculation, we saw similar patterns (electronic supplementary material, figure S2B). There appears to be no correlation between transcriptional regulator gene expression and AMP gene expression, as all treatment groups had similarly elevated AMP expression regardless of dorsal or relish expression (figure 2a,b). Interestingly, while S. alvi contains components sufficient for immune regulator upregulation, live cells do not trigger the same magnitude of response found in heat-killed treated groups.
(d). A mixture of live and heat-killed Snodgrassella alvi lowers the expression of Imd pathway genes while upregulating toll pathway components
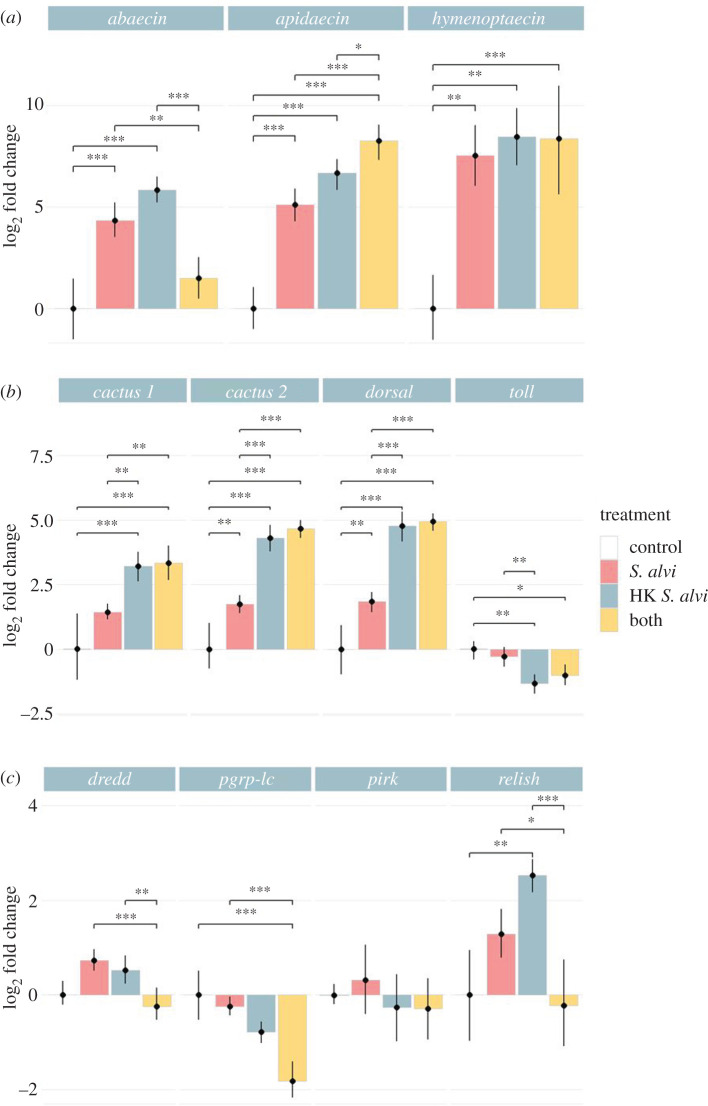
To further discern the impact of S. alvi on bee immune gene expression, we expanded our gene targets to include the receptor toll and pgrp-lc, as well as the Imd regulatory genes pirk and dredd. In Drosophila sp. Pirk is known to inhibit the function of the Imd pathway while Dredd modifies Relish, triggering its translocation to the nucleus [16,36]. Additionally, we added a treatment containing an equal mixture of live and heat-killed S. alvi. If live S. alvi alone fails to upregulate immune genes, we would expect signals generated by heat-killed S. alvi to dominate. However, if live S. alvi actively suppresses host immune gene upregulation, we would expect a gene expression pattern more comparable to live S. alvi alone. While live and heat-killed S. alvi treatments on their own confirmed previous results, our mixed treatment did not favour one or the other (figure 3).
Figure 3.
Loss of host Imd pathway gene expression post-inoculation with both live and heat-killed S. alvi. Bee gene expression relative to the housekeeping gene RPS18 measured using qPCR from cDNA derived from bee whole abdomens 5 days post-treatment. HK = heat-killed. (a) Bees treated with live or heat-killed S. alvi trigger AMP upregulation. Mixed treatment of both live and heat-killed S. alvi triggers upregulation of apidaecin and hymenoptaecin, but not abaecin, compared to the control group. (b) Bees treated with live S. alvi, heat-killed S. alvi, or both show upregulation of Toll pathway genes cactus and dorsal compared to the control group. Bees treated with heat-killed S. alvi or both live and heat-killed S. alvi trigger downregulation of the gene toll. (c) Bees treated with both live and heat-killed S. alvi show lower expression of Imd pathway genes dredd, pgrp-lc, and relish compared to other groups. Total N = 48 bees from one hive. *** = p < 0.001, ** = p < 0.01, * = p < 0.05. p-values obtained using Tukey honest significant difference method. (Online version in colour.)
We saw considerable upregulation of AMPs apidaecin and hymenoptaecin but lower levels of abaecin in the mixed treatment group (figure 3a). For the Toll pathway genes toll, cactus and dorsal, the mixed treatment group favoured expression similar to treatment with heat-killed S. alvi alone (figure 3b). While we found no change in pirk levels, the expression of the Imd pathway genes dredd, pgrp-lc, and relish in bees treated with both live and heat-killed S. alvi was reduced compared to live or heat-killed treatments alone (figure 3c). These results show a context-dependent loss of Imd pathway gene expression when bees were treated with a mixture of S. alvi and heat-killed S. alvi. Additionally, these results suggest that live S. alvi can reduce the expression of Imd pathway genes activated by heat-killed S. alvi.
(e). Live Snodgrassella alvi aids in the clearance of the pathogen S. marcescens while heat-killed Snodgrassella alvi does not
Both live and heat-killed S. alvi increase host survival following subsequent challenge with S. marcescens, and both trigger differential immune gene expression. To determine if these treatments could aid in S. marcescens clearance, microbiota-free bees were inoculated with live S. alvi, heat-killed S. alvi or a mixture of the two, then challenged with S. marcescens. Treatment with live S. alvi reduced S. marcescens CFUs both 1 day and 3 days after pathogen challenge (figure 4). Bees treated with a mixture of live and heat-killed S. alvi showed a similar reduction (figure 4). However, bees fed only heat-killed S. alvi showed no significant reduction in pathogen CFUs (figure 4). These findings indicate that colonization by live S. alvi boosts pathogen resistance, increasing bee survival and suppressing pathogen proliferation. However, pre-treatment with heat killed S. alvi supports tolerance, increasing bee survival but failing to reduce pathogen numbers.
Figure 4.
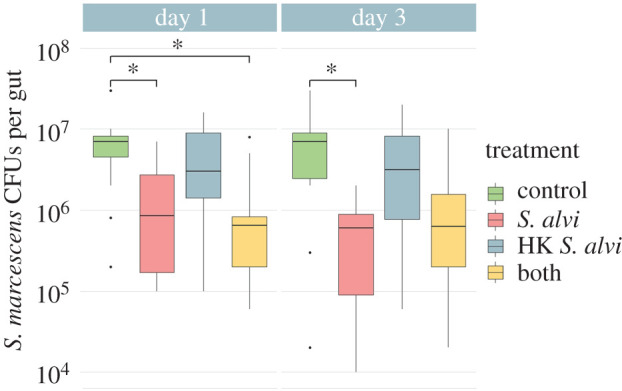
Live S. alvi aids in the clearance of the pathogen S. marcescens while heat-killed S. alvi does not. S. marcescens CFUs of different treatment groups 1 and 3 days post-inoculation with 5 μl of S. marcescens at OD600 = 0.5. HK = heat-killed. Bees previously treated with live S. alvi and a mixture of live and heat-killed S. alvi showed significant reduction of S. marcescens CFUs on Day 1 and bees treated with live S. alvi continued this trend at Day 3. Bees treated with only heat-killed S. alvi showed no significant reduction of S. marcescens CFUs compared to controls. Total N = 91 bees from one hive. * = p < 0.05. p-values obtained using Tukey honest significant difference method. (Online version in colour.)
4. Discussion
Increased survival after pathogen challenge in honeybees treated with heat-killed S. alvi suggests that immune priming underlies at least part of this symbiont's protective effect (figure 1). Inoculation with live S. alvi, however, leads to higher survival after pathogen challenge, similar AMP upregulation, and increased pathogen clearance. These results together suggest that S. alvi's colonization of the gut ileum's epithelial wall plays a key role in colonization resistance (figures 1, 2a and 4). These results reinforce earlier findings that S. alvi induces the upregulation of immune genes [23,24]. We observed greater upregulation of AMP genes than documented in previous studies, possibly due to our inclusion of bee fat bodies in our assays. Additionally, our results confirm previous studies showing immune priming by heat-killed bacterial cells in insects [37]. In our case, a commensal species provides priming against a pathogenic species, whereas in most studies priming is achieved through exposure to heat-killed pathogenic cells [37].
Comparing bees treated with heat-killed versus live S. alvi, the former had the greater upregulation of immune regulatory genes but similar AMP expression (figure 2). However, mRNA levels do not always correspond to protein levels, due to post-transcriptional and post-translational regulatory processes [38]. While little is known about Dorsal and Relish in bees, post-translational regulation of these proteins has been extensively documented in Drosophila.
Binding of the inhibitor Cactus prevents the transcription factor Dorsal from translocating to the nucleus [13]. Activation of the Toll pathway causes phosphorylation, ubiquitination and degradation of Cactus [13]. The observed similarities in dorsal and cactus upregulation may signify that higher expression of dorsal is matched by an increase in Cactus inhibitors preventing increases in AMP expression (figures 2b and 3b).
In the Drosophila Imd pathway, Relish activation requires both cleavage and phosphorylation, carried out by other enzymes. Cleavage and phosphorylation are required for translocation to the nucleus and recruitment of RNA polymerase II [14,39]. Therefore, the transcriptional upregulation of relish may not lead to a heightened host immune response if these other regulatory processes are hindered. Future studies should investigate how post-transcriptional processes affect expression and should quantify immune gene protein levels in response to these treatments.
Both resistance and tolerance play roles in pathogen defence in animals [40–42]. Resistance reduces pathogen abundance, while tolerance only limits the impact of the pathogen [40–42]. Live and heat-killed S. alvi both enhance survival following S. marcescens exposure and upregulate immune genes, but only live S. alvi or a mixed treatment aids in pathogen clearance (figures 1 and 4). These results suggest that S. alvi plays a role in resistance beyond immune gene upregulation, potentially by direct antagonism or by out-competing invading pathogens for space and resources. However, a protective host response does not imply a fitness benefit.
In Drosophila, exposure to Salmonella typhimurium or Listeria monocytogenes triggers anorexia and altered immune responses, increasing tolerance to the former while decreasing resistance to the latter [43]. Similarly, Mycobacterium marinum activates the Drosophila immune system, causing the dysregulation of metabolic pathways leading to host wasting and mortality [44]. In bees, lower survivorship following treatment with heat-killed bacteria may indicate a cost for high immune activation (figure 1). Furthermore, lower gene expression in bees treated with live S. alvi could reflect reduced investment in costly immune responses made superfluous by the presence of this defensive symbiont.
Bacterial biofilms can prevent the immune system from recognizing bacterial components and clearing an infection [45]. However, we know little about host–biofilm interactions in insects. In tsetse flies, the commensal symbiont Sodalis glossinidius requires the outer membrane protein OmpA to form a biofilm and colonize the fly gut [46]. The knockout of OmpA causes the clearance of Sodalis by the host immune system [46]. Similarly, S. alvi requires outer membrane proteins for its colonization of the honeybee, forming a biofilm directly on the gut epithelia of the ileum [6,47]. Like Sodalis, this biofilm may prevent targeting of S. alvi by immune effectors.
The honeybee gut harbours a diverse community of lytic phages [48,49]. It is likely that S. alvi lyses during colonization and growth, however, the dynamics of S. alvi and phage interactions remains unclear. Regardless, the biofilm produced by S. alvi may prevent lysed bacterial components and host immune receptors from coming into contact. Heat-killed cells cannot form a biofilm, and their components likely spread throughout the gut, interacting with membrane receptors in regions not typically associated with S. alvi. These features of heat-killed cells could explain the higher induction by heat-killed S. alvi and the mixed treatment for Toll pathway genes.
In the mixed treatment group, bees were fed both live and heat-killed cells simultaneously. Under this simultaneous exposure, heat-killed bacterial components may interact with immune receptors before S. alvi can form a biofilm or in places where S. alvi does not colonize. Future experiments should compare priority effects between live and heat-killed treatments to determine the impact of S. alvi colonization prior to feeding with heat-killed cells and vice versa. Additionally, using S. alvi mutants deficient for biofilm production would help to study the biofilm's significance in immune activation [50].
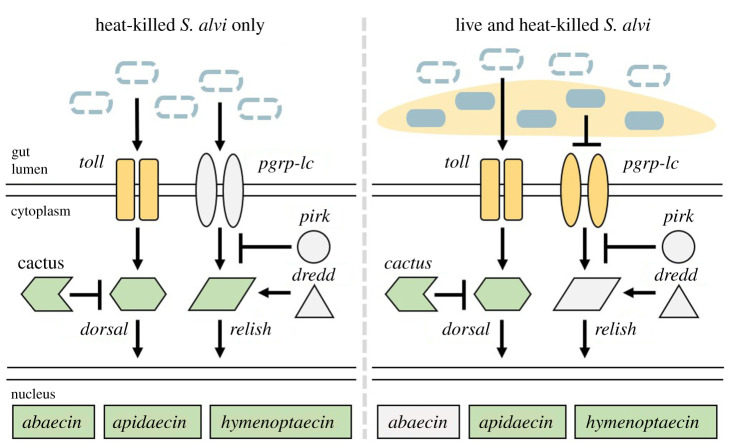
In Drosophila, Gram-negative bacteria typically trigger the Imd pathway, while Gram-positive bacteria trigger the Toll pathway [51,52]. Bees given either live or heat-killed S. alvi show similar expression patterns for Imd pathway genes, while live S. alvi treated bees have lower expression of Toll pathway genes (figure 3). Once live S. alvi is mixed with heat-killed cells, Toll pathway gene expression favours that of bees given heat-killed S. alvi alone. However, we find a sharp drop in Imd pathway gene expression (figures 3c and 5). Therefore, live S. alvi appears capable of reducing expression of Imd pathway genes (figure 5). Knockdown of immune gene levels by RNAi could clarify the roles of Imd and Toll components as well as possibly expose avenues for host–symbiont crosstalk [50].
Figure 5.
Proposed effect of live S. alvi on host gene expression. Bees treated with heat-killed S. alvi (dashed blue ovals) show high upregulation of Toll and Imd pathway components. Addition of live S. alvi (filled blue ovals) to heat-killed S. alvi triggers a reduction in Imd pathway expression while leaving Toll pathway expression unaffected. Pale yellow denotes S. alvi-produced biofilm. Pathway proteins and their localizations inferred from Drosophila melanogaster. Toll pathway genes: toll, cactus, dorsal. Imd pathway genes: pgrp-lc, pirk, dredd, relish. Antimicrobial peptides: abaecin, apidaecin, hymenoptaecin. Yellow = lower gene expression relative to control bees. Green = higher gene expression relative to control bees. Grey = no differential gene expression relative to control bees. (Online version in colour.)
Coevolution of bees and S. alvi over an approximately 80 million-year period has likely resulted in highly specific host–symbiont interaction networks [2]. For example, S. alvi is highly resistant to apidaecin, which is present in the gut lumen after inoculation with gut microbes [24]. This antimicrobial resistance may represent a coevolutionary response, allowing the host to manipulate its microbiome while leaving beneficial symbionts unharmed. Limitations in our knowledge of honeybee immune pathways have made investigations of these networks difficult. Future studies will require clarification of molecular mechanisms underlying honeybee immune responses. While this study raises new questions, we show replicable differential responses to the symbiont S. alvi by the host immune system. We propose that the gut symbiont S. alvi can possibly modulate the host immune system.
Supplementary Material
Supplementary Material
Acknowledgements
We thank J. Elijah Powell for protocols and experimental assistance, Kim Hammond for laboratory support and bee hive maintenance, and Margaret I. Steele for bacterial OD600 enumeration curves as well as assistance with S. marcescens clearance assay design.
Data accessibility
All data used in this study can be found in the electronic supplementary material, dataset provided.
Authors' contributions
All authors assisted in study design and manuscript revisions. Manuscript draft written by R.D.H. Experiments performed by R.D.H. Data analysis performed by R.D.H. and S.P.L.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by National Institutes of Health award R35GM131738 to N.A.M. and by a University of Texas Undergraduate Research Fellowship to R.H.
References
- 1.Engel P, Moran NA. 2013. The gut microbiota of insects — diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. ( 10.1111/1574-6976.12025) [DOI] [PubMed] [Google Scholar]
- 2.Kwong WK, Moran NA. 2012. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 2008–2018. ( 10.1099/ijs.0.044875-0) [DOI] [PubMed] [Google Scholar]
- 3.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. ( 10.1038/nrmicro.2016.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong WK, Medina LA, Koch H, Sing K-W, Soh EJY, Ascher JS, Jaffé R, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci. Adv. 3, e1600513 ( 10.1126/sciadv.1600513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Steele MI, Leonard SP, Motta EVS, Moran NA. 2018. Honey bees as models for gut microbiota research. Lab. Anim. (NY) 47, 317–325. ( 10.1038/s41684-018-0173-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840. ( 10.1128/aem.07810-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymann K, Shaffer Z, Moran NA. 2017. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15, e2001861 ( 10.1371/journal.pbio.2001861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spees AM, Lopez CA, Kingsbury DD, Winter SE, Bäumler AJ. 2013. Colonization resistance: battle of the bugs or ménage à trois with the host? PLoS Pathog. 9, e1003730 ( 10.1371/journal.ppat.1003730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 279, 90–105. ( 10.1111/imr.12563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steele MI, Kwong WK, Whiteley M, Moran NA. 2017. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 8, e01630-17 ( 10.1128/mbio.01630-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl Acad. Sci. USA 114, 4775–4780. ( 10.1073/pnas.1701819114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickard JM, Zeng MY, Caruso R, Núñez G. 2017. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. ( 10.1111/imr.12567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanji T, Ip YT. 2005. Regulators of the toll and imd pathways in the Drosophila innate immune response. Trends Immunol. 26, 193–198. ( 10.1016/j.it.2005.02.006) [DOI] [PubMed] [Google Scholar]
- 14.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. ( 10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 15.Brennan CA, Anderson KV. 2004. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22, 457–483. ( 10.1146/annurev.immunol.22.012703.104626) [DOI] [PubMed] [Google Scholar]
- 16.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. 2003. Caspase-mediated processing of the Drosophila NF-B factor relish. Proc. Natl Acad. Sci. USA 100, 5991–5996. ( 10.1073/pnas.1035902100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe K-M. 2002. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362. ( 10.1126/science.1070216) [DOI] [PubMed] [Google Scholar]
- 18.Evans JD, et al. 2006. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect. Mol. Biol. 15, 645–656. ( 10.1111/j.1365-2583.2006.00682.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danihlík J, Aronstein K, Petřivalský M. 2015. Antimicrobial peptides: a key component of honey bee innate immunity. J. Apicultural Res. 54, 123–136. ( 10.1080/00218839.2015.1109919) [DOI] [Google Scholar]
- 20.Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. 1989. Apidaecins: antibacterial peptides from honeybees. EMBO J. 8, 2387–2391. ( 10.1002/j.1460-2075.1989.tb08368.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casteels P, Ampe C, Riviere L, Damme J, Elicone C, Fleming M, Jacobs F, Tempst P. 1990. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 187, 381–386. ( 10.1111/j.1432-1033.1990.tb15315.x) [DOI] [PubMed] [Google Scholar]
- 22.Casteels P, Ampe C, Jacobs F, Tempst P. 1993. Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J. Biol. Chem. 268, 7044–7054. [PubMed] [Google Scholar]
- 23.Emery O, Schmidt K, Engel P. 2017. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 26, 2576–2590. ( 10.1111/mec.14058) [DOI] [PubMed] [Google Scholar]
- 24.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4, 170003 ( 10.1098/rsos.170003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans JD, Chen YP, di Prisco G, Pettis J, Williams V. 2009. Bee cups: single-use cages for honey bee experiments. J. Apicult. Res. 48, 300–302. ( 10.1080/00218839.2009.11101548) [DOI] [Google Scholar]
- 26.Powell JE, Martinson VG, Urban-Mead K, Moran NA. 2014. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387. ( 10.1128/aem.01861-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 28.Kassambara A, Kosinski M, Biecek P.. 2019. survminer: Drawing survival curves using ‘ggplot2’. R package version 0.4.6. https://CRAN.R-project.org/package=survminer.
- 29.Therneau TM, Grambsch PM. 2000. Modeling survival data: extending the Cox model. New York, NY: Springer. [Google Scholar]
- 30.Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457–481. ( 10.1080/01621459.1958.10501452) [DOI] [Google Scholar]
- 31.Cox DR. 1992. Regression models and life-tables. In Springer series in statistics, pp. 527–541. New York, NY: Springer. [Google Scholar]
- 32.Mantel N. 1966. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 50, 163–170. [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. ( 10.1111/j.2517-6161.1995.tb02031.x) [DOI] [Google Scholar]
- 34.Wickham H. 2016. Ggplot2 elegant graphics for data analysis. New York, NY: Springer International Publishing. [Google Scholar]
- 35.Tukey J. W. 1977. Exploratory data analysis. Reading, MA: Addison-Wesley Publishing Company. [Google Scholar]
- 36.Kleino A, Myllymäki H, Kallio J, Vanha-aho L-M, Oksanen K, Ulvila J, Hultmark D, Valanne S, Rämet M. 2008. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 180, 5413–5422. ( 10.4049/jimmunol.180.8.5413) [DOI] [PubMed] [Google Scholar]
- 37.Cooper D, Eleftherianos I. 2017. Memory and specificity in the insect immune system: current perspectives and future challenges. Front. Immunol 8, 539 ( 10.3389/fimmu.2017.00539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Beyer A, Aebersold R. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550. ( 10.1016/j.cell.2016.03.014) [DOI] [PubMed] [Google Scholar]
- 39.Erturk-Hasdemir D, et al. 2009. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl Acad. Sci. USA 106, 9779–9784. ( 10.1073/pnas.0812022106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raberg L, Sim D, Read AF. 2007. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 318, 812–814. ( 10.1126/science.1148526) [DOI] [PubMed] [Google Scholar]
- 41.Read AF, Graham AL, Råberg L. 2008. Animal defenses against infectious agents: is damage control more important than pathogen control. PLoS Biol. 6, e1000004 ( 10.1371/journal.pbio.1000004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider DS, Ayres JS. 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8, 889–895. ( 10.1038/nri2432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayres JS, Schneider DS. 2009. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7, e1000150 ( 10.1371/journal.pbio.1000150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. 2006. Akt and foxo dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 16, 1977–1985. ( 10.1016/j.cub.2006.08.052) [DOI] [PubMed] [Google Scholar]
- 45.Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. 2015. How biofilms evade host defenses. Microbiol. Spectrum 3, 1–10. ( 10.1128/microbiolspec.mb-0012-2014) [DOI] [PubMed] [Google Scholar]
- 46.Maltz MA, Weiss BL, O'Neill M, Wu Y, Aksoy S. 2012. OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl. Environ. Microbiol. 78, 7760–7768. ( 10.1128/aem.01858-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl Acad. Sci. USA 113, 13 887–13 892. ( 10.1073/pnas.1610856113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deboutte W, Beller L, Yinda CK, Maes P, de Graaf DC, Matthijnssens J.. 2020. Honey-bee-associated prokaryotic viral communities reveal wide viral diversity and a profound metabolic coding potential. Proc. Natl Acad. Sci. USA 117, 10 511–10 519. ( 10.1073/pnas.1921859117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonilla-Rosso G, Steiner T, Wichmann F, Bexkens E, Engel P. 2020. Honey bees harbor a diverse gut virome engaging in nested strain-level interactions with the microbiota. Proc. Natl Acad. Sci. USA 117, 7355–7362. ( 10.1073/pnas.2000228117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leonard SP, et al. 2018. Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7, 1279–1290. ( 10.1021/acssynbio.7b00399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. 1995. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl Acad. Sci. USA 92, 9465–9469. ( 10.1073/pnas.92.21.9465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemaitre B, Reichhart J-M, Hoffmann JA. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl Acad. Sci. USA 94, 14 614–14 619. ( 10.1073/pnas.94.26.14614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leonard SP, et al. 2020. Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576. ( 10.1126/science.aax9039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tesovnik T, Cizelj I, Zorc M, Čitar M, Božič J, Glavan G, Narat M. 2017. Immune related gene expression in worker honey bee (Apis mellifera carnica) pupae exposed to neonicotinoid thiamethoxam and Varroa mites (Varroa destructor). PLoS ONE 12, e0187079 ( 10.1371/journal.pone.0187079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lourenço AP, Guidugli-Lazzarini KR, Freitas FCP, Bitondi MMG, Simões ZLP. 2013. Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect. Biochem. Mol. Biol. 43, 474–482. ( 10.1016/j.ibmb.2013.03.001) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study can be found in the electronic supplementary material, dataset provided.