Abstract
A subset of patients with Covid-19 presents with negative RT-PCR screening but suspect CT findings. Using four commercially available anti-SARS-CoV-2 IgG immuno-assays, we found this subset constituted 9.2% of all consecutively admitted outpatients with Covid-19 in our hospital. Clinical specificity for Covid-19 of some N protein-based immuno-assays was suboptimal, as positive results were observed in control patients with recent common human coronavirus, influenza B and adenovirus infections.
Electronic supplementary material
The online version of this article (10.1007/s15010-020-01523-3) contains supplementary material, which is available to authorized users.
Keywords: ELISA, Covid-19, SARS-CoV-2, Serology
Introduction
Coronavirus disease 19 (Covid-19) is a viral illness caused by the recently emerged novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In contrast to the common human coronaviruses (NL63, 229E, OC43 and HKU1), which are associated with mild respiratory syndromes, SARS-CoV-2 may cause a severe lower respiratory tract infection.
The first available diagnostic test for Covid-19, moved upfront by the WHO, was RT-PCR with virus-specific primers and probes on upper respiratory tract samples. Two or more genetic targets should be included to obtain optimal specificity, certainly in regions where the prevalence is low. Its sensitivity dependents on the disease course, as viral loads are decreasing fast in the naso- and oropharynx after 5–7 days of illness [1]. RT-PCR testing of deeper respiratory samples might be useful in the second stage of the disease. However, bronchoalveolar lavage (BAL) procedures are avoided because of the risk of aerosol generation, putting the operator at risk for nosocomial transmission. CT thorax has proven to be an additional diagnostic tool, with high sensitivity during the second phase of the disease (day 7–14). Although typical radiologic features have been described, certain overlap is present with other respiratory pathogens or drug-related lung disease [2]. More recently, anti-IgM, anti-IgA and anti-IgG SARS-CoV-2 responses against the Nucleocapsid (N) and Spike (S) viral structural proteins have been detected after already 4 days of illness [3, 4]. Thus, the detection of these antibodies has emerged as a third diagnostic tool.
Since CT features of Covid-19 lack optimal specificity and BAL procedures impose the operator at significant risk, we hypothesized a specific serologic test would be a useful test in patients which present with a suspect CT image but negative RT-PCR screening. Here, we describe IgG SARS-CoV-2 responses measured by four commercial immuno-assays in a consecutive series of these patients. To determine the specificity of our test strategy, we tested sera from patients with common respiratory infections.
Methods
Patients
Patients were included in the AZ Sint-Jan Brugge-Oostende, a 1182-bed acute- and tertiary-care hospital in Belgium consisting of three separate campuses. From the beginning of week 13 (March 9 2020) to the end of week 16 (April 5 2020) 127 consecutive outpatients (median age 68 years, range 1–91, 61% male) were admitted because of high clinical suspicion of moderate or severe Covid-19 to a designated ward in attendance of their tests results. One-hundred eight patients had a positive screening with RT-PCR on a nasopharyngeal swab. One patient of this series, 10 years of age, tested positive for human coronavirus HKU1. The 18 remaining patients with negative RT-PCR test on the first nasopharyngeal swab were included in this study. According to local medical procedures, these patients underwent CT scan of the thorax and a serologic test for Covid-19 was ordered after the negative result of the RT-PCR was known. Additional serum samples from different time points were tested for SARS-CoV-2 IgG in case these were available in the local serum bank. CT scans were reviewed by one observer (K.V.D.M.) and scored according to the CO-RADS (COVID-19 Reporting and Data System). The day of self-reported onset of symptoms was determined upon review of the clinical file.
To evaluate the specificity of the immuno-assays, we used a set of serum samples from control patients with recent respiratory viral or atypical bacterial infections. The set included infections with coronavirus OC43 (n = 6), NL-63 (n = 1), HKU1 (n = 5), 229E (n = 2), OC43 and HKU1 (n = 1), RSV-A (n = 4), RSV-B (n = 2), adenovirus (n = 5), Influenza A (n = 5), Influenza B (n = 5), PIV-1 (n = 4), PIV-4 (n = 2), Mycoplasma pneumoniae (n = 5) and Chlamydia pneumoniae (n = 5). This study was approved by the ethical committee of AZ Sint-Jan Brugge-Oostende AV (numbers 2658 and 2665).
RT-PCR for SARS-Cov-2 and multiplex testing for other respiratory viruses
Specific RT-PCR testing for semi-quantitative determination of SARS-CoV-2 was performed by a laboratory-developed assay. Nucleic acids were extracted from clinical samples using a DNA Mini Kit on QiaSymphony (Qiagen, Hilden, Germany). As target genes for amplification, we used the N and RdRp (RNAse-dependent RNA-polymerase) genes. Additionally, as an extraction and inhibition control a phoque distemper virus was used. Multiparameter testing for 35 respiratory pathogens was performed using an in house Taqman Array Card as described previously [5].
Serologic immuno-assays
SARS-CoV-2 IgG was measured by 3 ELISA kits and 1 chemiluminescent microparticle immuno-assay (CMIA) kit according to the instructions of the manufacturer (respectively NovaTec Immundiagnostica GmbH, Dietzenbach, Germany; Vircell S.L., Granada, Spain and Euroimmun AG, Lübeck, Germany; and CMIA on Architect-I System from Abbott, Sligo, Ireland). These assays contain different antigens: recombinant structural S protein (Euroimmun), recombinant structural N protein (Novatec and Abbott), and a combination of recombinant structural S and N proteins (Vircell).
Results were interpreted as positive, negative of borderline according to the instructions of the manufacturer.
Results
From the beginning of week 13 (March 9 2020) to the end of week 16 (April 5 2020) 18 consecutive patients with CT finding suspect for Covid-19 but negative nasopharyngeal RT-PCR screening were admitted to our hospital. The clinical and radiological characteristics of these patients are shown in Supplementary Table 1. No other respiratory pathogens were found in these patients upon admission. Since the screening of the nasopharynx may yield false-negative results due to technical reasons, nearly all patients (17/18) were retested at least once, generating negative results. We also screened stool samples with RT-PCR in 7 patients, one of which had a positive test.
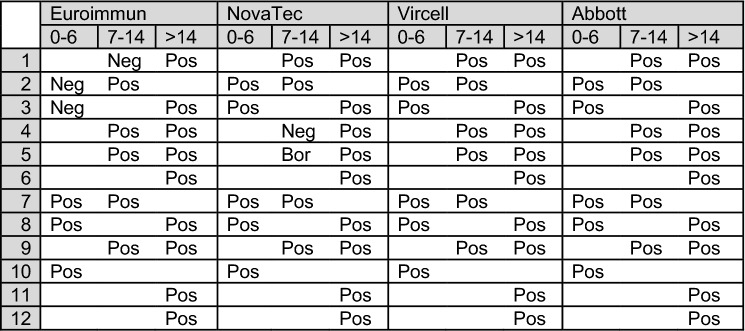
To evaluate the diagnostic value of anti-SARS-Cov-2 IgG in our study population, we used four different immuno-assays, three of which were ELISA-based. We included assays which use recombinant structural N protein, S protein or both as antigen, as previous studies have shown analytical sensitivity and specificity of ELISA assays may depend on the nature of the used antigen [4]. One patient died soon after admission and its serologic response could not be evaluated. Analyzing the available serum samples with the longest disease duration, 12 out of the 17 remaining patients tested positive with all 4 immuno-assays, including the patient with a positive RT-PCR screening on a stool sample. Figure 1 shows the results of these assays per patient, in the function of the day of self-reported onset of symptoms. The CO-RADS score was not significantly different between patients with a positive or negative test. Upon review of the clinical files of the 5 patients testing negative with all assays, we found an alternative diagnosis for the CT findings in 2 patients (recent bleeding from oropharyngeal tumor and cardiac decompensation). In 2 other patients, the rapid beneficial clinical course, and the absence of raised serum ferritin, raised troponin and lymphopenia strongly argued against Covid-19 [6]. Finally, one patient with a doubtful diagnosis underwent bronchoscopy and BAL, generating a negative RT-PCR test result for SARS-CoV-2. Thus, determination of SARS-CoV-2 IgG was essential for diagnosis of Covid-19 in 11 out of 120 (9.2%, 95% CI 5.2–16) consecutively admitted patients in our hospital. Concerning clinical sensitivity, we observed a delay of the seroconversion of 1 week in 3 out of 11 patients (number 1, 2 and 3 in Fig. 1) with the Euroimmun kit, and in 2 out of 11 patients (number 4 and 5 in Fig. 1) with the Novatec kit in comparison to the 2 other evaluated assays.
Fig. 1.
Serological results of patients with negative RT-PCR nasopharyngeal screening suspect CT findings and positive serology testing, in function of the day of self-reported symptom onset. The second line denotes the day of sampling in function of onset of symptoms. Each line represents an individual patient. Pos denotes a positive, Neg a negative and Bor a borderline test result. A blank cell denotes no serum sample was available for testing
Finally, we aimed to determine the clinical specificity of the immunoassays. To this end, we tested for cross-reactivity with sera of patients with other respiratory viral or atypical bacterial infections, including common coronavirus infections, since these may present with clinical and radiological findings similar to Covid-19. Furthermore, significant structural homology is present between the S and N structural proteins of different Coronaviridae [4]. Using the sera of 15 patients with recent human common coronavirus infections, we observed no positive results with the Euroimmun and Abbott kits (see Supplementary Table 2). In contrast, 3 samples tested positive with Novatec (OC43 n = 2, and OC43 and HKU1, n = 1), and 1 sample tested positive (HKU1) with Vircell. Concerning other respiratory viruses, 1 sample (Influenza B) tested positive with the Vircell kit, and 1 sample (adenovirus) with the Novatec kit, whereas no positive results were observed with the Euroimmun and Abbott kit. Finally, 1 serum sample of C. pneumoniae generated a borderline result with the Vircell and Novatec kits.
Discussion
Since the beginning of the pandemic, RT-PCR on upper respiratory samples for SARS-CoV-2 and CT scans of the lungs have been the major diagnostic tools for Covid-19. However, both the suboptimal sensitivity of RT-PCR (71–95%) and specificity of CT have precluded one of these to become the gold standard for diagnosis of Covid-19 [2, 7]. Recently, studies have shown that anti-SARS-Cov-2 IgG can be detected with high sensitivity by ELISA, already from 4 days after onset of symptoms [3]. High clinical sensitivity was found for patients with RT-PCR confirmed Covid-19 as well as clinically suspected cases [3, 8]. Our study focused on the clinical sensitivity and specificity of four commercial immuno-assays for anti-SARS-COV-2 IgG in the subset of patients presenting with negative RT-PCR and suspect CT findings.
RT-PCR on a nasopharyngeal swab was used in our hospital as a first screening assay during the Covid-19 epidemic. Zheng et al. found the highest analytic sensitivity with nasopharyngeal screening in comparison to other upper respiratory samples [9]. In addition, we screened stool samples from a selected group of patients with negative nasopharyngeal RT-PCR, as PCR positivity of stool was reported in 57% of patients in one study, remaining positive beyond the nasopharyngeal swab [10]. In our study, however, this test was of limited additional diagnostic value given only 1 out of 7 patients with negative nasopharyngeal screening tested positive. In contrast, determination of anti-SARS-CoV-2 IgG added significant diagnostic value and proved to be essential for the diagnosis in nearly 10% consecutively admitted outpatients. The highest clinical sensitivity was found for all 4 evaluated immuno-assays if disease duration was over 14 days, which is in line with previous reports [3, 8]. We found the clinical sensitivity to be lower in the Novatec and Euroimmun kits versus the Abbott and Vircell kit for disease duration under 14 days.
Since we used a serologic assay as the final test of a test cascade, we aimed to determine the clinical specificity of the different kits. Indeed, the misdiagnosis of admitted patients as Covid-19 may have serious consequences since these patients are isolated in a specific ward. We observed that some immunoassays containing recombinant N antigen showed suboptimal specificity, with cross-reactivity with other coronaviruses (OC43 and HKU1) and other common respiratory pathogens (Influenza, adenovirus and C. pneumoniae). Others have reported similar findings [4].
Discordance between serological SARS-CoV-2 IgG assays can be expected because of the lack of assay similarity. The 4 assays in this study include different antigenic targets. Furthermore, differences in performance between assays using the same target can still appear, resulting from differences in antigen purification or production, and methodology (e.g. ELISA, CMIA, …).
This study has several limitations. First, RT-PCR testing of lower respiratory samples was not performed in 4 cases with negative anti-SARS-Cov-2 IgG and RT-PCR testing on stool samples was not done in every patient, which might have underestimated its sensitivity. Furthermore, we could not test serum samples from included patients on all time points. Finally, absolute numbers were too small to allow for statistical testing.
In summary, we found good clinical sensitivity of anti-SARS-Cov-2 IgG immunoassays for Covid-19 in the subset of patients with negative RT-PCR 14 days after onset of symptoms. Clinical specificity was suboptimal in some N-based ELISA kits. Our findings imply a crucial role for serology testing in patients with negative RT-PCR screening and disease duration of over 14 days.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Manon Verhulst, Jessica Van Besien and Ewout Laureys for excellent technical or logistical assistance.
Author contributions
JTVP and MR: conceived and designed the study; JTVP, AC, KV and MR: acquired and analyzed the data; JTVP and MR: wrote the manuscript. All authors revised and approved the final version of the manuscript.
Funding
None.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 2.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing in Coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2019 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, et al. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome Coronavirus 2-specific antibody responses in Coronavirus disease 2019 patients. Emerg Infect Dis. 2020 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steensels D, Reynders M, Descheemaeker P, Curran MD, Jacobs F, Denis O, et al. Performance evaluation of direct fluorescent antibody, Focus Diagnostics Simplexa Flu A/B and RSV and multi-parameter customized respiratory Taqman(R) array card in immunocompromised patients. J Virol Methods. 2017;245:61–65. doi: 10.1016/j.jviromet.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020 doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang Province, China, January–March 2020: retrospective cohort study. BMJ. 2020 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.