Abstract
Background
Targeted therapies and immunotherapies are first-line treatments for patients with advanced melanoma. Serine–threonine protein kinase B-RAF (BRAF) and mitogen-activated protein kinase (MEK) inhibition leads to a 70% response rate in patients with advanced melanoma with a BRAFV600E/K mutation. However, acquired resistance occurs in the majority of patients, leading to relapse. Immunotherapies that activate immune cytotoxic effectors induce long-lasting responses in 30% of patients. In that context, combination of targeted therapies with immunotherapy (IT) is a promising approach. We considered boosting natural killer (NK) cell tumor immunosurveillance, as melanoma cells express stress-induced molecules and activate NK cell lysis.
Methods
Here we have generated vemurafenib (a BRAF inihibitor)-resistant (R) cells from BRAFV600E SK28 and M14-sensitive (S) melanoma cell lines and investigated how resistance interferes with immunogenicity to NK cells. We determined the levels of several soluble molecules including NK ligands in 61 melanoma patients at baseline and 6 months M post-treatment with targeted therapies or immunotherapies.
Results
Vemurafenib resistance involved activation of p-AKT in SK28R and of p-MEK/p-ERK in M14R cells and was accompanied by modulation of NK ligands. Compared with S cells, SK28R displayed an increased expression of natural killer group 2 D (NKG2D) receptor ligands (major histocompatibility complex class (MHC) I chain-related protein A (MICA) and UL16-binding protein 2 (ULBP2)) whereas M14R exhibited decreased ULBP2. SK28R and M14R cells induced higher NK degranulation and interferon gamma secretion and were more efficiently lysed by donor and patient NK cells. SK28R showed increased tumor necrosis factor-related apoptosis-inducing ligand receptor II (TRAIL-RII) expression and TRAIL-induced apoptosis, and TRAIL-induced apoptosis of M14R was decreased. Combined BRAF/MEK inhibitors abrogated the growth of SK28S, M14S, and M14R cells, while growth of SK28R was maintained. BRAF/MEK inhibition attenuated NK activity but R cell lines activated polyfunctional NK cells and were lysed with high efficiency. We investigated the relationship of soluble NK ligands and response to treatment in a series of melanoma patients. Soluble NKG2D ligands known to regulate the receptor function have been associated to cancer progression. Serum analysis of patients treated with target therapies or IT indicates that soluble forms of NK ligands (MICA, B7H6, programmed cell death ligand 1, and carcinoembryonic antigen cell adhesion molecule 1) may correlate with clinical response.
Conclusion
These results support strategies combining targeted therapies and NK-based immunotherapies.
Keywords: killer cells, natural, melanoma, immunotherapy, drug therapy, combination, immunity, cellular
Introduction
Over the past 7 years, immunotherapy (IT) has become a reference treatment for a growing number of solid tumors. Most immunomodulatory strategies focus on enhancing T cell responses, but there has been a recent surge of interest in harnessing the underexplored natural killer (NK) cell compartment for therapeutic interventions. NK cells show cytotoxic activity against diverse tumor cell types, and some of the clinical approaches originally developed to increase T cell cytotoxicity may also activate NK cells.
NK cells are potent cytolytic effectors involved in tumor immunosurveillance. NK cells represent a major source of interferon gamma (IFNγ) in vivo and NK-derived IFNγ is crucial in priming T helper 1 responses.1 Human CD3−CD56+ NK cells represent 5%–20% of peripheral blood mononuclear cells (PBMCs) and comprise CD56dim and CD56bright subsets. The cytotoxic CD56dim NK cell subset predominates in blood (90% of NK cells) while CD56bright NK cells, considered as precursors of CD56dim NK cells and producing cytokines, represent 5%–10% of blood NK cells.2 NK activation depends on an intricate balance between activating and inhibitory signals that determine whether the target will be susceptible to NK-mediated lysis.3 Three natural cytotoxicity receptors (NCRs) involved in NK cell activation were identified: NKp46 and NKp30 that bind to B7H6, expressed by resting NK cells, and NKp44, induced after stimulation by cytokines. Activation of NK cells is triggered by additional receptors. Natural killer group 2 D (NKG2D), expressed by most NK cells, binds major histocompatibility complex (MHC)-related antigens (MIC)-A/B molecules, and UL16-binding proteins (ULBP1–4), induced on stressed cells. NK cell activation is regulated by inhibitory receptors that recognize different human leukocyte antigen class I (HLA-I) molecules, including the killer immunoglobulin receptors, CD94/NKG2A, CD85j, and leukocyte-associated imunoglobulin-like receptors.4–6 Activated NK cells express tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and Fas-L and induce target cell apoptosis through engagement of death domain receptor Fas and TRAIL receptor II (TRAIL-RII).7 8
Tumor cells expressing stress-induced molecules and displaying frequent alterations of HLA-I expression trigger NK cell activation. However, cancer cells have the ability to escape from NK cell recognition through the modulation of NK activating receptors, the secretion of immunosuppressive factors, the expression of inhibitory checkpoint, or the altered migration/recruitment of NK cells to the tumor.9 NK cells infiltrate various tumors including melanoma10 and their frequencies have been related to favorable clinical outcome.11 Numbers of tumor infiltrating NK were positively correlated with regression of melanocytic lesions.12 Thus, NK cells enter the arsenal of immunotherapies and new approaches NK-based immunotherapies are evaluated.13 14
Metastatic melanoma is an aggressive disease with a high metastatic potential. For more than 20 years, therapies approved by the Food and Drug Administration for metastatic melanoma patients were high-dose interleukin-2 (IL-2) and dacarbazine (DTIC). These treatments were associated with response rates of 10%–20%, rare complete responses, and no improved overall survival. Better understanding of the genetic changes associated with melanoma development identified activating mutations in the genes encoding serine/threonine kinases BRAF and NRAS,15 establishing mitogen-activated phosphokinase (MAPK) signaling pathways as drug targetable in melanoma. Up to 50% of melanomas carry an activating BRAF mutation and 90% of reported BRAF mutations result in a substitution of glutamic acid for valine at amino acid 600 (the V600E mutation). This BRAF mutation constitutively activates BRAF and downstream signal transduction in the MAPK pathway. In melanoma patients bearing a V600E/K mutation, BRAF inhibitors induce rapid responses, improved overall survival, and progression-free survival but responses lack durability.16 17 The efficiency of BRAF inhibitors is limited by resistance mechanisms leading to disease relapse or progression in a majority of treated patients after an initial documented clinical response. Several mechanisms of resistance to BRAF inhibitors have been identified as leading to the reactivation of the downstream kinases in the MAPK pathway and the phosphoinositide 3-kinase/protein kinase B (PI3K/PKB) pathway.18 Actual treatment relies on the combination of specific BRAFV600E inhibitors with mitogen-activated protein kinase (MEK) inhibitors (either vemurafenib plus cobimetinib or dabrafenib plus trametinib or encorafenib plus binimetinib) resulting in higher responses rates and improved survival than BRAF inhibitor alone.19 20
There is growing evidence that targeted therapies likely modulate the immunogenicity of cancer cells, indicating the interest of their combinations with immunotherapies. In a previous report, we showed that treatment with vemurafenib of sensible BRAF-mutated melanoma cell lines was associated with change in the expression of NK ligands and decreased susceptibility to NK-mediated lysis.21 In the present study, we have evaluated the susceptibility to NK-mediated lysis of BRAF-mutated melanoma cell lines resistant to vemurafenib (BRAF inhibitor) and/or cobimetinib (MEK inhibitor). We have determined how this resistance modulates the immunogenicity of melanoma cells and discuss the consequence for combined therapies.
Materials and methods
Cell culture and generation of vemurafenib resistant cell lines
Vemurafenib (PLX4032/RO5185426; Genentech, South San Francisco, CA, USA), obtained from Institut Roche (Roche Scientific Partnership), was dissolved in dimethyl sulfoxide (DMSO; Sigma Aldrich, Saint-Louis, Missouri, USA). DMSO was used as vehicle control. M14 and SK28 cell lines were provided by other laboratories (SK28 by F. Faure; M14 by H. Benlalam). Melanoma cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) or Roswell Park Memorial Institute (RPMI) media (Life Technologies, Bleiswijk, the Netherlands) supplemented with 10% Fetal calf serum (FCS) (Eurobio, Les Ulis, France) and 1% penicillin–streptomycin (Life Technologies). To generate vemurafenib-resistant variants (R), exponentially growing sensible (S) SK28 and M14 cells were initially treated with 0.1 µM of vemurafenib. At confluence, cells were subcultured and treated with an increased concentration of vemurafenib, until reaching a concentration of 10 µM. Batches of S and R cells were frozen. Tumor cells were thawed every 2 months and cultured 1 week before use in experiments. The vemurafenib-resistant cell lines, SK28R and M14R, were maintained in culture medium without vemurafenib.
Melanoma patients and serums
Sixty-one consecutive patients with advanced melanoma, receiving a first-line systemic treatment (table 1) between June 2013 and November 2017 in the Department of Dermatology of the Dijon University Hospital, and agreeing to be included in MelBase, a French multicentric cohort dedicated to the prospective follow-up of patients with advanced melanoma, were included. Serum samples were collected at baseline (T0) and 6 months after the onset of treatment and were stored at the Centre de Ressources Biologiques in Dijon. Patients were treated by either first-line targeted therapy (BRAF/MEK inhibitors) or IT (ipilimumab or anti-programmed cell death 1 (PD-1)), according to investigator’s choice. Soluble (s) intercellular adhesion molecule 1 (ICAM1), carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1), and PD ligand 1 (PD-L1) were measured in addition to sMICA/B, sULBP1–6 (NKG2D ligands), and sB7H6 (NCR3 ligand). PBMCs from six metastatic melanoma patients (stage IV M1c, three women and three men) before treatment with immunotherapies were obtained to assess NK cell degranulation.
Table 1.
Characteristics of the patients included and treatment (clinical stage evaluated to American Joint Committee on Cancer (AJCC) 7th edition)
| Patients | |||
| Men | 27 | 10 | 1 |
| Women | 13 | 8 | 2 |
| Median age (years) | 69 | 61 | 74 |
| Clinical stage | |||
| IIIB | 1 | 2 | 0 |
| IIIC | 13 | 4 | 0 |
| IV M1a | 4 | 0 | 2 |
| IV M1b | 11 | 3 | 0 |
| IV M1c | 11 | 9 | 1 |
| Therapeutic treatment | Immunotherapy | Targeted therapy | Chemotherapy |
| Vemurafenib | 0 | 2 | 0 |
| Dabrafenib | 0 | 3 | 0 |
| Vemurafenib+cobimetinib | 0 | 5 | 0 |
| Dabrafenib+trametinib | 0 | 8 | 0 |
| Ipilimumab | 4 | 0 | 0 |
| Nivolumab | 15 | 0 | 0 |
| Pembrolizumab | 15 | 0 | 0 |
| Ipilimumab+nivolumab | 6 | 0 | 0 |
| Deticene | 0 | 0 | 2 |
| Fotemustine | 0 | 0 | 1 |
| Clinical responses at the time of sampling (6M) | |||
| Complete response | 7 | 0 | 0 |
| Partial response | 13 | 6 | 0 |
| Stabilization | 7 | 6 | 1 |
| Progression | 13 | 6 | 2 |
Sequencing
DNA purification was performed with the DNA easy blood and tissue Kit (Qiagen, Hilden, Germany). DNA concentration was measured with a NanoDrop device (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The Department of Genetics and Molecular Biology at Cochin Hospital (Paris, France) performed sequencing of BRAF and 22 known genes in the cell lines, using Ion Torrent Next-Generation Sequencing (Life Technologies) and Ion AmpliSeq Colon and Lung Cancer Research Panel (v2).
Expression of p-MEK, p-ERK, and p-AKT by western blot
Cells treated 30 min with medium containing DMSO or vemurafenib (10 µM) were lysed and proteins were extracted using NP-40 Buffer (Invitrogen, Carlsbad, California, USA) supplemented with sodium orthovanadate and proteases and harvested by scraping. The amount of proteins was measured by Pierce Micro BCA Protein Assay Kit (Life Technologies). Ten micrograms of proteins of each sample were electrophoretically segregated and transferred onto a nitrocellulose membrane. The primary antibodies to detect total and phosphorylated forms of ERK, MEK, and AKT (online supplementary table S1) (Cell Signaling Technology, Leiden, the Netherlands), diluted following the manufacturer’s instructions, were incubated overnight at 4°C. Then the membrane was incubated with anti-rabbit IgG secondary antibody for 1 hour at room temperature. Pierce ECL Plus (Life Technologies) was used for visualization of the protein bands.
jitc-2019-000275supp001.pdf (506.2KB, pdf)
Cell proliferation using the xCELLigence system and MTS assay
Cell proliferation was evaluated with the xCELLigence System (ACEA Biosciences, San Diego, California, USA). After background measurement, cells were seeded for a 6-hour adhesion in E-plates in a volume of 100 µL/well at 1.3×104 cells/well (M14) or 1×104 cells/well (SK28). Then, increasing concentrations of vemurafenib and cobimetinib in medium with DMSO (0.0001%) were added. Cell proliferation was monitored every 15 min for up to 3 days by measuring impedance values, expressed as cell index (CI), normalized to the addition of the drug. CI are self-calibrated values generated by the system, calculated as CI = (Zi − Z0)/15 ohm, where Zi is the impedance at an individual time point during the experiment and Z0 is the impedance at the beginning of the experiment. Alternatively, cell proliferation was monitored by MTS assay. Briefly, 8×103 cells/well for SK28 and 104 cells/well for M14 were seeded in a 96-well plate for 6 hours and treated as above for 24–72 hours. Then, 20 µL/well of combined MTS/PMS solutions (Promega, Madison, Wisconsin, USA) were added and the plate was incubated for 4 hours at 37°C. Absorbance was recorded at 490 nm.
Transcriptomic analysis
Melanoma cells were stored as dry pellets at −80°C prior to transcription analysis by GeneChip Human Transcriptome Array (Affimetrix, Santa Clara, California, USA). Transcriptomic data were analyzed with the Gene Set Enrichment Analysis (GSEA) software.
PBMC isolation and NK cell immunoselection
Blood samples from healthy donors were obtained from EFS (Etablissement Français du Sang, Hôpital St Louis). PBMCs were isolated by Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) density gradient centrifugation. NK cells were purified by negative immunoselection using the NK Cell Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany), leading to >95% of CD3−CD56+ cells. Purified NK cells (1×106/mL) were cultured in RPMI 1640 medium (Life Technologies), supplemented with 10% human serum AB (Biowest, France) and activated with IL-2 (100 IU/mL, Miltenyi Biotec) for 6 days with medium renewal every other day.
Flow cytometry analyses
Multicolor flow cytometry assays were performed on a FACSCanto II cytometer (BD Biosciences, San Jose, California, USA) and analyzed with FlowJo software (V.10.6.2). Melanoma cells treated or not with vemurafenib and cobimetinib in medium containing DMSO (0.0001%) were labeled with antibodies listed in online supplementary table S1.
CD107a degranulation and IFNγ secretion by NK cells
Donor NK (1×105) cells were stimulated with 1×105 melanoma cells in the presence of the CD107a-FITC antibody in 200 µL of medium containing monensin (BD Biosciences). Patient PBMCs (106) activated overnight with IL-2 (100 IU/mL) were stimulated with 105 melanoma cells in the presence of CD107a-FITC antibody. After 5 hours of coculture, cells were labeled for 30 min at 4°C with CD45, CD16, CD3, and CD56 antibodies (online supplementary table S1) and washed. Cells were analyzed on a FACSCantoII flow cytometer and CD107a degranulation was evaluated on gated CD45+CD3−CD56+ NK cells. Baseline NK cell CD107a degranulation was determined in the absence of target cells. Additional cocultures of 5×104 donor NK cells and 5×104 K562 or melanoma cells were conducted for 24 hours to measure IFNγ secreted by NK cells. Cell-free supernatants were harvested and an ELISA assay was carried out following the manufacturer’s instructions (OptiEIA Set Human IFNγ; BD Biosciences). In some experiments, melanoma cells were treated for 20 hours with medium DMSO (0.0001%) alone or with vemurafenib (5 µM) and cobimetinib (10 nM) before the coculture for 5 hours with donor IL-2 +IL-12 (50 IU/mL)-activated NK cells. Then, CD107a degranulation and IFNγ production were measured by intracellular staining in the presence of brefeldin A (Sigma Aldrich), using the Foxp3 staining buffer set (Invitrogen) and IFNγ-APC antibody (Miltenyi Biotec).
NK cell-mediated lysis assay using the xCELLigence system
NK cell-mediated lysis of adherent targets was assessed using the xCELLigence system.21 SK28S and SK28R cells (1×104), and 1.3×104 M14S and M14R cells were seeded in the E-plate and their adhesion was monitored for 6 hours, before addition of cytokine-activated NK cells (at 1:1 ratio). In some experiments, NK cells were incubated for 20 min with mAbs to block NKG2D, NKp30, NKG2A receptors (10 µg/mL) or control isotypes before being added to targets. To assess the impact of inhibitors, adherent targets were treated for 20 hours with medium containing DMSO (0.0001%) alone or with vemurafenib (5 µM)+cobimetinib (10 nM) before the addition of NK cells (at 1:1 ratio). CI was measured every 15 min for 6 hours. Results are expressed as percentage of lysis determined from normalized CI (nCI, to the addition of NK cells) with the RTCA software, with the following equation: % of lysis = [nCI(no effector) – nCI(effector)]/nCI(no effector) × 100. Percentages of lysis were calculated taking as reference the CI from targets without NK cells.
Apoptosis assay
Apoptosis was measured by Annexin-V staining. SK28 and M14 cell lines (4×104 and 5×105 cells, respectively) were plated in 24-well plates, treated with TRAIL recombinant protein (200 µg/mL, R&D System, Minneapolis, Minnesota, USA) or Staurosporin (1 µM, Sigma) for 12 hours and 24 hours. Cells were washed with cold Phosphate Buffered Saline (PBS) and resuspended in staining buffer containing Annexin-V-PE (BD Biosciences), according to the manufacturer’s instructions. Cells were analyzed using a FACSCantoII flow cytometer.
Soluble ligands in supernatants and serum
Cells were seeded in 12-well plates and treated for 24 or 48 hours with medium DMSO (0.0001%) alone or with vemurafenib (5 µM) and cobimetinib (10 nM). Soluble (s) MICA, sULBP2, and sB7H6 were quantified in cell-free supernatants by ELISA following the manufacturer’s instructions (Human MICA, ULBP2 or B7H6 DuoSet ELISA; R&D Systems). For patient serum analysis, a magnetic Luminex assay (immunoassay kit, R&D systems) was devised to measure the concentrations of sICAM1, sCEACAM1, sMICA and sMICB, sULBPs (1, 2/5/6, 3, 4), and sPD-L1. The operations were performed according to the instructions of the manufacturer. The detection was done on a Luminex xMAP system (Luminex Corporation, Austin, Texas, USA) and analyzed using the ProcartaPlex software (Thermo Fisher Scientific). sB7H6 were quantified by ELISA.
Statistical analyses
Analyses of significance were performed with the Wilcoxon rank test, a non-parametric paired t test, and Mann-Whitney non-parametric unpaired t test for clinical data, using the GraphPad Prism V.7.0 software (*p<0.05, **p<0.01, and ***p<0.001).
Results
Characterization of the paired vemurafenib S and R melanoma cells
BRAF inhibitor-resistant variants were obtained after several months of continuous culture of the cells with increasing doses of vemurafenib up to 10 µM. The genotyping of the cell lines was performed by next-generation sequencing, analyzing 22 gene transcripts. M14 bore homozygous BRAFV600E, TP53, and PIK3CA mutations, and SK28 bore homozygous BRAFV600E, TP53, EGFR, and PTEN mutations. SK28R and M14R cells displayed the same mutational status than the corresponding SK28S and M14S cell lines (online supplementary table S2).
jitc-2019-000275supp002.pdf (460.7KB, pdf)
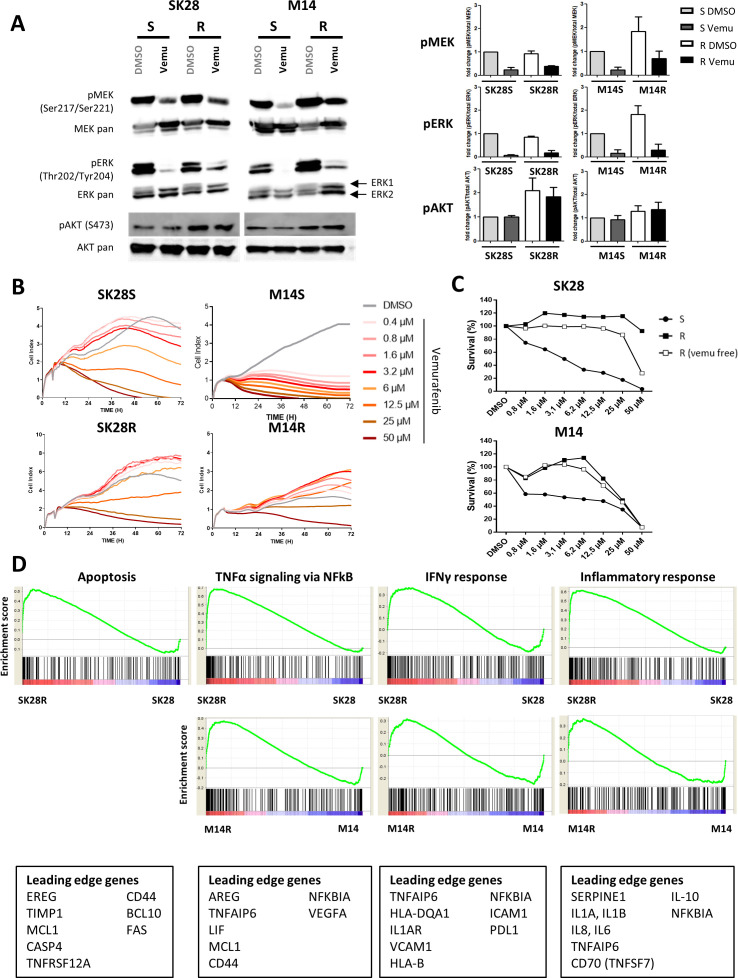
The acquisition of resistance to vemurafenib induced distinct modulation of ERK/MEK/AKT signaling pathway. Phosphorylated levels (p-) of MEK, ERK, and AKT molecules were analyzed by western blotting (figure 1A) in cells incubated for 30 min with medium containing 10 µM of vemurafenib or DMSO. The activation of each protein (phospho/total form) ratio was calculated and the results were expressed taking as reference S cells in DMSO. The modulation of the signal in R cells and in response to vemurafenib was determined (figure 1A, right panel). Levels of p-MEK and p-ERK were not affected in SK28R cells, but SK28R cells displayed high amounts of p-AKT compared with SK28S. In M14R, amounts of p-MEK and p-ERK were increased, and similar p-AKT levels were found M14S and M14R cells (figure 1A, right panel). Treatment with vemurafenib inhibited p-MEK and p-ERK in all cells.
Figure 1.
Characterization of vemurafenib-sensitive (S) and vemurafenib-resistant (R) cells. Activation of MAPK (ERK, MEK) and AKT pathways was determined by western blotting. Cells were incubated in medium with DMSO alone or with vemurafenib (10 µM) for 30 min to assess basal and drug-induced kinase activation (A). Signals were quantified and ratio to basal condition (S cells in medium DMSO) was calculated to express variations in R cells and/or following treatment (A, right panel). Proliferation and survival of S and R cell lines treated by increased concentrations of vemurafenib (0.4 µM–50 µM) for 72 hours. Cell growth was evaluated by dynamic measure of cell index by the xCELLigence technique: one representative experiment of three (B). R cells were cultured 2 months with or without drug and cell survival and IC50 values were measured by MTS assay (C). GSEA in transcription analysis of R versus S cells. GSEA enrichment plots in R cells for selected pathways related to NK immunogenicity. Each plot shows the ES of upregulated leading edge genes of each pathway. Each black bar depicts one individual gene and green line the ES of this gene (D). AKT, protein kinase B; DMSO, dimethyl sulfoxide; ERK, extracellular signal-regulated kinases; ES, enrichment score; GSEA, Gene Set Enrichment Analysis; IFNγ, interferon gamma; MEK, mitogen-activated protein kinase; NK, natural killer.
Cell growth of S and R cells was monitored in the presence of different concentrations of vemurafenib (0.4–50 µM). In a dose-dependent manner, vemurafenib abrogated the growth of M14S and strongly inhibited that of SK28S (figure 1B) while growth of R cells was less affected. SK28R and M14R cells exhibited increased IC50 of at least 25 µM as determined with the MTS assay, and resistance was maintained in the absence of the inhibitor in the culture for 8 weeks (figure 1C).
The comparison of the transcription profiles in S and R cells suggested an increased immunogenicity to NK cells in R cells. The gene expression study included 30 905 genes. GSEA was performed to determine concordant differences between differentially expressed genes and selected gene sets from the MSigDB database. Based on comparison with curated genomic databases (Kyoto Encyclopedia of Genes and Genomes, Biocarta, Reactome, and Gene Ontology sets), a relative enrichment of pathways related to TNFα signaling, inflammatory responses, IFNγ response was significantly increased in R cells. R cell profiles were associated with higher expression of adhesion molecules like ICAM1 or VCAM1 and proinflammatory molecules (TNFAIP6, IL-1, and IL-6). Genes involved in migration and epithelial–mesenchymal transition, such as MCL1, SERPINE1, and TIMP1, were upregulated in R cells. Interestingly, genes related to apoptosis pathway were increased in SK28R but not in M14R cells (figure 1D, online supplementary figure S1).
jitc-2019-000275supp003.pdf (1.8MB, pdf)
Phenotypic modulation of NK ligands in vemurafenib R cells activates NK-mediated lysis
SK28R and M14R cells displayed a modified phenotype compared with their sensitive counterparts. As for signaling, the phenotype was distinctly modulated in SK28 and M14 cells. Compared with parental, SK28R exhibited strongly increased expression of MICA and ULBP2. M14R exhibited reduced ULBP2 and MICA was slightly increased but remained low. B7H6 expression maintained in SK28 S and R cells was slightly decreased in M14R cells (figure 2A). HLA-I expression was maintained and HLA-E molecules were increased in SK28R. In contrast, HLA-E was reduced in M14R, and class-II HLA-DR molecules expressed by M14S cells increased in M14R cells. SK28S and R cells exhibited high expression of PD-L1 and PD-L2. PD-L1 was induced in SK28R cells. Acquired resistance was associated with reduced expression of adhesion molecules ICAM1 and CEACAM1 by R cells (figure 2A). As shedding of MICA and ULBP molecules regulates NKG2D function, we quantified the levels of sMICA and sULBP in cell supernatants (figure 2B). Higher ULBP2 membrane expression by SK28R cells was not associated with significant increase in sULBP2 levels. In M14R cells, decreased ULBP2 and B7H6 membrane expression was associated with increased sULBP2 and sB7H6, indicating a shedding-dependent regulation of the molecule in M14R cells. Very low amounts of sMICA were detected. Strongly reduced sCEACAM1 levels accompanied the decreased membrane expression of the molecule in R cells (figure 2B, online supplementary figure S2).
Figure 2.
Phenotype of S and R melanoma cell lines is associated with increased NK cell activation. Percentages of expression of NKG2D, NKp30, and PD-1 ligands, HLA and adhesion molecules (mean values of 7–12 experiments) by S and R cells (A). Levels of sNK ligands (sMICA, sULBP2, and sB7H6) in supernatants were quantified by ELISA and sCEACAM levels by Luminex assay: levels are normalized for 105 cells (B). Functional capacities (degranulation, IFNγ production) of activated NK cells. IL-2-activated NK cells were labeled with CD107a and cocultured for 6 hours with melanoma cell lines (left panels). IFNγ production in supernatants of NK/target cocultures (24 hours) was quantified by ELISA (right panels) (C). NK-mediated lysis was assessed by dynamic measure of cell index by the xCELLigence system. Targets were seeded and let to adhere 6 hours before the addition of NK cells (ratio 1/1). Results are mean values of seven independent experiments (D). In some experiments, NK cells were incubated with anti-NKR mAbs (NKG2D, NKp30, NKG2A: 10 µg/mL) for 20 min before addition to the targets: results are expressed as percentage of lysis inhibition calculated at T=300 min in the presence NKR mAbs (E). Statistical analyses were performed with the Wilcoxon rank test (*p<0.05, **p<0.01, and ***p<0.001). CEACAM, carcinoembryonic antigen cell adhesion molecule; HLA, human leukocyte antigen; IFNγ, interferon gamma; NK, natural killer.
jitc-2019-000275supp004.pdf (594.9KB, pdf)
To determine the effect of drug resistance on NK cell activation, we measured degranulation (evaluated by CD107a membrane expression) and IFNγ production in response to stimulation with S and R cell lines. In eight independent experiments, percentages of NK cell degranulation were increased in response to SK28R and M14R compared with SK28S and M14S, respectively (figure 2C). The production of IFNγ by IL-2-activated NK cells was higher in response to stimulation by SK28R (mean value 350 pg/mL) than SK28S (mean value 150 pg/mL), and in M14R (mean value 600 pg/mL) compared with M14S (mean value 200 pg/mL) (figure 2C). Moreover, blood NK cells from six metastatic melanoma patients showed a higher activation toward SK28 R cells (figure 2C, right panels).
NK-mediated lysis was monitored by the dynamic decrease CI of adherent targets incubated with NK cells.21 22 The results from seven independent experiments showed that NK cells induced the lysis of 80% of SK28R cells 120 min post-addition of NK cells, compared with 40% of lysis of SK28S (figure 2D). There was a mean increase of 15% of M14R lysis compared with M14S at 180 min (figure 2D). Blocking of NKG2D induced a 20% reduction of lysis of SK28S, SK28R, and M14S while the inhibition of M14R NK lysis by NKG2D reached 10%. Blocking NKp30 also reduced NK-mediated lysis of targets by 15%–20%. Blocking NKG2A increased the lysis of M14S by 10%–15% and by 5%–10% for M14R but did not affect the lysis of SK28 cell lines. Resistance was accompanied by increased NK cell activation and higher NK-mediated lysis of BRAF-resistant melanoma cells involving NKG2D, NKp30, and NKG2A receptors (figure 2E).
Drug and TRAIL-induced apoptosis in S and R cell lines
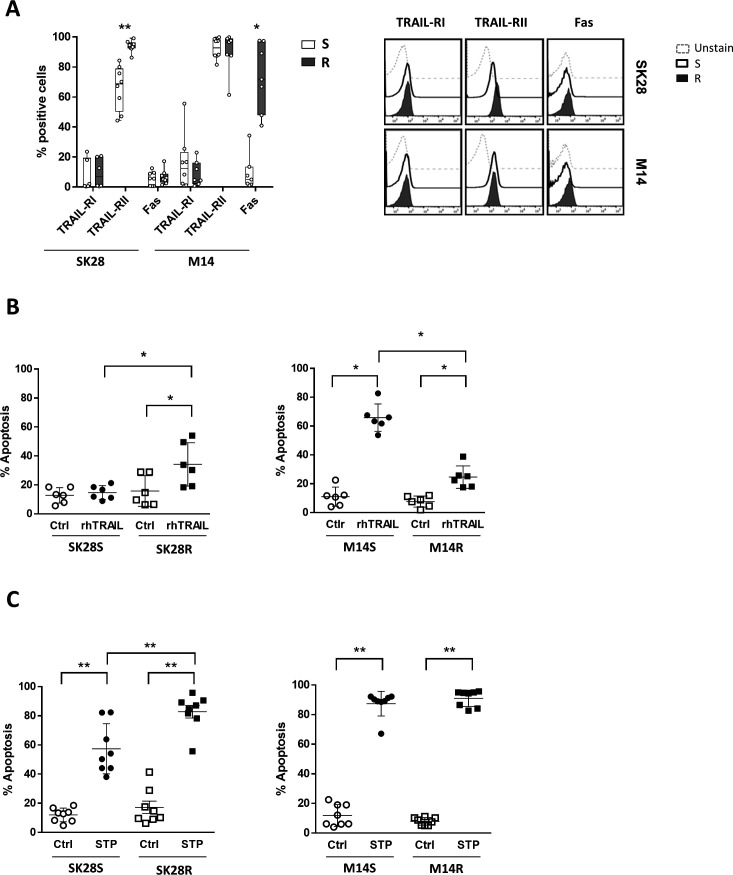
NK activation through death domain receptor engagement also contributes to tumor cell lysis. Investigating this pathway, we first showed that vemurafenib resistance was associated with increased TRAIL-RII expression by SK28R cells. Fas was not expressed. The low expression of Fas by M14S cells was increased in M14R cells and TRAIL-RII was strongly expressed by M14S and M14R cells (figure 3A).
Figure 3.
Apoptosis in S and R melanoma cell lines. Percentages of expression (mean values of eight experiments) and FACS plots (one representative experiment) of death domain receptors by S and R cell lines (A). TRAIL-triggered apoptosis in S and R cell lines incubated with TRAIL agonist (200 µg/mL) for 12 hours (B) and staurosporin (1 µM)-induced apoptosis (pool of 12 hours and 24 hours) (C). Apoptosis was assessed by Annexin-V staining. Statistical analyses were performed with the Wilcoxon rank test (*p<0.05 and **p<0.01). FACS, fluorescence-activated cell sorting; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; TRAIL-RI, TRAIL receptor I; TRAIL-RII, TRAIL receptor II.
To assess the death domain receptor functionality, receptors were engaged by agonist TRAIL and Fas-L molecules and apoptosis measured by Annexin-V staining. Low TRAIL-induced apoptosis of SK28S was increased in SK28R cells (mean 15%–35%, n=6). Staurosporin, activating caspase-independent apoptosis, induced higher apoptosis in SK28R than in SK28S cells. In contrast, the high TRAIL-induced apoptosis of M14S (mean 65%) dropped significantly to a mean value of 25% in M14R cells (n=6) (figure 3B). Moreover, Fas did not trigger apoptosis in M14R cells (data not shown) despite increased Fas expression and staurosporin apoptosis were maintained (figure 3C). SK28R cells exhibited higher susceptibility to apoptosis than SK28S cells while M14R acquired resistance to death domain receptors apoptosis.
Phenotype of S and R cell lines treated with combined vemurafenib and cobimetinib
As the treatment of melanoma patients with a BRAFV600E tumor now involves combined BRAF and MEK inhibitors, we treated S and R cell lines with 5 µM of vemurafenib in the presence of increasing doses of cobimetinib (5 nM–2.5 µM) for 72 hours. Combined inhibitors abrogated the growth of SK28S and M14S cells. The growth of M14R cells exhibiting high levels of p-ERK and p-MEK (see figure 1A) was abrogated with high doses of cobimetinib (0.1 and 2.5 µM) (figure 4A). SK28R cells in which p-ERK and p-MEK were not induced were resistant to the combination (figure 4A).
Figure 4.
Growth and phenotype of S and R cells treated by combined BRAF and MEK inhibitors. Cells were cultured in medium containing DMSO (5 µM) or vemurafenib (5 µM) combined with growing concentrations of cobimetinib (5 nM–2.5 µM) for 72 hours. Cell growth was assessed by the dynamic measure of cell index (A). Expression of ligands involved in NK cell/target interactions was determined in cells treated for 48 hours in medium containing DMSO (5 µM) or vemurafenib (5 µM)+cobimetinib (0.5 µM): box plots summarized four experiments (B). CEACAM, carcinoembryonic antigen cell adhesion molecule; DMSO, dimethyl sulfoxide; HLA, human leukocyte antigen; MEK, mitogen-activated protein kinase; MICA, major histocompatibility complex (MHC) class I chain-related protein A; PD-L, programmed cell death ligand; TRAIL-RII, tumor necrosis factor-related apoptosis-inducing ligand receptor II; ULBP, UL16-binding protein.
We determined the impact of combined inhibitors on phenotype of S and R cells. In SK28S cells, there was a decreased expression of MICA and ULBP2, whereas ULBP2 was induced and MICA was maintained in treated SK28R cells (figure 4B, online supplementary figure S3). Treatment abrogated TRAIL-RII in SK28S cells but not in SK28R cells. HLA-I maintained in SK28S was increased in SK28R, and HLA-E was augmented in SK28S and SK28R cells. PD-L1/PD-L2 expression was decreased in SK28S and augmented in SK28R cells. High CEACAM1 expression was maintained in SK28S and remained abrogated in treated SK28R.
jitc-2019-000275supp005.pdf (975.6KB, pdf)
BRAF+MEK inhibition decreased ULBP2 expression in M14S cells whereas it was slightly increased in M14R-treated cells (figure 4B, online supplementary figure S3). Combined inhibitors reduced TRAIL-RII expression in M14S and M14R cells. The expression of HLA-I and HLA-E was increased prominently in treated M14R cells (figure 4B). PD-1 ligands expression was not affected by the treatment (figure 4B, lower panels).
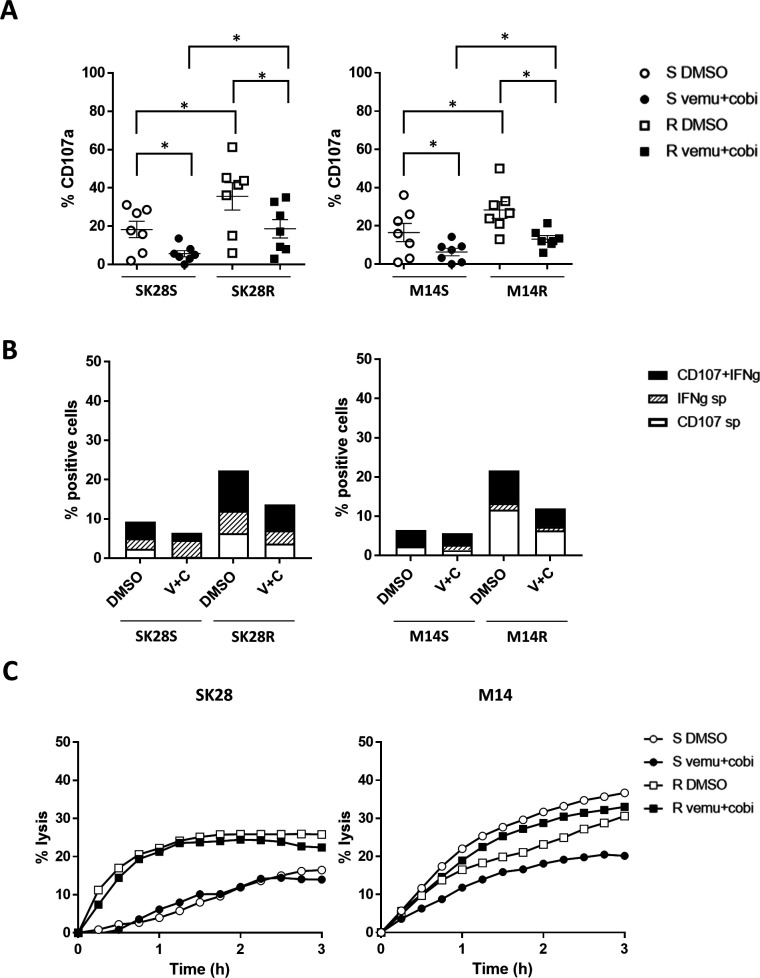
Resistant cells treated with both BRAF and MEK inhibitors maintained high NK cells immunogenicity
When the targets were treated with vemurafenib and cobimetinib (5 μM+10 nM) for 20 hours before their coculture with NK cells, the percentages of degranulating NK cells were reduced but still higher toward R than S cell lines (figure 5A). Furthermore, SK28R and M14R cells treated or not with inhibitors induced higher percentages of CD107+/IFNγ+ polyfunctional NK cells than SK28S and M14S cells, when cultured with IL-2 +IL-12-activated NK cells (figure 5B). Finally, the lysis of cells treated for 20 hours with BRAF+MEK inhibitors was assessed. Inhibitors reduced the NK-mediated lysis of M14S cells, while lysis of SK28S, SK28R, and M14R cells was not affected by the treatment further supporting NK immunotherapies for resistance to targeted therapies (figure 5C).
Figure 5.
Impact of vemurafenib and cobimetinib treatment of S and R targets on NK cell activation and NK-mediated lysis. Targets were incubated for 20 hours with medium containing DMSO (5 µM) or vemurafenib (5 µM) and cobimetinib (10 nM), washed, detached, and cocultured for 6 hours with IL-2-activated NK cells. Percentages of degranulating CD107a NK cells toward S and R targets treated or not with inhibitors (n=7) (A). Detection of polyfunctional (CD107a and IFNγ) NK cells in response to targets treated with DMSO or inhibitors: IL-2 +IL-12-activated NK cells were cultured for 6 hours with targets treated or not with inhibitors (B). NK cell-mediated lysis of targets treated or not with inhibitors measured by xCELLigence assay. Targets were cultured 20 hours in medium containing DMSO (0.0001%) alone or with vemurafenib (5 µM) and cobimetinib (10 nM). One representative experiment of three (C). Statistical analyses were performed with the Wilcoxon rank test (*p<0.05). DMSO, dimethyl sulfoxide; IFNγ, interferon gamma; IL, interleukin; NK, natural killer.
Soluble NKG2D and NKp30 ligands in melanoma patients treated by BRAF inhibitors or immune checkpoint blockers
Resistance to BRAF/MEK inhibition is accompanied by NK ligand modulations impacting NK cell functions. We investigated whether soluble NK ligands represent surrogate markers for clinical evolution and/or response to treatment. Sixty-one patients bearing advanced melanoma (IIIC to IVM1) were included: 18 BRAF-mutated (mut) patients received tyrosine kinase therapies (TKI), 40 BRAF wild-type (wt) patients received IT, and 3 received chemotherapy (table 1). Serum was harvested at baseline (T0) and 6 months (M6) after the beginning of the treatment. sMICA sMICB, sULBPs and sPD-L1, sICAM1 and sCEACAM1 were quantified in serum by Luminex and sB7H6 (NKp30 ligand) measured by ELISA. sMICA was detected in all but one patient, sMICB in 11 patients while sULBPs were present in few patients (<20). Levels of sB7H6 were detected at baseline in all but four patients showed high levels compared with donors (600 pg/mL vs 86 pg/mL). Serum sPD-L1 was detected in 52 patients, and sICAM1 and sCEACAM1 were present in all patients. At baseline, levels of sICAM1 (590 ng vs 727 ng), sCEACAM1 (54 ng vs 45 ng) were comparable in BRAFmut and BRAFwt patients (data not shown). A trend for higher sMICA was found in BRAFwt patients (192 pg vs 78 pg), while sPD-L1 and sB7H6 levels were comparable (figure 6A). We found increased levels of sCEACAM1 and sMICA levels at 6M in non-responder (stabilization and progression) patients but not in responder (complete or partial) patients (figure 6B). In BRAFmut-TKI-treated patients, lower sPD-L1 were found at 6M in non-responders (figure 6B, left panel). In IT-treated patients, higher baseline and M6 levels of sMICA and sB7H6 were found in non-responders (figure 6B, right panel) indicating that these sNK ligands may be prognostic in response to IT.
Figure 6.
Quantification of soluble markers in the serum of 61 melanoma patients received target therapies or IT. Serum samples at baseline (T0) and 6 months (M6) after the onset of treatment were obtained. Soluble molecules MICA and PD-L1 were measured by a magnetic Luminex immunoassay (R&D) and sB7H6 was measured by ELISA in BRAF-mutated (mut) patients receiving TKI and BRAF wild-type (wt) patients receiving IT. Baseline (T0) levels of sMICA, sPD-L1, and sB7H6 in BRAFmut and BRAFwt patients (A). Comparison of sCEACAM1, and sMICA measured in responders and non-responders at T0 and M6 in all patients (B). Comparison of sMICA, sPD-L1, sB7H6, and sCEACAM1 at T0 and M6 in TKI (left) and IT (right)-treated patients (C). Clinical response at 6M was evaluated according to RECIST V.1.1 criteria. Analyses of significance were performed with the Mann-Whitney rank test (*p<0.05). CEACAM, carcinoembryonic antigen cell adhesion molecule; IFNγ, interferon gamma; IT, immunotherapy; MICA, major histocompatibility complex (MHC) class I chain-related protein A; PD-L, programmed cell death ligand; TKI, tyrosine kinase inihibitors.
Discussion
Specific inhibitors of oncogene-driven mutations in cancer cells induce unprecedented high tumor response rates. BRAF and MEK inhibitors are widely used since BRAF mutation is present in almost 50% of cutaneous melanoma, and they induce clinical response in most of the patients. Unfortunately, resistance consequent from additional mutations or non-genomic changes provides alternate growth of tumor cells leading to patient relapse. A major goal, therefore, is to develop rational combinatorial approaches that exploit the specific benefits of oncogenic pathway disruption using targeted agents with the unique ability of IT to mediate long-term responses in metastatic cancers.23–25 Despite the fact that oncogene-driven therapeutic approaches does not take into account tumor immunology, emerging experiences suggest that targeted therapies can turn immune-resistant tumors into immune-sensitive ones.26 The impact of targeted therapies on the expression of immune regulators may directly influence antitumor immune responses. We here demonstrate that resistance of melanoma cells to BRAF (and MEK) inhibitors was accompanied by an increased immunogenicity to donor and metastatic patient NK cells. Modulation of NK ligands and sensitivity to death domain receptors favor activation of NK cells and tumor cell lysis. However, frequent relapses in patients indicate that tumor may escape NK cells in vivo and that boosting NK cells may be require in combination with targeted therapies to potentiate the antitumor response.
First, we show that vemurafenib-resistant variants from BRAFV600E SK28 and M14 cell lines, characterized by different additional mutations, exhibited distinct signaling. There was an increased MEK/ERK activation in M14R, and cobimetinib inhibited their growth. In contrast, the EGFR-mutated/BRAFV600E SK28R cells that displayed high activation of PI3K-AKT signaling (but no ERK/MEK activation) were resistant to combined BRAF/MEK inhibitor, confirming the role of EGFR mutation in MEK co-resistance.27–29 Moreover, EGFR mutation was associated with higher PD-L1 expression.30
Vemurafenib resistance is accompanied by a modulation of the NKG2D/NKG2DL interactions. This pathway, sensing oncogenic stress and cell transformation, is critical for tumor immunosurveillance.31 32 It is regulated by several mechanisms. ERK and AKT influence NKG2DLs expression on stressed cells.33 34 Furthermore, negative regulation of NKG2D function involves shedding of soluble NKG2DLs by tumor-derived matrix metalloproteases (MMP), leading to downregulation of NKG2D and hampering its signaling.35 In SK28R cells, resistance was associated with increased MICA and ULBP2 membrane expression that favored their NKG2D-mediated lysis by NK cells. Additionally, high expression of PD-1 ligands in SK28 cells suggests that immunotherapies targeting NKG2D and PD-1 for patients with BRAF/EGFR-mutant tumors represent an interesting option. In contrast, shedding of ULBP2 leading to increased sULBP2 may hamper NKG2D function in M14R cells, indicating that MMP may control proteolytic shedding of ligands involved in negative feedback signaling and affect NK receptors activity.36
Immunogenicity of M14R cells relied on additional pathways, with augmented HLA-DR and PD-L1 expression. While HLA-DR may be considered as a progression marker,37 38 it was reported that HLA-class II-positive melanoma cells presented antigenic peptides, stimulated CD4+ T cells, and associated with response to immune checkpoint blockade, increased progression-free and overall survival, as well as CD4+ and CD8+ T cells tumor infiltrates.39 Furthermore, PD-1/PD-L1 interaction score and IDO-1/HLA-DR co-expression were strongly associated with anti-PD-1 response.40 The increased PD-L1 in M14R is in agreement with previous reports41 and it suggests that immune checkpoint blockers may be relevant for such genetic profiles.
The data from the cellular models showed that changes in membrane and soluble forms of NKL accompanied resistance to treatment and may constitute useful prognostic biomarkers. Consequently, we have conducted analyses of soluble ligands in a series of melanoma patients. We observed that sMICA/B and sB7H6 and sCEACAM1 were negatively correlated with response in melanoma patients who received target therapies or immune checkpoint inhibitors. A meta-analysis of the literature42 indicated that serum sMICA/B represents a prognostic marker in various cancers. In melanoma patients, sULBP2 was associated with disease progression, and sMICA was correlated with reduced survival.43 Absence of sMICB and sULBP1 in baseline serum was correlated with improved survival in patients treated with IT.44 Targeting shedding of NKG2D and NKp30 ligands improved cell-based IT.45 Our results further indicate that baseline sMICA/B and sB7H6 values may predict response to IT.
NK cell antitumor function also involved caspase-dependent death domain receptor-induced apoptosis. Interestingly, we showed that resistance to vemurafenib promoted apoptosis in SK28R cells but not in M14R cells. Increased TRAIL-RII expression in SK28R cells correlated with higher TRAIL-induced apoptosis. Staurosporin-induced apoptosis was also augmented in SK28R, indicating that granzyme-mediated lysis and caspase-dependent apoptosis might be involved in their increased NK immunogenicity. In contrast, death domain receptor-induced apoptosis was reduced in M14R cells. High p-ERK and MCL-1 transcripts in M14R cells (online supplementary figure S4) may protect them from TRAIL-induced apoptosis by inhibition of SMAC/DIABLO release from mitochondria.46 The increase in anti-apoptotic molecules, XIAP and MCL-1, may be involved in their resistance to Fas-mediated apoptosis.47 Mutation of p53 in M14R cells may concur to their death domain receptor resistance.48 The comparative analysis of the apoptic pathway in SK28 and M14 cell models (online supplementary figure S4) outlines their distinct regulation of TNF receptor (TNFR) pathway: increased proapoptosis TRAF3 and NFKBIA in SK28R and TNFR/TRAF1 prosurvival signaling in M14R.
jitc-2019-000275supp006.pdf (1.5MB, pdf)
We described an additional impact on immunosurveillance induced by BRAF/MEK inhibitors, showing that resistance to inhibitors favored recognition and lysis by NK cells. The above observations confirm that the immune system exert a critical contribution to antitumor responses even in the context of non-immunotherapeutic treatments.49
Conclusions
We evidenced that beside the mainstream BRAFV600E-altered pathway, the different additional mutations present in melanoma cells interfere with immunogenicity to NK cells, acquisition of resistance to targeted therapies. Thus, it may be useful to take into account accompanying mutations in addition to oncogene-driven mutations to design appropriate combinations between target therapies and immunotherapies.
Combining BRAF/MEK inhibition with immunotherapies to increase response rates is actively investigated in preclinical models and clinical trials.50 51 The present results also favor the interest of NK-based immunotherapies that may offer a way to balance toward immunostimulation.
Acknowledgments
We thank Antonio Alberdi and the platform of Institut de Recherche Saint-Louis for support with molecular and FACS analyses.
Footnotes
AF and LR contributed equally.
Correction notice: This paper has been updated since first published to revise Figure 2.
Contributors: All authors participated in the preparation of the manuscript.
Funding: This work was supported by Roche Pharmaceuticals, Fondation ARC, Institut National du Cancer (PAIR Melanoma 2013-066), Cancéropôle Ile de France (Fondation Robert Debré pour la recherche Médicale for AF), and Société Française de Dermatologie (Grant to AC).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not required.
References
- 1.Fehniger TA, Cooper MA, Nuovo GJ, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood 2003;101:3052–7. 10.1182/blood-2002-09-2876 [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. Turning on natural killer cells. J Exp Med 2000;191:1259–62. 10.1084/jem.191.8.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. Embo J 2004;23:255–9. 10.1038/sj.emboj.7600019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long EO. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol 1999;17:875–904. 10.1146/annurev.immunol.17.1.875 [DOI] [PubMed] [Google Scholar]
- 5.López-Botet M, Carretero M, Bellón T, et al. The CD94/NKG2 C-type lectin receptor complex. Curr Top Microbiol Immunol 1998;230:41–52. 10.1007/978-3-642-46859-9_4 [DOI] [PubMed] [Google Scholar]
- 6.Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr Opin Immunol 2004;16:626–33. 10.1016/j.coi.2004.07.010 [DOI] [PubMed] [Google Scholar]
- 7.Screpanti V, Wallin RPA, Grandien A, et al. Impact of FasL-induced apoptosis in the elimination of tumor cells by NK cells. Mol Immunol 2005;42:495–9. 10.1016/j.molimm.2004.07.033 [DOI] [PubMed] [Google Scholar]
- 8.Sheard MA, Asgharzadeh S, Liu Y, et al. Membrane-Bound TRAIL supplements natural killer cell cytotoxicity against neuroblastoma cells. J Immunother 2013;36:319–29. 10.1097/CJI.0b013e31829b4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Vito C, Mikulak J, Zaghi E, et al. NK cells to cure cancer. Semin Immunol 2019;41:101272. 10.1016/j.smim.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 10.Kornstein MJ, Stewart R, Elder DE. Natural killer cells in the host response to melanoma. Cancer Res 1987;47:1411–2. [PubMed] [Google Scholar]
- 11.Schleypen JS, Baur N, Kammerer R, et al. Cytotoxic markers and frequency predict functional capacity of natural killer cells infiltrating renal cell carcinoma. Clin Cancer Res 2006;12:718–25. 10.1158/1078-0432.CCR-05-0857 [DOI] [PubMed] [Google Scholar]
- 12.McKay K, Moore PC, Smoller BR, et al. Association between natural killer cells and regression in melanocytic lesions. Hum Pathol 2011;42:1960–4. 10.1016/j.humpath.2011.02.019 [DOI] [PubMed] [Google Scholar]
- 13.Chiossone L, Vienne M, Kerdiles YM, et al. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol 2017;31:55–63. 10.1016/j.smim.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 14.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol 2016;17:1025–36. 10.1038/ni.3518 [DOI] [PubMed] [Google Scholar]
- 15.Tsao H, Zhang X, Fowlkes K, et al. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res 2000;60:1800–4. [PubMed] [Google Scholar]
- 16.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809–19. 10.1056/NEJMoa1002011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dummer R, Flaherty KT. Resistance patterns with tyrosine kinase inhibitors in melanoma: new insights. Curr Opin Oncol 2012;24:150–4. 10.1097/CCO.0b013e32834fca92 [DOI] [PubMed] [Google Scholar]
- 19.Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014;371:1877–88. 10.1056/NEJMoa1406037 [DOI] [PubMed] [Google Scholar]
- 20.Robert C, Grob JJ, Stroyakovskiy D, et al. Five-Year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med 2019;381:626–36. 10.1056/NEJMoa1904059 [DOI] [PubMed] [Google Scholar]
- 21.Frazao A, Colombo M, Fourmentraux-Neves E, et al. Shifting the Balance of Activating and Inhibitory Natural Killer Receptor Ligands on BRAFV600E Melanoma Lines with Vemurafenib. Cancer Immunol Res 2017;5:582–93. 10.1158/2326-6066.CIR-16-0380 [DOI] [PubMed] [Google Scholar]
- 22.Fregni G, Perier A, Pittari G, et al. Unique functional status of natural killer cells in metastatic stage IV melanoma patients and its modulation by chemotherapy. Clin Cancer Res 2011;17:2628–37. 10.1158/1078-0432.CCR-10-2084 [DOI] [PubMed] [Google Scholar]
- 23.Amaral T, Meraz-Torres F, Garbe C. Immunotherapy in managing metastatic melanoma: which treatment when? Expert Opin Biol Ther 2017;17:1523–38. 10.1080/14712598.2017.1378640 [DOI] [PubMed] [Google Scholar]
- 24.Keller HR, Zhang X, Li L, et al. Overcoming resistance to targeted therapy with immunotherapy and combination therapy for metastatic melanoma. Oncotarget 2017;8:75675–86. 10.18632/oncotarget.18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva IP, Long GV. Systemic therapy in advanced melanoma: integrating targeted therapy and immunotherapy into clinical practice. Curr Opin Oncol 2017;29:484–92. 10.1097/CCO.0000000000000405 [DOI] [PubMed] [Google Scholar]
- 26.Dushyanthen S, Teo ZL, Caramia F, et al. Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat Commun 2017;8:606. 10.1038/s41467-017-00728-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heppt MV, Tietze JK, Graf SA, et al. Combination therapy of melanoma using kinase inhibitors. Curr Opin Oncol 2015;27:134–40. 10.1097/CCO.0000000000000160 [DOI] [PubMed] [Google Scholar]
- 28.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. 10.1038/nature10868 [DOI] [PubMed] [Google Scholar]
- 29.Sun C, Wang L, Huang S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014;508:118–22. 10.1038/nature13121 [DOI] [PubMed] [Google Scholar]
- 30.Molnár E, Garay T, Donia M, et al. Long-Term vemurafenib exposure induced alterations of cell phenotypes in melanoma: increased cell migration and its association with EGFR expression. Int J Mol Sci 2019;20. 10.3390/ijms20184484. [Epub ahead of print: 11 Sep 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene 2008;27:5944–58. 10.1038/onc.2008.272 [DOI] [PubMed] [Google Scholar]
- 32.Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc Natl Acad Sci U S A 2008;105:1686–91. 10.1073/pnas.0701675105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borchers MT, Harris NL, Wesselkamper SC, et al. NKG2D ligands are expressed on stressed human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 2006;291:L222–31. 10.1152/ajplung.00327.2005 [DOI] [PubMed] [Google Scholar]
- 34.Ogbomo H, Michaelis M, Klassert D, et al. Resistance to cytarabine induces the up-regulation of NKG2D ligands and enhances natural killer cell lysis of leukemic cells. Neoplasia 2008;10:1402–10. 10.1593/neo.08972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmiedel D, Mandelboim O. NKG2D Ligands–Critical targets for cancer immune escape and therapy. Front Immunol 2018;9:9. 10.3389/fimmu.2018.02040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller MA, Oudin MJ, Sullivan RJ, et al. Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov 2016;6:382–99. 10.1158/2159-8290.CD-15-0933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostmeier H, Fuchs B, Otto F, et al. Can immunohistochemical markers and mitotic rate improve prognostic precision in patients with primary melanoma? Cancer 1999;85:2391–9. [DOI] [PubMed] [Google Scholar]
- 38.Costantini F, Barbieri G. The HLA-DR mediated signalling increases the migration and invasion of melanoma cells, the expression and lipid raft recruitment of adhesion receptors, PD-L1 and signal transduction proteins. Cell Signal 2017;36:189–203. 10.1016/j.cellsig.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 39.Zeng G, Touloukian CE, Wang X, et al. Identification of CD4+ T cell epitopes from NY-ESO-1 presented by HLA-DR molecules. J Immunol 2000;165:1153–9. 10.4049/jimmunol.165.2.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DB, Bordeaux J, Kim JY, et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin Cancer Res 2018;24:clincanres.0309.2018. 10.1158/1078-0432.CCR-18-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang X, Zhou J, Giobbie-Hurder A, et al. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res 2013;19:598–609. 10.1158/1078-0432.CCR-12-2731 [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Chen N, Yu Y, et al. Prognostic value of MICA/B in cancers: a systematic review and meta-analysis. Oncotarget 2017;8:96384–95. 10.18632/oncotarget.21466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maccalli C, Giannarelli D, Chiarucci C, et al. Soluble NKG2D ligands are biomarkers associated with the clinical outcome to immune checkpoint blockade therapy of metastatic melanoma patients. Oncoimmunology 2017;6:e1323618. 10.1080/2162402X.2017.1323618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paschen A, Sucker A, Hill B, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin Cancer Res 2009;15:5208–15. 10.1158/1078-0432.CCR-09-0886 [DOI] [PubMed] [Google Scholar]
- 45.Zingoni A, Vulpis E, Nardone I, et al. Targeting NKG2D and NKp30 ligands shedding to improve NK cell-based immunotherapy. Crit Rev Immunol 2016;36:445–60. 10.1615/CritRevImmunol.2017020166 [DOI] [PubMed] [Google Scholar]
- 46.Zhang XD, Borrow JM, Zhang XY, et al. Activation of ERK1/2 protects melanoma cells from TRAIL-induced apoptosis by inhibiting Smac/DIABLO release from mitochondria. Oncogene 2003;22:2869–81. 10.1038/sj.onc.1206427 [DOI] [PubMed] [Google Scholar]
- 47.Chetoui N, Sylla K, Gagnon-Houde J-V, et al. Down-Regulation of Mcl-1 by small interfering RNA sensitizes resistant melanoma cells to Fas-mediated apoptosis. Mol Cancer Res 2008;6:42–52. 10.1158/1541-7786.MCR-07-0080 [DOI] [PubMed] [Google Scholar]
- 48.Krayem M, Journe F, Wiedig M, et al. p53 Reactivation by PRIMA-1(Met) (APR-246) sensitises (V600E/K)BRAF melanoma to vemurafenib. Eur J Cancer 2016;55:98–110. 10.1016/j.ejca.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 49.Green DR, Ferguson T, Zitvogel L, et al. Immunogenic and tolerogenic cell death. Nat Rev Immunol 2009;9:353–63. 10.1038/nri2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boni A, Cogdill AP, Dang P, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res 2010;70:5213–9. 10.1158/0008-5472.CAN-10-0118 [DOI] [PubMed] [Google Scholar]
- 51.Ott PA, Adams S. Small-Molecule protein kinase inhibitors and their effects on the immune system: implications for cancer treatment. Immunotherapy 2011;3:213–27. 10.2217/imt.10.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2019-000275supp001.pdf (506.2KB, pdf)
jitc-2019-000275supp002.pdf (460.7KB, pdf)
jitc-2019-000275supp003.pdf (1.8MB, pdf)
jitc-2019-000275supp004.pdf (594.9KB, pdf)
jitc-2019-000275supp005.pdf (975.6KB, pdf)
jitc-2019-000275supp006.pdf (1.5MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.