Abstract
The first step of SARS-CoV-2 infection is binding of the spike protein’s receptor binding domain to the host cell’s ACE2 receptor on the plasma membrane. Here, we have generated a versatile imaging probe using recombinant Spike receptor binding domain conjugated to fluorescent quantum dots (QDs). This probe is capable of engaging in energy transfer quenching with ACE2-conjugated gold nanoparticles to enable monitoring of the binding event in solution. Neutralizing antibodies and recombinant human ACE2 blocked quenching, demonstrating a specific binding interaction. In cells transfected with ACE2-GFP, we observed immediate binding of the probe on the cell surface followed by endocytosis. Neutralizing antibodies and ACE2-Fc fully prevented binding and endocytosis with low nanomolar potency. Importantly, we will be able to use this QD nanoparticle probe to identify and validate inhibitors of the SARS-CoV-2 Spike and ACE2 receptor binding in human cells. This work enables facile, rapid, and high-throughput cell-based screening of inhibitors for coronavirus Spike-mediated cell recognition and entry.
Keywords: quantum dot, SARS-CoV-2, spike, receptor binding domain, angiotensin converting enzyme 2, fluorescence, endocytosis
The severe acute respiratory syndrome coronavirus of 2019 (SARS-CoV-2) is known to cause the coronavirus disease of 2019 (COVID-19).1 The global SARS-CoV-2 viral pandemic has resulted in millions of COVID-19 cases around the world. As of August 27, 2020, there have been over 24 million cases globally with over 828 000 confirmed deaths.2 This deadly virus has prompted a global mobilization of research activity. Because much of the biology and pathogenesis of COVID-19 is unknown, and there are few therapeutic options available to patients, the field of drug discovery and development for COVID-19 antivirals requires tools and reagents to study the viral mechanisms of infection in order to identify targets for therapeutic intervention.
The first step of SARS-CoV-2 infection involves the binding of the highly glycosylated Spike’s S1 subunit receptor binding domain (RBD) to the host cell’s angiotensin converting enzyme 2 (ACE2), a transmembrane enzyme expressed on the plasma membrane surface.3,4 Because the Spike and its binding to the ACE2 receptor play such a vital role in initiating viral infection, the development of probes to study this interaction is needed by virtually all researchers engaged in SARS-CoV-2 drug discovery efforts.5
An ideal tool would be able to (1) monitor Spike/ACE2 binding, (2) measure the cellular spatiotemporal dynamics of Spike/ACE2 binding and internalization, and (3) scale for high-throughput drug screening. To this end, we have developed versatile nanoparticle probes consisting of Spike subunits conjugated to quantum dots (QDs). Although considerably smaller than an average SARS-CoV-2 virion,3 these QD probes may approximate the virus particle shape and mimic its interactions with human cells. As such, they can be considered as model “pseudovirions” that facilitate the study of Spike protein–protein interactions and spatiotemporal dynamics.
The ideal QD includes features such as well-tailored emission characteristics and the ability to serve as a central anchor for multiple spike proteins. QDs have garnered significant attention over conventional organic fluorophores due to their unique photophysical properties that include (1) size- and composition-dependent tuning of fluorescence spectra, (2) broad excitation spectra, (3) high molar absorptivity, (4) high fluorescence quantum yield (QY), and (5) photochemical stability.6−9 Because QDs are photostable and relatively small in size and their surfaces can be easily functionalized with a series of biological molecules, there is great interest in developing QD-based Förster resonance energy transfer (FRET) biosensing systems with various energy transfer partners.10 One of the best energy acceptors for QDs is gold nanoparticles (AuNPs), due to their large absorptivity in the visible electromagnetic spectrum.11−15
In this work, we have combined the utility of fluorescent QDs, AuNPs, and ACE2-green fluorescent protein tagged (ACE2-GFP) cells to allow for facile monitoring of Spike-ACE2 interactions. Hereafter, we refer to the conjugates of Spike subunits bound to a central QD as “QD-[subunit]” (e.g., QD-RBD) and ACE2 receptors bound to a central AuNP as “AuNP-ACE2”. We designed an energy transfer system to monitor Spike-ACE2 binding in vitro where QD fluorescence is quenched by the nearby AuNP upon binding. This quenching can be disrupted by unlabeled ACE2 or neutralizing SARS-CoV-2 antibodies competing with or blocking QD-Spike binding to ACE2-AuNP, respectively. We further applied QD-RBD to ACE2-GFP to directly image Spike-ACE2 endocytosis [endo(RBD:ACE2)] using real-time confocal microscopy and high-resolution single-molecule tracking in living cells.
Our research supports the idea that endocytosis of Spike bound to ACE2 is one potential mechanism for viral entry.16 These assays can identify SARS-CoV-2 antivirals and suggest that QD-RBD conjugates can be used for high-throughput screening (HTS) as well as nanoparticle-based diagnostics for the detection of viral particles.17,18
Results and Discussion
Biochemical Assays
Nanoparticle-Based Assay Design
We initially focused on developing an energy transfer system to monitor the interaction between Spike and ACE2, using QD-RBD (green QD514, fluorescence maximum at 514 nm) (Figure 1d) and AuNP-ACE2 that quenches QD fluorescence with close proximity facilitated by RBD-ACE2 binding.11 Photoluminescence (PL) quenching of QDs is dependent on the binding affinity, conjugation ratio, and the integral overlap of donor–acceptor pairs (details in Methods). For cellular assays, QD-RBD and ACE2-GFP internalization was monitored using orange-emitting QDs (QD608, fluorescence maximum at 608 nm) (Figure 1c) and GFP signal (fluorescence maximum at 509 nm). Using the pseudovirion QD-RBD, we studied RBD:ACE2 internalization and its inhibition by recombinant ACE2 with the fragment crystallizable (Fc) region of the human immunoglobulin IgG1 (ACE2-Fc) or neutralizing antibodies.
Figure 1.
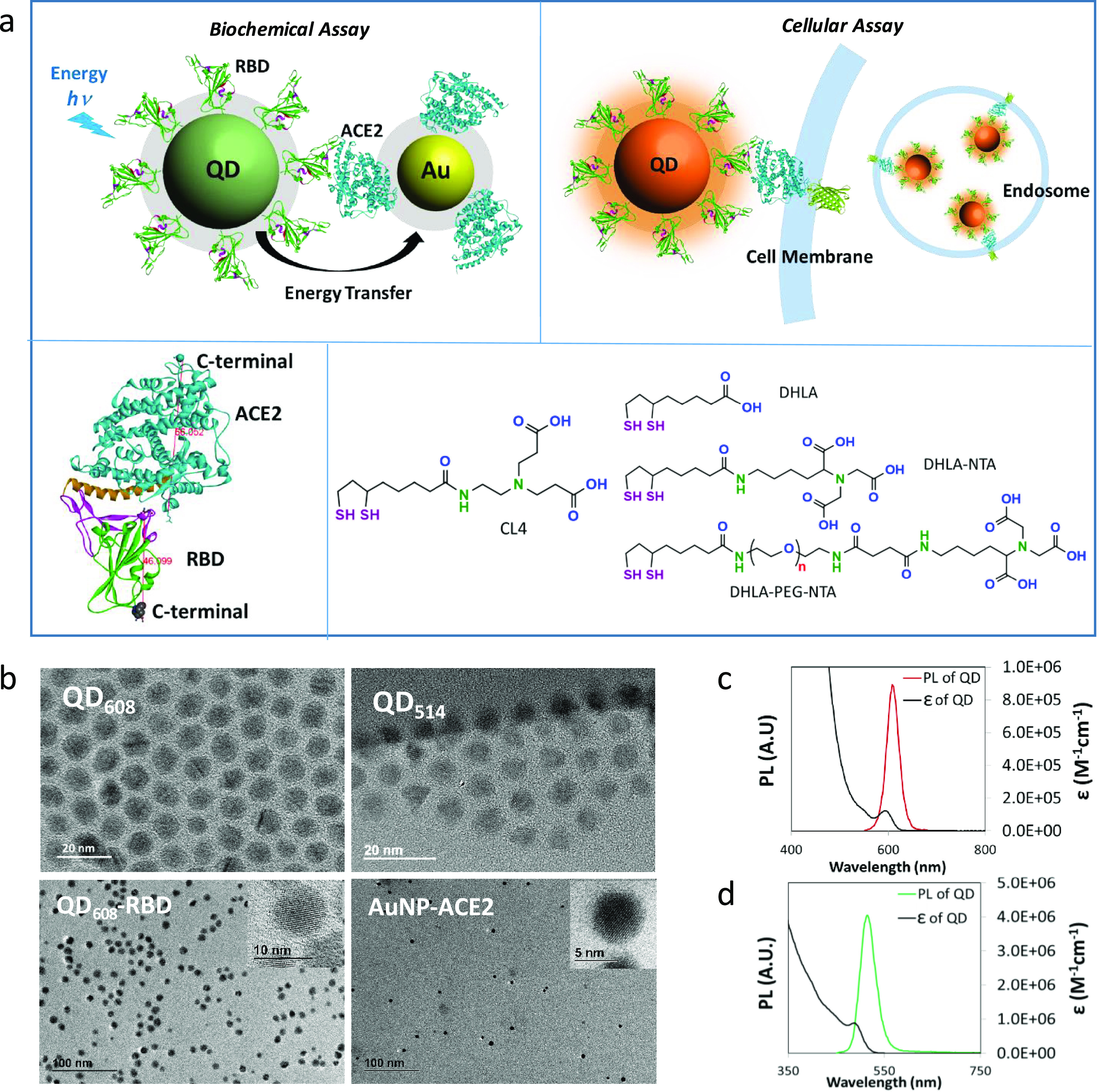
Assay design and physical properties of nanoparticles. (a) Schematic diagram of the biochemical assay using energy transfer from QD-RBD to AuNP-ACE2 (top left) and the cellular assay using QD-RBD interaction with ACE2 (with or without GFP modification at the end of the C-terminal) on the cell membrane (top right). The bottom image shows the binding of ACE2 and RBD (bottom left, the estimated size measured in Å) and the chemical structure of surface ligands for both QDs (CL4) and AuNPs (DHLA ligands) (bottom right). (b) TEM images of NPs. Top: As-synthesized QD608 (10.1 ± 0.94 nm) and QD514 (8.4 ± 0.84 nm). Bottom: QD608-RBD (10.1 ± 0.89 nm) and AuNP-ACE2 (5.8 ± 0.8 nm). (c) Absorption and fluorescence spectra of CL4-coated QD608 in water. (d) Absorption and fluorescence spectra of CL4-coated QD514 in water.
For this, QD surfaces were modified with compact ligands (CL4) and AuNPs with dihydrolipoic acid (DHLA) mixed with nitrilotriacetic acid-modified DHLA (DHLA-PEG-NTA, DHLA-NTA)19 (Figure 1a). QDs have narrow emission spectra, and measurements using transmission electron microscopy (TEM) determined their diameter to be 10.4 nm for orange QD608 and 8.4 nm for green QD514 (Figure 1b). TEM also confirmed that the QD sizes and shapes were not affected by ligand exchange nor protein conjugation and that the QDs were well dispersed (Table 1 and Supplementary Figure 1).
Table 1. Characteristics of Nanoparticles and Nanoparticle–Protein Conjugatesa.
| emission peak | extinction coefficient (M-1 cm-1) | TEM size (nm) | hydrodynamic size (intensity mode, nm) | hydrodynamic size (number mode, nm) | |
|---|---|---|---|---|---|
| QD608-CL4 | 608 nm (FWHM ∼26 nm) (QY∼30%) | 1.2 × 105 @592 | 10.0 ± 0.93 | 16.6 ± 0.6 | 12.2 ± 0.1 |
| QD608-RBD | 10.1 ± 0.89 | 23.8 ± 2.4 | 19.9 ± 0.9 | ||
| QD608-S1 | – | 28.2 ± 1.1 | 23.4 ± 0.7 | ||
| QD608-S1+S2 | – | 56.1 ± 0.9 | 53.1 ± 1.6 | ||
| QD514-CL4 | 514 nm (FWHM ∼34 nm) (QY∼40%) | 8.9 × 105 @488 | 8.4 ± 0.84 | 13.2 ± 0.5 | 10.8 ± 1.6 |
| QD514-RBD | 8.2 ± 0.72 | 21.8 ± 2.3 | 17.0 ± 1.3 | ||
| AuNP-NTA | – | 1.4 × 107@520 | 5.6 ± 0.7 | 15.6 ± 0.5 | 11.8 ± 0.4 |
| AuNP-ACE2 | 5.8 ± 0.8 | 21.7 ± 0.7 | 17.4 ± 1.7 |
Data shown as mean ± SD.
AuNP exhibited a surface resonance peak at 520 nm, and the size was 5.6 nm as measured by TEM (Figure 1b, Supplementary Figure 1). The hydrodynamic sizes of NPs in an aqueous solution were 13.2 and 16.6 nm for QD514-CL4 and QD608-CL4, respectively, and 15.6 nm for AuNP-NTA, which were larger than the TEM core sizes due to the hydration layer and ligand layer20 (Table 1). Next, histidine-tagged RBD (RBD-His) and histidine-tagged ACE2 (ACE2-His) were conjugated to NPs via coordination directly to the QD surface or NTA on the AuNP surface (details in Methods). After conjugation, the QD hydrodynamic size was increased by 7 nm with RBD (molar ratio of RBD/QD = 8, hereafter), 15 nm with S1 (S1/QD = 4), and 40 nm with S1 + S2 (S1 + S2/QD = 3), which were reasonable increases according to the protein sizes (Table 1). Similarly, the hydrodynamic size of AuNP was increased by 6 nm with ACE2 conjugation (ACE2/AuNP = 3). Gel electrophoresis revealed decreases in the mobility shifts as the conjugation ratio of RBD to QD increased from 0 to 16 and confirmed efficient conjugation of RBD-His to QD as well as ACE2-His to AuNP (Supplementary Figure 1).
NP-Based Energy Transfer Biosensor for RBD-ACE2 Binding
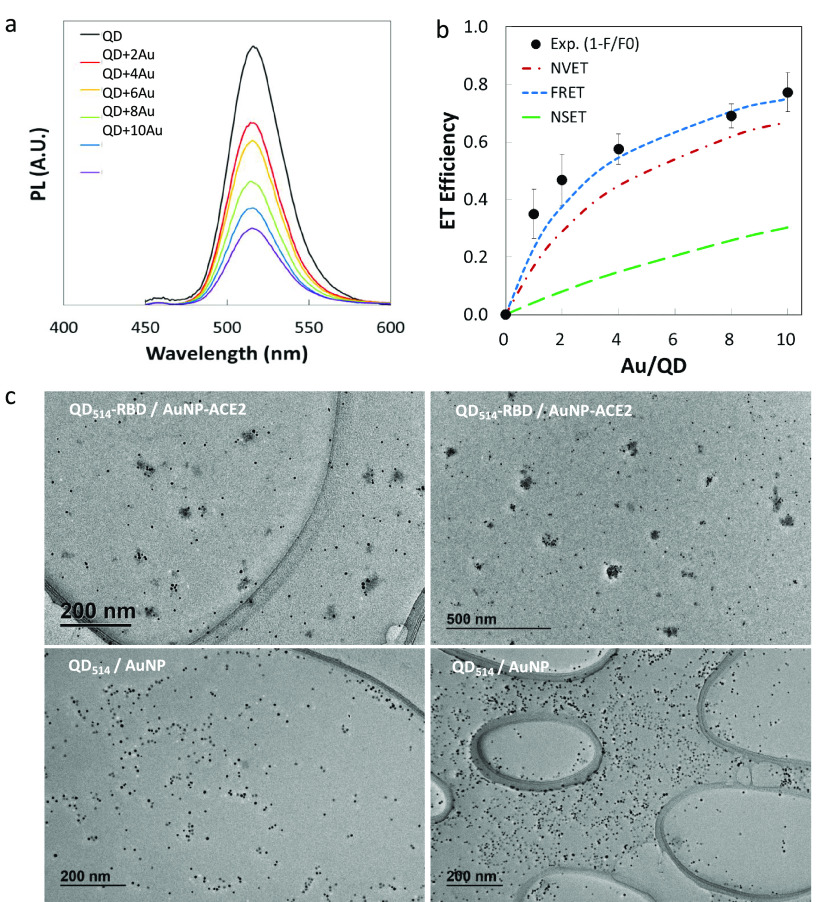
We chose QD514 as the energy transfer donor to achieve higher efficiency due to a better spectral overlap (J) with the AuNP absorption peak (520 nm) and its smaller core size (details in Methods). QD fluorescence decreased with increasing ratios of acceptor per donor (AuNP/QD = 0–10) (Figure 2a). The optimized biochemical assays exhibited 80% maximum energy transfer efficiency, and the control AuNP (without ACE2) did not quench the QD514-RBD PL, which confirmed the specificity of the RBD-ACE2 interaction (Supplementary Figure 2). The experimental energy transfer efficiency, E = 1 – (F/F0), was compared with three theoretical models: FRET (dipole–dipole interaction, E proportional to R6), NSET (nanosurface damping energy transfer, E proportional to R4), and NVET (nanovolume damping energy transfer, E proportional to R3). These models are described in the Supporting Information(21−23) (Figure 2b). In the FRET model, R is defined as the distance between the centers of the AuNP and QD. For NSET and NVET, R is the distance from the surface of the AuNP to the center of the QD. Here, F and F0 are the PL of QD with and without AuNP, respectively. The calculated distance with 50% energy transfer, R0, was 14.4 nm for FRET, which was longer than that for general organic dye acceptors (typically R0 < 10 nm), owing to the large extinction coefficient of AuNP (ε = 1.4 × 107 M–1 cm–1 at 520 nm).24 With QD514-AuNP, the FRET model provided the best fit to our experimental data with the specifically calculated and estimated distances, which was different from our previous study using QDs and ultrasmall AuNPs (1.5 nm) and other literature reports.15,23 Here, the calculated FRET model estimated higher efficiency than other models because the extinction coefficient of 5 nm AuNPs is 2–3 orders of magnitude higher than that of 1.5 nm AuNPs. The NVET model moderately estimated the experimental data, while the NSET model fit poorly because it does not account for acceptor size. While we observed consistent QD quenching using 3 nm AuNP-ACE2 and QD514-RBD, the FRET model underestimated the efficiency of smaller AuNPs due to the larger R-dependence (Methods, Supplementary Figure 2). QD514-S1 was also tested as the energy donor and exhibited much lower efficiency because the larger size of S1 compared to RBD (76.5 kDa vs 26.5 kDa) resulted in increased separation, R (Supplementary Figure 3).
Figure 2.
Energy transfer quenching assay using QD-RBD and AuNP-ACE2. (a) Changes in QD514-RBD photoluminescence (PL) with different ratios of AuNP-ACE2/QD514-RBD; from 0 (noted as “QD”) to 10 (noted as “QD + 10Au”). (b) Energy transfer efficiency as a function of the ratio of AuNP-ACE2 per QD514-RBD; the experimental plots and three different theoretical models. (c) TEM images of clustered AuNPs due to the binding between AuNP-ACE2 and QD514-RBD (ratio = 8).
The TEM images of the QD-RBD:AuNP-ACE2 complex captured clustering of QD and AuNP (Figure 2c, Supplementary Figure 2), and the decreased distance between NPs further corroborated specific RBD-ACE2 binding, while control samples without protein conjugates did not show any clustering. However, we could not completely exclude the inner-screening effect of AuNPs, the electron transfer, or the electrostatic interaction as additional contributions to quenching.23,24 Regardless of the fit to the models, the observed PL quenching indicated the binding between the Spike subunit and ACE2, demonstrating that QD-Spike is a viable method for the production of pseudovirions that can be monitored in real time by their emission characteristics.
Biologics Inhibit NP-Based Energy Transfer
After confirming that QD514-RBD quenching could be used to monitor RBD-ACE2 binding, we devised a method to test the inhibitory activity of biological molecules (Figure 3a). As a proof-of-concept, Fc-tagged recombinant ACE2 (ACE2-Fc) was used in a competition assay to block the interaction of QD514-RBD and AuNP-ACE2. Addition of 0.9 μM free ACE2-Fc resulted in 90% PL recovery and a half-maximal effective concentration (EC50) of 200 nM (Supplementary Figure 3). We then tested the efficacy of neutralizing antibodies Ab1 and Ab2, specific for SARS-CoV-2 S1 or RBD, respectively, and found that the PL of QD514-RBD was fully recovered in the presence of the neutralizing antibodies (Figure 3b and Supplementary Figure 3). As a control, we tested another non-neutralizing anti-spike antibody and observed no PL recovery (Figure 3c). The calculated EC50 using normalized PL was 60 nM and 125 nM with R2 > 99% for Ab1 and Ab2, respectively (Figure 3d). These results indicate that our pseudovirions can enable facile and rapid biochemical screening for repurposed or newly synthesized drugs in addition to neutralizing antibodies or other biologics to prevent SARS-CoV-2 infection.
Figure 3.
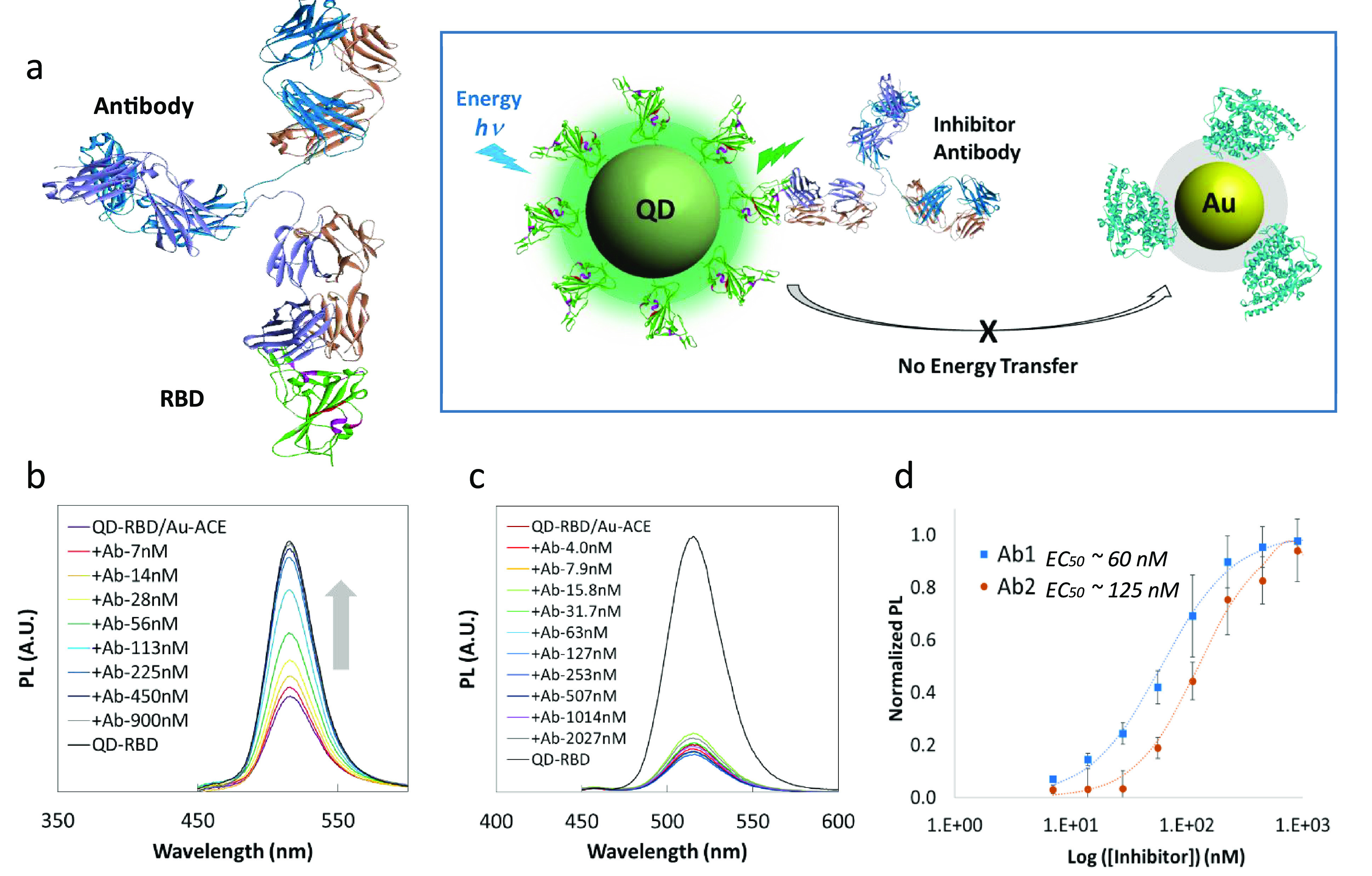
NP-based inhibition assay. (a) Left: The structure of neutralizing antibody (top) bound to SARS-CoV-2 Spike RBD (bottom, green). Right: Schematic diagram of the inhibition assay depicting blocking of the interaction between RBD and ACE2 and the resulting inhibition of energy transfer from QD to AuNP. (b) PL recovery of QD514-RBD in the presence of neutralizing antibody Ab1. (c) Inhibition test using anti-Spike antibody without neutralizing ability, showing almost no PL recovery of QD514-RBD. (d) Calculated EC50s for neutralizing antibodies Ab1 and Ab2 were 60 nM and 125 nM with R2 > 99%, respectively.
Cell-Based Assays
Quantum Dot Conjugates Induce ACE2-Spike Translocation
The biochemical assays described above demonstrated how QDs conjugated to SARS-CoV-2 Spike can act as a pseudovirion and bind to ACE2. To understand whether these nanoparticle probes were active in a cell-based system, we stably transfected a C-terminal GFP-tagged ACE2 fusion protein into HEK293T cells (ACE2-GFP HEK293T). This line propagated well, had a high transfection efficiency, and expressed high levels of ACE2 on the plasma membrane. ACE2-GFP clone 2 was treated with 100 nM QD514-RBD or QD608-RBD for 3 h, and the live cells were imaged using an Opera Phenix automated high-content confocal microscope (Figure 4).
Figure 4.

Quantum dot-conjugated Spike-RBD domain induces translocation of ACE2 and internalizes into cells. (a) Representative image montage of ACE2-GFP (yellow) HEK293T clone 2 treated with 100 nM QD514-RBD (magenta) and QD608-RBD (magenta). Digital phase contrast (cyan) was used during live-cell imaging to identify cell somas. (b) High-content analysis averages of spot count for QD514-RBD and QD608-RBD and ACE2-GFP. N ≥ 400 cells from duplicate wells. (c) Representative image montage of immunofluorescence staining for ACE2 in ACE2-GFP HEK293T cells. Cells were stained with Hoechst 33342 for nuclei (cyan), mouse anti-ACE2 antibody (yellow), and HCS Cell Mask Deep Red for whole cell fill (magenta). N = 9 fields each from 3 triplicate wells. (d) WT HEK293T cells were treated with 100 nM QD608-RBD. Digital phase contrast in cyan and QD608-RBD in magenta. (e) Representative image montage of ACE2-Expi293F and WT-HEK293T cells stained with Hoechst 33342 (cyan), mouse anti-ACE2 antibody (yellow), and HCS Cell Mask Deep Red (magenta). N = 3 triplicate wells. Optimem I alone used as control. Scale bar, 25 μm.
The control unconjugated QDs did not enter cells, nor did they induce any changes in the localization of ACE2-GFP (Figure 4a). QD514-RBD and QD608-RBD were both observed to internalize into cells and induced strong translocation of ACE2-GFP. Importantly, the separate ACE2-GFP and QD608-RBD signals were strongly colocalized, with little to no QD608 signal independent of ACE2-GFP fluorescence. The QD514 signal could not be discerned from the ACE2-GFP signal because of overlapping emission spectra and bleed-through as seen in stably transfected ACE2-Expi293F cells that do not have a GFP tag on ACE2 (Supplementary Figure 4). There was no bleed-through in signal when using QD608-RBD (Supplementary Figure 4); therefore it was selected for subsequent cell-based experiments. Furthermore, the QD-RBD fluorescence was only observed in ACE2-GFP cells as seen with ACE2-GFP HEK293T clone 1 that had a lower transfection efficiency. High-content analysis of fluorescent signals demonstrated a large assay window between cells treated with QD608-RBD and cells treated with unconjugated QD608 when analyzing spot counts, indicative of internalized QDs and ACE2 receptor (Figure 4b). We also generated QD528 during initial optimizations conjugated to the virtually full length recombinant SARS-CoV-2 S1+S2 ECD-His protein. QD528-S1+S2 bound to the cell surface, and subsequently intracellular puncta could be observed, although internalization was reduced compared to that observed with QD-RBD (Supplementary Figure 4). In addition, recombinant SARS-CoV-2 RBD alone was able to induce ACE2-GFP translocation (Supplementary Figure 5). Furthermore, QDs conjugated to the original SARS Spike, SARS-CoV S1, were internalized and strongly colocalized with ACE2-GFP (Supplementary Figure 5).
To verify that ACE2-GFP cells indeed expressed ACE2, fixed cells were immunostained with mouse anti-ACE2 antibody, and no independent yellow or magenta signal corresponding to GFP and QD, respectively, was observed (Figure 4c). Similarly, no uptake of QD608-RBD was observed in wild-type (WT) HEK293T cells (Figure 4d). In contrast to untagged ACE2-Expi293F, WT HEK239T did not express detectable levels of ACE2 as assessed by immunofluorescence staining (Figure 4e).
QD608-RBD Enters Cells and Induces ACE2-GFP Internalization through Endocytosis
We confirmed that QD608-RBD could be used at concentrations as low as 5 nM and still observe binding, internalization, and translocation of ACE2-GFP (Supplementary Figure 6). Concentrations of 10 and 20 nM were used in subsequent experiments to ensure sufficient amounts of QD-RBD. One potential mechanism of this translocation and internalization of ACE2-GFP bound to QD608-RBD is dynamin- and clathrin-dependent receptor endocytosis, a mechanism that has been proposed for viral entry in some cell types.25 To confirm this hypothesis, we conducted live-cell imaging of ACE2-GFP clone 2 cells treated with Optimem I as a control, 10 or 20 nM QD608-RBD, and 20 μM Dyngo-4a,26 a dynamin inhibitor (Figure 5a, Supplementary Videos 1–6).
Figure 5.
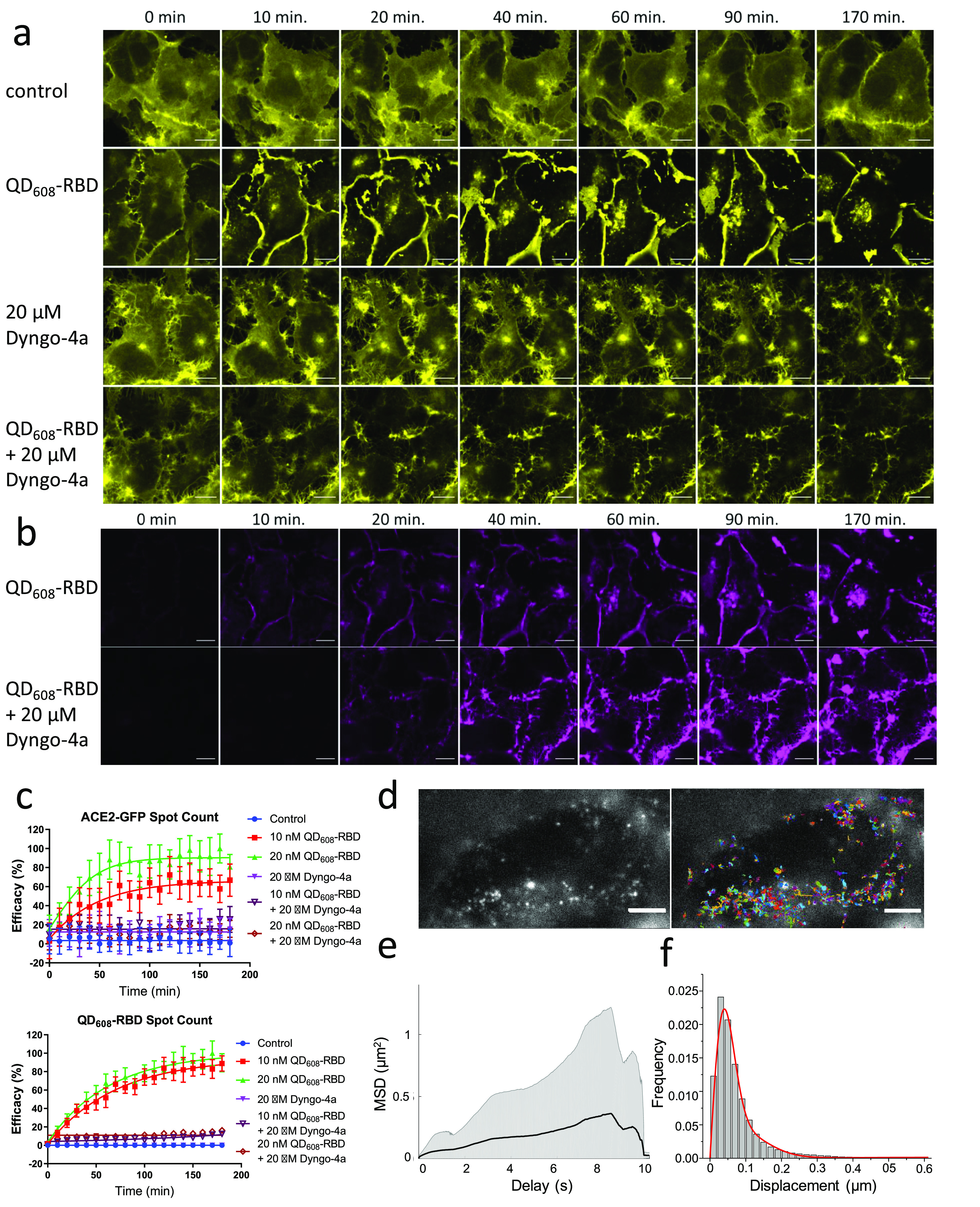
QD608-RBD-induced translocation of ACE2-GFP is blocked using endocytosis inhibitor Dyngo-4a. (a) Representative image montage of ACE2-GFP signal (yellow) in HEK293T clone 2 treated with 10 nM QD608-RBD. ACE2-GFP is represented by a yellow look-up table. (b) Same cells from (a) showing the QD608-RBD signal (magenta). Control cells were incubated with Optimem I alone. Dyngo-4a-treated cells were first preincubated with compound for 15 min. Time course spans 3 h and imaging began immediately after treating cells with QD608-RBD. Images were captured using a 63× objective. Scale bar, 25 μm. (c) High-content analysis averages of spot count for ACE2-GFP and QD608-RBD. Curves were fit using nonlinear regression. N ≥ 1100 cells from 4 fields each from 10 wells per condition, representative of three experiments. Error bars indicate SD. (d) Single-particle imaging of QD (left) and overlay with tracks (right) in ACE2-GFP HEK293T cells. Scale bar, 5 μm. (e) The ensemble mean of all mean square displacements (MSD) (1562 tracks) is shown as a black solid line. The grayed area represents the weighted standard deviation over all MSD curves. (f) Distribution of one-step jump distances with fitted curve (red solid line) of QDs in the ACE2-GFP HEK293T cells.
Signals from ACE2-GFP and QD608-RBD were captured for cells treated with and without QD608-RBD or Dyngo-4a. QD608-RBD rapidly bound to ACE2-GFP cells and began internalizing with ACE2-GFP within 10 min to form endo(RBD-ACE2). Dyngo-4a alone did not affect the ACE2-GFP localization (Supplementary Video 4), but treatment with Dyngo-4a prior to QD608-RBD treatment robustly blocked endo(RBD-ACE2) (Supplementary Videos 5,6). The inhibitory effect of Dyngo-4a was more apparent when quantifying the signal for QD608-RBD than for ACE2-GFP. The residual signal from the clustering of ACE2-GFP at the membrane was identified as “spots” during the high-content analysis (Figure 5b). However, QD608-RBD, while able to bind at the cell surface, was not able to enter cells in the presence of Dyngo-4a, and therefore the quantification revealed a strong inhibitory effect (Figure 5c).
Single-Molecule Tracking Confirms Endocytosis of QD608-RBD
To further study the spatiotemporal dynamics of QD608-RBD, we utilized inclined/total internal reflection fluorescence (TIRF) illumination microscopy,27 a high-resolution single-molecule microscopy method to measure the kinetics of individual quantum dots binding and internalizing into the ACE2-GFP HEK293T cell line (Figure 5d–f, Supplementary Video 7). In order to image single QDs, a very low concentration of QD608-RBD (200 pM) was incubated with ACE2-GFP HEK293T cells for 20 min to capture bound and endocytosing particles. We observed very fast binding (within minutes) of QD608-RBD to the surface of ACE2-GFP HEK293T cells. Furthermore, the fluorescence intensity fluctuation of QD608-RBD on the plasma membrane under TIRF mode was observed, suggesting single QD blinking (Supplementary Video 7). The localization of single QDs was determined through two-dimensional Gaussian fitting. The average mean square displacement (MSD) curve clearly indicated the nature of confined motion of QDs (Figure 5e). The ensemble MSD analysis revealed the average behavior of QD608-RBD. In order to better understand the mobility behavior in the population, we calculated the jump distance traveled by each QD track from one frame to the next and found three different mobilities of QD608-RBD in ACE2-GFP HEK293T cells: immobile (0.03 μm2/s), slow (0.13 μm2/s), and fast (0.84 μm2/s), occupying 32%, 52%, and 16% of the population, respectively (Figure 5f). Most of the tracks corresponded to immobile and slow population, demonstrating QD608-RBD interacted with the ACE2 receptor. The fast diffusion coefficient population likely reflects receptor-mediated endocytosis, as evidenced by active transport that was observed during imaging. Our results for the slow diffusion of QD608-RBD corresponded to those shown in the literature with regard to membrane compartments where confined diffusion was determined to be 0.12 μm2/s.28 These results indicate that the nanoparticle movement was vastly different from free diffusion29 and suggest that QD608-RBD interacted with the ACE2 receptor. While this experiment was conducted after 20 min of QD608-RBD incubation, longer-term incubations may reveal different endo(RBD:ACE2) mobility dynamics based on the endosomal location in the cell.
Inhibition of Spike Using Antibodies and Recombinant ACE2
The development of neutralizing antibodies and biologics as SARS-CoV-2 antivirals has garnered much attention because they can directly block viral entry.30−32 Using QD608-RBD, we demonstrated that neutralizing antibodies developed against SARS-CoV-2 S1 and RBD potently blocked the binding and internalization phenotype observed in ACE2-GFP HEK293T cells (Figure 6 and Supplementary Figure 7). While Ab1 was raised against SARS-CoV-2 S1-Fc, Ab2 was raised against SARS-CoV-2 RBD. However, our data showed Ab1 was more potent than Ab2 against RBD (Figure 6a,b). This result is consistent with the biochemical inhibitions carried out prior to the cell-based assays and ELISA assay data reported by the vendor. As with the biochemical inhibition assays, ACE2-Fc was less effective at blocking QD-RBD binding than were the antibodies; Ab1 was 8-fold more potent than ACE2-Fc (Figure 6c–e).
Figure 6.
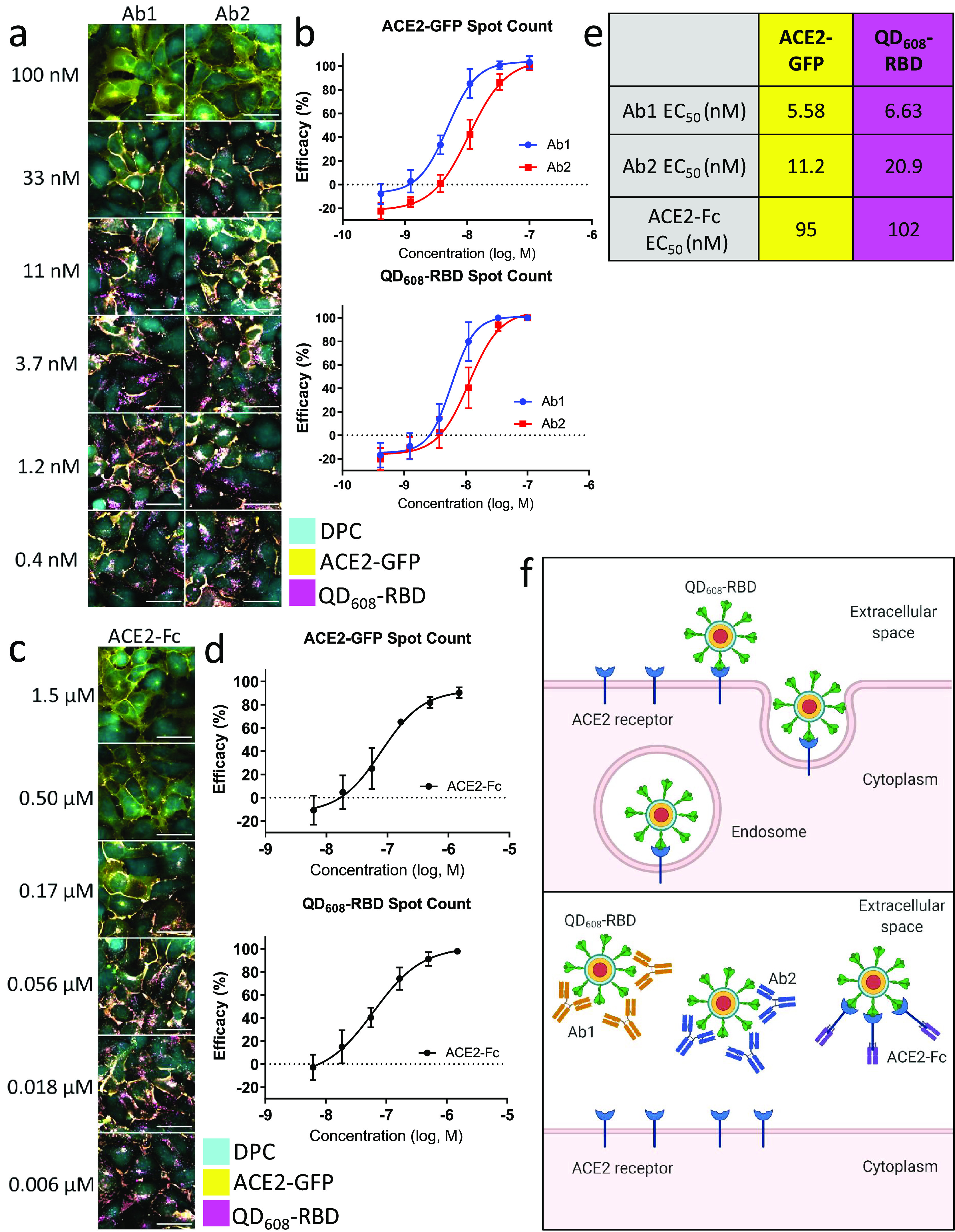
Neutralizing antibodies and ACE2-Fc block QD608-RBD-induced endocytosis. (a) Representative image montage of ACE2-GFP (yellow) HEK293T clone 2 treated with 10 nM QD608-RBD (magenta). Digital phase contrast (cyan) was used during live-cell imaging to identify cell bodies. Cells were treated with neutralizing antibodies Ab1 and Ab2 starting at 100 nM. (b) High-content analysis averages for ACE2-GFP and QD608-RBD spot count treated with neutralizing antibodies. (c) Representative image montage of ACE2-GFP (yellow) HEK293T clone 2 treated with 10 nM QD608-RBD (magenta). Digital phase contrast (cyan) was used during live-cell imaging to identify cell bodies. Cells were treated with ACE2-Fc starting at 1.5 μM. (d) High-content analysis averages for ACE2-GFP and QD608-RBD spot count treated with ACE2-Fc. N ≥ 2000 cells from triplicate wells each from three independent experiments. Error bars indicate SD. (e) Table showing the EC50 values for Ab1, Ab2, and ACE2-Fc based on spot count from (b) and (d). Images were captured using a 40× objective. (f) Illustration of QD608-RBD internalization via receptor-mediated endocytosis and the inhibition using Ab1, Ab2, and ACE2-Fc.
The addition of any exogenous material, whether small molecule or biologic, may have cytotoxic effects that can confound any observed experimental phenomenon. In order to assess the cytotoxicity of QD608-RBD, we conducted ATPlite cell viability assays following the biologics inhibition assays. The ATPlite luminescence signal is dependent upon the amount of adenosine triphosphate (ATP) in cells. Cells with low viability will have lower levels of ATP than cells with high viability. Neither QD608-RBD, Ab1, Ab2, nor ACE2-Fc exhibited any cytotoxicity after 3 h of treatment (Supplementary Figure 8). The negative control cells treated with QD608-RBD alone and the positive control cells treated with Optimem I alone both had equal levels of ATP as reported by the ATPlite luminescence-based reading. These data support the idea that QDs used in this study were not cytotoxic, as previously reported,33 nor was the RBD domain from SARS-CoV-2 itself.
Epithelial Lung Cancer Cell Line Calu-3 Can Uptake QD608-RBD
The permissiveness of different cell types and tissues for SARS-CoV-2 infection is a central question for the research community.34−36 It is important to understand how and whether cells are infected by SARS-CoV-2 and what those effects may be, cytopathic or otherwise. To shed some light on this question and to explore the utility of the QD-RBD reagent further, we cultured Calu-3 cells,38 a cancer cell line derived from lung epithelium and commonly used in coronavirus infection assays, and treated them with 20 nM QD608-RBD. A maximum intensity projection from a 28 μm confocal Z-stack demonstrated entry into some Calu-3 cells, particularly ones that were isolated as opposed to clustered (Supplementary Figure 9). We further immunostained Calu-3 cells for ACE2 expression using the mouse anti-ACE2 antibody and found some level of expression, although weaker than the ACE2-Expi293F cells shown above (Supplementary Figure 9). We hypothesize that even low levels of ACE2 expression in Calu-3 can facilitate QD608-RBD binding and cell entry.
Conclusion
In this work we demonstrate that QD nanoparticles labeled with SARS-CoV-2 RBD can act as pseudovirions that effectively bind ACE2, resulting in an efficient and facile biosensor for biochemical and cell-based assays. Importantly, the QD-RBD constructs and ACE2 enter cells together via dyamin/clathrin-dependent receptor-mediated endocytosis, bound together by the RBD’s high affinity to the ACE2 extracellular domain.
We have explored the utility of this NP-based sensing probe in multiple ways and demonstrate that biologics such as neutralizing antibodies and recombinant protein can act as very potent inhibitors of the viral Spike. Extrapolating to live virus infection assays, our data support the idea that the biologics bind the Spike on the surface of the viral particle, preventing its recognition by the ACE2 receptor, and blocking the downstream effects such as membrane fusion37 and viral endocytosis.25,37,40 The stably transfected ACE2-GFP cell line has proven an invaluable tool in this approach and suggests that some appreciable level of ACE2 is required for recognition of the viral particle. However, there may be other viral receptors that participate in viral entry and infection,25,41 and they could be investigated with our QD probes.
Future work involving advanced human airway epithelial tissue models42 will allow us to probe the spatiotemporal dynamics and features of Spike-ACE2 interactions. Our probes can also be used for HTS of potent antivirals for drug repurposing.43 Additional studies using full-length Spike with cells expressing the host cell protease TMPRSS2 will shed further light on virus–host cell interactions.38 Altogether, we have effectively established a platform technology not only for this SARS-CoV-2 viral pandemic but also for other viruses that have a Spike-mediated cell recognition and entry step as the first step in viral infection.44 We further postulate that the QD-Spike conjugates may act as highly specific and potent delivery vehicles for drugs and other molecules of therapeutic interest.
Methods
Reagents and Materials
CdO (99%) and tri-n-octylphosphine (TOP; min. 97%) were purchased from Strem Chemicals. Behenic acid (99%), 1,2-hexadecanediol (technical grade, 90%), oleylamine (technical grade, 70%), n-octanethiol (98.5+%), and LiOH (≥98%) were purchased from Sigma-Aldrich. 1-Octadecene (ODE; technical grade, 90%) was purchased from Acros Organics. Selenium dioxide (≥97%) was purchased from Fluka. Oleic acid (technical grade, 90%) and 2-(2-aminoethoxy)ethanol (98%) were purchased from Alfa Aesar. All other chemicals, including solvents, were purchased from Sigma-Aldrich or Acros Organics and were used as received.
Dulbecco’s modified essential media (DMEM) (10313021), tetrachloroauric(III) acid, sodium hydroxide, ascorbic acid, sodium citrate, boric acid, Optimem I (11058021), penicillin/streptomycin (15140122), 7.5% bovine serum albumin (BSA) fraction V (15260037), goat-anti-mouse AlexaFluor 488 (A32723; RRID:AB_2633275), High Content Screening Cell Mask Deep Red (H32721), Hoechst 33342 (H3570), and Lipofectamine 3000 (L3000001) were purchased from ThermoFisher Scientific. Mouse anti-ACE2 antibody (E-11): sc-390851 was purchased from Santa Cruz. Hyclone fetal bovine serum (FBS) (SH30071.03) was purchased from General Electric Healthcare. Paraformaldehyde (15714S, 35%) was purchased from Electron Microscopy Sciences. Greiner 96-well poly-d-lysine-coated clear-bottom black microplates (655946) were purchased from Greiner Bio-One. SARS-CoV S1-His (40150-V08B1), SARS-CoV-2 S1S2 ECD-His (40589-V08B1), SARS-CoV-2 S1-His (40591-V08H), SARS-CoV-2 RBD-His (40592-V08B), anti-SARS-CoV-2 S1 neutralizing antibody mouse mAb Ab1 (40591-MM43), and anti-SARS-CoV-2 RBD neutralizing antibody mouse mAb Ab2 (40592-MM57) were purchased from Sino Biological. ACE2-Fc (Z03484) was purchased from Genscript. pCMV6-AC-ACE2-GFP (RG208442) plasmid was purchased from Origene Technologies. Dyngo-4a (ab12068) was purchased from Abcam. ACE2-GFP HEK293T (CB-97100-203) and ACE2-untagged Expi293F cells were purchased from Codex Biosolutions.
QD Synthesis
The 514 nm emitting ZnSe/Cd0.4Zn0.6S/ZnS core–shell QDs and 528 nm emitting CdSe/CdS/ZnS core/shell QDs were synthesized as previously described.45,46 The 608 nm emitting CdSe/CdS/ZnS core/shell QDs were synthesized via modification of published procedures. (i) The CdSe core was synthesized following the published procedure with some modifications.47 CdO (77 mg, 0.60 mmol), behenic acid (0.613 g, 1.80 mmol), and ODE (5.0 mL) were loaded in a 50 mL three-neck flask. The mixture was heated to 260 °C under N2 to dissolve the Cd precursor. The mixture was cooled to 50 °C, and ODE (15 mL) and 1,2-hexadecanediol (0.155 g, 0.60 mmol) were further added. The mixture was degassed at 100 °C for 30 min, then cooled to room temperature. SeO2 (66.6 mg, 0.60 mmol) was added, and the reaction mixture was heated to 240 °C at a rate of ∼25 °C/min under N2. In 3 min after the temperature reached 240 °C, oleic acid (0.60 mL) was added dropwise, the heating mantle was removed, and the reaction mixture was cooled below 50 °C. TOP (0.6 mL), oleylamine (0.6 mL), hexane (9 mL), and methanol (18 mL) were added to the reaction mixture, and the methanol layer was discarded after vigorous stirring for a few minutes. An identical washing procedure was repeated a few more times. The QD solution was transferred to 40 mL vials, and excess isopropanol and ethanol were added to flocculate the QDs. The mixture was centrifuged at 3800 rpm for 5 min. The supernatant was discarded, and the QD pellet was dissolved in CHCl3. The final CdSe QD concentration was estimated following the literature method.48 (ii) Precursors for 0.2 M Cd oleate, 0.2 M Zn oleate, and 0.2 M n-octanethiol solutions for the overcoating procedure were prepared as previously described.45 (iii) For overcoating of CdSe core with CdS and ZnS shells, ODE (5.0 mL), oleylamine (5.0 mL), TOP (1.5 mL), and the CdSe QD core (0.15 μmol in 0.54 mL of CHCl3 solution) were loaded into a 100 mL four-neck round-bottom flask. The reaction mixture was degassed under vacuum at 100 °C to remove CHCl3 and other volatiles and backfilled with N2. The amount of shell precursors used for the overcoating was calculated following the literature procedure.49 For the coating of CdS layers, 0.2 M n-octanethiol in ODE (0.20 mL) was added to the reaction mixture at 100 °C. Then the reaction mixture was heated to 300 °C. Cd oleate (0.2 M) and 0.2 M n-octanethiol in ODE were separately added dropwise using syringe pumps starting at 200 °C. A 1.2-fold excess of n-octanethiol to Cd oleate was used during the CdS overcoating. After the precursor addition was done, the reaction mixture was left for 5 min, then cooled to 200 °C, and annealed for 30 min. The reaction mixture was further cooled to 100 °C and degassed for 30 min to remove volatiles. After backfilling with N2, a coating of ZnS layers was further performed in a similar fashion. The reaction mixture was heated to 290 °C. Zn oleate (0.2 M) and 0.2 M n-octanethiol in ODE were separately added dropwise starting at 250 °C. A 1.4-fold excess of n-octanethiol to Zn oleate was used during the ZnS overcoating. After the precursor addition was done, the reaction mixture was left for 5 min, then cooled to 240 °C, and annealed for 30 min.
QD Ligand Exchange
Typical procedures for the ligand exchange are as follows: QDs coated with native hydrophobic ligands (8.0 nmol in stock solution) were flocculated by mixing with isopropanol and methanol in a 20 mL vial. The mixture was centrifuged at 3800 rpm for 5 min. The clear supernatant was discarded. The QD pellet was mixed with 2-(2-aminoethoxy)ethanol (0.5 mL), CHCl3 (0.8 mL), and methanol (0.8 mL). The reaction mixture was stirred at 45 °C overnight under N2. Excess ethyl acetate was added to the mixture to flocculate the QDs. The mixture was centrifuged at 3800 rpm for 5 min, and the supernatant was discarded. The QD pellet was mixed with CHCl3 (1.0 mL) and methanol (0.5 mL). For the ligand preparation, LiOH (10.2 mg, 4.3 × 10–4 mol) was added to a mixture of CL4 methyl ester precursor50 (76 mg, 1.8 × 10–4 mol), methanol (0.8 mL), and DI water (0.7 mL). The reaction mixture was stirred at room temperature for 30 min. HCl (4 M) was then added dropwise to the reaction mixture to adjust the pH to approximately 7, and NaBH4 (20.4 mg, 5.4 × 10–4 mol) was added to the ligand solution, which was further stirred at room temperature for 1 h under N2. Then, 4 M HCl was added dropwise to the reaction mixture to adjust the pH to approximately 7. The ligand solution was injected by a syringe into the QD solution prepared above with vigorous stirring, and DI water (∼0.7 mL) was further mixed in. The biphasic mixture was stirred at 45 °C overnight under N2. After cooling, the CHCl3 layer was collected by a syringe and discarded. The residual CHCl3 in the aqueous layer was removed by evaporation. The aqueous layer was then filtered through a Millex-LCR membrane filter (pore size 0.45 μm, Millipore) and transferred to a centrifugal spin dialyzer (Amicon Ultra 50K, Millipore). The mixture was diluted with DI water and centrifuged at 3800 rpm for 5 to 10 min, and the clear, filtered solution was discarded. To remove excess unbound ligands and other byproducts, the QD dispersion was subject to a few additional rounds of centrifugation with DI water, followed by filtration through a Millex-LG membrane filter (pore size 0.20 μm, Millipore).
Synthesis of 5 nm AuNPs
AuNPs were synthesized as previously described with slight modification.50 The 5 nm AuNPs were synthesized by a seeded growth method using 3.2 nm seed AuNPs. First 3.2 nm seed NPs were synthesized with sodium citrate and NaBH4. Then 125 μL (1.25 × 10–5 mol) of a 100 mM tetrachloroauric(III) acid (HAuCl4·3H2O) aqueous stock solution and 125 μL (2.5 × 10–5 mol) of 200 mM sodium citrate stock solution were dissolved in 50 mL of deionized H2O; the mixture was then stirred at room temperature for 5 min. A 125 μL (1.0 × 10–4 mol) amount of 1 M sodium borohydride (NaBH4) stock solution in deionized water was added with vigorous stirring. For 5 nm AuNP, the growth solution was prepared with 100 μL (1.00 × 10–5 mol) of a 100 mM tetrachloroauric(III) acid (HAuCl4·3H2O) aqueous stock solution and 100 μL (2.0 × 10–5 mol) of a 200 mM sodium citrate stock solution that were dissolved in 50 mL of deionized H2O. The desired amount of seed NPs, calculated based on the target size of AuNPs and seed size, was added to the growth solution followed by addition of l-ascorbic acid (2 mM final concentration). The reaction mixture was stirred for 30 min at room temperature and kept without stirring for an additional 24 h for the complete reaction. Reaction completion was confirmed by the red shift of the AuNP surface plasmon band peak and the corresponding decrease of the ascorbic acid and aurate peaks in the near-UV region (<300 nm) using UV–vis absorption spectroscopy. The final sizes were confirmed by TEM measurement.
Ligand Exchange of AuNPs with DHLA-NTA/DHLA-PEG-NTA Ligands
Synthesis of the nitrilotriacetic acid-modified thioctic acid (TA-NTA/TA-PEG-NTA, disulfide ring in closed form; DHLA-NTA/DHLA-PEG-NTA, ring in open form) was as previously described.19,52 For ligand exchange, the presynthesized larger AuNPs were added to an excess ligand mixture.38 Briefly, 10 mL of as-synthesized citrate-modified AuNPs was mixed with an excess amount of mixed ligand stock solution containing 50% DHLA, 45% DHLA-NTA, and 5% DHLA-PEG-NTA, which had been deprotected from the ester derivative with an equivalent molar concentration of NaOH for 1 h. The solution was stirred for 8 h and adjusted to pH 8 by adding NaOH, and the dispersion was purified from free ligands by three cycles of centrifugation using a membrane filtration device (Amicon). For Ni coordination for NTA ligand, an excess amount of NiCl2 (500 times of 5 nm AuNP) was directly added to the as-prepared NTA-modified AuNPs and gently stirred for 30 min to promote the interaction between the Ni2+ and NTA on the AuNP surface. The Ni2+-NTA-modified AuNPs were purified using a centrifugal membrane filter (Amicon) and kept at 4 °C until further required.
Protein Conjugation to NPs: QD-Spike and AuNP-ACE2
Histidine-tagged RBD (RBD-His) was conjugated to the QD surface through coordination between the imidazole units of histidine and the ZnS QD shell. The ACE2-His was conjugated to the NTA on the AuNP after activation with the nickel ion that simultaneously coordinates the imidazole units of histidine and NTA.17 For QD-Spike conjugates, the prepared QDs were mixed with stock solution of the histidine-tagged Spike at targeted ratios of protein per QD, and the reaction mixture was adjusted to pH 8 by addition of borate solution (20 mM). After 1 h at room temperature with gentle agitation, BSA (20 μM final concentration) was added to the reaction mixture to prohibit potential nonspecific binding. The prepared QD-Spike conjugates were washed using a centrifuge membrane filter (Amicon Ultra) (100 kDa molecular cut-off, Millipore.Inc.) to remove small chemicals, and the mixture was redispersed in BSA buffer and stored at 4 °C until further use.
For AuNP-ACE2 conjugates, histidine-tagged ACE2 protein was directly added to the Ni-coordinated AuNPs at targeted ratios of ACE2 per AuNP, and the mixture was kept at 4 °C for at least 8 h to complete the reaction (see Results section for AuNP/ACE2 ratios studied in this work). BSA (20 μM final concentration) was added to the reaction mixture to prohibit nonspecific binding. The prepared AuNP-ACE2 conjugates were washed using a centrifuge membrane filter (100 kDa molecular cut-off, Millipore Inc.) to remove low molecular weight impurities, and the mixtures were redispersed in borate buffer (with BSA) and stored at 4 °C until further use.
Unconjugated NP Characterization
Three different techniques were used to characterize the QDs and AuNP used in this study: (1) Electronic absorption and PL emission spectra were recorded using a Shimadzu UV-1800 UV–vis spectrophotometer and a Horiba, Inc. fluorometer (excitation at λ = 395 nm), respectively. (2) Dynamic light scattering (DLS) was used to measure hydrodynamic size. The samples were transferred into a square-shaped capillary, and measurements were recorded on a ZetaSizer Ultra instrument equipped with a HeNe laser source (λ = 633 nm) (Malvern Instruments Ltd., Worcestershire, UK) and analyzed using Dispersion Technology Software (Malvern Instruments Ltd.) as previously described.20 (3) Structural characterization and elemental analysis of the as-prepared NPs were carried out using a JEOL 2200-FX analytical high-resolution transmission electron microscope with a 200 kV accelerating voltage. TEM samples were prepared by spreading a drop (5–10 μL) containing the NPs onto an ultrathin carbon/holey support film on a 300 mesh Au grid (Ted Pella, Inc.) and letting it dry. The concentration of NPs used for TEM was 50–100 nM. Individual particle sizes were measured using a Gatan digital micrograph (Pleasanton, CA, USA); average sizes along with standard deviations were extracted from analysis of at least 50–100 nanoparticles.
Gel Electrophoresis
Conjugation of proteins to QD608 or AuNPs was confirmed using an electrophoretic mobility shift assay with a 1% Agarose gel and 1× TBE buffer at 90 mV.5 Gel images were taken every 5 min utilizing a Bio-Rad ChemiDoc XRS+ gel imager under fluorescent light for QDs or Epi-white light for AuNPs. Ratios of RBD to QD608 varied from 0 to 16, and those of ACE2 to AuNP from 1 to 3. The retardation of migration through the gel as the ratio of protein to NP increased confirmed conjugation of the protein to NPs.
NP-Based Energy Transfer Assay
QD-RBD conjugates (or QD-S1) were mixed with AuNP-ACE2 conjugates with targeted ratios of AuNP-ACE2 to QD-RBD ranging from 0 to 10. The reaction mixtures were incubated for 2 h at room temperature. The general concentration of QD was approximately 3 to 10 nM. A basic buffer containing 20 mM borate and 20 μM BSA was used to stabilize all reactions, unless described separately. The QD fluorescence spectra were obtained at each ratio with 395 nm excitation. The fluorescence images of a series of solutions at increasing AuNP-ACE2 to QD-RBD ratios were taken under excitation with a hand-held UV lamp at 375 nm. For inhibition assays, the desired amount of inhibitor was incubated with QD-RBD for 3 h at room temperature, followed by adding AuNP-ACE2 and incubating for 2 h at room temperature. Fluorescence spectra were obtained in an identical manner to that described above.
Quantum Yield Measurements
Fluorescence quantum yields (Φ) were measured at room temperature with fluorescein in 0.1 N NaOH (Φ = 0.93)53 for QD514 and QD528 or Rhodamine 101 in ethanol (Φ = 1.0)54 for QD608 as standards. The obtained fluorescence spectra were corrected using the spectral output of a calibrated light source supplied by the National Bureau of Standards. The parameters in eq 1 include the integrated PL intensities of the QD and standard in arbitrary units (a.u.), PLQD and PLst, their optical density at excitation wavelength, ODQD and ODst, and the refractive indices of their media, nQD and nst, respectively.51
| 1 |
Generation of Stably Transfected Cell Lines
ACE2-GFP HEK293T
HEK293T cells were seeded into cells in a six-well plate with 70–80% confluency. For each well, the cells were transfected with 2.5 μg of pCMV6-AC-ACE2-GFP plasmid using Lipofectamine 3000 (ThermoFisher). Twenty-four hours later, the cells were disassociated with trypsin and transferred into 100 mm dishes. The cells were selected with 1.0 mg/mL G418 for 2–3 weeks. Single colonies were picked into 24-well plates containing 1.0 mL of DMEM with 10% FBS supplemented with 1.0 mg/mL G418. The clones with the brightest GFP signals were picked for propagation.
ACE2-Expi293F
Expi293F cells (ThermoFisher) were seeded into cells in a six-well plate with 70–80% confluency. For each well, the cells were transfected with 2.5 μg of pCMV-ACE2-IRES-Puromycin plasmid (Codex BioSolutions) using Lipofectamine 3000. Twenty-four hours later, the cells were disassociated with trypsin and transferred into 100 mm dishes. The cells were selected with 1 μg/mL puromycin for 2–3 weeks. Single colonies were picked into 24-well plates containing 1 mL of DMEM and 10% FBS supplemented with 1 μg/mL puromycin. Western blot was performed to screen the ACE2 expression clones with an ACE2-specific antibody (sc-390851, Santa Cruz).
Cell Culture
ACE2-GFP and ACE2-Expi293 cells were cultured using DMEM complete with 10% FBS and 1% Pen/Strep in large T175 flasks until 80% to 90% confluence prior to seeding in 96-well plates at 25 000 cells per well. Cells were incubated overnight at 37 °C and 5% CO2.
Calu-3 cells were cultured using EMEM complete with 10% FBS and 1% Pen/Strep in large T175 flasks until 80% to 90% confluence prior to seeding in 96-well plates at 20 000 cells per well. Cells were incubated overnight at 37 °C and 5% CO2.
Immunofluorescence Staining
Cells were washed three times with PBS prior to fixation using 4% PFA in PBS with 0.1% BSA for 30 min. Cells were washed three times followed by permeabilization with 0.5% saponin in Cell Staining Buffer for 15 min followed by blocking in Cell Staining Buffer for an additional 45 min. Then, cells were incubated with 1:200 mouse anti-ACE2 antibody overnight at 4 °C. The next day, cells were washed three times with PBS and incubated with 1:1000 goat anti-mouse AlexaFluor 488 for 1 h followed by 3× PBS washes. Finally, cells were incubated with Hoechst 33342 to stain the nuclei and HCS Cell Mask Deep Red when required. Cells were washed three final times in PBS prior to sealing of the plates for imaging.
QD and Spike Treatment
Prior to treatment with QD-Spike conjugates, cells were washed once with prewarmed Optimem I. Stock QD or recombinant protein solution was diluted directly in Optimem I, and 50 μL of QD working solution was added to cells for the indicated amount of time at 37 °C and 5% CO2.
High-Content Imaging and Analysis
Cells were placed into the Opera Phenix (PerkinElmer) automated confocal imaging system that was preheated to 37 °C. A 40× or 63× water immersion objective was used to capture multiple fields per well at a single Z-position. Cells were not washed further prior to imaging. Images were captured with digital phase contrast, green and orange channels. QDs were first exposed to UV light prior to capturing emission using the orange (λ = 570–630 nm) emission bandpass. Images were uploaded into the Columbus analyzer (PerkinElmer) and analyzed using custom protocols. Where applicable, the digital phase contrast channel was used to identify the cell bodies, and the spots were identified in the green (Cam1: λ = 435–550 nm) and orange (Cam2: λ = 570–630 nm) channels for ACE2-GFP and QD608, respectively. Data were exported into Microsoft Excel, and graphs were plotted using Graphpad Prism V8.4.3. For inhibition experiments using neutralizing antibodies or ACE2-Fc, data were normalized to the Optimem I only treated cells (100%, positive control) or QD608-RBD treated cells (0%, negative control).
For Dyngo-4a endocytosis inhibition experiments, cells were preincubated with 20 μM Dyngo-4a in Optimem I for 15 min. Afterward, 2× concentrated solutions of Dyngo-4a and QD608-RBD was added to an equal volume of Optimem I for a final concentration of 20 μM Dyngo-4a and 10 or 20 nM QD608-RBD. Imaging began immediately after the addition of QD608-RBD with minimal delay. Images were captured every 10 min for 3 h. For endocytosis experiments using Dyngo-4a, data were normalized to the Optimem I only treated cells (100%, positive control) or QD608-RBD treated cells (0%, negative control).
Image montages were constructed using Fiji (NIH). All images for each channel were first stacked before using the auto feature to equally adjust the brightness and contrast across the conditions. For time-lapse videos, images were registered using the StackReg plugin in Fiji, and stacks were saved as .avi files. Videos were subsequently slowed in Windows 10 video editor (Microsoft) and saved as .mpg files.
Single-Molecule Fluorescence Microscopy
Single-molecule imaging experiments were conducted on a custom-built Nikon Ti microscope coupled with a 100× oil-immersion objective lens (NA = 1.49), a multiband dichroic (405/488/561/633 BrightLine quad-band bandpass filter, Semrock, USA), and a piezo z-stage (ASI, USA). The lasers were focused into the back pupil plane of the objective to generate wide-field illumination. A Nikon N-STORM module was used to control the angle of the laser beam for generating inclined illumination. The emission was collected by the same objective passing through a quadband bandpass emission filter (FF01-446/523/600/677-25, Semrock, USA) in front of an sCMOS camera (Prime 95B, Teledyne Photometrics). The microscope, lasers, and the camera were controlled through NIS-Elements (Nikon, USA). A 488 nm laser was used excite the QDs.
Single-Molecule Tracking and Analysis
Single-molecule tracking was performed with custom-written MATLAB software.55 The MATLAB scripts, SLIMFAST/evalSPT, were used to localize and track single molecules. The positions of the diffraction-limited spots in the trajectories were determined with a 2D Gaussian fit. A maximal expected diffusion constant was set to connect localizations between consecutive frames.
Mean square displacements were calculated from x,y positions as previously described.56 We determined the instantaneous diffusion coefficients from a linear fit of the initial points of the MSD (between time lag 1 and 5). The MSD curves for all the tracks were computed with @msdanalyzer script.57
For jump distance analysis, the probability that a particle located at position r at time t in two dimension, will be found at position r′ at time t+tau is given by58
where D is the diffusion constant.
In the case of 2D diffusion, the displacement probability was obtained through integrating the above equation over the circular shell of width (dr):
Experimentally, this probability distribution can be approximated by counting the jump distances within respective intervals (r, r + dr) traveled by a single QD during a given time (camera exposure time).
The diffusion coefficient of different species was determined through nonlinear fitting the jump distance histogram with multicomponents. The F-test was performed to compare the single-, two-, and three-component fitting models.
Statistical Analysis and Illustration
For biochemical assays, all experiments were performed with at least three independent experiments, and the TEM size was analyzed with 50–100 randomly chosen nanoparticles in different images. For cell-based assays, all experiments where statistical analysis was performed included three independent experiments with three independent wells unless otherwise noted. Data shown as mean ± standard deviation (SD). Concentration–response curves and EC50 values were generated using nonlinear regression. The illustration in Figure 6f was created using Biorender.
Acknowledgments
We acknowledge Dr. Chiou and Dr. Liou at AIMLab at the University of Maryland, College Park, for their help in operating the TEM facility. We also thank NCATS scientists Dr. Matthew Hall and Dr. Catherine Chen for discussions as well as NRL scientists Dr. Michael Stewart and Dr. Alan Huston for discussions. We thank Dr. Zeenat Asghar Shyr for critical reading of the manuscript. We also appreciate the initial suggestion from Dr. Craig Thomas at NCATS for making the connection between NRL and NCATS. This research was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH. Naval Research Laboratory provided funding via its internal Nanoscience Institute. Reagent preparation was supported via the NRL COVID-19 base fund.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c05975.
Video 1. Confocal time lapse of ACE2-GFP HEK293T treated with Optimem I in the ACE2-GFP channel (yellow) (MP4)
Video 2. Confocal time lapse of ACE2-GFP HEK293T treated with 20 nM QD608-RBD in the QD608-RBD channel (magenta) (MP4)
Video 3. Confocal time lapse of ACE2-GFP HEK293T treated with 20 nM QD608-RBD in the ACE2-GFP channel (yellow) (MP4)
Video 4. Confocal time lapse of ACE2-GFP HEK293T treated with 20 μM Dyngo-4a in the ACE2-GFP channel (yellow).(MP4)
Video 5. Confocal time lapse of ACE2-GFP HEK293T treated with 20 nM QD608-RBD and 20 μM Dyngo-4a in the QD608-RBD channel (magenta) (MP4)
Supplementary Video 6. Confocal time lapse of ACE2-GFP HEK293T treated with 20 nM QD608-RBD and 20 μM Dyngo-4a in the ACE2-GFP channel (yellow) (MP4)
Video 7. Single-molecule imaging time lapse of ACE2-GFP HEK293T treated with 200 pM QD608-RBD in the QD608-RBD channel (AVI)
Supplemental discussion section describing in more detail the impetus for biochemical and cell-based assays and the rationale for choosing the methods that were used in the study; detailed description of the experiments and their implications; methods for the energy transfer models (PDF)
Author Contributions
Experimental: KG, KS, MX, MP, WZ, JCB, EO. Data analysis: KG, JCB, EO. Manuscript conception: KG, MW, EO. Manuscript writing: KG, JCB, MW, EO. Preparation of figures: KG, JCB, EO, XH. Critical manuscript editing and discussion: KG, KS, MW, EO.
The authors declare no competing financial interest.
Supplementary Material
References
- Rothan H. A.; Byrareddy S. N. The Epidemiology and Pathogenesis of Coronavirus Disease (COVID-19) Outbreak. J. Autoimmun. 2020, 109, 102433. 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E.; Du H.; Gardner L. An Interactive Web-Based Dashboard to Track COVID-19 in Real Time. Lancet Infect. Dis. 2020, 20, 533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; Mcguire A. T.; Veesler D. Structure, Function, and Antigenicity of The SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K.; Imai Y.; Rao S.; Gao H.; Guo F.; Guan B.; Huan Y.; Yang P.; Zhang Y.; Deng W. A Crucial Role of Angiotensin Converting Enzyme 2 (ACE2) in SARS Coronavirus–Induced Lung Injury. Nat. Med. 2005, 11, 875–879. 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Hou Y.; Shen J.; Huang Y.; Martin W.; Cheng F. Network-Based Drug Repurposing for Novel Coronavirus 2019-nCov/SARS-CoV-2. Cell Discov 2020, 6, 1–18. 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchez M. Jr.; Moronne M.; Gin P.; Weiss S.; Alivisatos A. P. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 1998, 281, 2013–2016. 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Alivisatos P. The Use of Nanocrystals in Biological Detection. Nat. Biotechnol. 2004, 22, 47–52. 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- Sapsford K. E.; Pons T.; Medintz I. L.; Mattoussi H. Biosensing with Luminescent Semiconductor Quantum Dots. Sensors 2006, 6, 925–953. 10.3390/s6080925. [DOI] [Google Scholar]
- Algar W. R.; Susumu K.; Delehanty J. B.; Medintz I. L. Semiconductor Quantum Dots in Bioanalysis: Crossing the Valley of Death. Anal. Chem. 2011, 83, 8826–8837. 10.1021/ac201331r. [DOI] [PubMed] [Google Scholar]
- Hildebrandt N.; Spillmann C. M.; Algar W. R.; Pons T.; Stewart M. H.; Oh E.; Susumu K.; Díaz S. A.; Delehanty J. B.; Medintz I. L. Energy Transfer with Semiconductor Quantum Dot Bioconjugates: A Versatile Platform for Biosensing, Energy Harvesting, and Other Developing Applications. Chem. Rev. 2017, 117, 536–711. 10.1021/acs.chemrev.6b00030. [DOI] [PubMed] [Google Scholar]
- Oh E.; Hong M.-Y.; Lee D.; Nam S.-H.; Yoon H. C.; Kim H.-S. Inhibition Assay of Biomolecules Based on Fluorescence Resonance Energy Transfer (FRET) between Quantum Dots and Gold Nanoparticles. J. Am. Chem. Soc. 2005, 127, 3270–3271. 10.1021/ja0433323. [DOI] [PubMed] [Google Scholar]
- Dyadyusha L.; Yin H.; Jaiswal S.; Brown T.; Baumberg J. J.; Booy F. P.; Melvin T. Quenching of CdSe Quantum Dot Emission, A New Approach for Biosensing. Chem. Commun. 2005, 3201–3203. 10.1039/b500664c. [DOI] [PubMed] [Google Scholar]
- Pons T.; Medintz I. L.; Sapsford K. E.; Higashiya S.; Grimes A. F.; English D. S.; Mattoussi H. On the Quenching of Semiconductor Quantum Dot Photoluminescence by Proximal Gold Nanoparticles. Nano Lett. 2007, 7, 3157–3164. 10.1021/nl071729+. [DOI] [PubMed] [Google Scholar]
- Kim Y.-P.; Oh Y.-H.; Oh E.; Ko S.; Han M.-K.; Kim H.-S. Energy Transfer-Based Multiplexed Assay of Proteases by Using Gold Nanoparticle and Quantum Dot Conjugates on a Surface. Anal. Chem. 2008, 80, 4634–4641. 10.1021/ac702416e. [DOI] [PubMed] [Google Scholar]
- Chen C.; Hildebrandt N. Resonance Energy Transfer to Gold Nanoparticles: NSET Defeats FRET. TrAC, Trends Anal. Chem. 2020, 123, 115748. 10.1016/j.trac.2019.115748. [DOI] [Google Scholar]
- Wang H.; Yang P.; Liu K.; Guo F.; Zhang Y.; Zhang G.; Jiang C. SARS Coronavirus Entry into Host Cells through a Novel Clathrin- and Caveolae-Independent Endocytic Pathway. Cell Res. 2008, 18, 290–301. 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B.; Kadhiresan P.; Kozlowski H. N.; Malekjahani A.; Osborne M.; Li V. Y. C.; Chen H.; Mubareka S.; Gubbay J. B.; Chan W. C. W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Weiss C.; Carriere M.; Fusco L.; Capua I.; Regla-Nava J. A.; Pasquali M.; Scott J. A.; Vitale F.; Unal M. A.; Mattevi C.; Bedognetti D.; Merkoçi A.; Tasciotti E.; Yilmazer A.; Gogotsi Y.; Stellacci F.; Delogu L. G. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14, 6383–6406. 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- Breger J. C.; Oh E.; Susumu K.; Klein W. P.; Walper S. A.; Ancona M. G.; Medintz I. L. Nanoparticle Size Influences Localized Enzymatic Enhancement—A Case Study with Phosphotriesterase. Bioconjugate Chem. 2019, 30, 2060–2074. 10.1021/acs.bioconjchem.9b00362. [DOI] [PubMed] [Google Scholar]
- Oh E.; Susumu K.; Goswami R.; Mattoussi H. One-Phase Synthesis of Water-Soluble Gold Nanoparticles with Control Over Size and Surface Functionalities. Langmuir 2010, 26, 7604–7613. 10.1021/la904438s. [DOI] [PubMed] [Google Scholar]
- Förster T. Zwischenmolekulare Energiewanderung Und Fluoreszenz. Ann. Phys. 1948, 437, 55–75. 10.1002/andp.19484370105. [DOI] [Google Scholar]
- Jennings T. L.; Singh M. P.; Strouse G. F. Fluorescent Lifetime Quenching near d = 1.5 nm Gold Nanoparticles: Probing NSET Validity. J. Am. Chem. Soc. 2006, 128, 5462–5467. 10.1021/ja0583665. [DOI] [PubMed] [Google Scholar]
- Oh E.; Huston A. L.; Shabaev A.; Efros A.; Currie M.; Susumu K.; Bussmann K.; Goswami R.; Fatemi F. K.; Medintz I. L. Energy Transfer Sensitization of Luminescent Gold Nanoclusters: More Than Just the Classical Förster Mechanism. Sci. Rep. 2016, 6, 35538. 10.1038/srep35538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddayasankar U.; Krull U. J. Energy Transfer Assays Using Quantum Dot–Gold Nanoparticle Complexes: Optimizing Oligonucleotide Assay Configuration Using Monovalently Conjugated Quantum Dots. Langmuir 2015, 31, 8194–8204. 10.1021/acs.langmuir.5b01932. [DOI] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M. J.; Deane F. M.; Robinson P. J.; Mccluskey A. Synthesis of Dynole 34–2, Dynole 2–24 and Dyngo 4a for Investigating Dynamin Gtpase. Nat. Protoc. 2014, 9, 851–870. 10.1038/nprot.2014.046. [DOI] [PubMed] [Google Scholar]
- Young L. J.; Ströhl F.; Kaminski C. F. A Guide to Structured Illumination TIRF Microscopy at High Speed with Multiple Colors. J. Visualized Exp. 2016, 53988. 10.3791/53988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia D.; Arumugam S.; Nasilowski M.; Joshi H.; Wunder C.; Chambon V.; Prakash V.; Grazon C.; Nadal B.; Maiti P. K.; Johannes L.; Dubertret B.; Krishnan Y. Quantum Dot-Loaded Monofunctionalized DNA Icosahedra for Single-Particle Tracking of Endocytic Pathways. Nat. Nanotechnol. 2016, 11, 1112–1119. 10.1038/nnano.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Shao L.; Chen B.-C.; Zhang X.; Zhang M.; Moses B.; Milkie D. E.; Beach J. R.; Hammer J. A.; Pasham M.; Kirchhausen T.; Baird M. A.; Davidson M. W.; Xu P.; Betzig E. Extended-Resolution Structured Illumination Imaging of Endocytic and Cytoskeletal Dynamics. Science 2015, 349, Aab3500. 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.; Hillyer C.; Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 335–359. 10.1016/j.it.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Li W.; Drabek D.; Okba N. M.; Van Haperen R.; Osterhaus A. D.; Van Kuppeveld F. J.; Haagmans B. L.; Grosveld F.; Bosch B.-J. A Human Monoclonal Antibody Blocking SARS-CoV-2 Infection. Nat. Commun. 2020, 11, 1–6. 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B.; Zhang Q.; Ge J.; Wang R.; Sun J.; Xiangyang G.; Jiazhen Y.; Shan S.; Zhou B.; Song S.; Tang X.; Yu J.; Lan J.; Yuan J.; Wang H.; Zhao J.; Zhang S.; Wang Y.; Shi X.; Liu L.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Maksoudian C.; Soenen S. J.; Susumu K.; Oh E.; Medintz I. L.; Manshian B. B. A Multiparametric Evaluation of Quantum Dot Size and Surface-Grafted Peptide Density on Cellular Uptake and Cytotoxicity. Bioconjugate Chem. 2020, 31, 1077–1087. 10.1021/acs.bioconjchem.0c00078. [DOI] [PubMed] [Google Scholar]
- Letko M.; Marzi A.; Munster V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Li X.; Chen M.; Feng Y.; Xiong C. The ACE2 Expression in Human Heart Indicates New Potential Mechanism of Heart Injury among Patients Infected with SARS-CoV-2. Cardiovasc. Res. 2020, 116, 1097–1100. 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S.; Chua R. L.; Trefzer T.; Kahn N. C.; Schneider M. A.; Muley T.; Winter H.; Meister M.; Veith C.; Boots A. W. SARS-CoV-2 Receptor ACE 2 and TMPRSS2 Are Primarily Expressed in Bronchial Transient Secretory Cells. EMBO J. 2020, 39, E105114 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov K.; Chen C. Z.; Bostwick R.; Rasmussen L.; Xu M.; Pradhan M.; Tran B. N.; Zhu W.; Shamim K.; Huang W., The SARS-CoV-2 Cytopathic Effect is Blocked with Autophagy Modulators. Biorxiv; 2020 10.1101/2020.05.16.091520 (accessed July 13, 2020). [DOI] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N.; Shen H.-M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Sun H.; Bu X.; Wan G. New Strategy for COVID-19: An Evolutionary Role for RGD Motif in SARS-CoV-2 and Potential Inhibitors for Virus Infection. Front. Pharmacol. 2020, 11, 912. 10.3389/fphar.2020.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V.; Kwon H.; Prado P.; Hagelkrüys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Del Pozo C. H.; Prosper F. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov O. O. Understanding SARS-CoV-2 Endocytosis for COVID-19 Drug Repurposing. FEBS J. 2020, 1–8. 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T. M.; Buchmeier M. J. Coronavirus Spike Proteins in Viral Entry and Pathogenesis. Virology 2001, 279, 371–374. 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susumu K.; Field L. D.; Oh E.; Hunt M.; Delehanty J. B.; Palomo V.; Dawson P. E.; Huston A. L.; Medintz I. L. Purple-, Blue-, and Green-Emitting Multishell Alloyed Quantum Dots: Synthesis, Characterization, and Application for Ratiometric Extracellular pH Sensing. Chem. Mater. 2017, 29, 7330–7344. 10.1021/acs.chemmater.7b02174. [DOI] [Google Scholar]
- Susumu K.; Oh E.; Delehanty J. B.; Pinaud F.; Gemmill K. B.; Walper S.; Breger J.; Schroeder M. J.; Stewart M. H.; Jain V. A New Family of Pyridine-Appended Multidentate Polymers as Hydrophilic Surface Ligands for Preparing Stable Biocompatible Quantum Dots. Chem. Mater. 2014, 26, 5327–5344. 10.1021/cm502386f. [DOI] [Google Scholar]
- Chen O.; Chen X.; Yang Y.; Lynch J.; Wu H.; Zhuang J.; Cao Y. C. Synthesis of Metal–Selenide Nanocrystals Using Selenium Dioxide as the Selenium Precursor. Angew. Chem., Int. Ed. 2008, 47, 8638–8641. 10.1002/anie.200804266. [DOI] [PubMed] [Google Scholar]
- Jasieniak J.; Smith L.; Van Embden J.; Mulvaney P.; Califano M. Re-Examination of the Size-Dependent Absorption Properties of CdSe Quantum Dots. J. Phys. Chem. C 2009, 113, 19468–19474. 10.1021/jp906827m. [DOI] [Google Scholar]
- Chen D.; Zhao F.; Qi H.; Rutherford M.; Peng X. Bright and Stable Purple/Blue Emitting Cds/Zns Core/Shell Nanocrystals Grown by Thermal Cycling Using a Single-Source Precursor. Chem. Mater. 2010, 22, 1437–1444. 10.1021/cm902516f. [DOI] [Google Scholar]
- Susumu K.; Oh E.; Delehanty J. B.; Blanco-Canosa J. B.; Johnson B. J.; Jain V.; Hervey IV W. J.; Algar W. R.; Boeneman K.; Dawson P. E. Multifunctional Compact Zwitterionic Ligands for Preparing Robust Biocompatible Semiconductor Quantum Dots and Gold Nanoparticles. J. Am. Chem. Soc. 2011, 133, 9480–9496. 10.1021/ja201919s. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R.2.10 Quantum Yield Standards, Principles of Fluorescence Spectroscopy, 2nd ed.; Springer Science & Business Media: New York, 2013; pp 52–53. [Google Scholar]
- Dwyer C. L.; Díaz S. N. A.; Walper S. A.; Samanta A.; Susumu K.; Oh E.; Buckhout-White S.; Medintz I. L. Chemoenzymatic Sensitization of DNA Photonic Wires Mediated through Quantum Dot Energy Transfer Relays. Chem. Mater. 2015, 27, 6490–6494. 10.1021/acs.chemmater.5b02870. [DOI] [Google Scholar]
- Magde D.; Wong R.; Seybold P. G. Fluorescence Quantum Yields and Their Relation to Lifetimes of Rhodamine 6G and Fluorescein in Nine Solvents: Improved Absolute Standards for Quantum Yields. Photochem. Photobiol. 2002, 75, 327–334. . [DOI] [PubMed] [Google Scholar]
- Karstens T.; Kobs K. Rhodamine B and Rhodamine 101 as Reference Substances for Fluorescence Quantum Yield Measurements. J. Phys. Chem. 1980, 84, 1871–1872. 10.1021/j100451a030. [DOI] [Google Scholar]
- Chen J.; Zhang Z.; Li L.; Chen B.-C.; Revyakin A.; Hajj B.; Legant W.; Dahan M.; Lionnet T.; Betzig E.; Tjian R.; Liu Z. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell 2014, 156, 1274–1285. 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M. J.; Jacobson K. Single-Particle Tracking: Applications to Membrane Dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 373–99. 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Tarantino N.; Tinevez J. Y.; Crowell E. F.; Boisson B.; Henriques R.; Mhlanga M.; Agou F.; Israël A.; Laplantine E. TNF And IL-1 Exhibit Distinct Ubiquitin Requirements for Inducing NEMO-IKK Supramolecular Structures. J. Cell Biol. 2014, 204, 231–45. 10.1083/jcb.201307172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D.; Abernathy A.; Golob N.; Morisaki T.; Mcnally J. G. A Benchmark for Chromatin Binding Measurements in Live Cells. Nucleic Acids Res. 2012, 40, E119 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.