Abstract
Genetically identical individuals can be phenotypically variable, even in constant environmental conditions. The ubiquity of this phenomenon, known as ‘intra-genotypic variability’ is increasingly evident, and the relevant mechanistic underpinnings are beginning to be understood. In parallel, theory has delineated a number of formal expectations for contexts in which such a feature would be adaptive. Here, we review empirical evidence across biological systems, and theoretical expectations, including nonlinear averaging and bet hedging. We synthesize existing results to illustrate the dependence of selection outcomes both on trait characteristics, features of environmental variability, and species’ demographic context. We conclude by discussing ways to bridge the gap between empirical evidence of intra-genotypic variability, studies demonstrating its genetic component, and evidence that it is adaptive.
Keywords: Intra-genotypic variability, vQTL, nonlinear averaging, bet hedging, micro-environmental plasticity, phenotypic variance
Responses to environmental variability
No environment on this planet is stable. Most organisms experience short-term fluctuations (such as seasonal variation, or stochastic fluctuations in temperature, rainfall and resources) as well as longer-term trends (such as those driven by global change). Such environmental variation drives the eco-evolutionary dynamics that contribute to maintaining genetic variation between individuals, and shapes phenotypic change. In turn, the fate of populations is determined in part by whether they harbor sufficient phenotypic variation to adapt to changing environments [1].
Perhaps the most frequently discussed source of phenotypic variation is genetic variation between individuals. Where sufficient genetic variation exists, a shift in the genetic makeup of the population will allow some individuals to persist in new conditions. This process can result in adaptive tracking (see Glossary), where the phenotypic mean of a population tracks a fitness optimum. Alternatively, adaptive phenotypic plasticity may allow populations to keep pace with environmental change, as specific genotypes modulate their phenotype in response to experienced environmental conditions [2]. These two components of population persistence in variable environments have been extensively studied both theoretically and empirically. An alternative strategy has received far less attention: changes in the level of phenotypic variance (rather than the mean) may be a key element in allowing populations to cope with changing environments.
The evolution of intra-genotypic variability, phenotypic variation observed across genetically identical individuals, leads to phenotypic variation even in a constant environment [3] (Box 1). This is in contrast to phenotypic plasticity, in which phenotypic variation is correlated with the environment. A life history strategy that maintains intra-genotypic variability will increase the probability that at least some individuals are well suited to particular environmental conditions [4]. Beyond genetic variation and phenotypic plasticity, intra-genotypic variability can thus be an important third axis by which populations can persist across changing environmental conditions, and is our focus here.
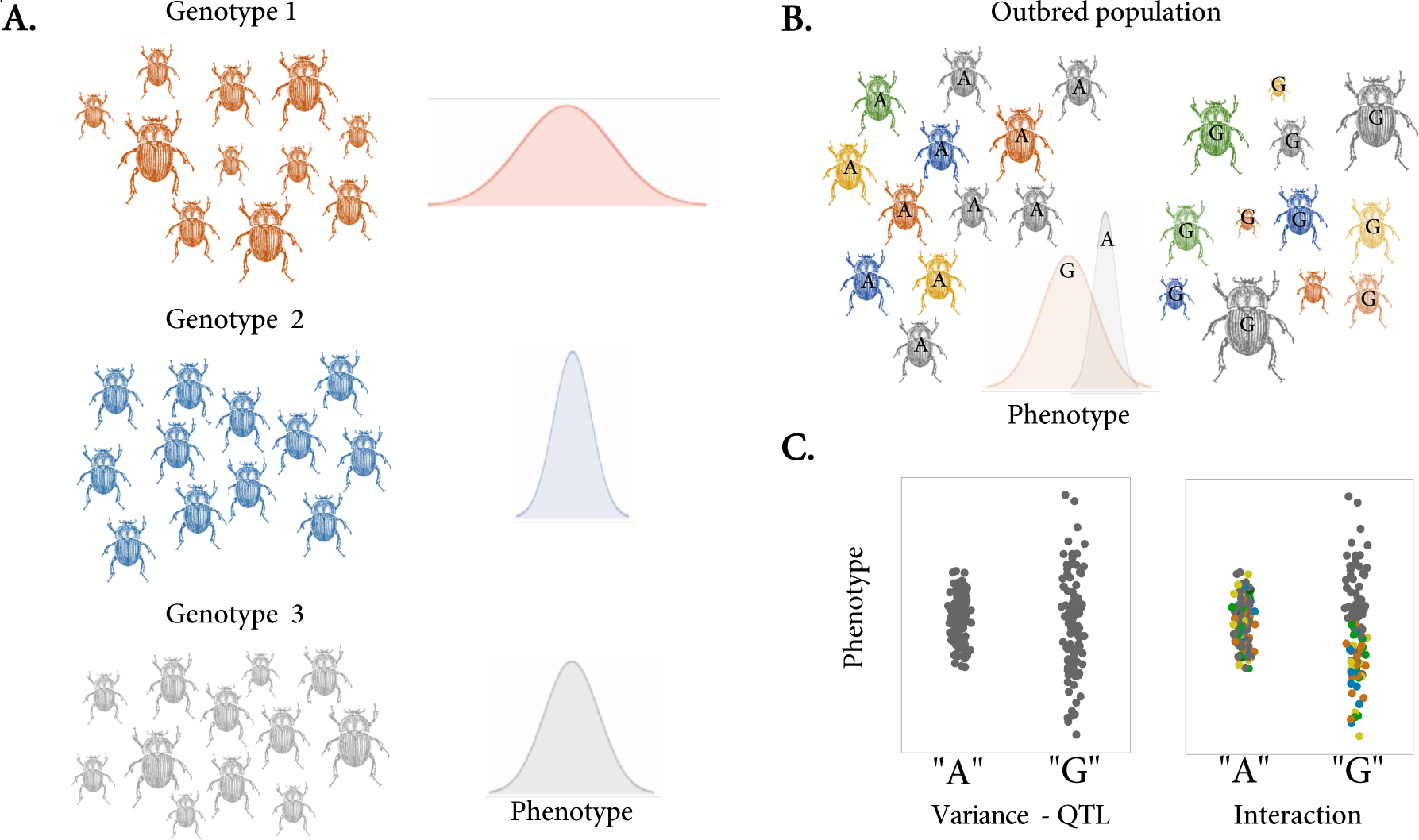
Box 1: Intra-genotypic variability in inbred and outbred populations.
Intra-genotypic variability is often described in the context of genetically identical individuals (e.g., parthenogenic species or isogenic lines). As illustrated in Fig I, panel A, when several isogenic individuals can be reared in the same nominal environment they often display different trait values. Here three genetic backgrounds (different colors) show different degrees of phenotypic variability for size, and the estimated variance across genotypes can be used directly to estimate heritability or map variance-QTLs. However, intra-genotypic variability also contributes to individual variation in outbred populations. Fig I, panel B represents a population of outbred individuals segregated by genotypes at a given locus (e.g., A or G) showing variance heterogeneity for size (the colors represent diverse genetic backgrounds). In both inbred and outbred populations, a given genotypic class is associated with different degrees of phenotypic variability. The key difference is that in an outbred population the observation of variance heterogeneity can either be driven by the presence of a variance controlling allele (e.g., A or G in this figure; the focus of this manuscript) or could result from mean-controlling loci interacting epistatically with another locus, leading to variance heterogeneity across genotypes [95,96]. Please note that an instance for inbred populations is not shown here. Fig I, panel C illustrates this point - on the left, the variance effect is driven by a variance QTL where a polymorphism is directly associated with a difference in variance across these alleles. On the right, the variance effect results from loci interacting epistatically. The phenotype of the “A” harboring individuals has low variance (regardless of the genetic background), while the phenotype of the “G” harboring individuals is a function of the genotype at a different locus (illustrated by the two distributions within the “G” allele). This distinction has important implications for the evolution of variance control and how it is studied. Here, we focus on the evolution of variance control from the perspective of variance controlling alleles, not variance heterogeneity driven by genetic interactions. While such heterogeneity could technically be maintained by epistasis under balancing selection, such cases have yet to be documented.
What is known about intra-genotypic variability?
Although there are surprisingly few studies investigating the contribution of intra-genotypic variability to phenotypic variation between individuals, various lines of evidence indicate that intra-genotypic variability is ubiquitous. Such variance in the variance commonly emerges in artificial breeding programs, as a nuisance to farmers who seek uniformity in their crops or livestock (e.g., observed in birth weight of rabbits (family: Leporidae) [5], mice (Mus musculus) [6] and rainbow trout (Oncorhynchus mykiss) [7]). Moreover, breeders have recently started using intra-genotypic variability as a resilience indicator [8,9]. Intra-genotypic variability has also been commonly observed for a wide range of traits in experimental model systems such as vinegar flies (Drosophila melanogaster), yeast (Saccharomyces cerevisiae) or nematodes (Caenorhabditis elegans) [10–12]. Finally, it has been observed in natural populations of plants (e.g., in seed banks [13,14]), mammals (e.g., in docility behavior in yellow-bellied marmots [Marmota flaviventris] [15]) and birds (in fledgling weight in great tits [Parus major] [16]), to list a few. However, the mechanisms generating intra-genotypic variability are still poorly understood. The phenotypic differences observed among genetically identical individuals are believed to result from both 1) the existence of stochastic micro-environmental variation affecting development (known as “developmental instability” [17]); and 2) the ability of an individual’s genotype to buffer development against these micro-environmental perturbations (and so limiting its effects on phenotype and thus phenotypic variability) [18].
For intra-genotypic variability to be of relevance for evolutionary ecology, it must be under genetic control. There is now ample evidence that this is the case, and heritabilities for intra-genotypic variability for a range of traits are reviewed by Hill & Mulder [19]. Moreover, recently it has become possible to map individual loci that regulate the degree of variability of a trait rather than its mean [20–22]. Such loci have been mapped in a wide range of organisms and for a variety of traits, for example locomotor handedness [12] and bristle numbers [23] in Drosophila, flowering time in Arabidopsis thaliana [24,25], morphological traits in maize (Zea mays) [26], calcium excretion in rats (Rattus norvegicus) [27], litter size in pigs (Sus scrofa) [28] and body mass index in humans [29]. The number of loci associated with variance may be on par with those associated with the mean [24,30] and there seems little overlap between loci affecting the mean of a trait and its variance [12,30] (although [31] provide a counter-example where the mean and variance are under the same genetic control).
When do we expect selection for increased intra-genotypic variability?
Given that intra-genotypic variability is under genetic control, we may expect that it can respond to selection. Results from basic theory allow us to predict selection for intra-genotypic variability and its direction across a range of contexts. If the relationship between traits and fitness is linear, e.g., in plants, the relation between vegetative and reproductive mass (as a measure for fitness) [32,33], there is no selection on the degree of variability (Fig. 1a,e). However, the relationship between traits and fitness is often nonlinear. If the shape of the relationship is convex (accelerating), this means that trait values some magnitude larger than the mean will result in large increases in fitness, while trait values equivalently smaller than the mean will result in relatively small reductions in fitness (Fig. 1b,f). Measures of plant size other than weight can yield such nonlinearities with reproductive output [33], also reported for the relationship between body size and egg number in mites (Sancassania berlesei) [34] and antler size and lifetime breeding success in red deer (Cervus elaphus) [35]. On average, then, if the trait mean is fixed, fitness can nonetheless be increased by increasing trait variance, as a result of ‘Jensen’s inequality’, a mathematical property of nonlinear functions [36,37] (Fig. 2a). The consequences of such nonlinear averaging have been described with respect to species coexistence [38], and may also be important in the evolution of variance control [39]. For example, Mulder et al. [16] found that stabilizing selection acts on variance in fledging weight in great tits due to the sigmoidal curve describing the relationship between fledging weight and recruitment - for such a concave (decelerating) relationship, an increase in the trait value (here, fledgling weight) yields smaller increases in fitness than the symmetrical decline.
Figure 1.
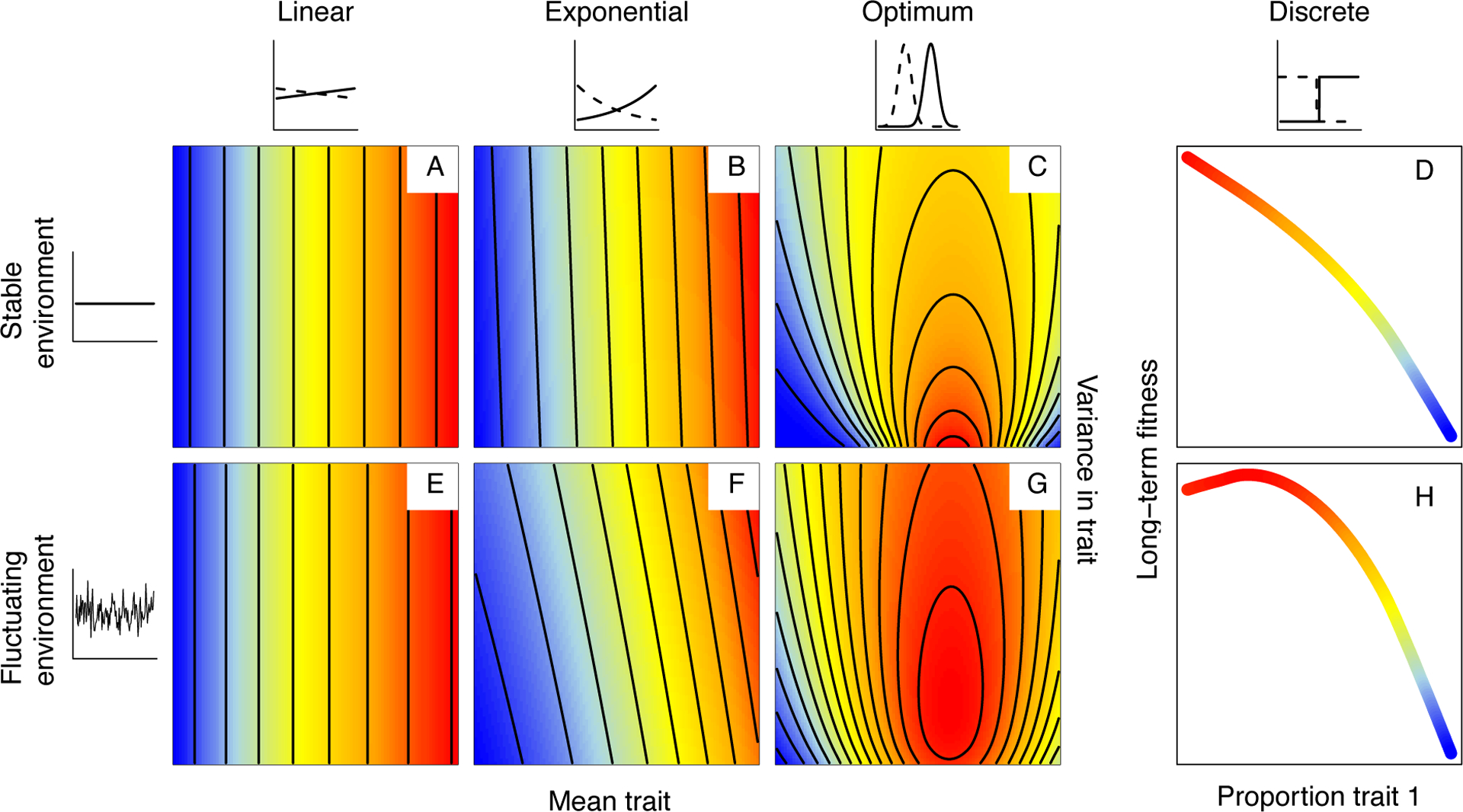
Long-term adaptive landscapes for different combinations of a mean phenotypic trait and variance in that trait to illustrate the conditions that might selection for intra-genotypic variability (Appendix S1–S3 for more information on how these landscapes were obtained). Upper row shows results for a stable environment, bottom row shows results for a fluctuating environment. Different columns show results for different relationships between the phenotypic trait and reproduction, as depicted in the small graphs. The environment modulates the relation between phenotype and fitness, illustrated by the solid and dotted lines. In a-c and e-g fitness landscapes for continuous phenotypic traits are shown. In d,h results for a discrete phenotypic trait are shown. Here, the x-axis shows the proportion of individuals expressing one trait, and is plotted against long-term fitness (y-axis). Red colors indicate higher long-term fitness, blue colors indicate lower values, and values are scaled for each graph separately. Variance has no effect in the case of the linear relation between phenotypic trait and fitness (a,e), while it results in a higher long-term fitness in the case of a convex relation, due to nonlinear averaging (b,f). When there is a phenotypic optimum that changes through time, for either a continuous (g) or discrete (h) trait, a bet hedging strategy results in the highest long-term fitness.
Figure 2.
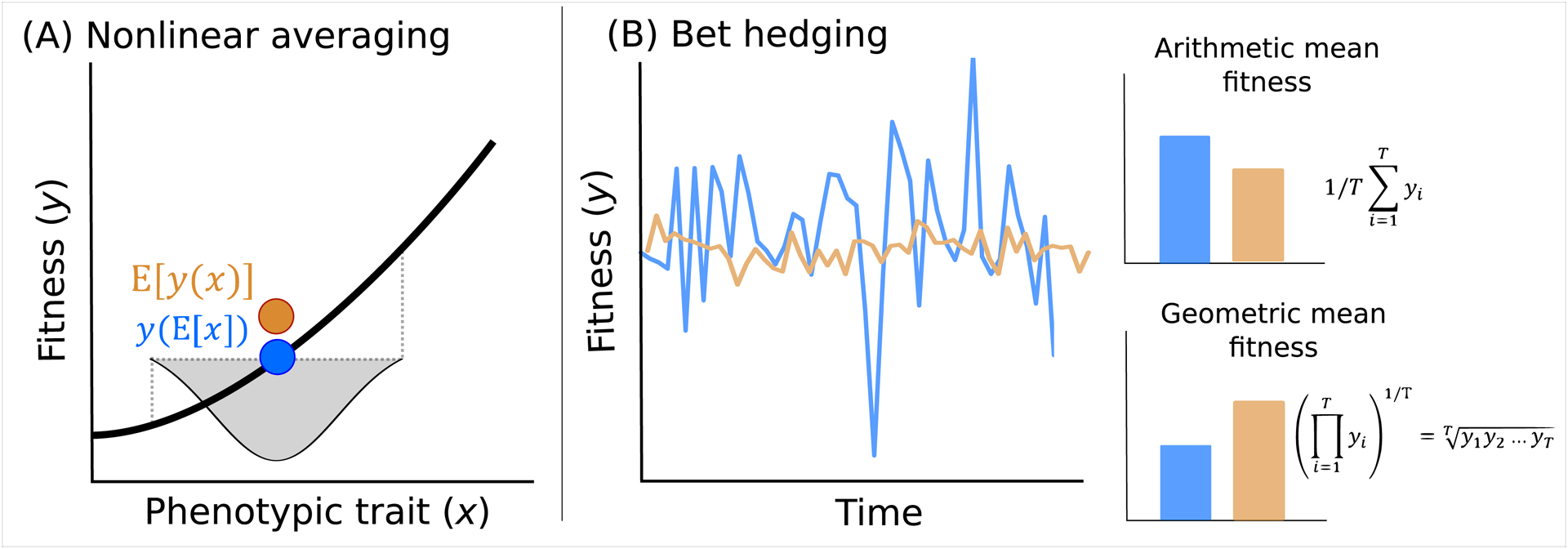
Two conditions that can result in selection favoring intra-genotypic variability: a) nonlinear averaging and b) bet hedging. a) The graph shows fitness (y) as a function of phenotype (x). If the relation is convex, an increase in the variance in x (indicated by the grey shading) results in a higher expected fitness value (E[y(x)]) (orange) than the fitness value of the average phenotype (y(E[x])) (blue), as a result of Jensen’s inequality. b) A bet hedging strategy (orange) reduces variance in fitness across generations compared to a non bet hedging strategy (blue). Despite a decrease in arithmetic mean fitness, bet hedging leads to an increase in geometric mean fitness, and is thus expected to be favored by natural selection.
Moving from constant environments to environments that vary (spatially or temporally), a next core theoretical driver of the evolution of intra-genotypic variability is bet hedging. Bet hedging is formally defined as a strategy that leads to a reduction in fitness variance across generations, thereby increasing geometric mean fitness (long-term fitness), at the cost of arithmetic mean fitness [4] (Fig. 2b). In other words, while year-to-year a bet hedging strategy might produce, on average, fewer offspring, over the long term its fitness will be larger. Germination delays in annual plants provide a classic example of diversified bet hedging, which is our focus here. This in contrast to conservative bet hedging, in which selection favors a single conservative phenotype (these two types of bet hedging may represent two extremes of one continuum, see [40]). A non-bet hedging strategy, where every year all seeds present germinate, leaves itself vulnerable to years of adverse environmental conditions where no or few germinants survive, and thus the population is at risk of extinction. A bet hedging strategy guarantees some seeds will have delayed germination, and thus persist in the seed bank during harsh years. Bet hedging involves a reduction in arithmetic mean fitness, accompanied by a reduction in fitness variance. In order to properly distinguish between bet hedging and nonlinear averaging, the fitness consequences on both the arithmetic mean and the variance of fitness must therefore be considered [41–43].
Research into bet hedging has often focused on traits related to timing, for example, seed germination [14,44,45] or diapause duration [46–51]. For these timing-related examples, the process underpinning bet hedging is often framed as a probability (e.g., probability of germinating). This implicitly defines the core trait underpinning fitness as discrete (e.g., being a seed or a germinant), with variance emerging purely from the binomial variance associated with the probability (implying that variance is maximal for a probability p=0.5, all else being equal). Interestingly, a large majority of both empirical and theoretical studies describing bet hedging in other traits (not related to timing) have also mostly considered discrete traits. Examples include whether or not aphids develop wings [52], left or right handedness in Drosophila [12], investment in spores or vegetative cells in slime molds (Dictyostelium discoideum) [53], stochastic on- and off switching of gene expression in yeast cells [10] as well as an array of theoretical models [54,55]. On consideration, a clear reason emerges for the observation that studies of bet hedging tend to predominantly focus on discrete traits: where the optimal trait value fluctuates unpredictably due to a changing environment and there is no ‘intermediate’ trait value possible, it is beneficial to express both discrete traits every time step [56] (Fig. 1d,h). However, bet hedging has also been observed in continuous traits, including thermal and phototactic preference in Drosophila [57], egg size in gypsy moths (Lymantria dispar) [58] and macrophyte (Scirpus maritimus) offspring size [59]. For continuous traits, theory indicates that bet hedging can be beneficial in traits for which the phenotypic optimum varies between years, provided that the magnitude of environmental fluctuation is sufficiently large [56] (Fig. 1c,g) and being a function of how much the strength of stabilizing selection varies through time [60].
Interaction between environmental fluctuations and lifespan on the evolution of variability
In Figure 1, we describe the range of selection scenarios for intra-genotypic variability depending on both characteristics of the trait: its mapping to fitness, and scope of environmental variation. The broader life history context will also be relevant. This is because fitness emerges from survival and fertility across the life cycle [61], and other life stages will thus define selective pressures on intra-genotypic variability, for example for traits affecting reproduction. Increasing survival probabilities decrease the optimal variance for two relationships between phenotype and reproduction we explored (Fig. 3; see Appendix S4 for more details on the performed simulations). This demonstrates that modifying survival can alter the optimum variance in another, uncorrelated, fitness component (reproduction) that together determine population fitness, and suggests that the more adverse the habitat is to survival, the more important it might be to invest in variable offspring. Furthermore, it implies that intra-genotypic variability, when affecting a trait related to reproduction, may be more adaptive in short-lived species (e.g., annual plants, Drosophila), as also indicated in results from Koons et al. [39]. For short-lived species, individuals are at a higher risk of dying without having reproduced, while for long-lived life cycles (e.g., trees, blue whales), individuals can afford to have one year of zero reproduction, because the yearly fitness is balanced by the high survival [62].
Figure 3.
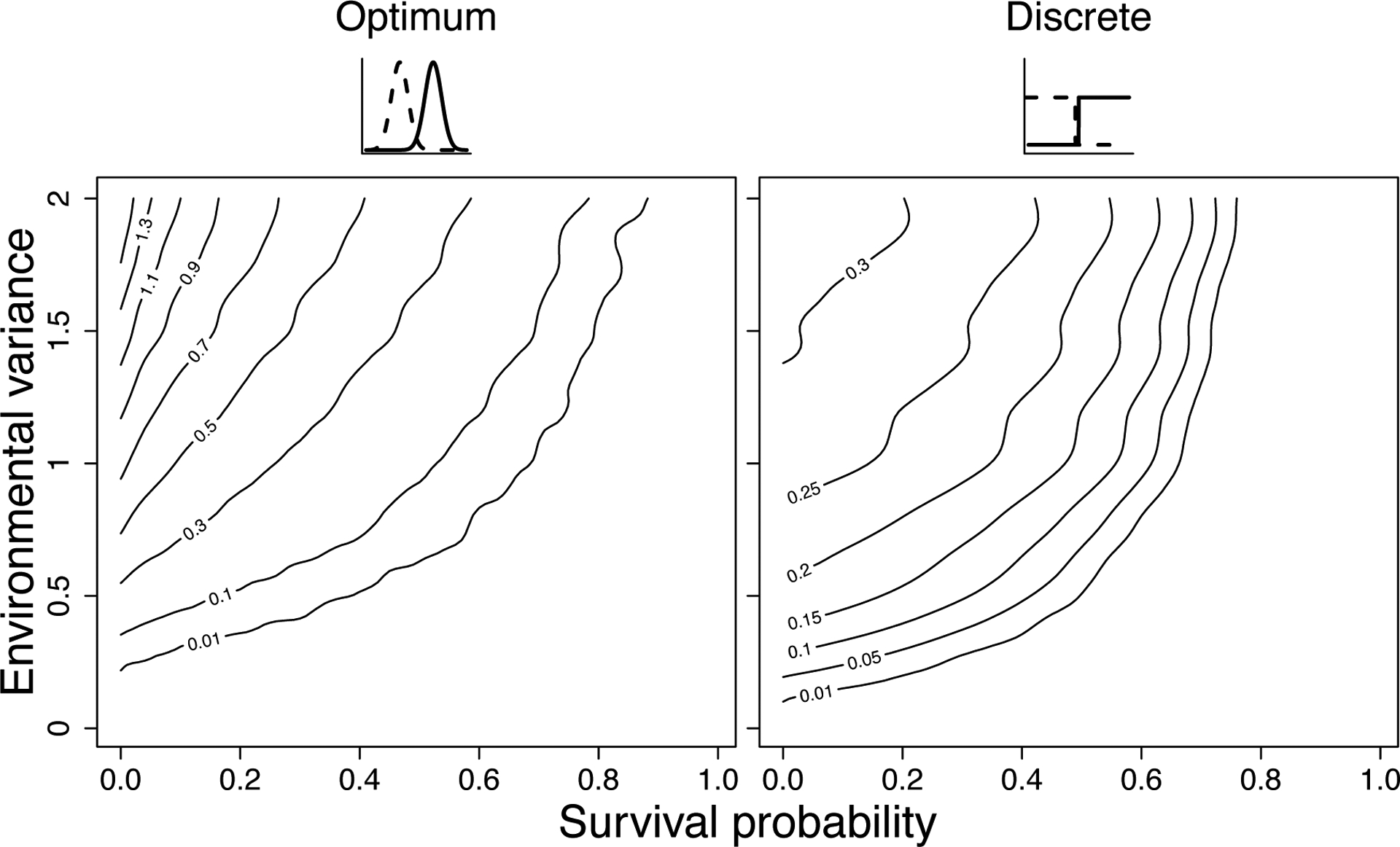
Graphs depict the optimal variance in a phenotypic trait affecting reproduction, as a function of survival probability per time-step (x-axis) and environmental variance (y-axis). Here we vary it between 0 and 2 while in Fig. 1, we set environmental variance at 0 (stable environment) and at 1 (fluctuating environment). We do so to further explore how environmental variance affects the optimal phenotypic variance for two phenotype-reproduction relations, where selection favored intermediate phenotypic variance (left: continuous, Fig. 1g; right: discrete, Fig. 1h). Like in Fig. 1, the environment modulates the relationship between phenotype and fitness, illustrated by the solid and dotted lines (Appendix S4 for more details on these simulations). The graphs show that higher environmental variance favors higher phenotypic variance. For example, when survival probability is set at 0.5, increasing the environmental variance from 0 to 2, increases the optimal phenotypic variance from 0 to 0.4 (left) and from 0 to 0.23 (right). Higher survival probabilities favor lower phenotypic variance. For example, when the environmental variance is set at 1, increasing survival from 0 to 1, decreases the optimal variance from 0.77 to 0 (left), and from 0.28 to 0 (right).
Evolution of intra-genotypic variability in natural populations
An array of demographic models suggests that, at the phenotypic level, non-zero variance is adaptive under commonly encountered environmental conditions and trait-phenotype relations (Figs. 1, 3) [10,16,54,56], but what drives such phenotypic variability? This is largely an open question, and it may, in some cases, reflect non-biological processes such as measurement error or incomplete statistical models [63]. More interestingly, however, it may also be due to stochastic variation in gene expression (‘gene expression noise’) [64]. Work on yeast cells has shown that the average gene expression noise differs between genes, and the degree of variability is suggested to be dependent on the gene function [65]. For instance, essential genes tend to have low stochastic variance in expression [65,66], while genes related to stress and plasma-membrane transporters show elevated noise levels [65,67,68]. Whether increased gene expression noise is adaptive is under debate, with some studies showing selection against variance-increasing alleles [69,70], while others suggest fitness benefits in response to environmental stress [71,72]. Optimal levels of expression noise will depend on the relationship between gene expression and emergent phenotype (both functioning and variance) following the principles linking phenotypic variance to fitness that we outline above. Defining these relationships will help predict for which genes we expect stochastic variation in expression to be beneficial, under which conditions, potentially helping explain why we observe gene expression noise in some genes, but less so in others. For example, for dosage-sensitive genes (in which changing expression levels is harmful), most genes indeed show low expression noise [69]. Whenever the relationship between expression level and performance is convex, we expect variance in gene expression to be beneficial [68]. Further, whenever an unpredictable environment changes the optimal gene expression level, bet hedging might be a beneficial strategy. Such an optimum relation has for instance been observed between lactose operon protein expression and fitness of Escherichia coli, whereby the optimum changes with lactose level [73].
Evidence available to date suggests that heritability for intra-genotypic variability is quite low [7,15,19]. Assuming that estimates are reasonably unbiased, we have three possible explanations for a low heritability of intra-genotypic variability. First, it may be a relatively constrained trait, with little variance. This will be modulated in part by the architecture of the gene networks constraining the degree of gene expression noise, and the degree to which its architecture is sensitive to mutations [74,75]. Second, populations may be close to their fitness optima, such that additional additive genetic variance has been eroded, though this scenario might be unlikely [76]. Evidence on whether populations are close to their optima, is mixed: theoretical predictions on optimal germination and flowering probabilities in plants [14,45,77], have been shown to closely match observations. In contrast, Philippi [78], also focusing on germination probabilities, showed that predicted optimal strategies did not match the observations. An apparent weak selection towards optimal bet hedging strategies in specific cases can be due to: i) selection for bet hedging being of recent origin, simply resulting in insufficient time for optimal strategies to have evolved, or ii) selection on bet hedging being infrequent, implying that bet hedging promoting alleles disappear due to drift or short-term selection against them, despite being beneficial on the long term [79]. Typically, theoretical predictions assume infinite large populations (e.g., Figs. 1,3 or [80]). However, real-world populations consist of individuals that are subject to demographic stochasticity, resulting in genetic drift. Whether a variance-increasing mutation appearing at one time point will get fixed in the population, depends on the population size, in addition to the selective advantage and the experienced environment [79]. Moreover, responses to selection are additionally shaped by genetic (co)variances, which have the potential to create a wide range of evolutionary trajectories towards the optimum (Box 2). Third, there may be standing genetic variation associated with phenotypic variability, but these alleles may appear neutral until particular environmental circumstances occur, and thus their contribution to heritability would be cryptic (Boxes 1 & 2). Research studies on this topic have primarily focused on fitness outcomes across environments that fluctuate, but in a stationary manner (i.e., around a consistent mean) over the long term (Figs. 1, 3) and generally do not allow for context-dependent manifestation of intra-genotypic variability [81,82]. Transient and directional changes in environmental conditions that co-occurred with expression of intra-genotypic variability could help sustain populations across challenging periods [81]. Characterizing these links empirically would require sufficient knowledge of the system to identify the condition-dependence of expression of intra-genotypic variability, as well as its demographic consequences as outlined above.
Box 2: Intra-genotypic variance in a Quantitative Genetic framework.
In a quantitative genetic model, assuming no dominance or epistasis, an individual’s phenotypic deviation from the population mean can be written as the sum of its breeding value (A) and an environmental (residual) effect (E), thus: P = A + E. In the absence of any covariance between A and E, the total phenotypic variance (VP) can be written as VP = VA + VE, where VA) is the additive genetic variance and VE is the environmental variance. Intra-genotypic variability involves a genetic component in the residual variance, where different genotypes can differ in VE. Different approaches to model this genetic component in the residual variance exist [19], for example according to an additive model [76,97], where the genetic component of the variance is modelled as an additive effect:
Here, χ is a standard normal deviate, distributed as N(0,1), scaled by Each individual is now characterized by two breeding values: one determining its average phenotype (Amean), and one determining the variance around this mean (Avariance). The evolutionary response in a population depends on both additive genetic variances VA-mean and VA-variance, as well as its covariance (covA(mean, variance) (which can be summarized in a so-called G-matrix) (Fig. I). Note that here Amean and Avariance affect the same phenotypic trait; when affecting different phenotypic traits, nonlinear relations between traits, on a phenotypic level, may emerge [98].
Figs. 1 and 3 (see main text) illustrate the fitness consequences of intra-genotypic variability in a phenotypic trait affecting reproduction, obtained using a demographic model (Appendices S1–3). Combining such adaptive landscapes with a quantification of the underlying genetic variances and covariances (Fig. II) will shed light on an array of interesting research questions on the evolution of variance control (see Outstanding Questions).
Figure I.
Evolutionary trajectories of the mean and variance in a trait. Contour lines depict the fitness landscape, corresponding to Fig. 1g (see main text). For clarity, we only show contour lines here. Green dots indicate the trait combination resulting in the highest fitness. Starting from a population with a low mean trait value and no variance (black dots), trajectories towards the fitness optimum (green dots) are shown, for varying genetic variances and covariances, using the multivariate breeder’s equation [99]. Additive genetic variances for the mean (VA-mean) and variance (VA-variance) in phenotypic trait were both set at 0.1, and their covariance (covA(mean, variance)) at 0. We then, one by one, varied these variance components, while keeping the rest of the G-matrix constant. We assessed how the phenotypic mean and variance evolve over the course of 200 time steps. Graph on the left shows the effect of varying values for VA-mean′ middle graph shows the effect of varying VA-variance′ and graph on the right the effect of changes in covA)(mean,variable). Different colors correspond to the range of values for the variable (see legend). Note that we considered a constant genetic variance-covariance matrix within each scenario, an assumption which might be often violated in natural population [100,101] and/or when environmental conditions change through time [102]
Figure II.

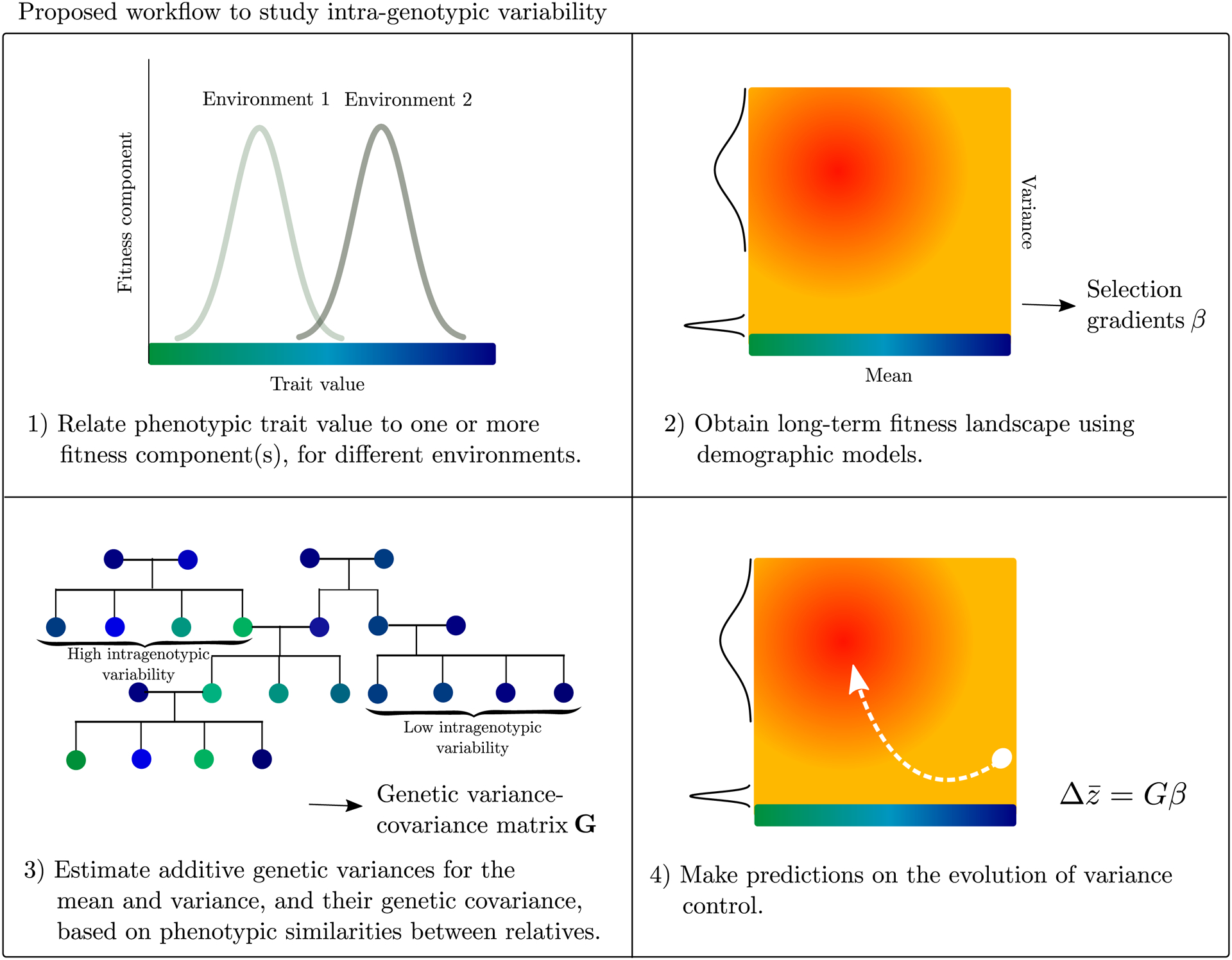
Proposed workflow to study the evolution of intra-genotypic variability, focusing on both the fitness consequences as well as on the genetics underlying the traits. 1–2) By using a demographic model (Appendices S1–3) to define the adaptive landscape, partial selection gradients at a given point in the landscape can be obtained. 3) The additive genetic variance462 covariance matrix (G) can be estimated using linear mixed effects models, based on knowledge on phenotypic similarities between relatives [7,15,16,19]. 4) The evolutionary change in multiple traits (), in this case phenotype mean and variance, can be written as the multivariate breeder’s equation [99], as the product of the genetic variance-covariance matrix (G) and the partial selection gradients for each trait (β).
Going forward
A growing number of studies on intra-genotypic variability are providing heritability estimates and are enriching a catalogue of loci associated with variability [7,15,19,23,24], however few studies have connected the degree of variability to fitness [16]. Observing heritable variation for variability tells us nothing about whether variability is adaptive or not, just that it can evolve. On the other hand, empirical or theoretical studies quantifying fitness gains from variability (for instance through bet hedging) generally lack knowledge of the underlying genetics basis. One notable exception is a recent study by Mulder et al. [16]. Here, the degree of variability in fledgling weight, in a wild great tit population, was related to fitness, showing stabilizing selection on intermediate phenotypic variance. Such studies are needed to understand when we expect intra-genotypic variation to evolve (Box 2).
A key priority is now to use theoretical expectations to guide the experiments that will provide empirical evidence. This remains challenging for a number of reasons. First, some of the best empirical examples have relied on humans, laboratory or domestic animals, making direct application of evolutionary predictions complicated (as these populations may not reflect the outcome of natural selection). Second, inferring the degree to which intra-genotypic variability is adaptive requires quantifying the impacts of a changing environment on (components of) fitness which is intractable unless a life history model can be constructed. This in turn requires measurement of core individual demographic rates (survival, growth, reproduction) as well as measurements of how variation in both the trait and the environment affect these demographic rates. Such information is rarely available (with the notable exception of plants with seed banks [13,14]). Third, evolutionary outcomes will be modulated by the degree to which genetic variation is available for selection to act upon, e.g., both additive genetic variance, and covariances with other traits (Box 2). Estimating genetic variances (e.g., using mixed effects models [83]) already demand large data resources for mean trait values, and the situation only worsens when attempting to do the same for trait variance. Estimating these components (see e.g., [84,85]) requires data on the relatedness between individuals, which generally calls for genetic analyses. Recent advances in genomic technology and the developments of powerful analytical tools are enabling the construction of high quality pedigree on an unprecedented large scale [86,87].
Concluding remarks
Making robust quantitative predictions on the evolution of variance control in natural populations require long-term data on i) temporal changes in one or more environmental variables, ii) individual phenotypes and demographic rates, interacting with the environment, and iii) the relatedness between individuals. Gathering such comprehensive datasets is a considerable undertaking, yet recognizing the unparalleled value of long-term field studies, a growing number of consortiums have risen to the challenge. For example, the Soay sheep (Ovis aries) in Scotland have become a key model to study disease ecology [88], the Amboseli baboon (Papio cynocephalus) project has transformed the field of behavioral ecology [89], other long-term studies in birds [90,91] and in plants [92] promise to be just as valuable. Such studies will provide estimates for key parameters such as the genetic component of phenotypic variance, and its genetic correlation with the mean trait [7,15,16] that are necessary to refine theoretical models and better understand the evolution of variance control (see Outstanding Questions). Model systems in laboratory settings remain an important tool to study the evolution of intra-genotypic variability. They afford the ability to combine genomics tools with quantitative genetics and creative experimental design, using for example experimental evolution under a range of conditions [93,94]. Although the catalog of variance controlling loci (vQTLs) that have been mapped to date remains relatively limited, a population genetics analysis of these loci would shed critical light on the dynamic driving their evolution. Finally, combining the adaptive landscapes with the evolutionary potential of intra-genotypic variability will be essential for making predictions about factors driving its evolution. Future studies applying demographic models, such as the one proposed here, to empirical data, will help to explain the ubiquity of the genetic control of phenotypic variation and to understand its relevance for natural populations facing environmental changes.
OUTSTANDING QUESTIONS.
How do life history parameters (from reproductive rate to lifespan) impact the adaptive advantage of intra-genotypic variability? In other words, is variance control more likely to evolve in fruit flies than in elephants? And, do we observe smaller coefficients of variation, and thus potentially higher canalization in long living species versus short-living species?
The relationship between canalization, plasticity and intra-genotypic variability, remains poorly understood. For example, does intra-genotypic variability favor the evolution of phenotypic plasticity and are they genetically correlated?
What is the genetic correlation between the mean and the variance, either affecting the same phenotypic trait or different phenotypic traits? Are their categories of traits (e.g., life history traits) for which such a correlation is more likely than others?
What is contribution of cryptic genetic variation to intra-genotypic variability? Is a significant fraction of variance controlling alleles only associated with variance control under certain (e.g., stressful) environments?
How often do predicted optimal strategies match the observed mean and variance?
What is the allele frequency spectrum of alleles association with variation in phenotypic variability? And what role does balancing selection play in shaping this spectrum?
Supplementary Material
HIGHLIGHTS.
Recent developments in the field of genetics show that the degree of phenotypic variability among genetically identical individuals can be under genetic control and can thus be tuned by natural selection.
To understand when the degree of intra-genotypic variability for a trait is the outcome of adaptive evolution, it is essential to map the focal trait to patterns of reproduction and survival across the life cycle.
Here, we evaluate the fitness consequences of phenotypic variance for commonly encountered relationships between traits, fitness, and environmental conditions.
ACKNOWLEDGEMENTS
We thank Luisa Pallares and Simon Forsberg for helpful comments and discussions. This research was funded by grants from the Nation Institute of Health to JFA R35 GM124881 and R01ES029929.
GLOSSARY
- Adaptive landscape
(fitness landscape) Visualization of the relation between fitness and one or more phenotypic characters or genes.
- Adaptive tracking
Evolutionary change in the average phenotypic trait in a population, where the phenotypic mean tracks a (changing) fitness optimum.
- Bet hedging
Selection for increased geometric mean fitness, despite a reduction in arithmetic mean fitness, due to a decrease in the variance in fitness over generations.
- Conservative bet hedging
Bet hedging where selection favors a single conservative phenotype, thus reducing fitness variance but without an increase in the phenotypic variance.
- Diversified bet hedging
Bet hedging where selection favors phenotypic variance within genotypes.
- G-matrix
Additive genetic variance-covariance matrix summarizing inheritance patterns of multiple traits.
- Intra-genotypic variability
Phenotypic variation between genetically identical individuals reared in a constant environment, most likely due to differences in an individual’s ability to buffer micro-environmental perturbations, leading to stochastic fluctuations in development.
- Jensen’s inequality
Mathematical property of a convex function. The expected value of a convex function (where the 2nd derivative is positive), is higher than the value at the average x-value, due to nonlinear averaging. In contrast, the expected value of a concave function (2nd derivative is negative), is lower than the value at the average x-value.
- Long-term fitness
The geometric mean population growth rate in a variable environment.
- Nonlinear averaging
The expected value of a nonlinear function (E[f(x)]) can differ from the value at the average x-value (f(E[x])). For convex functions, Jensen’s inequality is the result.
- Phenotypic plasticity
The ability of an organism to change its phenotype in response to the environment.
REFERENCES
- 1.Lande R (1976) Natural selection and random genetic drift in phenotype evolution. Evolution 30, 314–334 [DOI] [PubMed] [Google Scholar]
- 2.Piersma T and Drent J (2003) Phenotypic flexibility and the evolution of organismal design. Trends Ecol. Evol 18, 228–233 [Google Scholar]
- 3.Bradshaw AD (1965) Evolutionary significance of phenotypic plasticity in plants. Adv. Genet 13, 116–155 [Google Scholar]
- 4.Philippi T and Seger J (1989) Hedging one’s evolutionary bets, revisited. Trends Ecol. Evol 4, 41–44 [DOI] [PubMed] [Google Scholar]
- 5.Garreau H et al. (2008) Results of four generations of a canalising selection for rabbit birth weight. Livest. Sci 119, 55–62 [Google Scholar]
- 6.Formoso-Rafferty N et al. (2016) Genetic control of the environmental variance for birth weight in seven generations of a divergent selection experiment in mice. J. Anim. Breed. Genet 133, 227–237 [DOI] [PubMed] [Google Scholar]
- 7.Sae-Lim P et al. (2015) Genetic (co)variance of rainbow trout (Oncorhynchus mykiss) body weight and its uniformity across production environments. Genet. Sel. Evol 47, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgersma GG et al. (2018) Fluctuations in milk yield are heritable and can be used as a resilience indicator to breed healthy cows. J. Dairy Sci 101, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 9.Berghof TVL et al. (2019) Opportunities to improve resilience in animal breeding programs. Front. Genet 9, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acar M et al. (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet 40, 471–475 [DOI] [PubMed] [Google Scholar]
- 11.Diaz SA and Viney M (2014) Genotypic-specific variance in Caenorhabditis elegans lifetime fecundity. Ecol. Evol 4, 2058–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayroles JF et al. (2015) Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc. Natl. Acad. Sci. USA 112, 6706–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Childs DZ et al. (2010) Evolutionary bet-hedging in the real world: Empirical evidence and challenges revealed by plants. Proc. R. Soc. B Biol. Sci 277, 3055–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gremer JR et al. (2016) Within-and among-year germination in Sonoran Desert winter annuals: bet hedging and predictive germination in a variable environment. Ecol. Lett 19, 1209–1218 [DOI] [PubMed] [Google Scholar]
- 15.Martin JGA et al. (2017) Genetic basis of between-individual and within-individual variance of docility. J. Evol. Biol 30, 796–805 [DOI] [PubMed] [Google Scholar]
- 16.Mulder HA et al. (2016) Genetic variation in variability: Phenotypic variability of fledging weight and its evolution in a songbird population. Evolution 70, 2004–2016 [DOI] [PubMed] [Google Scholar]
- 17.Simons AM and Johnston MO (1997) Developmental instability as a bet-hedging strategy. Oikos 80, 401–406 [Google Scholar]
- 18.Willmore KE et al. (2007) Phenotypic variability: Its components, measurement and underlying developmental processes. Evol. Biol 34, 99–120 [Google Scholar]
- 19.Hill WG and Mulder HA (2010) Genetic analysis of environmental variation. Genet. Res 92, 381–395 [DOI] [PubMed] [Google Scholar]
- 20.Ansel J et al. (2008) Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 4, e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rönnegård L and Valdar W (2011) Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 188, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rönnegård L and Valdar W (2012) Recent developments in statistical methods for detecting genetic loci affecting phenotypic variability. BMC Genet. 13, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay TFC and Lyman RF (2005) Drosophila bristles and the nature of quantitative genetic variation. Philos. Trans. R. Soc. B Biol. Sci 360, 1513–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimenez-Gomez JM et al. (2011) Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet. 7, e1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X et al. (2012) Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 8, e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ordas B et al. (2008) Genetic variation and quantitative trait loci associated with developmental stability and the environmental correlation between traits in maize. Genet. Res 90, 385–395 [DOI] [PubMed] [Google Scholar]
- 27.Perry GML et al. (2012) Sex modifies genetic effects on residual variance in urinary calcium excretion in rat (Rattus norvegicus). Genetics 191, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sell-Kubiak E et al. (2015) Genome-wide association study reveals novel loci for litter size and its variability in a Large White pig population. BMC Genomics 16, 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J et al. (2012) FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgante F et al. (2015) Genetic architecture of micro-environmental plasticity in Drosophila melanogaster. Sci. Rep 5, 9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ros M et al. (2004) Evidence for genetic control of adult weight plasticity in the snail Helix aspersa. Genetics 168, 2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samson DA and Werk KS (1986) Size-dependent effects in the analysis of reproductive effort in plants. Am. Nat 127, 667–680 [Google Scholar]
- 33.Klinkhamer PGL et al. (1992) On the analysis of size-dependent reproductive output in plants. Funct. Ecol 6, 308–316 [Google Scholar]
- 34.Ozgul A et al. (2012) Population responses to perturbations: the importance of trait-based analysis illustrated through a microcosm experiment. Am. Nat 179, 582–594 [DOI] [PubMed] [Google Scholar]
- 35.Kruuk LEB et al. (2002) Antler size in red deer: hertability and selection but no evolution. Evolution 56, 1683–1695 [DOI] [PubMed] [Google Scholar]
- 36.Jensen JLWV (1906) Sur les fonctions convexes et les inegalites entre les valeurs moyennes. Acta Math. 30, 175–193 [Google Scholar]
- 37.Ruel JJ and Ayres MP (1999) Jensen’s inequality predicts effects of environmental variation. Trends Ecol. Evol 14, 361–366 [DOI] [PubMed] [Google Scholar]
- 38.Hart SP et al. (2016) How variation between individuals affects species coexistence. Ecol. Lett 19, 825–838 [DOI] [PubMed] [Google Scholar]
- 39.Koons DN et al. (2009) Is life-history buffering or lability adaptive in stochastic environments? Oikos 118, 972–980 [Google Scholar]
- 40.Starrfelt J and Kokko H (2012) Bet-hedging—a triple trade-off between means, variances and correlations. Biol. Rev 87, 742–755 [DOI] [PubMed] [Google Scholar]
- 41.Olofsson H et al. (2009) Bet-hedging as an evolutionary game: The trade-off between egg size and number. Proc. R. Soc. B Biol. Sci 276, 2963–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees M et al. (2009) Bet-hedging as an evolutionary game: The trade-off between egg size and number. Proc. R. Soc. B Biol. Sci 277, 1149–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ripa J et al. (2010) What is bet-hedging, really? Proc. R. Soc. B Biol. Sci 277, 1153–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans Margaret E. K. et al. (2017) Bet hedging via seed banking in desert evening primroses (Oenothera, Onagraceae): demographic evidence from natural populations. Am. Nat 169, 184. [DOI] [PubMed] [Google Scholar]
- 45.Gremer JR and Venable DL (2014) Bet hedging in desert winter annual plants: optimal germination strategies in a variable environment. Ecol. Lett 17, 380–387 [DOI] [PubMed] [Google Scholar]
- 46.Menu F et al. (2000) Bet-hedging diapause strategies in stochastic environments. Am. Nat 155, 724–734 [DOI] [PubMed] [Google Scholar]
- 47.Graham JK et al. (2014) Experimental evolution of bet hedging under manipulated environmental uncertainty in Neurospora Crassa. Proc. R. Soc. B Biol. Sci 281, 20140706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajon E et al. (2014) The evolution of bet hedging in response to local ecological conditions. Am. Nat 184, E1–E15 [DOI] [PubMed] [Google Scholar]
- 49.García-Roger EM et al. (2014) Bet-hedging in diapausing egg hatching of temporary rotifer populations - A review of models and new insights. Int. Rev. Hydrobiol 99, 96–106 [Google Scholar]
- 50.Furness AI et al. (2015) Adaptation in a variable environment: Phenotypic plasticity and bet-hedging during egg diapause and hatching in an annual killifish. Evolution 69, 1461–1475 [DOI] [PubMed] [Google Scholar]
- 51.Tarazona E et al. (2017) Experimental evolution of bet hedging in rotifer diapause traits as a response to environmental unpredictability. Oikos 126, 1162–1172 [Google Scholar]
- 52.Grantham ME et al. (2016) A case for a joint strategy of diversified bet hedging and plasticity in the pea aphid wing polyphenism. Biol. Lett 12, 20160654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martínez-García R and Tarnita CE (2017) Seasonality can induce coexistence of multiple bet-hedging strategies in Dictyostelium discoideum via storage effect. J. Theor. Biol 426, 1339–1351 [DOI] [PubMed] [Google Scholar]
- 54.Botero CA et al. (2014) Evolutionary tipping points in the capacity to adapt to environmental change. Proc. Natl. Acad. Sci. USA 112, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crowley PH et al. (2016) Dealing with stochastic environmental variation in space and time: bet hedging by generalist, specialist, and diversified strategies. Theor. Ecol 9, 149–161 [Google Scholar]
- 56.Bull JJ (1987) Evolution of phenotypic variance. Evolution 41, 303–315 [DOI] [PubMed] [Google Scholar]
- 57.Kain JS et al. (2015) Variability in thermal and phototactic preferences in Drosophila may reflect an adaptive bet-hedging strategy. Evolution 69, 3171–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossiter MC (1991) Maternal effects generate variation in life history: consequences of egg weight plasticity in the gypsy moth. Funct. Ecol 5, 386 [Google Scholar]
- 59.Charpentier A et al. (2012) Variable offspring size as an adaptation to environmental heterogeneity in a clonal plant species: integrating experimental and modelling approaches. J. Ecol 100, 184–195 [Google Scholar]
- 60.Zhang X-S and Hill WG (2005) Evolution of the environmental component of the phenotypic variance: stabilizing selection in changing environments and the cost of homogeneity. Evolution 59, 1237–1244 [PubMed] [Google Scholar]
- 61.Metcalf CJE and Pavard S (2007) Why evolutionary biologists should be demographers. Trends Ecol. Evol 22, 205–212 [DOI] [PubMed] [Google Scholar]
- 62.Visser MD et al. (2011) Strict mast fruiting for a tropical dipterocarp tree: a demographic cost-benefit analysis of delayed reproduction and seed predation. J. Ecol 99, 1033–1044 [Google Scholar]
- 63.Westneat DF et al. (2015) The biology hidden inside residual within-individual phenotypic variation. Biol. Rev 90, 729–743 [DOI] [PubMed] [Google Scholar]
- 64.Raser JM and O’Shea EK (2005) Noise in gene expression: Origins, consequences, and control. Science 309, 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newman JR et al. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840. [DOI] [PubMed] [Google Scholar]
- 66.Fraser HB et al. (2004) Noise minimization in eukaryotic gene expression. PLoS Biol. 2, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bar-Even A et al. (2006) Noise in protein expression scales with natural protein abundance. Nat. Genet 38, 636–643 [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z et al. (2009) Positive selection for elevated gene expression noise in yeast. Mol. Syst. Biol 5, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehner B (2008) Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol. Syst. Biol 4, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Metzger BPH et al. (2015) Selection on noise constrains variation in a eukaryotic promoter. Nature 521, 344–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blake WJ et al. (2006) Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24, 853–865 [DOI] [PubMed] [Google Scholar]
- 72.Viney M and Reece SE (2013) Adaptive noise. Proc. R. Soc. B Biol. Sci 280, 20131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dekel E and Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436, 588–592 [DOI] [PubMed] [Google Scholar]
- 74.Chalancon G et al. (2012) Interplay between gene expression noise and regulatory network architecture. Trends Genet. 28, 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shreif Z and Periwal V (2014) A network characteristic that correlates environmental and genetic robustness. PLoS Comput. Biol 10, e1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hill WG and Zhang X (2004) Effects on phenotypic variability of directional selection arising through genetic differences in residual variability. Genet. Res 83, 121–132 [DOI] [PubMed] [Google Scholar]
- 77.Rees M et al. (2006) Seed dormancy and delayed flowering in monocarpic plants: selective interactions in a stochastic environment. Am. Nat 168, E53–E71 [DOI] [PubMed] [Google Scholar]
- 78.Philippi T (1993) Bet-hedging germination of destert annuals: Beyond the first year. Am. Nat 142, 474–487 [DOI] [PubMed] [Google Scholar]
- 79.King OD and Masel J (2007) The evolution of bet-hedging adaptations to rare scenarios. Theor. Popul. Biol 72, 560–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rees M and Ellner SP (2016) Evolving integral projection models: Evolutionary demography meets eco-evolutionary dynamics. Methods Ecol. Evol 7, 157–170 [Google Scholar]
- 81.Bishop AL et al. (2007) Phenotypic heterogeneity can enhance rare-cell survival in ‘stress-sensitive’ yeast populations. Mol. Microbiol 63, 507–520 [DOI] [PubMed] [Google Scholar]
- 82.Raj A et al. (2010) Variability in gene expression underlies partial penetrance in Caenorhabditis elegans. Nature 12, 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cleasby IR et al. (2015) Quantifying the predictability of behaviour: statistical approaches for the study of between-individual variation in the within-individual variance. Methods Ecol. Evol 6, 27–37 [Google Scholar]
- 84.Mulder HA et al. (2013) Estimation of genetic variance for macro-and micro-environmental sensitivity using double hierarchical generalized linear models. Genet. Sel. Evol 45, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sae-Lim P et al. (2017) Estimation of breeding values for uniformity of growth in Atlantic salmon (Salmo salar) using pedigree relationships or single-step genomic evaluation. Genet. Sel. Evol 49, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Barba M et al. (2017) High-throughput microsatellite genotyping in ecology: improved accuracy, efficiency, standardization and success with low-quantity and degraded DNA. Mol. Ecol. Resour 17, 492–507 [DOI] [PubMed] [Google Scholar]
- 87.Chen N et al. (2019) Allele frequency dynamics in a pedigreed natural population. Proc. Natl. Acad. Sci. USA 116, 2158–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nussey DH et al. (2014) Multivariate immune defences and fitness in the wild: complex antibodies, health and survival. Proc. R. Soc. B Biol. Sci 281, 20132931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Altmann J (2001) Baboon Mothers and Infants, University of Chicago Press. [Google Scholar]
- 90.Grant PR and Grant BR (2014) 40 Years of Evolution: Darwin’s Finches on Daphne Major Island, Princeton University Press. [Google Scholar]
- 91.Chen N et al. (2016) Genomic consequences of population decline in the endangered Florida scrub-jay. Curr. Biol 26, 2974–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson-Teixeira KJ et al. (2015) CTFS-Forest GEO: a worldwide network monitoring forests in an era of global change. Glob. Chang. Biol 21, 528–549 [DOI] [PubMed] [Google Scholar]
- 93.Bradley BP (1980) Developmental stability of Drosophila melanogaster under artificial and natural selection in constant and fluctuating environments. Genetics 95, 1033–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kubrak OI et al. (2017) Adaptation to fluctuating environments in a selection experiment with Drosophila melanogaster. Ecol. Evol 7, 3796–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Forsberg SKG and Carlborg Ö (2017) On the relationship between epistasis and genetic variance heterogeneity. J. Exp. Bot 68, 5431–5438 [DOI] [PubMed] [Google Scholar]
- 96.Corty RW and Valdar W (2018) QTL mapping on a background of variance heterogeneity. G3 Genes, Genomes, Genet. 8, 3767–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mulder HA et al. (2007) Prediction of breeding values and selection responses with genetic heterogeneity of environmental variance. Genetics 175, 1895–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mulder HA et al. (2015) Heritable environmental variance causes nonlinear relationships between traits: application to birth weight and stillbirth of pigs. Genetics 199, 1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lande R (1979) Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33, 402. [DOI] [PubMed] [Google Scholar]
- 100.Steppan SJ et al. (2002) Comparative quantitative genetics: evolution of the G matrix. Trends Ecol. Evol 17, 320–327 [Google Scholar]
- 101.Arnold SJ et al. (2008) Understanding the evolution and stability of the G-matrix. Evolution 62, 2451–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoffmann AA and Merilä J (1999) Heritable variation and evolution under favourable and unfavourable conditions. Trends Ecol. Evol 14, 96–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


