Sir,
The coronavirus disease 2019 (COVID-19) pandemic has afected millions of people and poses a global health emergency. The disease is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), from the Betacoronavirus genus. Transmission of SARS-CoV-2 takes place through close contact from person-to-person, via fomites or through inhalation of viral particles [1]. SARS-CoV-2 has been shown to retain infectivity for up to 16 h in respiratory aerosols [2], and can be viable on various surfaces for several days [3]. An effective strategy for medical workers to protect themselves from infection is to wear personal protective equipment (PPE), including scrubs, which are often used over day-long shifts. Scrubs and other clothing used for long periods of time during the care of patients shedding SARS-CoV-2 may accumulate virus on their external surfaces and, without decontamination, could pose a risk to healthcare workers [4,5]. Self-decontaminating PPE is an elegant solution to this problem as this may reduce the risk of infection from PPE without effort from the user. Duritex™ is a natural biopolymer and disinfectant of complex chemical composition that can be bonded to fabrics by a thermal process, which establishes ionic or covalent bonds, depending on the composition of the fibre used. We tested whether fabric treated with this biopolymer disinfects effectively against SARS-CoV-2.
We used pieces of 80/20 polycotton fabric [6] made from combining dimethyl ester dimethyl terephthalate and monoethylene glycol (polyesther) to cotton. These pieces of fabric were bonded with Duritex, cut into 1 x 1-cm squares and added to six-well cell culture plates before testing. We used SARS-CoV-2 strain USA_WA1/2020 [7], obtained from the World Reference Center for Emerging Viruses and Arboviruses at passage 4, and all manipulations of the virus were performed in a biosafety level 3 laboratory. The virus was amplified in Vero cells at multiplicity of infection of 0.001 using Dulbecco's minimal essential medium (DMEM) containing 2% fetal bovine serum (FBS), 200 mg/mL streptomycin and 200 U/mL penicillin, and 100 μL of virus stock containing 5 x 105 plaque-forming units (PFU) was dripped on to and absorbed by the treated fabric, or untreated control fabric, and held at room temperature (22–25˚C) at approximately 60% relative humidity. After each timepoint (15 min, 30 min, 45 min, 1 h, 2 h), the fabric was transferred to a microcentrifuge tube containing 500 μL DMEM using sterilized forceps. The tube was vortexed for 30 s, and eluted samples were serially diluted in DMEM supplemented with 2% FBS, penicillin and streptomycin as above. Each dilution was added to six-well plates containing a monolayer of susceptible Vero E6 cells for virus adsorption for 1 h, incubated at 37˚C in an atmosphere of 5% CO2, and rocked every 15 min. After adsorption, cells were overlaid with DMEM containing 0.8% agarose. Two days later, the cells were stained with neutral red for 6 h and viral plaques were counted.
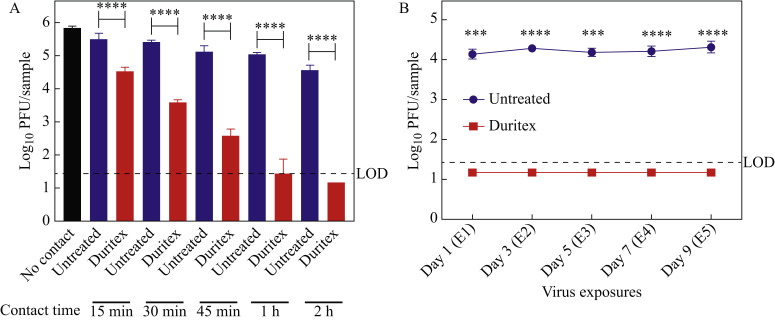
After adding virus to the treated fabric, we observed reductions in the titres compared with the untreated fabric of 1 log10 after 15 min of contact, 1.8 log10 after 30 min of contact and 2.5 log10 after 45 min of contact (Figure 1 A). After 1 h of exposure, the decrease was approximately 3.5 log10, and after 2 h of exposure, the virus was decreased to titres below the detection limit of 30 PFU/sample (Figure 1A). To determine whether residual virus from multiple exposures could saturate the self-disinfecting property of the treated fabric and affect long-term re-usage, we performed five serial applications of virus, once every 2 days. For each application, 20 μL of virus containing 1 x 105 PFU was placed on to to the fabric for 2 h at room temperature before virus titration. This volume was chosen to avoid saturating the fabric with liquid and risking spillover on to the six-well plate. Other parameters were the same as for the single application experiment described above. At all timepoints, we measured a reduction of ≥ 3 log10 of infectious virus (below the limit of detection) in the treated fabric compared with the untreated control fabric (Figure 1B).
Figure 1.
Fabric treated with Duritex™ is self-decontaminating against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). (A) SARS-CoV-2 titres are decreased when in contact with treated fabric. SARS-CoV-2 [100 μL containing 5 x 105 plaque-forming units (PFU)] was added to the treated or untreated fabric and timepoints of contact were tested. (B) Treated fabric can reduce viral titres over multiple exposures to the virus. SARS-CoV-2 (20 μL containing 1 x 105 PFU) was added to the fabric and allowed to incubate at room temperature for 2 h before the plaque assay. The fabric was exposed every 2 days. For both panels, error bars represent the standard deviation of triplicate samples. The limit of detection (LOD) of the assays was 30 PFU/sample. One-way analysis of variance with Sidak's multiple comparison test was used to determine significance for these assays; ∗∗∗∗P≤0.0001.
These results demonstrate that treatment of fabric with Duritex via thermal bonding can be used to render it self-disinfecting against SARS-CoV-2. This represents a potentially important application for cloth PPE used over long periods of time in settings where the virus is likely to be present, such as hospitals and other healthcare facilities. Compared with other methods of disinfection, such as heat, ultraviolet radiation and chemical disinfection, Duritex self-disinfection has the advantage that decontamination starts before doffing of PPE, a major safety feature. This technology may also have the potential for use in laboratory PPE such as masks, shoe covers and laboratory coats, or for making self-disinfecting clothing and face coverings for the general population during the COVID-19 pandemic.
Conflict of interest statement
This work was supported by Ghost Tactical which developed Duritex.
Funding source
Ghost Tactical.
References
- 1.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fears A.C., Klimstra W.B., Duprex P., Hartman A., Weaver S.C., Plante K.S. Persistence of severe acute respiratory syndrome coronavirus 2 in aerosol suspensions. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2609.201806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Maio P., Iocca O., Cavallero A., Giudice M. Performing the nasopharyngeal and oropharyngeal swab for 2019-novel coronavirus (SARS-CoV-2) safely: how to dress, undress, and technical notes. Head Neck. 2020;42:1548–1551. doi: 10.1002/hed.26230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans H.L., Thomas C.S., Bell L.H., Hink A.B., O'Driscoll S., Tobin C.D. Development of a sterile personal protective equipment donning and doffing procedure to protect surgical teams from SARS-CoV-2 exposure during the COVID-19 pandemic. Surg Infect (Larchmt) 2020 doi: 10.1089/sur.2020.140. [DOI] [PubMed] [Google Scholar]
- 6.Moiz A., Padhye R., Wang X. Coating of TPU-PDMS-TMS on polycotton fabrics for versatile protection. Polymers (Basel) 2017;9:660. doi: 10.3390/polym9120660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv. 2020 doi: 10.1101/2020.03.02.972935. [DOI] [Google Scholar]