Abstract
Antibody-dependent enhancement (ADE) exists in several kinds of virus. It has a negative influence on antibody therapy for viral infection. This effect was first identified in dengue virus and has since also been described for coronavirus. To date, the rapid spread of the newly emerged coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), has affected over 3.8 million people across the globe. The novel coronavirus poses a great challenge and has caused a wave of panic. In this review, antibody-dependent enhancements in dengue virus and two kinds of coronavirus are summarized. Possible solutions for the effects are reported. We also speculate that ADE may exist in SARS-CoV-2.
Keywords: Antibody-dependent enhancement (ADE), Coronavirus disease 2019 (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV)
Introduction
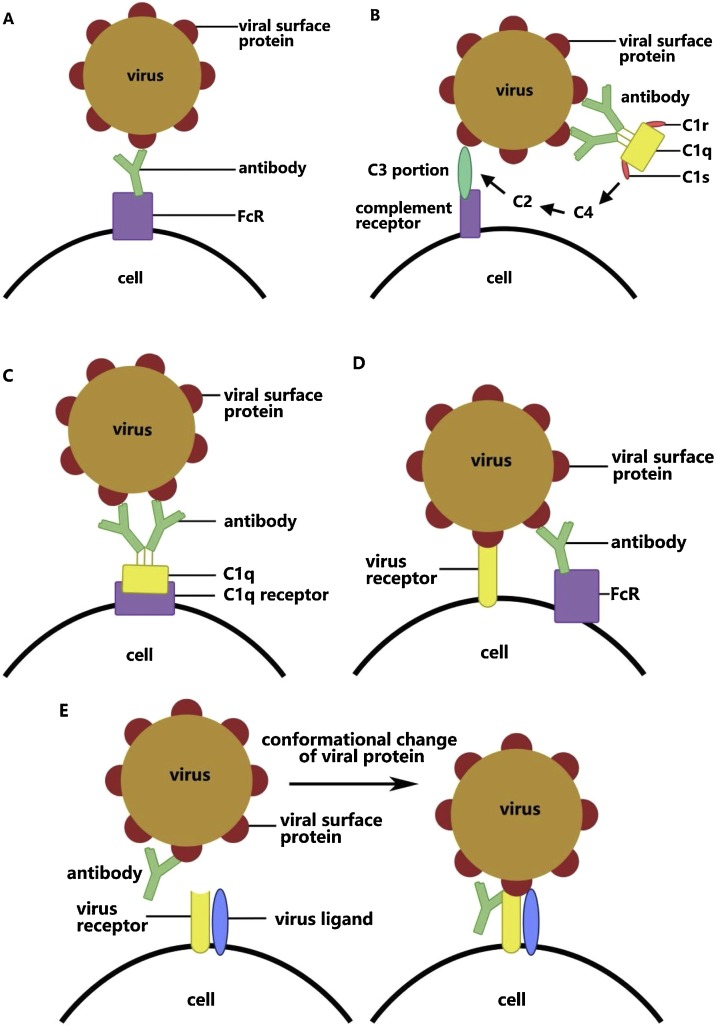
Viral infection initiates with attachment of the virus particle to the cytoplasm membrane on the cell surface, a process in which viral surface protein binds uniquely to specific receptors on the host cell. To block this viral attachment to target cells, antibodies that target the viral surface proteins specifically are secreted, which bind and neutralize the viruses, weakening their infective ability. However, in some viruses, the binding of specific antibodies to viral surface proteins can promote viral invasion into certain types of cell instead, and enhance viral infection. This effect is called antibody-dependent enhancement (ADE) (see Glossary) (Taylor et al., 2015). ADE happens in two main cases: (1) when viral-specific antibody promotes viral entry into host monocytes/macrophages and granulocytes, and (2) when it enhances viral infection in cells via interplay with the Fc receptor (FcR) and/or complement receptor. Enhancement of the attachment between viruses and target cells play important role in most cases. ADE has been identified in over 40 kinds of virus. These viruses have several different antigenic epitopes, some of which induce neutralizing antibodies, while some stimulate enhanced antibodies. Conventional vaccine has shown a weak preventive and therapeutic effect on these viruses (Wang et al., 2016), and in some cases has also been shown to increase the susceptibility of those who are vaccinated.
Mechanisms of antibody-dependent enhancement
Studies so far have presumed that there are five mechanisms that underlie ADE and that various viruses work under different mechanisms and are not necessarily facilitated by a single mechanism.
The first mechanism of ADE is dependent on FcR (Figure 1 A). FcRs are mainly distributed on immune cells and are receptors targeting the Fc portions on antibodies (Dustin, 2016). In this FcR-mediated ADE, the viral surface protein combines with the antibody to form a virus–antibody complex. The complex strengthens viral adhesion through interaction of the Fc portion on the antibody and its receptor on the surface of particular cells. This mechanism has been found in West Nile virus, dengue virus, and human immunodeficiency virus (HIV). Among these, dengue is the disease most prominently affected by ADE (de Alwis et al., 2020). It is one of the diseases observed to have ADE earliest (Dustin, 2016), and is also the fastest growing global epidemic.
Figure 1.
Mechanisms of antibody-dependent enhancement.
Dengue is generally a self-limiting disease; however, secondary dengue may present as a more severe disease. There are four serotypes of dengue virus, DENV1, DENV2, DENV3, and DENV4. There is no cross-immune protection between the four serotypes, which means that the antibodies induced by one serotype do not work on the others. When it comes to secondary infection, if the individual is infected by virus of the same serotype, the antibodies produced before can neutralize the virus quickly. However, if the individual is infected by virus of a different serotype, these antibodies will not only fail to neutralize the virus, but may even facilitate viral entry via antibody Fc portions and increase viremia in vivo. This was confirmed in research using DENV-4 and DENV-2 to infect Macaca mulatta (rhesus monkeys) in succession. In ADE of dengue virus, after viral surface proteins have attached to antibodies, the Fc portions on the antibodies bind to cells bearing FcγR. Then virus–antibody complexes gather on the cell surface with the assistance of FcγRIIA, thereby promoting viral entry and enhancing infectivity. The suppression of antiviral genes also occurs during this process. The interplay between the host cell and virus during viral replication helps virus to escape the antiviral and immune responses of the host (Morrone and Lok, 2019). The monocytes and T lymphocytes are excessively activated and secrete more cytokines, leading to increases in vascular permeability and triggering dengue hemorrhagic fever and dengue shock syndrome. Nevertheless, not all secondary dengue virus infections are at risk of ADE (Wilder-Smith et al., 2019); the proportion of antibody to virus determines whether this occurs.
Only under the titer at which FcγR-mediated viral entry is inhibited can the isogenic serotype of dengue virus be neutralized by antibodies. Also, modification of the FcγR binding domain on the antibody Fc portion eliminates its ability to bind to FcγR on cells without changing its half-life period, and this can effectively lower the viral load and improve the survival rate. Cross-linking of FcγRIIB has also been shown to inhibit the ADE in dengue virus infection (Chan et al., 2011).
The second mechanism of ADE is C3-dependent. Complement C3 is activated in the classical pathway through the binding of antibody to viral surface protein, following which the interaction between complement C3 and corresponding receptor enhances viral adhesion in the form of virus–antibody–complement complex (Figure 1B). Complement receptor has a wider distribution than FcR, including immune cells, follicular dendritic cells, and smooth muscle cells (Dustin, 2016). In this type of ADE, C1q molecules as part of complement C1, work together with serine protease proenzyme, C1r and C1s. This is dependent on calcium ions. Then, when C1q binds to antibody or antibody complex, C1r and C1s depart from C1q under the induction of the C1 inhibitor in blood plasma. C1s then cleaves complement C2 and C4, allowing C1q to activate the combination between distant effector complement C3 and its receptors on cells. In this way, the viruses bind to complement receptors. This mechanism underlies ADE in both West Nile virus and HIV.
The third mechanism of ADE is C1q-dependent (Figure 1C). Virus–antibody complexes are combined by C1q, promoting fusion between the viral capsule and cell membrane through the deposition of the combination of C1q and its receptor (von Kietzell et al., 2014). Tight binding of two or more monomer IgG antibodies and specific epitopes allows C1q to bind with antibody Fc portions, which causes the formation of virus–antibody–C1q complex. The complex binds to the C1q receptor on cells, initiates the intracellular signaling pathway, and then promotes binding of the virus and its specific receptor, as well as endocytosis of the target cells. In some cases, C1q binds directly to gp41, one of the glycoproteins on the viral outer membrane during HIV infection. C1q receptors are found not only on inflammatory monocytes/macrophages, but are also distributed on many different cell types, including neutrophils, B cells, fibroblasts, smooth muscle cells, and endothelial cells. Hence this C1q-mediated ADE explains why early-stage antiviral serum can enhance the infection in non-monocytes. This mechanism is also exploited by Ebola virus.
The fourth mechanism of ADE is the suppression of the expression of antiviral genes via the stimulation and enhancement of certain target cell effects, such as endocytosis (von Kietzell et al., 2014) (Figure 1D). This mechanism was identified in Ross River virus. In the course of this mechanism, viruses rely mainly on Fc receptors to enter cells in the form of ADE, while normal viral entry through binding with viral receptor is decreased. This occurs in the replication of viruses, leading to the suppression of the antiviral genes, such as those for tumor necrosis factor (TNF) and induced nitric oxide synthase, hence helping the virus with immunological escape (Mahalingam and Lidbury, 2002). Similar pathways have been observed in dengue virus as well.
The fifth mechanism of ADE is the enhancement of the fusion of viruses and cells via a change in the conformation of viral protein through its binding with antibody (Figure 1E). This was found in HIV infection by Sullivan and co-workers. Monoclonal antibodies recognize the glycoprotein gp120 on the outer membrane of HIV and combine with one of its subunits under a sub-neutralizing concentration. This then induces a conformational change in the residual subunits and promotes membrane fusion of viruses and target cells via the activation of viral glycoprotein. The specific antibody towards gp120 will also regulate the interaction between gp120 and virus ligand CCR5 (Guillon et al., 2002).
ADE in coronavirus
ADE and its mechanisms in coronavirus infection
Several different kinds of coronavirus have been shown to cause disease in mammals and birds. Among these, seven are known to infect humans (Table 1 ), including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), which has overwhelmed the whole world this year. Four of the remaining six just elicit common cold symptoms; these are human coronavirus 229E, NL63, OC43, HKU1 (Fung and Liu, 2019). All of these are known to be endemic. The final two are well known and highly pathogenic betacoronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Both of these may cause a lethal infection in humans, with symptoms including acute respiratory distress syndrome (Takano et al., 2019). Early in the last century, researchers discovered coronaviruses in animals, like feline enteric coronavirus. This is exploited by ADE because of ineffective antibodies (Takano et al., 2019), leading to an exacerbation of disease symptoms. Antibodies to feline infectious peritonitis virus also enhance infection of monocytes. It has been sequentially confirmed in subsequent studies that ADE of SARS-CoV and MERS-CoV also occur, with different mechanisms. Whether ADE works in other kinds of coronavirus infections remains to be investigated. Unlike dengue virus, ADE in SARS and MERS is not triggered by a heterovirus strain, however it is clear that the effects of both have negative consequences for the human body and are probably an obstacle to the development of viral vaccines (Yong et al., 2019).
Table 1.
Coronaviruses that infect humans.
| Virus name | Hierarchy | ADE or not | Mechanism | Reference |
|---|---|---|---|---|
| Human coronavirus 229E | Alphacoronavirus | Unsure | – | |
| Human coronavirus OC43 | Betacoronavirus lineage A | Unsure | – | |
| Human coronavirus NL63 | Alphacoronavirus | Unsure | – | |
| Human coronavirus HKU1 | Betacoronavirus lineage A | Unsure | – | |
| Severe acute respiratory syndrome coronavirus (SARS-CoV) | Betacoronavirus lineage B | Yes | Viruses enter macrophages through antibody Fc portion and alter function | Liu et al. (2019), Luo et al. (2018), Yip et al. (2016) |
| Middle East respiratory syndrome coronavirus (MERS-CoV) | Betacoronavirus lineage C | Yes | Mersmab1 induces a formational change of spike and causes viral entry through Fc portion | Wan et al. (2020), Du et al. (2014) |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2019 novel coronavirus) | Betacoronavirus | Unsure | – |
According to previous studies on human coronaviruses SARS-CoV and MERS-CoV, the mechanisms of ADE are different to those in dengue virus, the one more closely related to the existence of different subtypes of virus strain. Both kinds of human coronavirus enter cells under the assistance of FcR. SARS-CoV causes enhanced lung injury by inducing hyperimmunity through the interactions of antibody and FcR, which alters the functions of macrophages (Liu et al., 2019). Regarding MERS-CoV, this promotes viral entry under the induction of binding between antibodies and FcR, which is similar to the traditional route of viral entry (Wan et al., 2020). Otherwise, these processes in the two coronaviruses are related to the titers and compositions of antibody.
ADE in severe acute respiratory syndrome
SARS-CoV enters host cells by recognizing and binding to its viral receptor angiotensin-converting enzyme 2 (ACE2) in the classical pathway, while antibodies neutralize the viruses by blocking the interaction of viral spike proteins and ACE2. This block was confirmed in a type of human antibody extracted from the antibody library of non-immune people early in 2004 (Sui et al., 2004). Then in 2007, Yiu Wing Kam et al. explored whether antibody against SARS-CoV viral spike protein can induce viral entry into FcR-bearing cells and evoke ADE (Kam et al., 2007). The results showed that these antibodies increase the affinity of SARS-CoV towards FcγRII-bearing cells. This increase is mediated by the Fc portion of anti-spike antibody and FcγRII on cells, while ACE2 is not required in the process. Later research by Jaume et al. in 2014 showed that antibody against SARS-CoV viral spike protein strengthened the infection towards monocytes and lymphocytes, both of which do not express viral receptors (Yip et al., 2014). This was in agreement with the results of Yiu Wing Kam’s team. In the same year, studies conducted by Chen and Huang’s team indicated that ADE is mainly induced by diluted antibodies against spike proteins rather than nucleocapsid protein(Wang et al., 2014). These studies further demonstrated that anti-spike antibodies induce ADE during SARS-CoV infection, and this effect mainly works in immune cells. In 2018, the rhesus monkey was used as an animal model to study the relationship between ADE and the antibody titer induced by vaccine. The results demonstrated that those vaccines that elicit low titers of antibody may not induce ADE after infection with SARS-CoV (Luo et al., 2018), while highly diluted serum may in turn promote the infectivity of the virus.
Up until 2019, the mechanism of ADE in SARS-CoV remained unclear. Then Chen et al. studied the mechanism of the SARS-CoV viral spike induced effect in detail, using the Chinese rhesus monkey as an animal model. This team developed the first SARS vaccine in 2005, which encodes the complete SARS-CoV viral spike in the modified attenuated poxvirus vector. A study was performed in 2019 to investigate the vaccine. It was found to induce the production of large amounts of neutralizing antibodies (S-IgG) soon after injection. Although these antibodies can effectively reduce the viral load in the upper respiratory tract, they also enhance lung injury. A positive correlation has been found between the amount of neutralizing antibody in serum and the degree of pathological injury in the lung. Further studies found that the virus enters macrophages with the help of FcR during ADE (Liu et al., 2019).
There are two main types of macrophage. One is classically activated macrophage (M1), whose main function includes secreting proinflammatory factors and mediating the host defense against varieties of bacteria, protozoa, and viruses. This type of macrophage has a strong ability to kill microorganisms. However, this property is also prone to cause tissue injury. Another type of macrophage is the alternatively activated macrophage (M2), which has anti-inflammatory functions and regulates wound healing in most cases. Antibody against SARS-CoV spike alters the function of M2 macrophages through binding with FcR. Endocytosis of glycoprotein and immunodepression in macrophages are then weakened while the enrichment of cytokines increases. M1 macrophages, which should repair pathological injury in the lung, then convert into cells that promote inflammation. This conversion partially relies on the role of FcγR (Liu et al., 2019).
The application of an FcR blocker effectively inhibits the secretion of proinflammatory factors. Comparison of the pathological damage and concentration of neutralizing antibody in serum between patients who died and those who recovered led to conclusions consistent with those reported for animal models (Liu et al., 2019). However, the findings of Jaume et al. in 2016 differ; they suggested that the inflammatory response in macrophages changes mildly during ADE of SARS-CoV (Yip et al., 2016). These studies confirmed the existence of the effect in the virus and provided ideas for resolving the negative consequences of ADE during the treatment of viral infection.
ADE in Middle East respiratory syndrome
Spike protein anchored on the cytomembrane induces coronavirus to enter host cells. The ectodomain of spike protein consists of receptor-binding subunit S1, S1/S2 cleavage site, and membrane-fusion subunit S2. Receptor-binding domains on each S1 subunit induce the recognition of receptor, while the S2 subunit possesses an S2′ cleavage site, which allows its hydrophobic amino acid to insert into cells and mediates fusion of the viral envelope and the cell membrane. The viral receptor of MERS-CoV on cells is dipeptidyl peptidase 4 (DPP4). During MERS-CoV viral packaging, spike protein is cleaved. The S1 subunit on the spike protein binds with DPP4 and stabilizes the receptor-binding site, which promotes a conformational change in the spike protein. Successive cleavage of the S1/S2 and S2′ enzymatic cleavage sites by host protease will lead to the S1 subunit departing and a change in the conformation of the S2 subunit, which gradually induces membrane fusion of the virus and host cells. Neutralizing monoclonal antibody that acts specifically against the receptor binding site on the MERS-CoV viral spike protein (Mersmab1) can block classical viral invasion by contesting the binding site for viral spike against DPP4 (Du et al., 2014).
MERS-CoV was first discovered in 2012, so there have not been many studies on ADE of MERS-CoV up to now. The rhesus macaque was the first animal model adopted for the development of MERS-CoV vaccine. Research by Tseng et al. in 2016 first reported that the MERS inactivated vaccine could lead to a hypersensitive lung pathology similar to that in SARS. Although the vaccine induced the production of neutralizing antibodies and also reduced the virus in the lung, it caused the enhancement of mononuclear infiltration. An increase in eosinophil granulocytes promoted the secretion of interleukins IL-5 and IL-13. This only occurred under the mediation of inactivated vaccine. Thus, the cause of ADE during vaccine treatment may be viral components or contaminants. Another important support for ADE in MERS-CoV is the finding that the inflammatory response caused by the virus aggravates symptoms (Prescott et al., 2018). Only in 2020 did researchers first uncover the mechanism of ADE of MERS-CoV in vitro comprehensively. Only Mersmab1 with a complete structure can induce the indirect interaction between viral spike protein and FcR on cells. This helps the virus enter FcR-bearing cells while inhibiting its entry into DPP4-expressing cells, whereas the residual Fc portion of antibody induces viral entry into DPP4-expressing cells. Viral entry under ADE similar to that under the classical pathway has also been demonstrated in MERS-CoV. Both viral entry pathways are induced through the exposure of the S2′ site after a conformational change in the viral spike protein (Wan et al., 2020).
Moreover, the dose of antibody affects the two types of MERS-CoV viral entry mentioned above in diverse ways. With the increase in Mersmab1 dose, fewer virus particles enter DPP4-expressing cells, while the amount of virus entering FcR-bearing cells increases first before it decreases. Researchers have provided a reasonable explanation for this result. Although antibody induces an indirect interaction between viral spike protein and FcR at a low dose, after it reaches a certain concentration, it blocks FcRs before binding to viral spike proteins, hence blocking the indirect interaction (Wan et al., 2020). Therefore, high doses of antibody may help to reduce the risk brought by ADE during the use of antibody. Other factors that determine the dose of antibody to be used include the expression level of DPP4 and FcR in specific tissues. The affinity between antibodies and FcR is also important.
The membrane fusion during invasion of coronavirus needs the activation of host protease, especially lysosomal proteases. Several types of protease inhibitor were found to affect both DPP4 and Mersmab1-dependent MERS viral entry during an investigation of the impact of proprotein invertase inhibitors. These inhibitors promote virus entry into DPP4-bearing cells in the absence of Mersmab1, whereas they promote virus entry into FcR-bearing cells (Wan et al., 2020). All of these studies provided good suggestions to avoid ADE during the treatment of MERS-CoV infection.
ADE may exist in SARS-CoV-2
The 2019 novel coronavirus SARS-CoV-2 emerged this year and has caused high numbers of deaths. The most recent sequencing results have shown that SARS-CoV-2 shares a similar genome sequence of up to 79.5% with SARS-CoV, and the viral receptor for both is ACE2. A team from the University of Texas found that SARS-CoV-2 has affinity for the ACE2 receptor 10–20 times that of SARS-CoV (Wrapp et al., 2020), which explains why it has a higher basic reproduction number. These results also indicate a pathogenic similarity between the two viruses.
Studies on SARS-CoV have highlighted the complexity of the role of antibodies in the pathogenesis of highly pathogenic coronaviruses (Fleming and Raabe, 2020). Not long after the outbreak of COVID-19 was declared, the heterogeneity of severe cases in Hubei Province, China and in other areas was noted and this was attributed to ADE. Before a vaccine or specific therapy is available, convalescent plasma therapy is considered to be a useful tool for research. Golden Syrian hamsters are considered to be a good model for the study of SARS-CoV-2 as they can be consistently infected with the virus (Chan et al., 2020). Zhiwei Chen et al. used this model for research, and in a recent interview they reported that in their most recent study on convalescent plasma and the transmission of SARS-CoV-2, antibodies played no role in pathological injury in the lung (Li, 2020). However, hamsters and human beings belong to different species after all, so results may differ greatly between them.
The latest publication on convalescent plasma transmission in humans showed satisfactory results without any aggravation of symptoms; however only 10 adults with severe disease were involved (Duan et al., 2020). The most recent research published on May 6, 2020 tested a purified inactivated SARS-CoV-2 virus vaccine candidate. Although inactivated virus vaccines are thought to have ADE, the results of this newest study performed in mice, rats, and non-human primates showed good neutralization of 10 representative strains, without ADE (Gao et al., 2020). Nevertheless, use of the vaccine in humans has not yet been reported in a research paper, so whether SARS-CoV-2 will induce ADE in patients still needs further verification. As stated by Jiang (who has contributed towards vaccines and treatments for coronaviruses), safety testing matters most during the counterattack against the new coronavirus (Jiang, 2020).
To better reveal the mechanism of SARS-CoV-2 and provide new diagnostic tools, the One Health approach should be considered (Tilocca et al., 2020a). We should work at the local, regional, national, and global levels, with the goal of achieving optimal health outcomes while recognizing the interconnection between people, animals, plants, and their shared environment. An immunoinformatics approach is one that will play a great role. By comparing the sequence of spike protein or envelope protein, two proteins with high immunogenicity and multifunctional properties, at the epitope level among different coronaviruses with tropism towards various species, new avenues towards the biological mechanism of viral infection may be opened. In the studies by Tilocca et al., high similarity of spike protein between SARS-CoV-2 and bovine coronavirus/canine virus partially supported the possibility of ADE occurring during this epidemic event, which may explain the diversity of clinical cases inside and outside Wuhan Province (Tilocca et al., 2020a, 2020b).
The solutions for ADE in coronavirus
According to previous studies, a solution for ADE in coronavirus infection can be approached in several ways. The first is to control the dose. A high dose of antibodies can inhibit the ADE in MERS-CoV without influencing its antiviral ability (Wan et al., 2020). The second way is to change the antibody target. Although blocking the binding of coronavirus spike proteins is a good therapeutic approach because of its high efficiency in reducing the viral load, the binding with antibody against spikes makes it easier to mediate ADE. The third approach is to take advantage of some inhibitors. For example, protease inhibitors and Fc inhibitors play roles in the inhibition of ADE in MERS-CoV and SARS-CoV, respectively (Liu et al., 2019; Wan et al., 2020). Previous studies on SARS-CoV showed that an adjuvant promoted Th2-type immunity and reduced the immunopathology, thereby suggesting the latent importance of adjuvant (Hotez et al., 2020). In addition, the case of dengue virus can also be of reference; for example, reduce the risk of ADE by modifying the FcγR binding site on the antibody Fc portion. Another difficulty in these cases is to guarantee the inhibition of classical viral entry via antibodies while solving ADE. A potential solution is to combine Cyclospora A and Chinese drugs pharmaceutics that have an immunodepression function with colloidal subparticles, which can enhance the targeting to macrophages and promote an immunosuppressive effect. This could act not only by inhibiting the immune-injury inflammation, but also against the virus and bacteria.
Conclusions and future perspectives
Our understanding of ADE has been relatively thorough up to now. However, the detailed mechanism of ADE and how to resolve this in coronavirus infections is not yet totally clear. From previous research on ADE in other coronaviruses, in particular SARS-CoV and MERS-CoV, it appears that the existence of ADE will elicit more severe body injury, while actually reducing the viral load at the same time. This may affect the results of vaccine therapy. The presence of this phenomenon in these two coronaviruses indicates a potential risk in the vaccine therapy for the novel coronavirus SARS-CoV-2, as it shares the same viral receptor and similar genome sequence with SARS-CoV. SARS-CoV-2 may have a similar mechanism of viral entry and thus may share similar mechanisms of ADE. This novel coronavirus has not long been known, so studies in this field have not yet led to any conclusions.
Previous studies have shown that different teams have sometimes given different explanations for ADE in the same virus. A possible important factor may be the late emergence of human coronaviruses that cause severe symptoms. Understanding of ADE in SARS-CoV and MERS-CoV infections has taken several years and was relatively clear till these two years. From this, we speculate that studies on ADE in this newly emerged coronavirus will take some time, but the elucidation of this effect is of great importance.
Funding source
This work was supported by Shenzhen Peacock Plan Technological Innovation (QJSCX2010170728150303243) and by the Shenzhen Branch Committee on Technology and Innovation(JCYJ2018030512481244444).
Ethical approval
All analyses were based on previously published studies, thus ethical approval and patient consent were not required.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Glossary
- Angiotensin-converting enzyme II
A potent negative regulator of the renin–angiotensin system. Viral receptor for both SARS-CoV and SARS-CoV-2.
- A
- ntibody-dependent enhancement
ADE is an effect mediated by antibodies. In some viruses, the binding of specific antibodies to viral surface proteins will promote viral invasion into certain types of cell instead of blocking viral attachment.
- Antiviral genes
Genes that reduce the infectivity or invasive ability of virus.
- Antiviral serum
Immune serum obtained by immunizing horses or other large animals with bacteria or viruses themselves.
- C1rType
of β globin, can activate C1s.
- C1sType
of α2 globin, has enzyme-like activity, activates C4 and C2 in the presence of Mg
- C2
Intrinsic component of the classical activation pathway, belongs to serine.
- C4
Plays a role in complement activation, promotion of phagocytosis, prevention of immune complex’s formation and viral neutralization after hydrolyzation.
- CCR5
An important HIV-1 co-receptor involved in membrane fusion.
- Conventional vaccines
Consist of whole pathogenic organisms, which may either be killed or live vaccines.
- Dipeptidyl peptidase 4
A serine protease on the surface of a cell, viral receptor for MERS-CoV.
- FcγRIIAFcγRII
(CD32) widely expressed on almost all bone marrow cells. FcγRⅡA elicits cell excitation through intracellular signaling.
- FcγRIIB
Mainly conduct inhibitory signals.
- Fc receptor
Receptor for C terminal of the Fc portion on immunoglobulin, induces the interaction between the antigen–antibody complex and the cell, plays an important role in the immune response and its regulation.
- gp120
A glycoprotein that binds to CD4 receptor on T cells bearing such receptors before infection of the cell can occur.
- IL-5
Participates in cytokine activity and interleukin-5 receptor binding.
- IL-13
Participates in cytokine activity and interleukin-13 receptor binding.
- Induced nitric oxide synthase
A class of enzyme that
- Neutralizing antibodies
Antibody produced by B lymphocytes, can bind with antigen on the surface of pathogenic microorganisms and then block the attachment and invasion to cells.
- Serotype
In microbiology, it can be used to identify different types of the same virus.
- Tumor necrosis factor
A cytokine released predominantly from macrophages or a variety of other immune cells. As a pyrogen, it is important in the acute phase of inflammation and infection, with signaling through nuclear factor kappa B.
- Viral load
The number of viruses in each milliliter of blood shown by measurement.
- Viral specific antibody
Host cell membrane components that can specifically bind to the virus, mediate viral invasion, and promote viral infection. Most of them are proteins, some are glycoproteins, proteoglycans, lipids, or glycolipids.
References
- Chan K.R., Zhang S.L.-X., Tan H.C., Chan Y.K., Chow A., Lim A.P.C., et al. Ligation of Fc gamma receptor IIB inhibits antibody-dependent enhancement of dengue virus infection. Proc Natl Acad Sci USA. 2011;108:12479–12484. doi: 10.1073/pnas.1106568108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa325. pii: ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R., Chen S., Gan E.S., Ooi E.E. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z., et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L. Complement receptors in myeloid cell adhesion and phagocytosis. Microbiol Spectr. 2016;4(November (6)) doi: 10.1128/microbiolspec.MCHD-0034-2016. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A.B., Raabe V. Current studies of convalescent plasma therapy for COVID-19 may underestimate risk of antibody-dependent enhancement. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yan M., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon C., Schutten M., Boers P.H.M., Gruters R.A., Osterhaus A.D.M.E., et al. Antibody-mediated enhancement of human immunodeficiency virus type 1 infectivity is determined by the structure of gp120 and depends on modulation of the gp120-CCR5 interaction. J Virol. 2002;76:2827–2834. doi: 10.1128/JVI.76.6.2827-2834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P.J., Corry D.B., Bottazzi M.E., et al. COVID-19 vaccine design: the Janus face of immune enhancement. Nat Rev Immunol. 2020;20(6):1–2. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature. 2020;579(7799):321. doi: 10.1038/d41586-020-00751-9. [DOI] [PubMed] [Google Scholar]
- Kam Y.W., Kien F., Roberts A., Cheung Y.C., Lamirande E.W., Vogel L., et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine. 2007;25:729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. 2020. Southern Weekly.http://infzm.com/contents/181390 [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.123158. pii: 123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Liao F.-L., Wang H., Tang H.-B., Yang Z.-Q., Hou W. Evaluation of antibody-dependent enhancement of SARS-CoV infection in rhesus macaques immunized with an inactivated SARS-CoV vaccine. Virol Sin. 2018;33:201–204. doi: 10.1007/s12250-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam S., Lidbury B.A. Suppression of lipopolysaccharide-induced antiviral transcription factor (STAT-1 and NF-kappa B) complexes by antibody-dependent enhancement of macrophage infection by Ross River virus. Proc Natl Acad Sci USA. 2002;99:13819–13824. doi: 10.1073/pnas.202415999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone S.R., Lok S.M. Structural perspectives of antibody-dependent enhancement of infection of dengue virus. Curr Opin Virol. 2019;36:1–8. doi: 10.1016/j.coviro.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Prescott J., Falzarano D., de Wit E., Hardcastle K., Feldmann F., Haddock E., et al. Pathogenicity and viral shedding of MERS-CoV in immunocompromised rhesus macaques. Front Immunol. 2018;9:205. doi: 10.3389/fimmu.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Yamada S., Doki T., Hohdatsu T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J Vet Med Sci. 2019;81(6):911–915. doi: 10.1292/jvms.18-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilocca B., Soggiu A., Musella V., Britti D., Sanguinett M., Urbani A., et al. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microbe Infect. 2020;22(4–5):218–220. doi: 10.1016/j.micinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A., Foo S.-S., Bruzzone R., Vu Dinh L., King N.J.C., Mahalingam S., et al. Fc receptors in antibody-dependent enhancement of viral infections. Immunol Rev. 2015;268(1):340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilocca B., Soggiu A., Sanguinetti M., Babini G., De Maio F., Britti D., et al. Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess crossprotection against COVID-19. Microbe Infect. 2020;22(4–5):182–187. doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kietzell K., Pozzuto T., Heilbronn R., Grössl T., Fechner H., Weger S. Antibody-mediated enhancement of parvovirus B19 uptake into endothelial cells mediated by a receptor for complement factor C1q. J Virol. 2014;88(14):8102–8115. doi: 10.1128/JVI.00649-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Shihui S., Wanbo T., Jing C., Qibin G., et al. Molecular mechanism for antibodydependent enhancement of coronavirus entry. J Virol. 2020;94(5) doi: 10.1128/JVI.02015-19. pii: e02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis. 2016;2(5):361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.F., Tseng S.-P., Yen C.-H., Yang J.-Y., Tsao C.-H., Shen C.-W., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A., Ooi E.-E., Ol. Horstick, Wills B. Dengue. Lancet. 2019;393(10169):350–363. doi: 10.1016/S0140-6736(18)32560-1. doi: 10.1016. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.S., Leung N.H.L., Cheung C.Y., Li P.H., Lee H.H.Y., Daëron M., et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.S., Leung H.L., Li P.H., Cheung C.Y., Dutry I., Li D., et al. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;(3 Suppl. 4):25–31. [PubMed] [Google Scholar]
- Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]