Abstract
SARS-CoV-2 uses −1 programmed ribosomal frameshifting (−1 PRF) to control expression of key viral proteins. Because modulating −1 PRF can attenuate the virus, ligands binding to the RNA pseudoknot that stimulates −1 PRF may have therapeutic potential. Mutations in the pseudoknot have occurred during the pandemic, but how they affect −1 PRF efficiency and ligand activity is unknown. Studying a panel of six mutations in key regions of the pseudoknot, we found that most did not change −1 PRF levels, even when base-pairing was disrupted, but one led to a striking 3-fold decrease, suggesting SARS-CoV-2 may be less sensitive to −1 PRF modulation than expected. Examining the effects of a small-molecule −1 PRF inhibitor active against SARS-CoV-2, it had a similar effect on all mutants tested, regardless of basal −1 PRF efficiency, indicating that anti-frameshifting activity can be resistant to natural pseudoknot mutations. These results have important implications for therapeutic strategies targeting SARS-CoV-2 through modulation of −1 PRF.
Keywords: COVID-19, programmed −1 ribosomal frameshifting, translation, small-molecule inhibitor
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; −1 PRF, −1 programmed ribosomal frameshifting
Graphical Abstract
Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causing the COVID-19 pandemic that is currently sweeping the globe features a −1 programmed ribosomal frameshifting (−1 PRF) site [1]. −1 PRF, which involves a shift in the reading frame of the ribosome at a specific location in the RNA message to generate an alternate gene product, is stimulated by a structure in the mRNA, typically a pseudoknot, that is located 5–7 nucleotides downstream of the “slippery” sequence where the reading-frame shift occurs [2,3]. The −1 PRF signal is essential to SARS-CoV-2: the frameshift gene products include the RNA-dependent RNA polymerase that is required for viral replication. Previous work on SARS-CoV showed that mutations suppressing −1 PRF significantly attenuated viral propagation in cell culture, by up to several orders of magnitude [[4], [5], [6]]. Indeed, work on other viruses has found more generally that −1 PRF levels must typically be held within a relatively narrow range to avoid attenuation [7,8]. As a result, frameshift-stimulatory structures are potential targets for anti-viral drugs [[9], [10], [11], [12], [13], [14], [15], [16], [17], [18]].
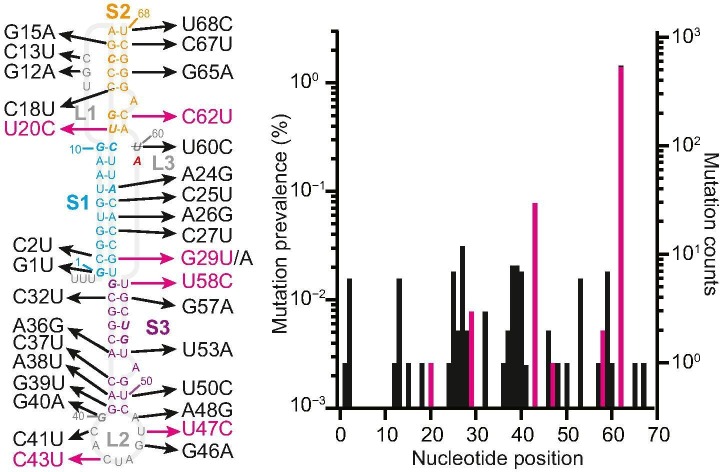
The pseudoknot stimulating −1 PRF in SARS-CoV-2 has a three-stem architecture that is characteristic of coronaviruses, in contrast to the more common two-stem architecture of most frameshift-stimulatory pseudoknots in viruses [19]. Although the pseudoknot sequence is highly conserved among coronaviruses [20], several mutations have been identified in the pseudoknot from viral samples isolated from COVID-19 patients during the pandemic, tracked in the GISAID database [21]. Some of these mutations have been seen in only single patients; others have been found in patients from many different regions of the world (Figure 1 , right). Understanding how these mutations affect −1 PRF in SARS-CoV-2 can provide insight into issues of central relevance to developing therapeutic −1 PRF modulators. Such issues include the range of −1 PRF levels over which the virus can survive and propagate (defining the degree of modulation needed to achieve a therapeutic effect), the regions of the pseudoknot that are most sensitive to disruption (and hence most profitable to target therapeutically), and the extent to which mutations in the pseudoknot may be able to induce drug resistance. However, the effects of naturally occurring mutations in the SARS-CoV-2 pseudoknot on the levels of −1 PRF in SARS-CoV-2 and any changes they may induce in the activity of anti-frameshifting ligands have not yet been investigated. Here we do so, focusing on a ligand previously shown to inhibit −1 PRF in SARS-CoV.
Figure 1.
Natural mutations in SARS-CoV-2 pseudoknot. Left: Mutations identified from COVID-19 patients occur in all regions of the pseudoknot structure. Bases shown in italic are protected against nuclease digestion [4]. Right: Most mutations have low occurrence. Mutations studied herein shown in magenta.
Results
Surveying over 40,000 patient-derived sequences deposited in the GISAID database as of June 9, 2020 to identify mutations in the frameshift-stimulatory pseudoknot, we found that ~ 1.8% of all sequences contained mutations, affecting 33 of the 68 nucleotides in the pseudoknot. In all cases but one, mutations involved single-nucleotide polymorphisms, without insertions or deletions; the exception involved the mutation of three adjacent nucleotides (A38–G40). These mutations occurred in all regions of the secondary structure (Figure 1), distributed relatively evenly except within stem 1, which featured few mutations in the 5′ strand and many on the 3′ strand. The vast majority involved transitions (purine–purine or pyrimidine–pyrimidine conversions) rather than transversions. Most (62%) of the mutations in the stems also preserved the wild-type base-pairing by converting G:C pairs to G:U, A:U to G:U, or G:U to G:C. Finally, 5 of the 13 positions (Figure 1 left, italic) that are protected from nuclease digestion in the SARS-CoV pseudoknot [4] experienced mutation in SARS-CoV-2, close to the same rate of mutation as in the pseudoknot as a whole (38% versus 48%), indicating that the protected residues are not significantly more resistant to undergoing mutation.
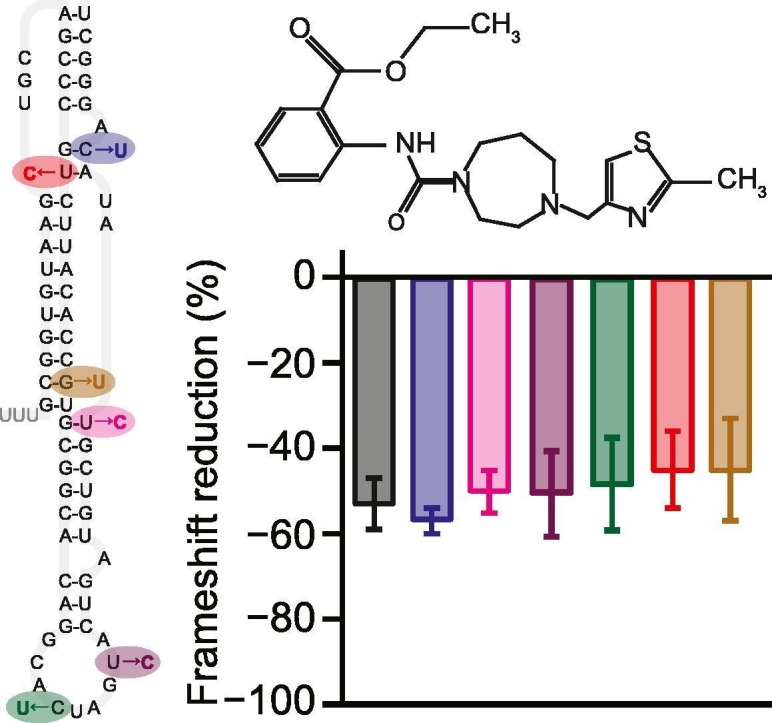
To examine the functional effects of these mutations on frameshifting, we selected a panel of six mutations (Figure 1, magenta), consisting of U20C, G29U, C43U, U47C, U58C, and C62U. These mutations were chosen for their prevalence or their potential functional and/or structural importance: C62U and C43U were the two most common mutations; C62U is also located adjacent to an adenine bulge (A63) critical to −1 PRF in SARS-CoV [4]; C43U and U47C are part of a domain in loop 2 that promotes pseudoknot dimerization [22]; U20C disrupts the cross-junction base-pair stacking between stems 1 and 2 that is important for −1 PRF in many pseudoknots [23], and U20 is also protected against nuclease digestion in SARS-CoV [4]; G29U destabilizes the end of stem 1 near the junction with stem 3, which may facilitate threading of the 5′ end of the RNA through the stem 1/stem 3 junction as proposed in structural models [24,25]; and finally, U58C pairs with G31 (which is nuclease-protected in SARS-CoV) and stabilizes the end of stem 3 near the stem 1/stem 3 junction, again possibly affecting the structure. Note that we included in this study only single-nucleotide polymorphisms found from patient samples, rather than any of the engineered mutations found to suppress −1 PRF in SARS-CoV [4,22], because the former are most relevant from a therapeutic perspective. For each mutant, we measured the −1 PRF efficiency induced by the pseudoknot using cell-free translation of a dual-luciferase reporter system consisting of the Renilla luciferase gene in the 0 frame upstream of the firefly luciferase gene in the −1 frame and separated by the SARS-CoV-2 frameshift signal [1]. The −1 PRF efficiency was obtained from the ratio of luminescence emitted by the two enzymes, compared to controls with 100% and 0% firefly luciferase read-through.
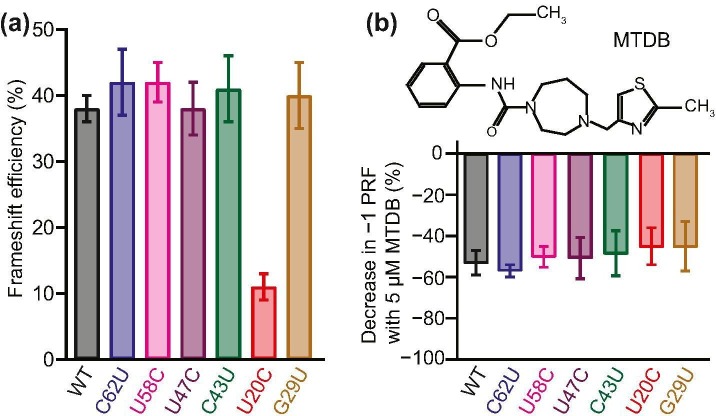
Comparing the results for each mutant to the −1 PRF efficiency seen for the consensus (wild-type) pseudoknot sequence, we found that five of the six mutations left the −1 PRF efficiency effectively unchanged within error (Figure 2(a)). Mutations that disrupted base-pairing in a stem (e.g. G29U) or the dimerization domain (C43U, U47C) as well as those that left all base-pairing intact (U58C, C62U) were observed to have the same lack of effect, suggesting that most natural mutations do not alter the −1 PRF efficiency characteristic of the virus. Such a result is consistent with previous work suggesting that −1 PRF levels are regulated in a narrow range, outside of which the virus is significantly attenuated [[5], [6], [7], [8]], and that they are not determined directly by characteristics such as stability or specific static structures but rather are most closely related to the conformational heterogeneity of the RNA [26,27]. Remarkably, however, the U20C mutation, which disrupted the A:U base-pair at the end of stem 2 in the stem 1/stem 2 junction, caused a very significant decrease in −1 PRF efficiency, suppressing it over 3-fold (Figure 2(a), red). This result is particularly striking because in SARS-CoV, a ~ 3.5-fold reduction of −1 PRF efficiency was shown to cause an over 1000-fold attenuation of the virus [5,6]. The fact that the U20C mutant was sampled from a COVID-19 patient implies that SARS-CoV-2 can survive at a wider range of −1 PRF levels than expected based on work on other viruses.
Figure 2.
Effects of mutations on −1 PRF efficiency and anti-frameshifting activity of MTDB. (a) Most mutations left the −1 PRF efficiency unchanged, with the notable exception of U20C. (b) The anti-frameshifting activity of the small-molecule ligand MTDB was not affected by any of the mutations. Error bars represent standard error of the mean from 5 to 16 replicates.
Finally, we explored if these natural mutations might lead to resistance to the effects of an anti-frameshifting ligand, 2-{[4-(2-methylthiazol-4-ylmethyl)-[1,4]diazepane-1-carbonyl]-amino}-benzoic acid ethyl ester, denoted as MTDB (Figure 2(b), inset). MTDB was previously found to bind to the pseudoknot from SARS-CoV and inhibit −1 PRF [9,11]. MTDB was also recently shown to have a similar effect on −1 PRF in wild-type SARS-CoV-2 [1]. We repeated the dual-luciferase assays of −1 PRF for all six mutants described above while adding 5 μM MTDB, to quantify the reduction in −1 PRF caused by the ligand. Comparing to the effect of MTDB on −1 PRF when using the wild-type pseudoknot (Figure 2(b), black), we found that for all of the mutants, 5 μM MTDB reduced −1 PRF efficiency by roughly half. This result was obtained even for the U20C mutant, which already stimulated −1 PRF at a substantially lower efficiency. Hence, none of mutations significantly altered the inhibition of −1 PRF by MTDB, regardless of whether the mutations altered the basal −1 PRF efficiency.
Discussion
Considering how the mutations affected −1 PRF levels, the surprising effect of the U20C mutation on −1 PRF may arise from a combination of key properties of U20 in the SARS-CoV-2 pseudoknot. For one thing, this mutation destabilizes an inter-stem junction; stacking of paired bases across junctions is known to play an important role in the stimulatory power of pseudoknots [23]. Structural modeling also indicates that U20 plays an important role in triplex-like interactions with loop 1 that organize the stem 1/stem 2 interface [24], and previous work showed that removing triples in pseudoknots can reduce −1 PRF efficiency [28]. The relatively sparse network of triples predicted in this pseudoknot [24] compared to others, in combination with the effects on the stem junction, could explain the sensitivity of −1 PRF to mutation of U20. Interestingly, G29U also disrupts an inter-stem junction, yet it leaves −1 PRF unchanged, suggesting that the stem 1/stem 3 interface is less important than the stem 1/stem 2 junction, consistent with work on SARS-CoV showing that stem 3 is not essential for frameshifting [4].
The results presented here have important implications for therapeutic strategies targeting −1 PRF modulation. First, the example provided by U20C suggests that SARS-CoV-2 can tolerate a substantial reduction in −1 PRF levels. Hence any putative drug suppressing −1 PRF may need to do so at quite dramatic levels to be effective therapeutically. In fact, 5 μM MTDB induces a smaller decrease in −1 PRF than does the U20C mutation; significantly higher concentrations would be needed to abolish −1 PRF, and MTDB is thus unlikely to be effective as a drug for treating COVID-19. Just as important, however, is the evidence that the effects of an anti-frameshifting ligand can be insensitive to a wide range of natural mutations. The fact that the details of how the mutations affected the pseudoknot (whether they stabilized or destabilized the structure, disrupted base-pairing, affected one region of the structure or another, or even perturbed the pseudoknot enough to alter the basal −1 PRF rate significantly) were effectively immaterial to the activity of MTDB suggests that the inhibitory effect of the ligand is not easily evaded by simple mutations. Even though such an insensitivity to mutation was seen for only a single anti-frameshifting ligand, this behavior shows the promise of the pseudoknot as a therapeutic target, and holds out hope that a ligand with higher anti-frameshifting activity may be found in future work.
Methods
Preparation of mRNA constructs
A dual luciferase reporting system was created by cloning the sequence corresponding to Renilla luciferase and the multiple cloning site from the plasmid pMLuc-1 (Novagen) upstream of the firefly luciferase sequence in the plasmid pISO (addgene), as described previously [1]. The frameshift signal from SARS-CoV-2 was then cloned into multiple cloning site between the restriction sites PstI and SpeI. Three different types of constructs were made. First, a construct for measuring −1 PRF stimulation was made, containing the frameshift signal with consensus (wild-type) slippery sequence (U UUA AAC) and consensus or mutant pseudoknot sequence, placing the downstream firefly luciferase gene in the −1 frame so that its expression was dependent on −1 PRF. Next, two controls were derived from this construct: (1) a negative control for measuring the background firefly luciferase luminescence (0% firefly luciferase read-through), in which the slippery sequence was mutated to include a stop codon (U UGA AAC), and (2) a positive control for measuring 100% firefly luciferase read-through, in which the slippery sequence was disrupted (U AGA AAC) and the firefly luciferase gene was shifted into the 0 frame. Sequences for all constructs are listed in Table S1.
Transcription templates were amplified from these plasmids by PCR, using a forward primer that contained the T7 polymerase sequence as a 5′ extension to the primer sequence [1]. The mRNA for dual-luciferase measurements was produced from the transcription templates by in vitro transcription (MEGAclear).
Dual-luciferase frameshift assay
Frameshifting efficiency was measured by a cell-free dual-luciferase assay [29]. Briefly, for each construct, 1 μg of mRNA transcript was heated to 65 °C for 3 min and then incubated on ice for 2 min. The mRNA was added to a solution mixture containing amino acids (10 μM Leu and Met, 20 μM all other amino acids), 35 μl of nuclease-treated rabbit reticulocyte lysate (Promega), 5 U RNase inhibitor (Invitrogen), and brought up to a reaction volume of 50 μl with water. The reaction mixture was incubated for 90 min at 30 °C. Luciferase luminescence was then measured using a microplate reader (Turner Biosystems). First, 20 μl of the reaction mixture was mixed with 100 μl of Dual-Glo Luciferase reagent (Promega) and incubated for 10 min before reading firefly luminescence, then 100 μl of Dual-Glo Stop and Glo reagent (Promega) was added to the mixture to quench firefly luminescence, and the reaction was incubated for 10 min before reading Renilla luminescence. The −1 PRF efficiency was calculated from the ratio of firefly to Renilla luminescence (F:R), subtracting the background F:R measured from the negative control and then normalizing by F:R measured from the positive control.
To quantify the effects of MTDB on −1 PRF, 5 μM MTDB was added to the reaction volume for each construct (wild-type and mutant pseudoknots, positive and negative controls). At least five replicates were measured and the results averaged, as described previously [30].
Sequences of measurement and control constructs. DNA sequence of constructs with slippery site (green) and linker region (blue). Pseudoknot mutation sites shown in bold.
Accession numbers of mutant pseudoknot sequences in GISAID database.
CRediT authorship contribution statement
Krishna Neupane: Conceptualization, Resources, Investigation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing, Supervision. Sneha Munshi: Resources, Investigation, Formal analysis, Writing - review & editing. Meng Zhao: Investigation, Formal analysis, Visualization, Writing - review & editing. Dustin B. Ritchie: Resources, Investigation, Formal analysis, Writing - review & editing. Sandaru M. Ileperuma: Investigation, Writing - review & editing. Michael T. Woodside: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Funding acquisition.
Acknowledgments
Acknowledgments
We gratefully acknowledge the originating laboratories that obtained SARS-CoV-2 specimens and submitting laboratories that generated and shared genetic sequence data via the GISAID Initiative, and all associated authors. This work was supported by the Canadian Institutes of Health Research (grant reference no. OV3-170709), Alberta Innovates, and National Research Council Canada.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Edited by Sarah A. Woodson
References
- 1.Kelly J.A., Olson A.N., Neupane K., Munshi S., San Emeterio J., Pollack L., Woodside M.T., Dinman J.D. Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2) J. Biol. Chem. 2020;295:10741–10748. doi: 10.1074/jbc.AC120.013449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giedroc D.P., Cornish P.V. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley I., Gilbert R.J., Pennell S. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; 2010. Pseudoknot-dependent programmed −1 ribosomal frameshifting: structures, mechanisms and models; pp. 149–174. [Google Scholar]
- 4.Plant E.P., Pérez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plant E.P., Rakauskaitė R., Taylor D.R., Dinman J.D. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plant E.P., Sims A.C., Baric R.S., Dinman J.D., Taylor D.R. Altering SARS coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses. 2013;5:279–294. doi: 10.3390/v5010279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinman J.D., Wickner R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dulude D., Berchiche Y.A., Gendron K., Brakier-Gingras L., Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 9.Park S.-J., Kim Y.-G., Park H.-J. Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 10.Ahn D.-G., Lee W., Choi J.-K., Kim S.-J., Plant E.P., Almazán F., Taylor D.R., Enjuanes L. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 2011;91:1–10. doi: 10.1016/j.antiviral.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie D.B., Soong J., Sikkema W.K., Woodside M.T. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J. Am. Chem. Soc. 2014;136:2196–2199. doi: 10.1021/ja410344b. [DOI] [PubMed] [Google Scholar]
- 12.Belew A.T., Dinman J.D. Cell cycle control (and more) by programmed −1 ribosomal frameshifting: implications for disease and therapeutics. Cell Cycle. 2015;14:172–178. doi: 10.4161/15384101.2014.989123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofori L.O., Hilimire T.A., Bennett R.P., Brown N.W., Jr., Smith H.C., Miller B.L. High-affinity recognition of HIV-1 frameshift-stimulating RNA alters frameshifting in vitro and interferes with HIV-1 infectivity. J. Med. Chem. 2014;57:723–732. doi: 10.1021/jm401438g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilimire T.A., Bennett R.P., Stewart R.A., Garcia-Miranda P., Blume A., Becker J., Sherer N., Helms E.D. N-methylation as a strategy for enhancing the affinity and selectivity of RNA-binding peptides: application to the HIV-1 frameshift-stimulating RNA. ACS Chem. Biol. 2016;11:88–94. doi: 10.1021/acschembio.5b00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilimire T.A., Chamberlain J.M., Anokhina V., Bennett R.P., Swart O., Myers J.R., Ashton J.M., Stewart R.A. HIV-1 frameshift RNA-targeted triazoles inhibit propagation of replication-competent and multi-drug-resistant HIV in human cells. ACS Chem. Biol. 2017;12:1674–1682. doi: 10.1021/acschembio.7b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anokhina V.S., McAnany J.D., Ciesla J.H., Hilimire T.A., Santoso N., Miao H., Miller B.L. Enhancing the ligand efficiency of anti-HIV compounds targeting frameshift-stimulating RNA. Biorg. Med. Chem. 2019;27:2972–2977. doi: 10.1016/j.bmc.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brakier-Gingras L., Charbonneau J., Butcher S.E. Targeting frameshifting in the human immunodeficiency virus. Expert Opin. Ther. Targets. 2012;16:249–258. doi: 10.1517/14728222.2012.665879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcheschi R.J., Tonelli M., Kumar A., Butcher S.E. Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication. ACS Chem. Biol. 2011;6:857–864. doi: 10.1021/cb200082d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plant E.P., Dinman J.D. The role of programmed −1 ribosomal frameshifting in coronavirus propagation. Front. Biosci. 2008;13:4873. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangan R., Zheludev I.N., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimaru D., Plant E.P., Sims A.C., Yount B.L., Jr., Roth B.M., Eldho N.V., Perez-Alvarado G.C., Armbruster D.W. RNA dimerization plays a role in ribosomal frameshifting of the SARS coronavirus. Nucleic Acids Res. 2013;41:2594–2608. doi: 10.1093/nar/gks1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornish P.V., Stammler S.N., Giedroc D.P. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omar S.I., Zhao M., Sekar R.V., Moghadam S.A., Tuszynski J.A., Woodside M.T. Modeling the structure of the frameshift stimulatory pseudoknot in SARS-CoV-2 reveals multiple possible conformers. bioRxiv. 2020 doi: 10.1371/journal.pcbi.1008603. 2020.06.08.141150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, K., Zheludev, I. N., Hagey, R. J., Wu, M. T.-P., Haslecker, R., Hou, Y. J., Kretsch, R., Pintilie, G. D., Rangan, R., Kladwang, W., Li, S., Pham, E. A., Bernardin-Souibgui, C., Baric, R. S., Sheahean, T. P., D'Souza, V., Glenn, J. S., Chiu, W., and Das, R. (2020) Cryo-electron microscopy and exploratory antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. bioRxiv 2020.07.18.209270.
- 26.Halma M.T., Ritchie D.B., Cappellano T.R., Neupane K., Woodside M.T. Complex dynamics under tension in a high-efficiency frameshift stimulatory structure. Proc. Natl. Acad. Sci. U. S. A. 2019;116:19500–19505. doi: 10.1073/pnas.1905258116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie D.B., Foster D.A., Woodside M.T. Programmed −1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G., Chang K.-Y., Chou M.-Y., Bustamante C., Tinoco I. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs J.L., Dinman J.D. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of measurement and control constructs. DNA sequence of constructs with slippery site (green) and linker region (blue). Pseudoknot mutation sites shown in bold.
Accession numbers of mutant pseudoknot sequences in GISAID database.