Abstract
The human 16p11.2 gene locus is a hot-spot for copy number variations which predispose carriers to a range of neuropsychiatric phenotypes. Microduplications of 16p11.2 are associated with autism spectrum disorder (ASD), intellectual disability (ID) and schizophrenia (SZ). Despite the debilitating nature of 16p11.2 duplications, the underlying molecular mechanisms remain poorly understood. Here we performed a comprehensive behavioral characterization of 16p11.2 duplication mice (16p11.2dp/+) and identified social and cognitive deficits reminiscent of ASD and ID phenotypes. 16p11.2dp/+ mice did not exhibit the SZ-related sensorimotor gating deficits, psychostimulant-induced hypersensitivity or motor impairment. Electrophysiological recordings of 16p11.2dp/+ mice found the deficient GABAergic synaptic transmission and elevated neuronal excitability in the prefrontal cortex (PFC), a brain region critical for social and cognitive functions. RNA-sequencing identified genome-wide transcriptional aberrance in the PFC of 16p11.2dp/+ mice, including downregulation of the GABA synapse regulator Npas4. Restoring Npas4 expression in PFC of 16p11.2dp/+ mice ameliorated the social and cognitive deficits and reversed the GABAergic synaptic impairment and neuronal hyper-excitability. These findings suggest that prefrontal cortical GABAergic synaptic circuitry and Npas4 are strongly implicated in 16p11.2 duplication pathology, and may represent potential targets for therapeutic intervention in ASD.
Keywords: 16p11.2 duplication, GABA, Npas4, autism spectrum disorder, intellectual disability, prefrontal cortex
INTRODUCTION
The human 16p11.2 genetic locus (chromosome 16, position 11.2) constitutes a ~550 kb (26 gene) chromosomal region that is susceptible to copy number variations (CNVs; i.e. deletion or duplication), which confer risk for a range of neurodevelopmental conditions (1–3). Microduplications of 16p11.2 are estimated to affect 1 in every 4,216 live births (4), and often carry broad and multifaceted phenotypic consequences due to frequent comorbidity among psychiatric, physical/developmental and cognitive symptoms. 16p11.2 duplication carriers most commonly exhibit neurodevelopmental deficits characterized by intellectual disability (ID), speech & language deficits/autism spectrum disorder (ASD), and developmental/motor delays (1, 2, 5–10). 16p11.2 duplications are also associated with schizophrenia and bipolar disorder (2, 11–14). In addition, epilepsy, dysmorphic features, and microcephaly are often observed in 16p11.2 duplications (6, 7, 15).
Numerous clinical reports have substantiated the debilitating nature of 16p11.2 duplications. Mice carrying duplication of the genomic region homologous to 16p11.2 (mouse chromosome 7F3) exhibit neurocognitive and metabolic phenotypes (16, 17), however, it remains to be determined whether 16p11.2 duplication mice (16p11.2dp/+) thoroughly and accurately depict the clinical features present in human patients, and what molecular mechanisms are underlying these behavioral abnormalities. We thus performed a comprehensive behavioral examination of 16p11.2dp/+ mice, and report social and cognitive behavioral deficits reminiscent of ASD and ID phenotypes, respectively.
Dysfunction of inhibitory gamma-aminobutyric acid (GABA) neurotransmission is highly implicated in ASD (18), and the resulting imbalance of excitatory and inhibitory synaptic activity (E/I imbalance) has been theorized to underlie ASD pathology (19, 20). Moreover, brain GABA levels are significantly reduced in human ASD patients (21), and numerous mouse models of ASD exhibit disrupted E/I balance in cortical regions and specifically in the medial prefrontal cortex (mPFC) (22–26), a brain region critical for higher level executive functions and involved in social cognition (27). In the current study, we found that GABAergic synaptic transmission was disrupted and neuronal excitability was elevated in the mPFC of 16p11.2dp/+ mice, an electrophysiological profile consistent with existing explanations of ASD pathology, which may explain the social deficits in 16p11.2 duplication carriers.
Our genome-wide search for gene alterations associated with the disrupted GABA signaling in 16p11.2dp/+ mice led to the discovery of the downregulated gene Npas4, an activity-dependent transcription factor highly expressed in PFC (28). Npas4 is induced in response to neuronal excitation and subsequently regulates the formation of inhibitory GABAergic synapses onto pyramidal neurons (29–31). Npas4 expression in the PFC during adolescence appears to be critical for the proper establishment of GABAergic synapse markers (32), and Npas4 deficiency is associated with cognitive impairment and compromised memory formation (32–35) along with social deficits (34). Here, we found that restoring Npas4 expression in PFC of 16p11.2dp/+ mice was sufficient to reverse GABAergic synaptic deficits and ameliorate the observed social and cognitive phenotypes, implicating Npas4 and the prefrontal cortical GABA system in the pathogenesis of social and cognitive deficits in 16p11.2 duplication syndrome.
MATERIALS AND METHODS
Animals and Human Postmortem Tissue
16p11.2dp/+ mice carrying a heterozygous duplication of the 7F3 chromosomal region homologous to human 16p11.2 were generated as previously described (16). All animal studies were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of the State University of New York at Buffalo. Frozen human postmortem tissue (Brodmann’s Area 9) from autism patients and healthy controls (age- and gender-matched) were provided by NIH NeuroBioBank. Detailed information about the ASD human patients is included in Supplemental Table 1. Tissue was stored in a −80°C freezer. See Supplementary Methods for details.
Behavioral Testing
See Supplementary Methods for details.
Electrophysiological Recordings
See Supplementary Methods for details.
Immunohistochemistry
See Supplementary Methods for details.
RNA-Sequencing and Analysis
See Supplementary Methods for details.
Quantitative Real-time RT-PCR
Primers for all target genes are listed in Supplemental Table 2. See Supplementary Methods for details.
Western Blotting of Nuclear Proteins
See Supplementary Methods for details.
Viral Vectors and Animal Surgeries
See Supplementary Methods for details.
Statistical Analyses
All statistical analyses were performed with Graphpad Prism and Minitab 18. Sample sizes were determined based on power analyses and were similar to those reported in previous works (36). Experiments with more than two groups were subjected to one-way ANOVA, two-way ANOVA, or three-way ANOVA with Bonferroni correction for multiple post-hoc comparisons. Experiments with two groups were analyzed statistically using two-tailed unpaired t-tests, unless the data failed Shapiro-Wilk tests for normality, in which case the data were subjected to Mann-Whitney U tests. All data are presented as the mean ± s.e.m. Data points identified as statistically significant outliers (determined by Grubb’s test, p < 0.05) were removed from the analyses. The variance between groups being statistically compared was similar. Detailed statistical data for all data shown are presented in Supplemental Table 3.
RESULTS
16p11.2dp/+ Mice Exhibit Social and Cognitive Behavioral Deficits Reminiscent of ASD and ID
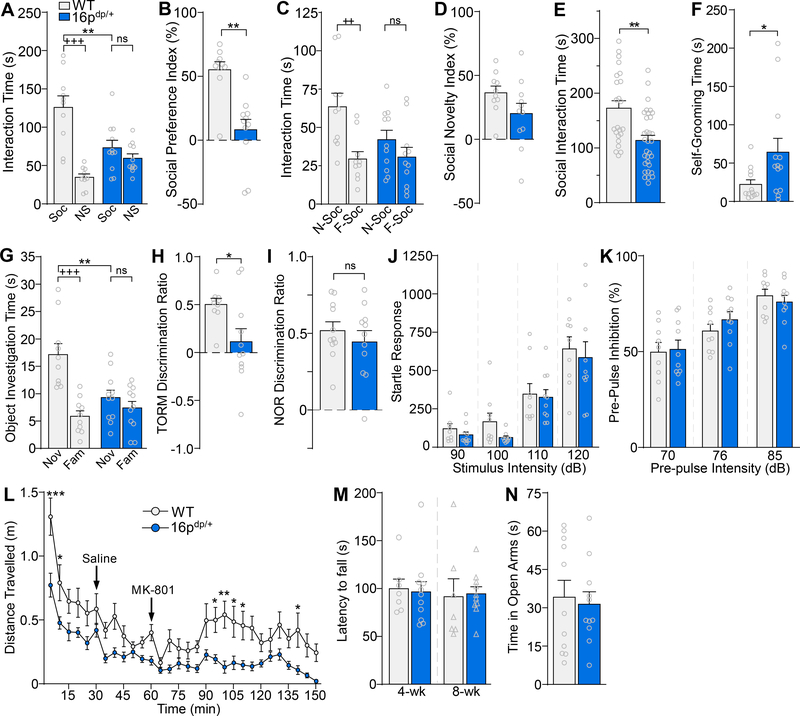
To determine whether mice carrying the 16p11.2 duplication (16p11.2dp/+) exhibit phenotypes resembling the clinical features present in human patients, we performed an array of behavioral tests on both male and female 7–9-week-old 16p11.2dp/+ mice and age-matched wild-type (WT) controls. Since human 16p11.2 duplication carriers are strongly predisposed to ASD (1, 2, 5–9, 37), we first evaluated social behavior in the three-chamber social preference test. When animals were exposed to a social stimulus and a non-social stimulus, 16p11.2dp/+ mice spent significantly less time than WT mice interacting with the social stimulus (Figure 1A, F1,38 (genotype x stimulus) = 16.7, p = 0.0002, two-way ANOVA), and correspondingly demonstrated a significantly lower social preference index (Figure 1B, U = 9, p = 0.0006, Mann-Whitney U test). When animals were exposed to a novel social stimulus and a familiar social stimulus, WT mice spent significantly more time interacting with the novel mouse, whereas 16p11.2dp/+ mice did not display a clear preference for the novel mouse (Figure 1C, F1,38 (genotype x stimulus) = 2.91, p = 0.10, two-way ANOVA), resulting in a trend toward a lower social novelty preference index in 16p11.2dp/+ mice (Figure 1D, t(19) = 1.67, p = 0.11, unpaired t-test). In the social approach test, 16p11.2dp/+ mice spent significantly less time than WT controls interacting with the social stimulus (Figure 1E, t(53) = 3.65, p = 0.0006, unpaired t-test). WT and 16p11.2dp/+ mice did not differ in the total distance travelled during the three-chamber social preference test (n = 9–14 mice/group, t(21) = 0.27, p = 0.79, unpaired t-test) or the social approach test (n = 8–11 mice/group, t(17) = 1.10, p = 0.29, unpaired t-test), suggesting that differences in locomotion are not contributing to the observed social phenotypes. Self-grooming, a rodent behavior thought to model repetitive behaviors observed in human ASD patients (38), was also assessed. Relative to WT animals, 16p11.2dp/+ mice spent significantly more time self-grooming (Figure 1F, U = 29, p = 0.02, Mann-Whitney U test). Collectively, these data indicate that 16p11.2dp/+ mice exhibit both social deficits and repetitive behaviors, the two core behavioral features of ASD.
Figure 1.
16p11.2dp/+ mice exhibit social deficits, repetitive behaviors, and cognitive impairment reminiscent of ASD and ID symptoms. A, B, Bar graphs comparing the amount of time spent interacting with the social (Soc) vs. non-social (NS) stimuli (A) and the social preference index (B) in the 3-chamber social preference test of WT and 16p11.2dp/+ mice. n = 10–11 mice/group. C, D, Bar graphs showing the amount of time spent exploring the novel social stimulus (N-Soc) vs. the familiar social stimulus (F-Soc) (C) and the social novelty index (D) in the 3-chamber preference test of WT and 16p11.2dp/+ mice. n = 10–11 mice/group. E, Bar graphs showing the amount of time spent interacting with the social stimulus in the social approach test of WT and 16p11.2dp/+ mice. n = 23–32 mice/group. F, Bar graphs showing self-grooming time for WT and 16p11.2dp/+ mice. n = 12–13 mice/group. G, H, Bar graphs showing the amount of time spent exploring the novel (Nov) vs. familiar (Fam) objects (G) and the discrimination ratio (H) in temporal order recognition memory (TORM) test of WT and 16p11.2dp/+ mice. n = 10–11 mice/group. I, Bar graphs showing the discrimination ratio in the novel object recognition (NOR) test of WT and 16p11.2dp/+ mice. n = 11 mice/group. J, K, Bar graphs showing startle responses at various stimulus intensities (J) and pre-pulse inhibition levels at various pre-pulse intensities (K) for WT and 16p11.2dp/+ mice. n = 9–10 mice/group. L, Plot showing the distance travelled (in 5-minute bins) by WT and 16p11.2dp/+ mice at baseline (0–30 min), after saline injection (30–60 min), and after injection of the NMDAR antagonist MK-801 (2 mg/kg, i.p., 60–150 min). n = 9–11 mice/group. M, Bar graphs showing the latency to fall in the rotarod test of WT and 16p11.2dp/+ mice at different ages. n = 7–11 mice/group. N, Bar graphs showing the total amount of time spent exploring the open arms in the elevated plus maze test of WT and 16p11.2dp/+ mice. n = 11 mice/group. All data are presented as mean ± SEM. In all figures, ns = not significant, * p < 0.05; **, ++ p < 0.01; ***, +++ p < 0.0001.
We next sought to assess whether 16p11.2dp/+ mice exhibit cognitive deficits reminiscent of ID, another phenotype strongly associated with 16p11.2 duplications (7–9, 37). Temporal Order Recognition Memory (TORM), a task testing the animal’s ability to remember which of two objects it was more recently exposed to, was used to assess cognitive processes mediated by the medial prefrontal cortex (mPFC) (39). In the TORM task, 16p11.2dp/+ mice spent significantly less time than WT controls interacting with the more novel (less recent) object (Figure 1G, F1,38 (genotype x object) = 10.62, p = 0.002, two-way ANOVA), and correspondingly exhibited a significantly lower discrimination ratio (Figure 1H, t(19) = 2.55, p = 0.02, unpaired t-test), indicating PFC-dependent cognitive impairment. However, in the Novel Object Recognition (NOR) task, which is mediated primarily by the perirhinal cortex (39, 40), 16p11.2dp/+ mice displayed unimpaired performance (Figure 1I, t(19) = 0.79, p = 0.44, unpaired t-test), suggesting that the cognitive deficits afflicting 16p11.2dp/+ mice may be driven by brain region-specific neurobiological changes.
Since several reports have linked 16p11.2 duplications to schizophrenia (SZ) (2, 11–14), we next examined SZ-related behaviors in 16p11.2dp/+ mice. Pre-pulse inhibition (PPI) is a measure of sensorimotor gating which is disrupted in human SZ patients and animal models of SZ (41–43). Abnormalities in startle-responses or PPI have also been reported in autism (44–46) and fragile X patients (47–49), as well as in mouse models of ASD and fragile X syndrome (48, 50). Compared to WT counterparts, 16p11.2dp/+ mice displayed normal startle responses at multiple stimulus intensities (Figure 1J, F1,17 (genotype) = 0.86, p = 0.36, two-way ANOVA), and intact pre-pulse inhibition at all pre-pulse intensities (Figure 1K, F1,17 (genotype) = 0.11, p = 0.75, two-way ANOVA), suggesting the lack of SZ-related sensorimotor gating deficits.
Based on the NMDAR hypofunction theory of SZ (51), NMDAR antagonists have been used to evoke psychosis-related behaviors, including hyperlocomotion (52–55). We tested whether a single administration of the NMDAR antagonist MK-801 (2.0 mg/kg) could induce enhanced hyperlocomotion in 16p11.2dp/+ mice. Prior to MK-801 injection, 16p11.2dp/+ mice exhibited significantly lower baseline locomotor activity relative to WT mice. In contrast to WT animals, 16p11.2dp/+ mice failed to display elevated locomotion after MK-801 injection (Figure 1L, F1,18 (genotype) = 20.41, p = 0.0003, two-way ANOVA). These data indicate that 16p11.2dp/+ mice do not exhibit SZ-related hypersensitivity to psychostimulants.
Motor deficits, which are associated with 16p11.2 duplications (1, 2, 5–9), were assessed in 16p11.2dp/+ mice via the rotarod test. At both 4- and 8-weeks of age, latency to fall did not differ between 16p11.2dp/+ and WT mice, suggesting a lack of motor coordination deficits (Figure 1M, 4 weeks: t(16) = 0.22, p = 0.83, unpaired t-test; 8 weeks: t(15) = 0.16, p = 0.87, unpaired t-test). General anxiety has also been reported in 16p11.2 duplication patients (9, 56). In the elevated plus maze test, 16p11.2dp/+ mice did not differ from WT animals in the amount of time spent exploring the open arms (Figure 1N, t(20) = 0.33, p = 0.74, unpaired t-test), indicating the lack of anxiety-like behaviors. Collectively, our behavioral characterization indicates that 16p11.2dp/+ mice exhibit many clinical features associated with human 16p11.2 duplications, including ASD-related social deficits and repetitive behaviors, along with cognitive deficits reminiscent of ID.
GABAergic Synaptic Transmission is Impaired in PFC of 16p11.2dp/+ mice
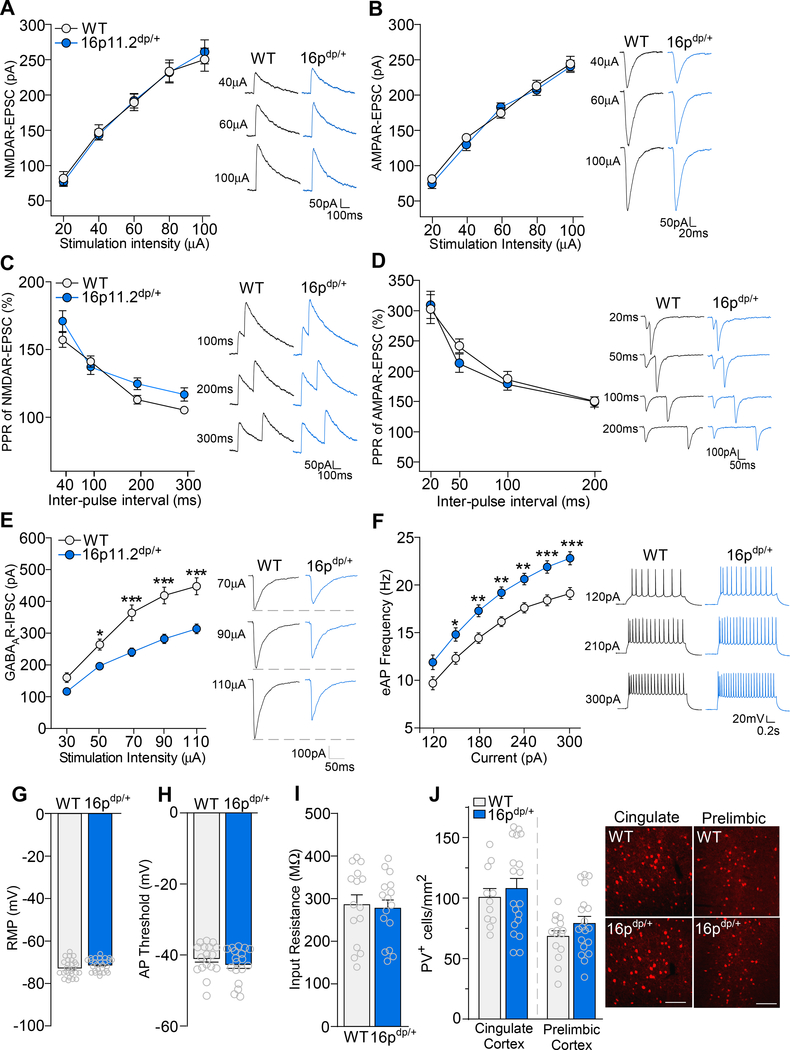
Considering that 16p11.2dp/+ mice exhibited impaired sociability and cognition, two major behavioral functions mediated by the PFC (27, 39), we next performed whole-cell patch clamp recordings on WT and 16p11.2dp/+ medial PFC (mPFC; prelimbic and infralimbic) layer V pyramidal neurons to identify synaptic transmission deficits which may underlie the observed behavioral phenotypes. NMDA receptor (NMDAR)-mediated excitatory postsynaptic current (EPSC) amplitudes did not differ between 16p11.2dp/+ and WT neurons at various stimulation intensities (Figure 2A, F1,29 (genotype) = 0.002, p = 0.96, two-way ANOVA). WT and 16p11.2dp/+ mPFC neurons also demonstrated comparable AMPA receptor (AMPAR)-mediated EPSC amplitudes (Figure 2B, F1,25 (genotype) = 0.22, p = 0.64, two-way ANOVA). In addition, 16p11.2dp/+ mPFC neurons exhibited normal paired-pulse ratios of NMDAR-EPSC (Figure 2C, F1,40 (genotype) = 0.01, p = 0.90, two-way ANOVA) and AMPAR-EPSC (Figure 2D, F1,14 (genotype) = 0.33, p = 0.57, two-way ANOVA). These data suggest that glutamatergic transmission is largely unchanged in 16p11.2dp/+ mPFC neurons.
Figure 2.
16p11.2dp/+ mPFC pyramidal neurons exhibit GABAergic synaptic deficits and elevated excitability. A, B, Summarized input-output curves of NMDAR-EPSC (A) and AMPAR-EPSC (B) in WT and 16p11.2dp/+ PFC neurons. Inset: representative NMDAR-EPSC and AMPAR-EPSC traces. NMDA: n = 14–17 cells, 3–4 mice/group; AMPA: n = 12–15 cells, 3 mice/group. C, D, Plot of paired-pulse ratio (PPR) of NMDAR-EPSC (C) and AMPAR-EPSC (D) evoked by double-pulses with various intervals in PFC pyramidal neurons from WT and 16p11.2dp/+ mice. Inset: representative traces. NMDA: n = 16–24 cells, 3–5 mice/group; AMPA: n = 8 cells, 2 mice/group. E, Summarized input-output curves of GABAAR-IPSC in WT and 16p11.2dp/+ mPFC pyramidal neurons. Inset: representative GABAR-IPSC traces. n = 28–31 cells, 7–8 mice/group. F, Plot of AP firing frequencies evoked by different depolarizing current injections in WT and 16p11.2dp/+ PFC neurons. Inset: representative eAP firing traces. n = 26–27 cells, 4 mice/group. G, Bar graph showing resting membrane potential (RMP) in PFC pyramidal neurons from WT and 16p11.2dp/+ mice. n = 26–27 cells, 4 mice/group. H, Bar graph showing action potential (AP) threshold in PFC pyramidal neurons from WT and 16p11.2dp/+ mice. n = 18 cells, 4 mice/group. I, Bar graph showing input resistance in PFC pyramidal neurons from WT and 16p11.2dp/+ mice. n = 15–16 cells, 4 mice/group. J, Bar graph showing the number of Parvalbumin expressing (PV+) cells in the cingulate cortex and prelimbic cortex of WT and 16p11.2dp/+ mice. Inset: representative immunostaining images; scale bars = 200 μM. Cingulate cortex: n = 11–19 slices, 4 mice/group; Prelimbic cortex: n = 15–19 slices, 4 mice/group. All data are presented as mean ± SEM. In all figures, * p < 0.05; ** p < 0.01; *** p < 0.0001.
We next recorded GABAA receptor (GABAAR)-mediated inhibitory postsynaptic currents (IPSCs). Relative to WT cells, 16p11.2dp/+ mPFC neurons displayed significantly reduced GABAAR-IPSC amplitudes at multiple stimulation intensities (Figure 2E, F1,57 (genotype) = 24.41, p < 0.0001, two-way ANOVA), indicating marked disruption of GABAergic synaptic transmission in 16p11.2dp/+ PFC. We then measured action potential (AP) firing to assess neuronal excitability, which could be influenced by the alteration of synaptic inhibition. Relative to WT cells, 16p11.2dp/+ mPFC neurons displayed significantly increased frequencies of APs evoked by multiple current intensities (Figure 2F, F1,51 (genotype) = 13.03, p = 0.0007, two-way ANOVA). However, no changes were observed between WT and 16p11.2dp/+ neurons in the resting membrane potential (Figure 2G, t(51) = 1.55, p = 0.13, unpaired t-test), action potential threshold (Figure 2H, t(34) = 1.12, p = 0.27, unpaired t-test), or input resistance (Figure 2I, t(29) = 0.28, p = 0.78, unpaired t-test), suggesting that the intrinsic membrane properties of mPFC neurons from 16p11.2dp/+ mice are unchanged.
To determine whether the diminished GABAergic synaptic responses in PFC pyramidal neurons was potentially caused by the loss of interneurons, we performed immunostaining for parvalbumin (PV) in two regions of the prefrontal cortex, the prelimbic and cingulate areas. WT and 16p11.2dp/+ mice did not differ in the number of PV-expressing (PV+) cells in the cingulate cortex or the prelimbic cortex (Figure 2J, Cingulate: t(28) = 0.59, p = 0.56, unpaired t-test; Prelimbic: t(32) = 1.38, p = 0.18, unpaired t-test), indicating that the observed GABAergic synaptic deficits are not due to the loss of parvalbumin-expressing interneurons in the PFC. Collectively, these data indicate that 16p11.2dp/+ PFC neurons exhibit selective impairments in synaptic inhibition, which may be mediated by the loss of GABAergic synapses.
Genome-wide Transcriptional Dysregulation in PFC of 16p11.2dp/+ mice
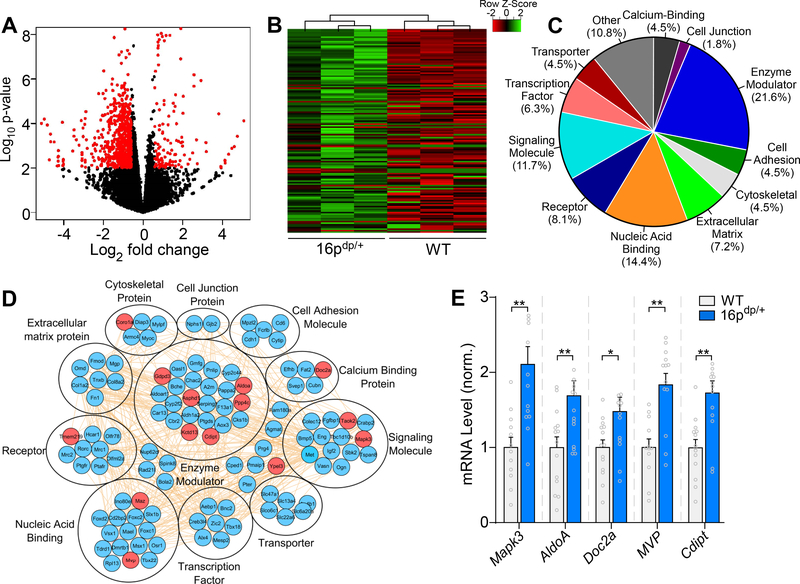
In order to determine the genome-wide transcriptional impact of the 16p11.2 duplication, we next performed RNA-sequencing (RNA-seq) with mPFC tissue. RNA-seq identified a total of 388 gene transcripts with significantly altered expression levels in 16p11.2dp/+ PFC (>1.5-fold increase or decrease, p < 0.05, and FDR < 0.3), with the majority of genes showing downregulation (Figure 3A), suggesting that 16p11.2 duplication has a predominantly repressive impact on genome-wide transcriptional levels in PFC. As shown in the heat map in Figure 3B, 111 gene transcripts demonstrated significant upregulation in 16p11.2dp/+ mPFC (Supplemental Table 4). Gene ontology (GO) analysis was performed to classify the upregulated genes into 11 categories based on biological functions (Figure 3C). Enrichment was observed in functional categories including enzyme modulator, nucleic acid binding and signaling molecule, suggesting that transcriptional upregulation in 16p11.2dp/+ PFC occurs in diverse gene classes. The interactome network demonstrated that the upregulated genes have rich interconnections (Figure 3D). Quantitative PCR (qPCR) analysis was performed on WT and 16p11.2dp/+ mPFC tissue, and verified the upregulation of several genes located in the duplicated 16p11.2 genomic region, including Mapk3, AldoA. Doc2a, Mvp, and Cdipt (Figure 3E).
Figure 3.
RNA-sequencing identifies numerous upregulated genes in PFC of 16p11.2dp/+ mice. A, Volcano plot illustrating gene distributions based on expression levels in 16p11.2dp/+ mice relative to WT animals; black dots represent genes not significantly altered, red dots represent differentially expressed genes in 16p11.2dp/+ (>1.5-fold change, p < 0.05, FDR < 0.3). B, Heat map representing expression (row z-score) of 111 significantly upregulated genes in PFC from 16p11.2dp/+ mice relative to WT values. C, Pie chart displaying the biological function classification of the upregulated genes in 16p11.2dp/+ PFC based on Gene Ontology. D, Interactome network showing predicted interactions between the upregulated genes in various ontological classifications. Genes located within the duplicated 16p11.2 chromosomal region are designated in red. E, Bar graph comparing mRNA level of five upregulated genes located in the 16p11.2 region between WT and 16p11.2dp/+ PFC. n = 12–15 mice/group. All data are presented as mean ± SEM. In all figures, * p < 0.05; ** p < 0.01.
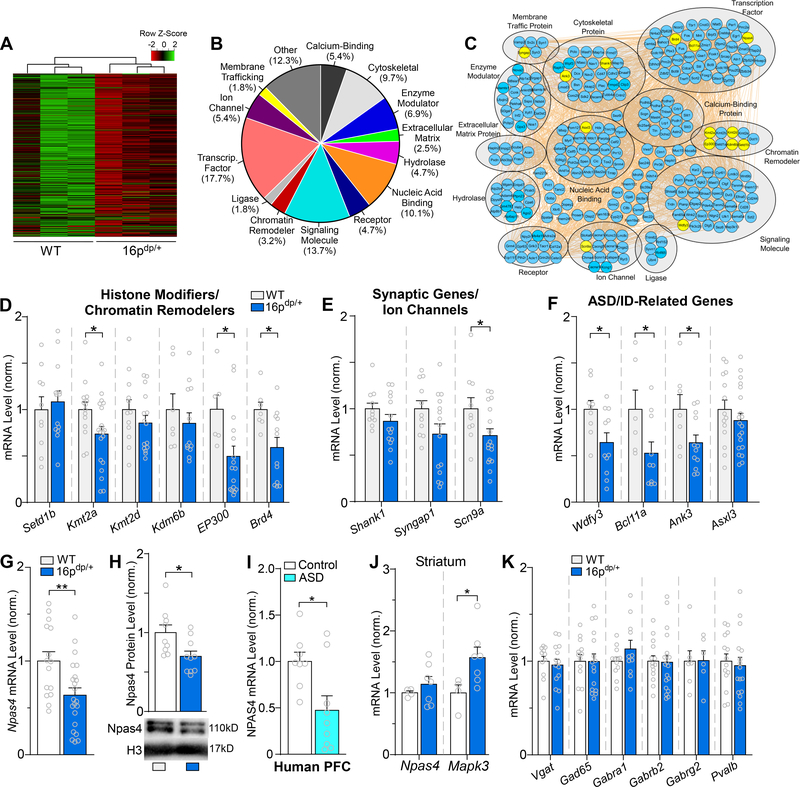
RNA-seq identified an additional 277 gene transcripts exhibiting significant downregulation in 16p11.2dp/+ PFC (Figure 4A, Supplemental Table 5). GO analysis was performed to classify significantly downregulated genes into 14 categories. Enrichment was observed in categories such as transcription factors, signaling molecules, nucleic acid binding and cytoskeletal genes (Figure 4B), indicating that transcriptionally repressed genes in 16p11.2dp/+ PFC assume a variety of functional roles. An interactome network was also built to illustrate predicted interactions between the downregulated genes, along with their respective ontological classifications (Figure 4C).
Figure 4.
RNA-sequencing identifies downregulated genes from diverse classes in 16p11.2dp/+ PFC, including the GABA-synapse regulator Npas4. A, Heat map representing expression (row z-score) of 277 significantly downregulated genes in PFC from 16p11.2dp/+ mice relative to WT values. B, Pie chart displaying the biological function classification of the downregulated genes in 16p11.2dp/+ PFC based on Gene Ontology. C, Interactome network showing predicted interactions between the downregulated genes in various ontological classifications. Genes assessed via qPCR are designated in yellow. D, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA expression level for several genes encoding chromatin remodelers identified as significantly downregulated via RNA-seq. n = 7–20 mice/group. E, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA level of genes encoding synaptic components/ion channels identified as significantly downregulated via RNA-seq. n = 10–17 mice/group. F, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA level of genes related to ASD/ID identified as significantly downregulated via RNA-seq. n = 6–20 mice/group. G, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA expression level of the GABA synapse regulator Npas4. n = 15–22 mice/group. H, Bar graph showing NPAS4 protein expression level in nuclear fractions isolated from WT and 16p11.2dp/+ PFC. Inset: representative immunoblot images. n = 9–10 mice/group. I, Bar graph showing Npas4 mRNA expression in human postmortem PFC tissue from healthy controls and ASD patients. n = 8–9/group. J, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA expression level of Npas4 and the 16p11.2 gene Mapk3 in striatum. n = 4–7 mice/group. K, Bar graph comparing WT and 16p11.2dp/+ PFC mRNA level of genes related to GABAergic synaptic transmission. n = 6–19 mice/group. All data are presented as mean ± SEM. In panel E, * p < 0.05; ** p < 0.01.
In order to verify the transcriptional reduction of the downregulated genes identified by our RNA-seq experiments, we next performed qPCR analysis of selected genes from various ontological classifications. Transcriptional levels were assessed for several histone modifiers/chromatin remodelers, and significant downregulation was confirmed for the epigenetic enzymes Kmt2a, EP300, and Brd4, while other genes such as Setd1b, Kmt2d, and Kdm6b failed to show significant reduction in mPFC of 16p11.2dp/+ mice (Figure 4D). Expression level of the synaptic genes Shank1 and Syngap1, both of which showed significant downregulation in RNA-seq, exhibited a trend of reduction in PFC of 16p11.2dp/+ mice, while the sodium ion channel Scn9a was significant downregulated (Figure 4E). Additionally, the mRNA level of other ASD- and/or ID risk genes identified by genomic screening, including Wdfy3, Bcl11a, Ank3, and Asxl3 (57–59), was significantly reduced in PFC of 16p11.2dp/+ mice (Figure 4F).
Among the top 20 most strongly downregulated genes in 16p11.2dp/+ PFC identified by RNA-seq, Npas4 (FC = −1.6, FDR = 0.0073, p < 0.0001, Supplemental Table 5), a gene encoding the neuron-specific transcription factor neuronal PAS domain-containing protein 4 (Npas4) (60), caught our attention. Npas4 is a neuronal activity-dependent immediate early gene, which promotes GABAergic synapse formation and plays a key role in maintaining homeostatic excitability (29–31). In agreement with RNA-seq data, qPCR found a significant reduction of Npas4 mRNA in 16p11.2dp/+ PFC (Figure 4G, t(35) = 2.92, p = 0.006, unpaired t-test). Western blotting revealed a significant loss of Npas4 protein expression in the nuclear fraction of PFC from 16p11.2dp/+ mice (Figure 4H, t(17) = 2.59, p = 0.019, unpaired t-test). Furthermore, qPCR analyses of human postmortem PFC tissue revealed that NPAS4 mRNA level was significantly reduced in idiopathic human ASD patients compared to healthy controls (Figure 4I, U = 14, p = 0.036, Mann-Whitney U test), suggesting that Npas4 dysregulation may be broadly involved in ASD.
Npas4 exhibits restricted regional expression in the brain, with the highest expression in cortical areas. However, Npas4 is also expressed at relatively high levels in other areas including the striatum (28). To determine whether the observed loss of Npas4 expression is ubiquitous throughout the brain or specific to PFC, we compared Npas4 mRNA in the striatum of WT and 16p11.2dp/+ mice. As shown in Figure 4J, Npas4 mRNA level was unchanged in striatum of 16p11.2dp/+ mice, whereas the Mapk3 gene which is located in the duplicated 16p11.2 region exhibited significant upregulation in striatum. This suggests that Npas4 dysregulation in 16p11.2dp/+ mice is region-specific.
Other than Npas4, we also evaluated the expression level of various genes encoding GABAergic synaptic components in PFC of WT and 16p11.2dp/+ mice. qPCR analyses indicated no change in mRNA levels of Vgat, Gad65, Gabra1, Gabrb2, Gabrg2, and Pvalb (Figure 4K), consistent with our RNA-seq data. This suggests that the observed GABAergic synaptic dysfunction in PFC of 16p11.2dp/+ mice is unlikely caused by the direct transcriptional changes of GABA transporters, enzymes or receptors, but may be due to dysregulation of GABA synapses by Npas4.
Restoring Npas4 Expression in 16p11.2dp/+ mPFC Ameliorates Synaptic and Behavioral Deficits
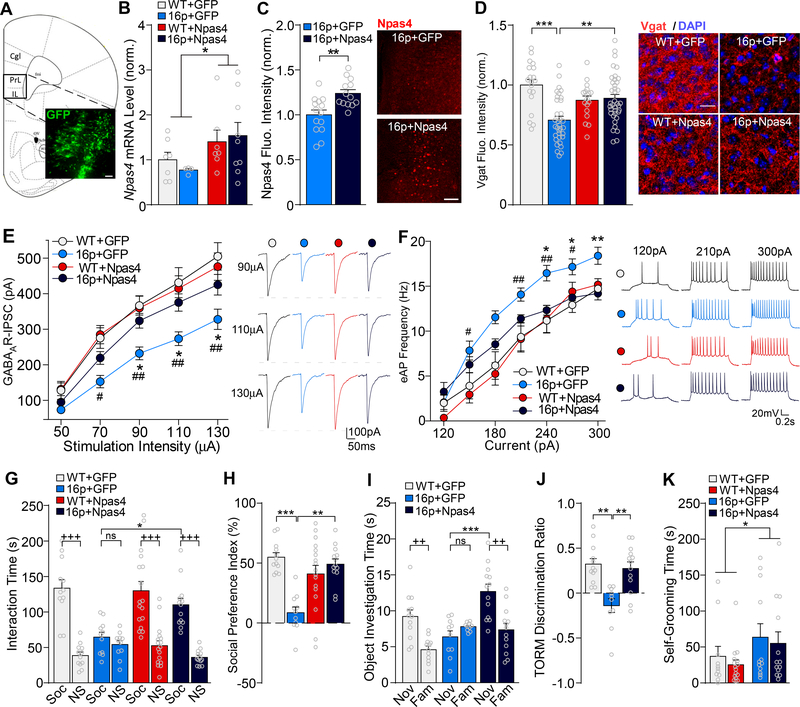
Considering the GABAergic deficits observed in 16p11.2dp/+ PFC, we sought to further investigate the role that Npas4 downregulation may play in 16p11.2dp/+ or ASD pathology. Since Npas4 plays a major role in regulating GABAergic synapse development (29, 31) and is implicated in neurodevelopmental disorders (32, 34, 61), we hypothesized that Npas4 downregulation in 16p11.2dp/+ PFC may underlie the observed GABAergic synaptic impairment and social/cognitive deficits. To test this, we examined whether restoring Npas4 expression in 16p11.2dp/+ PFC could ameliorate the synaptic and behavioral deficits. Either Npas4 CRISPR lentiviral activation particles or GFP control lentiviral particles were stereotaxically injected into mPFC of WT and 16p11.2dp/+ mice (Figure 5A). The significant upregulation of Npas4 mRNA level in Npas4-injected groups relative to GFP-injected groups was verified via qPCR (Figure 5B, F1,23 (treatment) = 4.69, p = 0.041, two-way ANOVA). Additionally, immunostaining of Npas4 revealed the significantly increased Npas4 expression in mPFC of Npas4-injected 16p11.2dp/+ mice, relative to GFP-injected 16p11.2dp/+ mice (Figure 5C, t(25) = 3.48, p = 0.002, unpaired t-test), authenticating the viral upregulation of Npas4. Viral upregulation of Npas4 was detected in both CaMKII-expressing pyramidal neurons and GAD67-positive interneurons (data not shown).
Figure 5.
Restoring Npas4 expression in PFC ameliorates the social and cognitive deficits and restores GABAergic synaptic transmission in 16p11.2dp/+ mice. A, Immunofluorescent image showing the location of GFP expression in a viral-injected mouse. Scale bar = 50 μm. B, Bar graph showing Npas4 mRNA levels in PFC of WT or 16p11.2dp/+ mice injected with GFP or Npas4 virus. n = 4–9 mice/group. C, Bar graph showing Npas4 fluorescence intensity in mPFC of GFP-injected and Npas4-injected 16p11.2dp/+ mice. Inset: representative images showing Npas4 expression in mPFC of both groups. Scale bar = 100 μm. n = 13–14 slices, 3–4 mice/group. D, Bar graph showing VGAT immunostaining fluorescence intensity in mPFC (prelimbic area) of WT and 16p11.2dp/+ mice injected with GFP or Npas4 virus. Inset: representative images showing VGAT (red) and DAPI (blue) staining. Scale bar = 20 μm. n = 18–38 slices, 2–3 mice/group. E, F, Plot of input-output curves of GABAAR-IPSC (E) and AP firing frequencies (F) in mPFC pyramidal neurons from WT or 16p11.2dp/+ mice injected with GFP or Npas4 virus. Insets: representative GABAAR-IPSC and AP firing traces. GABAAR-IPSC: n = 9–25 cells, 3–4 mice/group; eAP: 11–17 cells, 3–4 mice/group. G, H, Bar graphs showing the amount of time spent interacting with Soc vs. NS stimuli (G) and the social preference index (H) in the 3-chamber social preference test of WT or 16p11.2dp/+ mice injected with GFP or Npas4 virus. n = 11–17 mice/group. I, J, Bar graphs showing the amount of time spent interacting with the novel (Nov) vs. familiar (Fam) objects (I) and the discrimination ratio (J) in the TORM test of WT or 16p11.2dp/+ mice injected with GFP or Npas4 virus. n = 10–13 mice/group. K, Bar graphs showing self-grooming time in WT or 16p11.2dp/+ mice injected with GFP or Npas4 virus. n = 11–16 mice/group. All data are presented as mean ± SEM. In all figures, *, # p < 0.05; **, ## p < 0.01; ***, +++ p < 0.0001; ns = not significant. In figures E/F, *: 16p+GFP vs. 16p+Npas4; #: 16p+GFP vs. WT+GFP.
To determine whether Npas4 upregulation was driving GABA synapse formation in 16p11.2dp/+ mPFC, we next performed immunostaining for the vesicular GABA transporter VGAT. Relative to GFP-injected WT mice, GFP-injected 16p11.2dp/+ mice displayed a marked reduction of VGAT expression in PFC, and VGAT expression was rescued to the control level in PFC of Npas4-injected 16p11.2dp/+ mice (Figure 5D, F1,105 (genotype x treatment) = 16.16, p = 0.0001, two-way ANOVA). The cellular expression level of Npas4 was significantly correlated with the level of VGAT expression in the immediate proximity of the soma (n = 77 cells/4 mice, R2 = 0.25, p < 0.0001). This suggests that upregulating Npas4 expression in 16p11.2dp/+ PFC is sufficient to induce the pronounced restoration of GABAergic synaptic density.
We next performed whole-cell patch clamp electrophysiology on mPFC pyramidal neurons to assess whether the Npas4-driven induction of GABA synapse formation could reverse the observed synaptic deficits in 16p11.2dp/+ PFC. Compared to GFP-injected WT neurons, GABAAR-IPSC amplitudes were significantly diminished in GFP-injected 16p11.2dp/+ neurons, and this deficit was significantly reversed by Npas4 injection into the PFC of 16p11.2dp/+ mice (Figure 5E, F3,54 (group) = 7.41, p = 0.0003, two-way ANOVA). Furthermore, Npas4-injected 16p11.2dp/+ neurons exhibited significantly reduced AP firing frequencies relative to GFP-injected 16p11.2dp/+ neurons (Figure 5F, F3,52(group) = 5.70, p = 0.002, two-way ANOVA), collectively indicating that restoring Npas4 expression in 16p11.2dp/+ PFC is sufficient to reverse the GABAergic synaptic deficits and restore homeostatic neuronal excitability.
We next tested whether restoring Npas4 expression in 16p11.2dp/+ PFC could ameliorate the ASD- and ID-related behavioral phenotypes. In the 3-chamber social preference test, Npas4-injected 16p11.2dp/+ mice spent significantly more time than GFP-injected 16p11.2dp/+ mice interacting with the social stimulus (Figure 5G, F1,107 (interaction) = 9.1, p = 0.003, three-way ANOVA), and exhibited a significantly elevated preference for the social stimulus over the non-social stimulus (Figure 5H, F1,49 (interaction) = 21.78, p < 0.0001, two-way ANOVA). In the TORM task, Npas4-injected 16p11.2dp/+ mice spent significantly more time than GFP-injected 16p11.2dp/+ mice investigating the novel object (Figure 5I, F2,64 (object x group) = 9.56, p = 0.0002, two-way ANOVA), and displayed a significant preference for the more novel object over the more familiar object (Figure 5J, F2,32 (group) = 11.72, p = 0.0002, one-way ANOVA). However, viral upregulation of Npas4 did not affect self-grooming behavior in 16p11.2dp/+ mice (Figure 5K, F1,48 (genotype x treatment) = 0.01, p = 0.91, two-way ANOVA). Collectively, these data indicate that restoring Npas4 expression in 16p11.2dp/+ PFC is capable of ameliorating the social and cognitive deficits related to ASD and ID.
DISCUSSION
The phenotypic impact of the 16p11.2 duplication has been thoroughly characterized in human patients and the associated neurodevelopmental deficits are well-defined, though the underlying molecular mechanisms remain almost completely unknown. Here we have demonstrated that transgenic 16p11.2dp/+ mice exhibit ASD- and ID-related behavioral phenotypes resembling neurodevelopmental deficits in human 16p11.2 duplication patients, and discovered deficient GABAergic synaptic transmission in the PFC of 16p11.2dp/+ mice. Furthermore, we observed the pronounced downregulation of Npas4, a transcription factor responsible for the formation of GABAergic synapses in response to neuronal excitation (29). Restoring Npas4 expression in 16p11.2dp/+ PFC ameliorated the observed social and cognitive deficits and restored GABAergic synaptic function and normal neuronal excitability, suggesting a central role for Npas4 in 16p11.2 duplication pathology.
Our behavioral assays indicate that 16p11.2dp/+ mice exhibit social deficits and repetitive behaviors reminiscent of ASD, PFC-dependent cognitive impairment, and hypolocomotion, with the absence of schizophrenia-associated sensorimotor gating impairment, motor deficits, and anxiety. Thus, it is evident that the behavioral profile of 16p11.2dp/+ mice recapitulates many, but not all, neurodevelopmental deficits observed in human 16p11.2 duplication carriers. Importantly, the performance of 16p11.2dp/+ mice in certain behavioral assays such as social approach and self-grooming tests reflected heterogeneity within litters and specific batches, indicating that – like human 16p11.2 duplication carriers – individual 16p11.2dp/+ mice may present with variable behavioral phenotypes and at different degrees of severity. Our results have confirmed the hypolocomotion, elevated self-grooming and social deficits of 16p11.2dp/+ mice that were reported earlier (16, 17) and more comprehensively assessed behavioral phenotypes related to ASD/SZ.
In addition to 16p11.2 duplication mice, 16p11.2 deletion mice (16p11.2+/−) also exhibit deficits in sociability (17, 62, 63) and various cognitive impairments (17, 63, 64). While 16p11.2-deletion and 16p11.2-duplication mice share similar behavioral phenotypes, it is notable that the two models exhibit opposing electrophysiological profiles in PFC. Specifically, 16p11.2+/− PFC neurons exhibit hypoactivity (65), while 16p11.2dp/+ PFC neurons display abnormal hyper-excitability. Moreover, these divergent phenotypes appear to underlie the shared behavioral abnormalities, as elevating PFC activity ameliorated the social and cognitive deficits in 16p11.2+/− mice (65), whereas restoring inhibitory GABAergic transmission in PFC of 16p11.2dp/+ mice gave similar therapeutic effects. These divergent phenotypes offer an intriguing bidirectional explanation for the behavioral pathologies in 16p11.2 CNVs. The alteration of excitation and inhibition has also been reported in the hippocampus of 16p11.2+/− mice (66). Taken together, these findings suggest that E/I imbalances across several implicated brain regions likely contribute to the pathogenesis of neuropsychiatric phenotypes in mouse models of 16p11.2 CNVs.
Whole-cell patch clamp electrophysiology experiments revealed marked reductions in IPSC amplitudes and elevated action potential firing frequencies in 16p11.2dp/+ mPFC pyramidal neurons, indicating the disruption of GABAergic synaptic transmission and a potentially subsequent increase in neuronal excitability. The electrophysiological phenotype of 16p11.2dp/+ PFC is consistent with extensive evidence implicating GABAergic deficits and excitatory/inhibitory imbalance in both human ASD patients and animal models of ASD (18–24). Additionally, the elevated excitability of 16p11.2dp/+ PFC neurons could provide a mechanism driving the epileptic phenotypes reported in some human 16p11.2 duplication patients (6, 8, 67).
Our RNA-seq experiments identified Npas4, a transcription factor with a key role in GABA synapse formation, as one of the top 20 most strongly downregulated genes in 16p11.2dp/+ PFC. Consistently, RNA sequencing of mice and humans have found that 16p11.2 CNV is associated with altered expression of genes and networks that converge on synaptic function and transcriptional regulation (68). Npas4 knockout mice exhibit social anxiety (34) and impaired performance on various cognitive and contextual learning tasks (32–34). Considering the distinct role of Npas4 in GABAergic synapse formation, we hypothesized that disruption of Npas4 may underlie GABAergic synaptic deficits, which leads to social and cognitive deficits in 16p11.2 duplications and other forms of ASD. Indeed, we found that Npas4 mRNA expression was significantly reduced in postmortem PFC tissue from idiopathic ASD patients, suggesting that the dysregulation of Npas4 may be broadly implicated in ASD pathology. Furthermore, restoring Npas4 expression in 16p11.2dp/+ PFC significantly increased sociability in the 3-chamber social preference test and ameliorated the cognitive deficits in the temporal order recognition memory task, indicating that Npas4 expression is functionally linked to the observed behavioral phenotypes. In contrast, Npas4 upregulation in PFC did not affect self-grooming behavior in 16p11.2dp/+ mice, consistent with evidence suggesting that grooming behavior is controlled primarily by striatal circuits (38). Collectively, our findings suggest that PFC Npas4 expression is critical for the proper development of social and cognitive functions, and that Npas4 dysregulation may broadly underlie the behavioral features of ASD and ID.
It has been extensively shown that Npas4 plays a key role in the formation of GABAergic synapses (29–31). Knockdown of Npas4 reduces GABAergic synapse density and disrupts GABAergic synaptic transmission, whereas overexpressing Npas4 drives excessive GABA synapse formation (29). In the current study, we found that restoring Npas4 expression in 16p11.2dp/+ PFC significantly elevated GABAR-mediated IPSCs and normalized action potential firing frequencies in 16p11.2dp/+ mPFC pyramidal neurons. Furthermore, Npas4 upregulation restored the downregulated expression of the presynaptic GABA transporter VGAT in PFC of 16p11.2dp/+ mice, suggesting that Npas4 expression may directly rescue the density of presynaptic GABAergic synaptic terminals. Furthermore, since viral upregulation of Npas4 was observed in both pyramidal neurons and interneurons, and Npas4 expression in either cell type promotes GABAergic input onto pyramidal neurons (30), it is likely that the observed VGAT upregulation represents an Npas4-induced increase of GABAergic synaptic input to pyramidal neurons, which is mediated through both pre- and post-synaptic mechanisms.
The current study presents strong evidence for the involvement of Npas4 and prefrontal cortical GABA dysregulation in 16p11.2 duplication pathology. We propose that Npas4 dysregulation yields E/I imbalances in prefrontal cortical synaptic circuitry, resulting in social and cognitive deficits in 16p11.2 duplications, a mechanism that may be more broadly implicated in ASD and ID.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Xiaoqing Chen, Dr. Zi-Jun Wang and Dr. Luye Qin for excellent technical support. We acknowledge the support of University at Buffalo’s Genomics and Bioinformatics Core and the New York State Center of Excellence in Bioinformatics and Life Sciences. We are grateful for Dr. Michael Greenberg at Harvard University for providing Npas4 antibody. This work was supported by Nancy Lurie Marks Family Foundation and National Institutes of Health (MH112237; MH108842) to Z. Y.
Footnotes
DECLARATION OF INTERESTS
The authors report no competing financial or other interests.
REFERENCES
- 1.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England Journal of Medicine. 2008;358:667–75. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niarchou M, Chawner S, Doherty JL, Maillard AM, Jacquemont S, Chung WK, et al. Psychiatric disorders in children with 16p11.2 deletion and duplication. Transl Psychiatry. 2019;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillentine MA, Lupo PJ, Stankiewicz P, Schaaf CP. An estimation of the prevalence of genomic disorders using chromosomal microarray data. J Hum Genet. 2018;63:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez BA, Roberts W, Chung B, Weksberg R, Meyn S, Szatmari P, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. [DOI] [PubMed] [Google Scholar]
- 6.Shinawi M, Liu P, Kang S-HL, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. Journal of Medical Genetics. 2010;47:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber JC, Hall V, Maloney VK, Huang S, Roberts AM, Brady AF, et al. 16p11.2-p12.2 duplication syndrome; a genomic condition differentiated from euchromatic variation of 16p11.2. Eur J Hum Genet. 2013;21:182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Angelo D, Lebon S, Chen Q, Martin-Brevet S, Snyder LG, Hippolyte L, et al. Defining the Effect of the 16p11.2 Duplication on Cognition, Behavior, and Medical Comorbidities. JAMA Psychiatry. 2016;73:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green Snyder L, D’Angelo D, Chen Q, Bernier R, Goin-Kochel RP, Wallace AS, et al. Autism Spectrum Disorder, Developmental and Psychiatric Features in 16p11.2 Duplication. J Autism Dev Disord. 2016;46:2734–48. [DOI] [PubMed] [Google Scholar]
- 10.Bernier R, Hudac CM, Chen Q, Zeng C, Wallace AS, Gerdts J, et al. Developmental trajectories for young children with 16p11.2 copy number variation. Am J Med Genet B Neuropsychiatr Genet. 2017;174:367–80. [DOI] [PubMed] [Google Scholar]
- 11.Chang H, Li L, Li M, Xiao X. Rare and common variants at 16p11.2 are associated with schizophrenia. Schizophr Res. 2017;184:105–8. [DOI] [PubMed] [Google Scholar]
- 12.Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13:868–80. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg S, de Jong S, Mattheisen M, Costas J, Demontis D, Jamain S, et al. Common variant at 16p11.2 conferring risk of psychosis. Mol Psychiatry. 2014;19:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld JA, Coppinger J, Bejjani BA, Girirajan S, Eichler EE, Shaffer LG, et al. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. Journal of Neurodevelopmental Disorders. 2010;2:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horev G, Ellegood J, Lerch JP, Son Y-EE, Muthuswamy L, Vogel H, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. PNAS. 2011;108:17076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arbogast T, Ouagazzal AM, Chevalier C, Kopanitsa M, Afinowi N, Migliavacca E, et al. Reciprocal Effects on Neurocognitive and Metabolic Phenotypes in Mouse Models of 16p11.2 Deletion and Duplication Syndromes. PLoS Genet. 2016;12:e1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coghlan S, Horder J, Inkster B, Mendez MA, Murphy DG, Nutt DJ. GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci Biobehav Rev. 2012;36:2044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schur RR, Draisma LW, Wijnen JP, Boks MP, Koevoets MG, Joels M, et al. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Hum Brain Mapp. 2016;37:3337–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee E, Lee J, Kim E. Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biol Psychiatry. 2017;81:838–47. [DOI] [PubMed] [Google Scholar]
- 24.Antoine MW, Langberg T, Schnepel P, Feldman DE. Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron. 2019;101:648–61.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZJ, Zhong P, Ma K, Seo JS, Yang F, Hu Z, et al. Amelioration of autism-like social deficits by targeting histone methyltransferases EHMT1/2 in Shank3-deficient mice. Mol Psychiatry. 2019; [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapanelli M, Tan T, Wang W, Wang X, Wang ZJ, Zhong P, et al. Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry. 2019; [epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 27.Amodio DM, Frith CD. Meeting of minds: the medial frontal cortex and social cognition. Nature Reviews Neuroscience. 2006;7:268–77. [DOI] [PubMed] [Google Scholar]
- 28.Damborsky JC, Slaton GS, Winzer-Serhan UH. Expression of Npas4 mRNA in Telencephalic Areas of Adult and Postnatal Mouse Brain. Front Neuroanat. 2015;9:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shepard R, Heslin K, Coutellier L. The transcription factor Npas4 contributes to adolescent development of prefrontal inhibitory circuits, and to cognitive and emotional functions: Implications for neuropsychiatric disorders. Neurobiol Dis. 2017;99:36–46. [DOI] [PubMed] [Google Scholar]
- 33.Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS One. 2012;7:e46604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PLoS One. 2011;6:e23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan T, Wang W, Williams J, Ma K, Cao Q, Yan Z. Stress exposure in dopamine D4 receptor knockout mice induces schizophrenia-like behaviors via disruption of GABAergic transmission. Schizophr Bull. 2019;45:1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Molecular Psychiatry. 2015;20:140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, Fentress JC. Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci. 2016;17:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker GR, Bird F, Alexander V, Warburton EC. Recognition memory for objects, place, and temporal order: a disconnection analysis of the role of the medial prefrontal cortex and perirhinal cortex. J Neurosci. 2007;27:2948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clementz BA, Geyer MA, Braff DL. Poor P50 suppression among schizophrenia patients and their first-degree biological relatives. Am J Psychiatry. 1998;155:1691–4. [DOI] [PubMed] [Google Scholar]
- 42.Wong AH, Josselyn SA. Caution When Diagnosing Your Mouse With Schizophrenia: The Use and Misuse of Model Animals for Understanding Psychiatric Disorders. Biol Psychiatry. 2016;79:32–8. [DOI] [PubMed] [Google Scholar]
- 43.Braff DL, Grillon C, Geyer MA. Gating and habituation of the startle reflex in schizophrenic patients. Arch Gen Psychiatry. 1992;49:206–15. [DOI] [PubMed] [Google Scholar]
- 44.Perry W, Minassian A, Lopez B, Maron L, Lincoln A. Sensorimotor gating deficits in adults with autism. Biol Psychiatry. 2007;61:482–6. [DOI] [PubMed] [Google Scholar]
- 45.Kohl S, Wolters C, Gruendler TO, Vogeley K, Klosterkotter J, Kuhn J. Prepulse inhibition of the acoustic startle reflex in high functioning autism. PLoS One. 2014;9:e92372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madsen GF, Bilenberg N, Cantio C, Oranje B. Increased prepulse inhibition and sensitization of the startle reflex in autistic children. Autism Res. 2014;7:94–103. [DOI] [PubMed] [Google Scholar]
- 47.Hessl D, Berry-Kravis E, Cordeiro L, Yuhas J, Ornitz EM, Campbell A, et al. Prepulse inhibition in fragile X syndrome: feasibility, reliability, and implications for treatment. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankland PW, Wang Y, Rosner B, Shimizu T, Balleine BW, Dykens EM, et al. Sensorimotor gating abnormalities in young males with fragile X syndrome and Fmr1-knockout mice. Mol Psychiatry. 2004;9:417–25. [DOI] [PubMed] [Google Scholar]
- 49.Yuhas J, Cordeiro L, Tassone F, Ballinger E, Schneider A, Long JM, et al. Brief report: Sensorimotor gating in idiopathic autism and autism associated with fragile X syndrome. J Autism Dev Disord. 2011;41:248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunner D, Kabitzke P, He D, Cox K, Thiede L, Hanania T, et al. Comprehensive Analysis of the 16p11.2 Deletion and Null Cntnap2 Mouse Models of Autism Spectrum Disorder. PLoS One. 2015;10:e0134572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olney JW, Farber NB. Glutamate receptor dysfunction and schizophrenia. Arch Gen Psychiatry. 1995;52:998–1007. [DOI] [PubMed] [Google Scholar]
- 52.Carlsson M, Carlsson A. The NMDA antagonist MK-801 causes marked locomotor stimulation in monoamine-depleted mice. J Neural Transm. 1989;75:221–6. [DOI] [PubMed] [Google Scholar]
- 53.Neill JC, Barnes S, Cook S, Grayson B, Idris NF, McLean SL, et al. Animal models of cognitive dysfunction and negative symptoms of schizophrenia: focus on NMDA receptor antagonism. Pharmacol Ther. 2010;128:419–32. [DOI] [PubMed] [Google Scholar]
- 54.Bickel S and Javitt DC. Neurophysiological and neurochemical animal models of schizophrenia: focus on glutamate. Behav. Brain Res, 2009; 204:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bubenikova-Valesova V, Horacek J, Vrajova M, Hoschl C. Models of schizophrenia in humans and animals based on inhibition of NMDA receptors. Neurosci Biobehav Rev. 2008;32:1014–23. [DOI] [PubMed] [Google Scholar]
- 56.Filges I, Sparagana S, Sargent M, Selby K, Schlade-Bartusiak K, Lueder GT, et al. Brain MRI abnormalities and spectrum of neurological and clinical findings in three patients with proximal 16p11.2 microduplication. Am J Med Genet A. 2014;164A:2003–12. [DOI] [PubMed] [Google Scholar]
- 57.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeyabalan N, Clement JP. SYNGAP1: Mind the Gap. Front Cell Neurosci. 2016;10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maya-Vetencourt JF. Activity-dependent NPAS4 expression and the regulation of gene programs underlying plasticity in the central nervous system. Neural Plast. 2013;2013:683909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaehne EJ, Klaric TS, Koblar SA, Baune BT, Lewis MD. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behav Brain Res. 2015;281:276–82. [DOI] [PubMed] [Google Scholar]
- 62.Yang M, Mahrt EJ, Lewis F, Foley G, Portmann T, Dolmetsch RE, et al. 16p11.2 deletion syndrome mice display sensory and ultrasonic vocalization deficits during social interactions. Autism Research. 2015;8:507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoppel LJ, Kazdoba TM, Schaffler MD, Preza AR, Heynen A, Crawley JN, et al. R-Baclofen Reverses Cognitive Deficits and Improves Social Interactions in Two Lines of 16p11.2 Deletion Mice. Neuropsychopharmacology. 2018;43:513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang M, Lewis FC, Sarvi MS, Foley G, Crawley JN. 16p11.2 deletion mice display cognitive deficits in touchscreen learning and novelty recognition tasks. Learning and Memory. 2015;22:622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W, Rein B, Zhang F, Tan T, Zhong P, Qin L, et al. Chemogenetic Activation of Prefrontal Cortex Rescues Synaptic and Behavioral Deficits in a Mouse Model of 16p11.2 Deletion Syndrome. J Neurosci. 2018;38:5939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu HC, Mills AA, Tian D. Altered synaptic transmission and maturation of hippocampal CA1 neurons in a mouse model of human chr16p11.2 microdeletion. J Neurophysiol. 2018;119:1005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinman KJ, Spence SJ, Ramocki MB, Proud MB, Kessler SK, Marco EJ, et al. 16p11.2 deletion and duplication: Characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet A. 2016;170:2943–55. [DOI] [PubMed] [Google Scholar]
- 68.Blumenthal I, Ragavendran A, Erdin S, Klei L, Sugathan A, Guide JR, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94:870–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.