Abstract
Atopic dermatitis (AD) is a skin disease that results from a combination of skin barrier dysfunction and immune dysregulation. The immune dysregulation is often associated with IgE sensitivity. There is also evidence that autoallergens Hom s 1, 2, 3, and 4 play a role in AD; it is possible that patients with specific HLA subtypes are predisposed to autoreactivity due to increased presentation of autoallergen peptides. The goal of our study was to use in silico epitope prediction platforms as an approach to identify HLA subtypes that may preferentially bind autoallergen peptides and are thus candidates for further study. Considering the previously described association of DRB1 alleles with AD and progression of disease, emphasis was placed on DRB1. Certain DRB1 alleles (08:04, 11:01, and 11:04) were identified by both algorithms to bind a significant percent of the generated autoallergen peptides. Conversely, autoallergen core peptide sequences FRQLSHRFH and IRAKLRLQA (Hom s 1), IRKSKNILF (Hom s 2), FKWVPVTDS and MAAIEKVRK (Hom s 3), and FRYFATLKV (Hom s 4) were predicted to bind many DRB1 alleles and thus, may play a role in the pathogenesis of AD. Our findings provide candidate DRB1 alleles and autoallergen epitopes that will guide future studies exploring the relationship between DRB1 subtype and autoreactivity in AD. A similar approach can be used for any antigen that has been associated with an IgE respsonse and AD.
Keywords: atopic dermatitis, HLA alleles, autoreactivity
4. Introduction
Currently, existing in silico T-cell epitope discovery algorithms, given a full-length protein and a HLA allele, are able to predict likely peptide epitopes and their binding affinities to HLA molecules(1–5). Immune Epitope Database Analysis Resource (IEDB) and NetMHCIIpan 3.1 are epitope prediction platforms that have been developed based on large data sets of known peptide/MHC molecule interactions and previously validated against in vitro epitope binding data(4–13). These algorithms, in identifying likely interactions between HLA molecules and certain peptides, can be used to direct future binding experiments and genetic association studies, thus providing a unique approach to probing the molecular interactions that underlie T-cell mediated disease processes.
Atopy, the predisposition to develop allergic hypersensitivity reactions, is such a disease process, as it is generally characterized by a TH2-dominant cellular response and often IgE antibody production against specific allergens. In atopic individuals, the severity and specificity of the immune response to allergens are dependent on a multifactorial process that includes genetic and environmental factors, as well as the type of allergen exposure(14, 15). One, and often the first clinical, manifestation of atopy is atopic dermatitis (AD), a common inflammatory skin disease characterized by itchy, red patches occurring most frequently at flexural areas of the elbows and knees(16, 17). In the United States alone, AD affects 10–20% of children of all races and ethnic groups(18–20).
Immune dysregulation, characterized by T-cell mediated responses and elevated serum IgE antibodies, plays an important complementary role in the mechanism of AD(21, 22). The imbalanced activation and differentiation of TH2 and TH22 cells have been associated with acute and chronic AD(23–25). The production of TH2-associated cytokines (IL-3 and IL-4) can contribute to the impairment of the epidermal barrier function(26–28), which may account for the barrier dysfunction observed in AD patients who do not have known genetic defects in epidermal barrier proteins. The additional presence of IgE antibodies to an allergen can enhance specific T-cell responses to that allergen, including the release of pro-inflammatory cytokines, and contribute to disease symptoms(29–31). There are also hypotheses that the sensitization of AD patients has an autoimmune component(32). Severity of AD symptoms has been correlated with intensity of IgE autoreactivity in several studies(33).
Furthermore, sensitization to specific autoallergens, Hom s 1, Hom s 2, Hom s 3, and Hom s 4, has been reported in AD patients(34–36). More specifically, TH1 and TH2 responses to Hom s 1 and Hom s 4, as well as Hom s 2-specific autoreactive T-cells have been reported in AD patients(34, 37–39); previous studies have also identified specific IgE sensitization to Hom s 2 and Hom s 3 in AD patients(33). While the exact role of these autoallergens in the pathogenesis of AD remains unclear, in a model in which autoreactivity plays a role, it is possible that there exists preferential binding between specific HLA molecules and autoallergen peptides. Because HLAs plays a large role in immune system activation by presenting antigen peptides to T cells, an increased molecular affinity of specific HLA subtypes to bind autoallergen peptides could result in increased presentation of autoallergen peptides to T cells, and lead to sensitization and autoreactivity.
Therefore, the goal of this study is to use in silico epitope prediction platforms as an approach to explore the relationship between HLA subtype and autoallergen peptides in AD. We first perform validation of NetMHCIIpan 3.1 and IEDB using previously published epitopes and then utilize the epitope prediction platforms to identify HLA subtypes that may display preferential binding to specific peptide epitopes of the previously identified autoallergens, Hom s 1, Hom s 2, Hom s 3, and Hom s 4. We focus on these autoantigens in order to use them as a model for future exploration of the possible interaction between specific autoallergen peptides and HLA subtype in the context of AD; however, other antigens such as those shown to result in specific IgE elevation in individuals with AD can also be studied in a similar fashion.
5. Materials and Methods
8.1. T-cell Epitope Prediction
T-cell epitope prediction was performed using the Immune Epitope Database Analysis Resource (IEDB) MHC I and MHC II epitope prediction algorithms, both based on a combination of NN-align, NNM-align, Combinatorial Library, Sturniolo, and ARB prediction methods(4, 6, 12, 38, 40–42). The consensus method was used for epitope prediction. Within the IEDB algorithm, a binding percentile rank was determined by comparing the binding affinity of the given peptide to those of a large, randomly selected set of peptides from the SWISS-PROT database. According to IEDB recommendations, a percentile rank of ≤1% was used as the cutoff to predict peptide binders for MHC I and a percentile rank of ≤10% was used as the cutoff to predict peptide binders for MHC II(4). In the MHC I IEDB algorithm, overlapping 8-, 9-, 10-, and 11-mer peptides were generated from the full-length protein sequences. For MHC II, overlapping 15-mer peptides (default setting) were generated from the full-length protein sequences. The number of predicted epitopes was normalized based on the number of possible peptides and expressed as a proportion of all possible peptides.
Additional T-cell epitope prediction was performed using the MHC II-specific NetMHCIIpan 3.1 algorithm, whose prediction method is based on an artificial neural network(2, 5, 6, 13). The NetMHCIIpan 3.1 program determines a percentile rank for each epitope by comparing the binding affinity of the given peptide with a random selection of 200,000 peptides of the same length(5). The NetMHCIIpan recommended cutoffs of percentile rank ≤ 10% for weakly binding MHC II epitopes and percentile rank ≤ 2% for strongly binding MHC II epitopes were used(5). Unless the length of peptides was otherwise changed to match published epitopes during proof-of-concept testing, 15-mer peptides were generated from the full-length protein sequences for epitope prediction. The number of predicted epitopes was expressed as a proportion of all possible peptides.
5.2. Selection of HLA Alleles
The top ten most common HLA-A, -B, -C, and -DRB1 alleles were selected for the four different ethnicities (Caucasians, African Americans, Asians, and Hispanics), as defined by the National Marrow Donor Program (see Table S1 for full listing of all alleles used)(43).
8.3. Selection of Epitopes for Validation of IEDB and NetMHCIIpan
For proof of concept of the in silico epitope prediction algorithms, H1N1 nucleoprotein(44), peanut (Ara h 1)(9), and mugwort (Art v 1) epitopes(45, 46) that were previously validated during in vitro studies, were selected. The full-length proteins were entered into IEDB and NetMHCIIpan algorithms and the sets of predicted epitopes were compared to the peptides originating from these proteins and known to bind to HLA molecules from in vitro binding studies.
Control proteins were selected as a baseline comparison for the number of predicted epitopes (expressed as a proportion of all possible epitopes). We hypothesize that proteins known to elicit a strong T-cell response among individuals with different HLA phenotypes will have a relatively high proportion of predicted epitopes and low variability in the number of predicted epitopes across different HLA alleles. One such protein is H1N1 influenza hemagglutinin (HA), which is known to elicit a strong T-cell response and has significant cross-reactivity among different HLA class I subtypes(47–49). Hepatitis B surface antigen was also chosen as a control peptide; based on its high immunogenicity but variable reactivity among different HLA alleles, a relatively high proportion of predicted epitopes and high variability among different HLA subtypes is expected(50, 51). Tuberculin was chosen as a negative control, as there has been no association found between HLA class II alleles and reactivity, and M. tuberculosis naïve individuals do not react to the tuberculin skin test even upon repeated exposure(52, 53).
8.4. Selection of Allergens of Interest
The allergens of interest, Hom s 1, Hom s 2, Hom s 3, and Hom s 4, were selected based on previous studies that reported TH1 and TH2 responses to Hom s 1, Hom s 2, and Hom s 4 in AD patients, as well as specific IgE sensitization to Hom s 2 and Hom s 3 [34–38]. Based on previous sequence identity findings, Hom s 1 corresponds to the protein SART-1, Hom s 2 corresponds to the alpha chain of the human nascent polypeptide-associated complex, Hom s 3 corresponds to the protein BCL7B, and Hom s 4 corresponds to the mitochondrial calcium-binding protein MICU1. Hom s 1 is expressed in proliferating cells, and Hom s 2, Hom s 3, and Hom s 4 are expressed ubiquitously(54). Thus, all four allergens are widely available as potential allergens (Table S2).
8.5. Binding Pocket Analysis
Peptide binding to HLA class II molecules depends on the cumulative binding affinities of the different pockets formed along the length of the binding grove, as the side chains of a particular peptide bound to the groove interacts with these side chain amino acids. Specific residues involved in binding pockets on the antigen presenting domain of the HLA-DRB1 molecule have been previous described; all residues in HLA-DRB1 within a 5.0Å radius of the center of each binding pocket were aligned across the different HLA-DRB1 alleles(55, 56). The relevant residues were numbers 9, 11, 13, 26, 28, 47, 57, 60, 61, 67, 70, 71, 74, 78, 82, 85, 86, 89, and 90, comprising binding pockets 1, 4, 6, 7, and 9; binding pockets 4 and 9 were of particular interest, as we previously reported associations between binding pockets 4 and 9 of HLA-DRB1 and the persistence of AD in children(56). The HLA-DRB1 alleles were classified into groups based on sequence homology of binding pocket residues. The full amino acid sequence of each HLA-DRB1 allele in our set was obtained from the IMGT/HLA database(57).
9. Results
9.1. In Silico Prediction of Previously Published Epitopes
To establish the credibility of the approach, we first considered as to whether the predictive algorithms would successfully predict the binding of peptides already known to be physically bound, based on binding studies, to either class I or class II alleles. H1N1 nucleoprotein peptides that were previously discovered to bind in vitro to HLA-B*35:01 were also predicted as strongly binding epitopes in the IEDB MHC I prediction algorithm using the recommended 1% cutoff (Table 1). Of the 17 published peanut Ara h 1 epitopes, 15 were predicted using the IEDB algorithm at the recommended 10% cutoff; in NetMHCIIpan, only 2 epitopes were predicted using the recommended 2% cutoff for strong binders and 8 epitopes were predicted using the recommended 10% cutoff for weak binders (Table 1). All of the Ara h 1 peptides that NetMHCIIpan predicted to bind were also predicted to bind by IEDB. The published mugwort peptides were not predicted to bind DRB1*01:01 using either the IEDB or NetMHCIIpan algorithms (Table 1).
Table 1.
Proof-of-concept testing of IEDB and NetMHCIIpan algorithms.
| Protein | HLA Allele | Published Epitope | IEDB | NetMHCIIpan |
|---|---|---|---|---|
| H1N1 | B*35:01 | LPFDRTTIM | ✓ | |
| Nucleoprotein | B*35:01 | LPFDRPTIM | ✓ | |
| Peanut Ara h 1 | DRB1*01:01 | KEGDVFIMPAAHPVAINASS | ✓ | ✓ |
| DRB1*01:01 | QRSRQFQNLQNHRIVQIEAK | ✓ | ✓ | |
| DRB1*01:01 | DN1LVIQQGQATVTVANGNN | ✓ | ✓ | |
| DRB1*11:01 | TSRNNPFYFPSRRFSTRYGN | ✓ | ✓ | |
| DRB1*04:01 | QRSRQFQNLQNHRIVQIEAK | ✓ | ✓ | |
| DRB1*04:01 | KEGDVFIMPAAHPVAINASS | ✓ | ✓ | |
| DRB1*04:01 | QKESH FVSARPQSQSQSPSS | ✓ | ✓ | |
| DRB1*15:02 | QRSRQFQNLQNHRIVQIEAK | ✓ | ✓ | |
| DRB1*11:01 | LEAAFNAEFNEIRRVLLEEN | ✓ | ||
| DRB1*14:01 | LEAAFNAEFNEIRRVLLEEN | |||
| DRB1*04:04 | NSKAMVIVVVNKGTGNLELV | ✓ | ||
| DRB1*04:01 | FNEIRRVLLEENAGGEQEER | ✓ | ||
| DRB1*11:01 | VVNKGTGNLELVAVRKEQQQ | ✓ | ||
| DRB1*03:01 | NNFGKLFEVKPDKKNPQLQD | ✓ | ||
| DRB1*04:04 | FNEIRRVLLEENAGGEQEER | ✓ | ||
| DRB1*11:01 | LELVAVRKEQQQRGRREEEE | ✓ | ||
| DRB1*14:01 | FNEIRRVLLEENAGGEQEER | |||
| Mugwort Art v 1 | DRB1*01:01 | KCIEWEKAQHGA | ||
| DRB1*01:01 | DKKCIEWEKAQHGA | |||
| DRB1*01:01 | NKKCDKKCIEWEKAQHGA |
9.2. Cross-reactivity of Autoallergens across HLA Alleles
We explored the ten most common HLA alleles in each ethnicity. For HLA-A, these groups of 10 variants represent 94.9% of prevalent HLA variants for Caucasians, 84.6% of African Americans, 88% of Asians, and 88% of Hispanics. For HLA-B, 77.9% of Caucasians, 72.8% of African Americans, 64.5% of Asians and 59.9% of Hispanics are represented. For HLA-C, 97.1% of Caucasians, 92% of African Americans, 87.95 of Asians, and 92.4% of Hispanics are represented. For HLA-DRB1, 88.5% of Caucasians, 83% of African Americans, 78.9% of Asians, and 75.8% of Hispanics are represented (Table S3).
Particular emphasis is given on the epitope prediction results for HLA-DRB1 because previous genetic studies have indicated an association between loci in the HLA-DRB1 region and AD [56,58–64]. Full results for epitope prediction of HLA-DRB1 from IEDB and NetMHCIIpan are presented in Tables S4 and S5. Additionally, full results for epitope prediction for HLA-A, -B, and C- are presented in Tables S6, S7, and S8, respectively.
The number of predicted peptide binders is expressed as a percent of the total possible peptides generated (Table 2). H1N1 HA has a relatively large proportion of predicted epitopes among the HLA-DRB1 alleles with a median of 21.05% predicted binders in IEDB and 8.26% predicted binders in NetMHCIIpan. HBVSAg has a more variable proportion of predicted epitopes across the HLA-DRB1 alleles with a median of 38.21% and an interquartile range of 21.34% to 51.06% in IEDB, and a median of 11.79% and interquartile range of 5.78% to 14.98% in NetMHCIIpan (Table 2). For tuberculin, the algorithms predicted a relatively lower percent of binders, with a median of 1.33% (interquartile range 0%−18.00%) in IEDB and 0% (interquartile range 0%−0%) in NetMHCIIpan (Table 2). Among the four autoallergens, Hom s 4 had the largest median proportion of peptides predicted to bind HLA-DRB1 and Hom s 2 had the second largest median proportion of predicted binders; both findings were consistent across both epitope prediction algorithms (Table 2).
Table 2.
The median, minimum, first quartile, third quartile, and maximum percent of peptides predicted to bind among the set of HLA-DRB1 alleles.
| Hom s 1 | Hom s 2 | Hom s 3 | Hom s 4 | H1N1 HA | HBVSAg | Tuberculin | ||
|---|---|---|---|---|---|---|---|---|
| IEDB | median | 5.92% | 14.18% | 9.04% | 20.35% | 21.05% | 38.21% | 1.33% |
| min | 1.65% | 2.99% | 0.00% | 2.60% | 3.81% | 1.42% | 0.00% | |
| Q1 | 3.34% | 10.20% | 2.26% | 12.12% | 9.35% | 21.34% | 0.00% | |
| Q3 | 21.60% | 22.01% | 18.75% | 35.01% | 34.30% | 51.06% | 18.00% | |
| max | 33.84% | 35.82% | 28.19% | 47.40% | 45.37% | 61.79% | 34.67% | |
| NetMHCIIpan | median | 2.67% | 7.96% | 1.33% | 10.15% | 8.26% | 11.79% | 0.00% |
| min | 0.64% | 1.99% | 0.00% | 2.38% | 3.81% | 0.00% | 0.00% | |
| Q1 | 1.94% | 6.09% | 0.00% | 9.13% | 4.90% | 5.78% | 0.00% | |
| Q3 | 4.55% | 12.06% | 2.66% | 11.83% | 9.03% | 14.98% | 0.00% | |
| max | 9.16% | 15.92% | 7.98% | 14.04% | 11.62% | 22.64% | 4.00% |
We then ranked the HLA-DRB1 alleles according to the percent of predicted binders for each autoallergen and identified alleles that are in the top five according to percent of predicted binders for two or more autoallergens (Figure 1A). DRB1*11:04, DRB1*08:04, and DRB1*11:01 met these requirements in both the IEDB and NetMHCIIpan platforms. In IEDB, DRB1*13:01 and DRB1*15:02 were also in the top five alleles for all autoallergens and in NetMHCIIpan, DRB1*08:02 and DRB1*08:03 were in the top five alleles for all autoallergens (Figure 1A).
Figure 1. HLA-DRB1 alleles that have the greatest percent of predicted binders among all autoallergens.
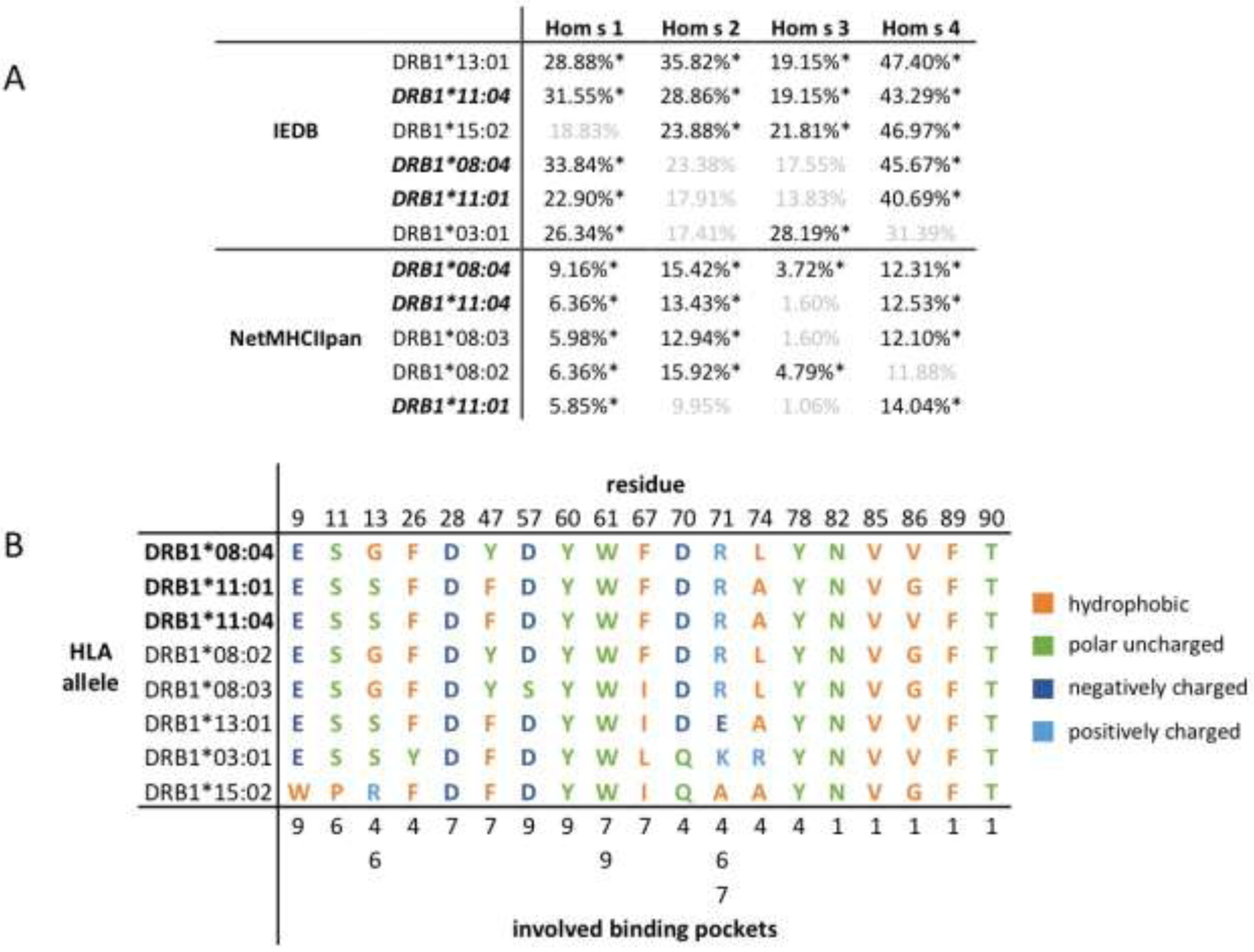
(A) Number of predicted binders for each alleles, expressed as a percent of all epitopes. (*) denotes that the allele was in the top five alleles ranked by percent of predicted binders for the given autoallergen; faded cells indicate that the allele was not in the top five alleles with the most predicted binders for the given autoallergen. Alleles that appeared in the top five for more than one autoallergen are listed, and alleles that overlap between the IEDB and NetMHCIIpan algorithms are bold and italicized. (B) Amino acid sequences at key positions implicated in binding pockets in the antigen presenting domain of the MHC molecule. Residues are categorized as hydrophobic, polar uncharged, negatively charged, and positively charged.
9.3. Binding Pocket Analysis
DRB1*08:04, DRB1*11:01, and DRB1*11:04, the three alleles that were predicted to bind a high number of epitopes by both IEDB and NetMHCIIpan algorithms, share the majority of residues that are in the antigen presenting domain of HLA-DRB1 and are thought to play a role in binding pocket functionality (Figure 1B). The only differences in residue composition between the three alleles are at positions 13, 47, 74, and 86. At positions 74 and 86, alleles DRB1*08:04 and DRB1*11:01, respectively, differ from the other two, but all residues in those positions remain hydrophobic (Figure 1B). At positions 13 and 47, DRB1*08:04 differs from the other two alleles, with less conservation of binding pocket structure (Figure 1B).
Of all the alleles that were predicted to bind many epitopes for two or more autoallergens (Figure 1A), most had a similar pattern of hydrophobic, polar, and charged residues in the key residue positions; allele DRB1*15:02 differed the most from the other alleles, with five positions in which the type of residue was not expressed at that position in at least three different alleles. A full listing of binding pocket residues for all HLA-DRB1 alleles can be found in Figure S1.
9.4. Allergen Epitope Prediction
For each of the four autoallergens, we also identified the core epitope sequences of each of the autoallergen that most commonly appeared to bind the DRB1 alleles. Sequences FRQLSHRFH (Hom s 1), IRAKLRLQA (Hom s 1), IRKSKNILF (Hom s 2), FKWVPVTDS (Hom s 3), MAAIEKVRK (Hom s 3), and FRYFATLKV (Hom s 4) were the most frequent core sequences identified in the predicted epitopes for their respective autoallergens (Table 3). Additionally, we identified the DRB1 allele associated with the most frequently occurring core sequences (Table 3). All Hom s 1, Hom s 2, Hom s 3, and Hom s 4 peptides predicted to bind HLA alleles (including HLA-A, -B, -C, and -DRB1) are presented in supplementary file S10.
Table 3.
Most common peptide core sequences among the predicted epitopes and associated HLA-DRB1 alleles for Hom s 1, Hom s 2, Hom s 3, and Hom s 4.
| Autoallergen | Core Sequence | Epitope Count IEDB | Epitope Count NetMHCIIpan | HLA-DRB1 Alleles (IEDB and NetMHCIIpan overlap) |
|---|---|---|---|---|
| Horn s 1 | FRQLSHRFH | 28 | 63 | 15:03, 12:02, 11:04, 11:01, 08:04, 03:02,01:02 |
| IRAKLRLQA | 38 | 60 | 15:03, 13:01, 12:02, 12:01, 11:04, 11:01, 08:04, 07:01, 03:02, 03:01,01:02 | |
| Horn s 2 | IRKSKNILF | 127 | 110 | All alleles except 01:01 |
| Horn s 3 | FKWVPVTDS | 34 | 24 | 08:02, 04:07, 04:05, 04:04, 04:01 |
| MAAIEKVRK | 36 | 21 | 12:02, 11:04, 11:01, 08:04, 08:03, 08:02, 03:02, 03:01 | |
| Horn s 4 | FRYFATLKV | 106 | 138 | All alleles except 13:02 |
10. Discussion
In this study, we used in silico epitope prediction algorithms to explore the potential binding patterns of four autoallergens, Hom s 1, Hom s 2, Hom s 3, and Hom s 4, and various HLA alleles. The goal was to explore whether specific allergen peptides (epitopes) bind preferentially to certain HLA alleles, which would identify alleles and peptides of interest for further study. The effort is complimentary to the approach we have taken in the past to evaluate the potential association of HLA class II alleles with atopic dermatitis and progression to disease, irrespectively of presumed autoallergens(56).
We first demonstrated that the algorithms could identify previously published and experimentally verified epitopes. The IEDB MHC I algorithm identified all of the H1N1 test case epitopes and the MHC II algorithm identified 15 of the 17 peanut allergen epitopes. The NetMHCIIpan algorithm identified 8 of the 17 epitopes under the weak binding (less stringent) cutoff. These findings suggest that the NetMHCIIpan algorithm is less sensitive than the IEDB MHC II platform, but perhaps more specific. However, both the IEDB and the NetMHCIIpan failed to identify the published mugwort epitope. This is not entirely unexpected given that the in silico algorithms are not perfectly sensitive, but is notable because the mugwort allergen Art v 1 has only a single known epitope thought to contribute to the allergic reaction(45, 46). Indeed, previous studies attempting to evaluate the sensitivity of MHC II epitope prediction algorithms have estimated a sensitivity of about 67% for the consensus method used in this study(12). The above results verified the employed predictive algorithms as credible, though not completely sensitive, means of assessing the generation of autoallergen peptides that bind to different HLA alleles and form potential T cell epitopes associated with AD.
We then used the algorithms to predict epitopes for the four autoallergens of interest binding across a set of HLA alleles that represents the most frequent alleles in a given population. Particular emphasis has been given in the analysis of the peptides bound to DRB1 since this locus has been previously reported to be associated with both susceptibility to AD and disease progression and also because of the well-established role of DRB1 in TH2 responses(56, 58–66). Additionally, although MHC II molecules generally present peptides derived from exogenous proteins, they can also present endogenous antigens within the antigen-presenting cells(67). Regarding the analysis of tuberculin peptides, there was a low number of predicted epitopes overall in both IEDB and NetMHCIIpan as expected but there was higher variability in the proportion of predicted epitopes for tuberculin in IEDB compared to NetMHCIIpan. This might be partly explained by the relatively short length of tuberculin (89 amino acids) and partly by IEDB’s apparent lower specificity than NetMHCIIpan. Across all of the tested proteins, NetMHCIIpan predicted fewer epitopes than IEDB, which is consistent with the findings in our initial proof of concept and again suggests that while IEDB is likely more sensitive, NetMHCIIpan may be more specific.
Notably, we found that certain DRB1 alleles, DRB1*08:04, DRB1*11:04, and DRB1*11:01, had a high number of predicted epitopes for each of the four autoallergens across both prediction algorithms. The overall similarity in residues at key binding pocket positions for DRB1*08:04, DRB1*11:01, and DRB1*11:04 suggests that these three alleles, overall, have very similar binding pocket structure in the antigen presenting domain. This similarity in the residue composition of major binding pockets suggests an underlying molecular characteristic of the three alleles, along with the others that were also predicted to bind a high number of predicted epitopes by one algorithm, which may explain why they were all predicted to bind a high number of epitopes.
There are currently limited data demonstrating the association between DRB1 alleles and AD. Our in silico approach in the context of previous association studies aids in providing supporting evidence for the potential significance of specific HLA subtypes in the AD disease process: in an AD disease model that includes autoreactivity as a catalyst, it is possible that there is a predisposition of certain T-cells to recognize autoallergens via preferential binding between specific HLA molecules and autoallergen peptides. This would result in an initial skin barrier dysfunction via the production of inflammatory cytokines. With or without a genetically determined barrier defect, the initial skin barrier disruption would allow immune cells to be exposed to external environmental antigens to which the patient could be further sensitized, resulting in the continued production of inflammatory cytokines and the propagation of a vicious circle.
Our previous study found a statistically significant association between the persistence of AD and residues at positions 26 and 78 of HLA-DRB1, which corresponded to alleles DRB1*07:01, DRB1*09:01, and DRB1*03:01(56). Although we did not find that DRB1*07:01 or DRB1*09:01 ranked in the top five alleles for number of predicted epitopes in two or more autoallergens, DRB1*03:01 did rank in the top five alleles on the IEDB platform for Hom s 1 and Hom s 3. In another study, DRB1*11:01, which had a high number of predicted epitopes in both prediction algorithms, was found to be associated with AD in Korean children(68).
The potential association of specific HLA alleles with a higher likelihood of autoreactivity has implications for different risk among different ethnic groups, as many of the alleles that are common in one ethnic group are less common in other ethnic groups. For example, in the National Marrow Donor Program, DRB1*08:04 is ranked 8th among African Americans (5.1%), 27th among Hispanics (1.13%), and 31st among Causcasians (0.04%); in Asian Americans, it is so rare that the prevalence is essentially 0%(43). Taking into consideration the results from both algorithms, we identify DRB1*08:04, DRB1*11:04, DRB1*08:03, DRB1*08:02, DRB1*11:01, DRB1*13:01, DRB1*15:02, and DRB1*03:01 as candidates for future study regarding their relationship to AD in the context of autoreactivity.
Among the four autoallergens, Hom s 4 and Hom s 2 had the largest number of predicted epitopes across all DRB1 alleles, perhaps suggesting that they are more likely than the other autoallergens to trigger autoreactivity. However, a limitation of the in silico epitope prediction algorithms is that it is unclear which of the predicted epitopes will actually bind in vivo if the correct immunodominant epitope is predicted at all. It is not necessarily the case that if more predicted epitopes exist for a protein, it is more likely that at least one of those predicted epitopes will bind, as only one strong binding interaction is necessary for autoreactivity to occur. This suggests the necessity of confirmatory in vitro binding assays for all of the proposed autoallergens.
Thus, for each of the autoallergens, we also identified several specific peptide sequences that appeared in a large number of predicted epitopes across the HLA-DRB1 alleles. Based on BLAST protein sequence queries(69), the identified core sequences are highly specific to their corresponding proteins (SART1, nascent polypeptide-associated complex, BCL7B, MICU1) and are unlikely to be found in other antigens. Therefore, the identified core sequences serve as candidates for further exploration with in vitro epitope binding assays. Because the additional presence of IgE antibodies to an allergen can enhance specific T-cell responses to that allergen, it may also be of interest in the future to determine if increased specific IgE binding to the proposed epitopes of interest exists in AD patients. A limitation of this study in providing guidance for specific epitopes of interest is that the epitope prediction algorithms are not perfectly sensitive. In the cases for which there are very few immunodominant epitopes, there is the possibility that the one or two clinically significant epitopes may not be predicted at all in the algorithms, as we discovered in the mugwort Art v 1 test case. Nevertheless, in cases for which associations have already been discovered that involve specific HLA alleles without a clear mechanism, these epitope discovery algorithms can be used to support genetic associations by further elucidating the underlying molecular relationship between specific HLA molecules and peptides of interest, guiding in vitro or clinical studies.
In conclusion, through use of two in silico epitope discovery algorithms, we have identified candidate HLA-DRB1 alleles that may be predisposed to autoreactivity with Hom s 1, Hom s 2, Hom s 3, and Hom s 4 in the context of AD. Additionally, we have identified specific peptide epitopes that may play a role in the pathogenesis of AD and warrant future exploration to determine their immunodominance. The proposed HLA alleles and sequences provide a focus for future translational studies exploring the relationship between AD, HLA subtype, and autoreactivity. A similar evaluation could be conducted for external antigens, such as epitopes common to peanuts, candida, Staph. aureus, and others that result in elevations of sIgE that have been associated with the onset of atopic dermatitis and atopic diatheses.
Supplementary Material
11. Acknowledgements
We thank Nandita Mitra and Ole Hoffstad for their helpful input. This work was supported by the University of Pennsylvania Center for Clinical Epidemiology and Biostatistics Summer Research Fellowship.
13. List of Abbreviations
- AD
atopic dermatitis
- IEDB
Immune Epitope Database Analysis Resource
- HA
hemagglutinin
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Competing Interests
The authors have declared no conflicts of interest.
References
- 1.Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S. NetMHCIIpan-2.0 - Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 2010;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4(7):e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul S, Kolla RV, Sidney J, Weiskopf D, Fleri W, Kim Y, et al. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clinical and Developmental Immunology. 2013;2013(ID 467852):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, et al. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008;36(Web Server issue):W513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013;65(10):711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185(2):943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, et al. Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production. J Immunol. 2012;189(2):679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramesh M, Yuenyongviwat A, Konstantinou GN, Lieberman J, Pascal M, Masilamani M, et al. Peanut T-cell epitope discovery: Ara h 1. J Allergy Clin Immunol. 2016;137(6):1764–71.e4. [DOI] [PubMed] [Google Scholar]
- 10.Schulten V, Greenbaum JA, Hauser M, McKinney DM, Sidney J, Kolla R, et al. Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci U S A. 2013;110(9):3459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tangri S, Mothe BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, et al. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174(6):3187–96. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160(7):3363–73. [PubMed] [Google Scholar]
- 14.Valenta R The future of antigen-specific immunotherapy of allergy. Nat Rev Immunol. 2002;2(6):446–53. [DOI] [PubMed] [Google Scholar]
- 15.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. [DOI] [PubMed] [Google Scholar]
- 16.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, et al. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. . Allergy. 2006;61(8):969–87. [DOI] [PubMed] [Google Scholar]
- 17.Leung DY, Bieber T. Atopic dermatitis. [Review] [100 refs]. Lancet. 2003;361(9352):151–60. [DOI] [PubMed] [Google Scholar]
- 18.Hay RJ, Johns NE, Williams HC, Bolliger IW, Dellavalle RP, Margolis DJ, et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. Journal of Investigative Dermatology. 2014;134(6):1527–34. [DOI] [PubMed] [Google Scholar]
- 19.Margolis JS, Abuabrar K, Bilker W, Hoffstad O, Margolis DJ. Persistance of mild of mild to moderate atopic dermatitis. JAMA Dermatology. 2014;150(6):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010.[Erratum appears in Lancet. 2013 Feb 23;381(9867):628 Note: AlMazroa, Mohammad A [added]; Memish, Ziad A [added]]. Lancet. 2012;380(9859):2197–223. [DOI] [PubMed] [Google Scholar]
- 21.Leung DY. Atopic dermatitis: new insights and opportunities for therapeutic intervention. J Allergy Clin Immunol. 2000;105(5):860–76. [DOI] [PubMed] [Google Scholar]
- 22.Leung DY. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol Int. 2013;62(2):151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eyerich K, Novak N. Immunology of atopic eczema: overcoming the Th1/Th2 paradigm. Allergy. 2013;68(8):974–82. [DOI] [PubMed] [Google Scholar]
- 24.Eyerich S, Onken AT, Weidinger S, Franke A, Nasorri F, Pennino D, et al. Mutual antagonism of T cells causing psoriasis and atopic eczema. N Engl J Med. 2011;365(3):231–8. [DOI] [PubMed] [Google Scholar]
- 25.Gittler JK, Shemer A, Suarez-Farinas M, Fuentes-Duculan J, Gulewicz KJ, Wang CQF, et al. Progressive activation of Th2/Th22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. Journal of Allergy & Clinical Immunology. 2012;130:1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandt EB, Sivaprasad U. Th2 cytokines and atopic dermatitis. J Clin Cell Immunol. 2011;2(3):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewe M, Walther S, Gyufko K, Czech W, Schopf E, Krutmann J. Analysis of the cytokine pattern expressed in situ in inhalant allergen patch test reactions of atopic dermatitis patients. J Invest Dermatol. 1995;105(3):407–10. [DOI] [PubMed] [Google Scholar]
- 28.Howell MD, Fairchild HR, Kim BE, Bin L, Boguniewicz M, Redzic JS, et al. Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J Invest Dermatol. 2008;128(9):2248–58. [DOI] [PubMed] [Google Scholar]
- 29.Jurgens M, Wollenberg A, Hanau D, de la Salle H, Bieber T. Activation of human epidermal Langerhans cells by engagement of the high affinity receptor for IgE, Fc epsilon RI. J Immunol. 1995;155(11):5184–9. [PubMed] [Google Scholar]
- 30.Maurer D, Ebner C, Reininger B, Fiebiger E, Kraft D, Kinet JP, et al. The high affinity IgE receptor (Fc epsilon RI) mediates IgE-dependent allergen presentation. J Immunol. 1995;154(12):6285–90. [PubMed] [Google Scholar]
- 31.Valenta R, Maurer D, Steiner R, Seiberler S, Sperr WR, Valent P, et al. Immunoglobulin E response to human proteins in atopic patients. J Invest Dermatol. 1996;107(2):203–8. [DOI] [PubMed] [Google Scholar]
- 32.Valenta R, Seiberler S, Natter S, Mahler V, Mossabeb R, Ring J, et al. Autoallergy: a pathogenetic factor in atopic dermatitis? J Allergy Clin Immunol. 2000;105(3):432–7. [DOI] [PubMed] [Google Scholar]
- 33.Natter S, Seiberler S, Hufnagl P, Binder BR, Hirschl AM, Ring J, et al. Isolation of cDNA clones coding for IgE autoantigens with serum IgE from atopic dermatitis patients. Faseb j. 1998;12(14):1559–69. [DOI] [PubMed] [Google Scholar]
- 34.Valenta R, Natter S, Seiberler S, Wichlas S, Maurer D, Hess M, et al. Molecular characterization of an autoallergen, Hom s 1, identified by serum IgE from atopic dermatitis patients. J Invest Dermatol. 1998;111(6):1178–83. [DOI] [PubMed] [Google Scholar]
- 35.Tang TS, Bieber T, Williams HC. Does “autoreactivity” play a role in atopic dermatitis? J Allergy Clin Immunol. 2012;129(5):1209–15.e2. [DOI] [PubMed] [Google Scholar]
- 36.Roesner LM, Werfel T. Autoimmunity (or Not) in Atopic Dermatitis. Front Immunol. 2019;10:2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aichberger KJ, Mittermann I, Reininger R, Seiberler S, Swoboda I, Spitzauer S, et al. Hom s 4, an IgE-reactive autoantigen belonging to a new subfamily of calcium-binding proteins, can induce Th cell type 1-mediated autoreactivity. J Immunol. 2005;175(2):1286–94. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roesner LM, Heratizadeh A, Wieschowski S, Mittermann I, Valenta R, Eiz-Vesper B, et al. alpha-NAC-Specific Autoreactive CD8+ T Cells in Atopic Dermatitis Are of an Effector Memory Type and Secrete IL-4 and IFN-gamma. J Immunol. 2016;196(8):3245–52. [DOI] [PubMed] [Google Scholar]
- 40.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57(5):304–14. [DOI] [PubMed] [Google Scholar]
- 41.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17(6):555–61. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maiers M, Gragert L, Klitz W. High resolution HLA alleles and haplotypes in the US population. Human Immunology. 2007;68:779–88. [DOI] [PubMed] [Google Scholar]
- 44.Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT, et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1–2009 and H1N1–1918 influenza A viruses. Proc Natl Acad Sci U S A. 2010;107(28):12599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jahn-Schmid B, Kelemen P, Himly M, Bohle B, Fischer G, Ferreira F, et al. The T cell response to Art v 1, the major mugwort pollen allergen, is dominated by one epitope. J Immunol. 2002;169(10):6005–11. [DOI] [PubMed] [Google Scholar]
- 46.Van Hemelen D, Mahler V, Fischer G, Fae I, Reichl-Leb V, Pickl W, et al. HLA class II peptide tetramers vs allergen-induced proliferation for identification of allergen-specific CD4 T cells. Allergy. 2015;70(1):49–58. [DOI] [PubMed] [Google Scholar]
- 47.Duvvuri VR, Duvvuri B, Jamnik V, Gubbay JB, Wu J, Wu GE. T cell memory to evolutionarily conserved and shared hemagglutinin epitopes of H1N1 viruses: a pilot scale study. BMC Infect Dis. 2013;13:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, et al. Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccin Immunother. 2013;9(7):1598–607. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, James E, Gates TJ, DeLong JH, LaFond RE, Malhotra U, et al. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol. 2013;25(8):447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Godkin A, Davenport M, Hill AV. Molecular analysis of HLA class II associations with hepatitis B virus clearance and vaccine nonresponsiveness. Hepatology. 2005;41(6):1383–90. [DOI] [PubMed] [Google Scholar]
- 51.Thio CL, Thomas DL, Karacki P, Gao X, Marti D, Kaslow RA, et al. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol. 2003;77(22):12083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menzies R, Vissandjee B, Rocher I, St Germain Y. The booster effect in two-step tuberculin testing among young adults in Montreal. Ann Intern Med. 1994;120(3):190–8. [DOI] [PubMed] [Google Scholar]
- 53.Selvaraj P, Reetha AM, Uma H, Xavier T, Janardhanam B, Prabhakar R, et al. Influence of HLA-DR and -DO phenotypes on tuberculin reactive status in pulmonary tuberculosis patients. Tuber Lung Dis. 1996;77(4):369–73. [DOI] [PubMed] [Google Scholar]
- 54.Brown GR, Hem V, Katz KS, Ovetsky M, Wallin C, Ermolaeva O, et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43(Database issue):D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Androulakis IP, Nayak NN, Ierapetritou MG, Mono DS, Floudas CA. A predictive method for th evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins: Structures, function, and Genetics. 1997;29:87–102. [PubMed] [Google Scholar]
- 56.Margolis DJ, Mitra N, Kim B, Gupta J, Hoffstad OJ, Papadopoulos M, et al. Association of HLA-DRB1 genetic variants with the persistence of atopic dermatitis. Human Immunology. 2015;76(8):571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodriguez E, Matanovic A, Marenholz I, et al. High-density genotyping study indentifies four new susceptibility loci for atopic dermatitis. Nature Genetics. 2013;45:808–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nature Genetics. 2009;41(5):596–601. [DOI] [PubMed] [Google Scholar]
- 60.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nature Genetics. 2012;44(11):1222–6. [DOI] [PubMed] [Google Scholar]
- 61.Mansur AH, Williams GA, Bishop DT, Markham AF, Lewis S, Britton J, et al. Evidence for a role of HLA DRB1 alleles in the control of IgE levels, strengthened by interacting TCR A/D marker alleles. Clin Exp Allergy. 2000;30(10):1371–8. [DOI] [PubMed] [Google Scholar]
- 62.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47(12):1449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Potaczek DP, Kabesxh M. Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy. 2012;42:852–71. [DOI] [PubMed] [Google Scholar]
- 64.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nature Genetics. 2011;43(7):690–4. [DOI] [PubMed] [Google Scholar]
- 65.Torres-Galvan MJ, Quiralte J, Blanco C, Castillo R, Carrillo T, Perez-Aciego P, et al. Pocket 4 in the HLA-DRB1 antigen-binding groove: an association with atopy. Allergy. 2000;55(4):398–401. [DOI] [PubMed] [Google Scholar]
- 66.Weidinger S, Willis-Owen SA, Kamatani Y, Baurecht H, Morar N, Liang L, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Human Molecular Genetics. 2013;22(23):4841–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leung CS. Endogenous Antigen Presentation of MHC Class II Epitopes through Non-Autophagic Pathways. Front Immunol. 2015;6:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park H, Ahn K, Park MH, Lee SI. The HLA-DRB1 polymorphism is associated with atopic dermatitis, but not egg allergy in Korean children. Allergy Asthma Immunol Res. 2012;4(3):143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST--a tool for discovery in protein databases. Trends Biochem Sci. 1998;23(11):444–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


