SUMMARY
Background
Autism typically presents with highly heterogeneous features, including frequent comorbidity with intellectual disability (ID). The overlap between these phenotypes has confounded diagnosis and discovery of genetic factors associated with autism. We analyzed pathogenicde novo genetic variants in individuals with autism who had either ID or normal cognitive function to determine whether genes associated with autism also contribute towards ID comorbidity.
Methods
We analyzed 2,290 individuals from the Simons Simplex Collection (SSC) for de novolikely-gene disruptive (LGD) variants and copy-number variants (CNVs), and determined their relevance towards IQ and Social Responsiveness Scale (SRS) measures.
Results
Individuals who carried de novo variants in a set of 173 autism-associated genes showed an average 12.8-point decrease in IQ scores(p=5.49×10−6) and 2.8-point increase in SRS scores(p=0.013) compared with individuals without such variants. Furthermore, individuals with high-functioning autism (IQ>100) had lower frequencies of de novo LGD variants (42/397 vs. 86/562, p=0.021) and CNVs (9/397 vs. 24/512, p=0.065) compared with individuals who manifested both autism and ID (IQ<70). Pathogenicvariants disrupting autism-associated genes conferred a 4.85-fold increased risk (p=0.011) for comorbid ID, while de novo variants observed in individuals with high-functioning autism disrupted genes with little functional relevance towards neurodevelopment.
Conclusions
Pathogenic de novo variants disrupting autism-associated genes contribute towards autism and ID comorbidity, while other genetic factors are likely to be causal for high-functioning autism.
Keywords: Autism, intellectual disability, comorbid features, genetic complexity
INTRODUCTION
Autism spectrum disorder, which presents in children with social communication difficulties, repetitive behavior, and restricted interests[1],is a highly heterogeneousneurodevelopmental disorder characterized by complex genetic etiology and strong comorbidity with other developmental disorders[2]. For example, approximately 30% of individuals with autism also manifestwith intellectual disability (ID)[3], defined by IQ scores <70[1]. The high degree of co-occurrence of autism with IDhas been shown to confound accurate diagnosis of autism.In fact, we recently showed that 69% of individuals diagnosed with ID are likely to be recategorized and diagnosed with autism[4]. The diagnostic overlap between autism and ID suggests that de novo gene disruptive variants and copy-number variants (CNVs) identified in individuals ascertained for autism in large-scale studies could also be confounded by ID comorbidity. Here, using genetic and phenotypic data from 2,290 individuals with autism from the Simons Simplex Collection (SSC)[5], we show that gene discoveries in autism are biased towards genes that contribute towards both autism and comorbid ID.
MATERIALS AND METHODS
We analyzed rarede novo likelygene-disruptive (LGD) variants derived from exome sequencing data[6, 7], 78disease-associated copy-number variants (CNVs)[8]derived from microarray data[9], and Full-scale IQ and Social Responsiveness Scale (SRS) T-scores for SSCprobands obtained from the Simons Foundation Autism Research Initiative[5]. As these data were de-identified, they were exempt from IRB review and conformed to the Helsinki Declaration. We identified 173 genes associated with autism (Supplementary File 1) from multiple database sources, including tier 1 genes (>2 de novo LGD variants) from the Developing Brain Disorders Gene Database [10], genes with >5 non-SSC de novo LGD variants from denovo-db[11], and SFARI Gene tiers 1 and 2 (https://gene.sfari.org/). Clinical case reports were reviewed for a subset of 22 genes that appeared in all three databases (Supplementary File 2). Expected frequencies of de novo variants were calculated from gene-specific probabilities of de novo nonsense and frameshift variants based on a sequence context-dependent model[12]. Phenotypic data for mouse knockout models were obtained from the Mouse Genome Informatics database[13]. Gene-set enrichment for specific expression in brain regions during development, based on expression data derived from the BrainSpan Atlas[14], was calculated using the Specific-Expression Analysis (SEA) online tool[15]. All statistics were calculated using R v.3.4.2 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
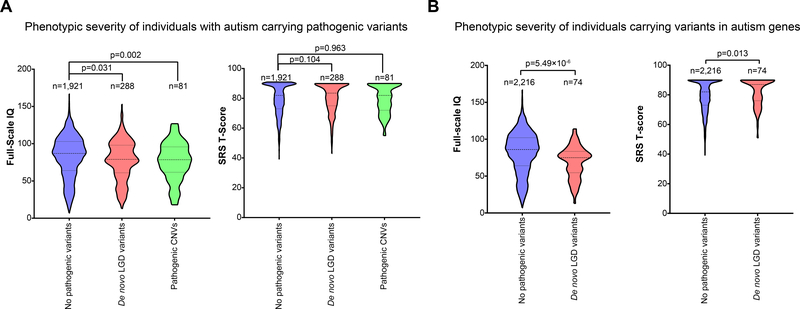
Wefirst compared the phenotypes of288individuals with de novoLGD variants and 81 individuals with pathogenic CNVs to 1,921 individuals without such variants obtained from the SSC cohort. Similar to previous autism studies thatidentified correlations between de novo variants and IQ scores[12, 16–18],we found that individuals with de novo LGD variants(average IQ=77.7, p=0.031, two-tailed Mann-Whitney test) orpathogenic CNVs (average IQ=76.3, p=0.002) hada significant decrease in IQ scores compared to individualswithoutsuchvariants (average IQ=82.3) (Figure 1A).However, nodifferencesin autism severity, measured using SRS T-scores, were observed between groups of individuals with and without pathogenicvariants(p=0.104 for de novo LGD variants and 0.963 for CNVs) (Figure 1A).This suggeststhat pathogenic variantsin general contribute to ID independent of autism severity, although this could also be due to an ascertainment bias in the SSC cohorttowards individuals with severe autism.We furtheridentifiedindividuals who carried de novoLGD variants in173autism-associated genes, defined as geneswith recurrent de novovariantsreported inmultiple databases of sequencing studies (Supplementary File 1). We found that74 individualscarrying de novoLGD variants in autism-associated genes had decreased IQ (average IQ=69.1, p=5.49×10−6, two-tailed Mann-Whitney test) and increased SRS T-scores (average SRS=82.4, p=0.013) compared with2,216 individualswithout de novo LGD variants in these genes (average IQ=81.9, average SRS=79.6), implying that candidate autism genes contribute to both autism and ID phenotypes (Figure 1B). To validate this finding, we examined 76 published case reports of affected individuals with pathogenic variants in a subset of 22autism-associatedgenes for ID comorbidity (Table 1, Supplementary File 2). For example, recent case studies have identifiedautism co-occurring with ID in 21 individuals with de novo SHANK3 variants[19], 19 individuals with NRXN1 variants[20], and 18 individuals with TCF20 variants[21]. Overall, 460/497(92.6%) individuals with autism described in these studies had ID, emphasizing that variants in these genes contribute to severe forms of autism with comorbid ID(Table 1). The remaining 37 individuals (7.4%) who manifested autism but not ID each carried variants in genes primarily contributing towards autism and comorbid ID, in particular CHD8 and NRXN1, suggesting that these genes exhibit incomplete penetrance or allelic heterogeneity towards ID phenotypes.
Figure 1.
Phenotypic comparison of individuals with autism from the SSC cohort with and without pathogenic variants.(A)Individuals with pathogenic variants (de novo LGD and CNV) had significantly lower IQ scores than individuals without pathogenic variants, but no change in autism severity (SRS T-score) was observed between the three groups. (B)Individuals with de novoLGD variants in candidate autism genes had lower IQ scores and more severe autism phenotypes (SRS T-score)than individuals without such variants. n indicates sample size, p-values were derived from two-tailed Mann-Whitney tests, and dotted lines within each violin plot indicate the median and first and third quartiles.
Table 1.
Individuals carrying variants in autism-associated genes with comorbid ID.
| Autism- associated genes | Cases with ID | Autism cases (with or without ID) | Autism cases with comorbid ID | Autism cases without ID |
|---|---|---|---|---|
| ADNP | 134/134 | 114/134 | 114/114 | 0/114 |
| ANK2 | 1/1 | 1/1 | 1/1 | 0/1 |
| ANKRD11 | 10/10 | 9/10 | 9/9 | 0/9 |
| ARID1B | 137/153 | 80/153 | 80/80 | 0/80 |
| ASH1L | 14/14 | 4/14 | 4/4 | 0/4 |
| ASXL3 | 18/19 | 16/19 | 15/16 | 1/16 |
| BCL11A | 11/16 | 4/16 | 3/4 | 1/4 |
| CHD2 | 3/3 | 3/3 | 3/3 | 0/3 |
| CHD8 | 56/75 | 61/75 | 46/61 | 15/61 |
| CUL3 | 1/1 | 1/1 | 1/1 | 0/1 |
| DDX3X | 97/97 | 33/97 | 33/33 | 0/33 |
| DYRK1A | 7/26 | 9/26 | 7/9 | 2/9 |
| KMT2A | 76/99 | 12/99 | 10/12 | 2/12 |
| MECP2 | 1/1 | 1/1 | 1/1 | 0/1 |
| MYT1L | 1/1 | 1/1 | 1/1 | 0/1 |
| NRXN1 | 45/60 | 34/60 | 23/34 | 11/34 |
| POGZ | 48/49 | 29/49 | 29/29 | 0/29 |
| SCN2A | 19/33 | 9/33 | 7/9 | 2/9 |
| SETD5 | 15/16 | 7/16 | 7/7 | 0/7 |
| SHANK3 | 35/37 | 26/37 | 24/26 | 2/26 |
| SYNGAP1 | 11/23 | 11/23 | 11/11 | 0/11 |
| TCF20 | 47/48 | 32/48 | 31/32 | 1/32 |
| Total | 787/916 (85.9%) | 497/916 (54.3%) | 460/497 (92.6%) | 37/497 (7.4%) |
We next comparedgenetic data from 397SSC individuals (17.3% of the SSC cohort) with “high-functioning autism”, defined as having severe autism (SRS>75) and average or above-average IQ scores(IQ>100), to 562 individuals (24.5% of the SSC cohort) with both autism and ID (SRS>75 and IQ<70).Individuals with high-functioning autism had a significantly lower (p=0.021,one-tailed Fisher’s Exact test) frequency ofde novo LGD variants (42/397, 10.6%) than individuals with autism and ID (86/562, 15.3%). Similarly, individuals with high-functioning autism were less likely (p=0.065) to carrypathogenic CNVs (9/397, 2.3%)than individuals with both autism and ID(24/562, 4.3%). In fact, de novo LGD variants conferred a 1.53-fold higher likelihoodofmanifesting ID among individuals with autism(p=0.035, 95% confidence interval 1.03–2.26), and pathogenic CNVs similarly conferred a 1.92-fold increased risk for co-occurrence of IDamong individuals with autism (p=0.099, 95% CI 0.88–4.18). We replicated these observations by analyzing an additional combined cohort of 2,357 individuals from both the SSC and the Autism Sequencing Collection[22]. Here, individuals with both autism and ID had a significantly higher rate (p=3.04×10−6, one-tailed Student’s t-test) of de novovariants in genes intolerant to variation, as measured by probability of Loss-of-function Intolerant (pLI) scores>0.9 (70/643, 10.8%), than individuals manifesting autism but not ID (114/1747, 6.65%). We also found that only3/397 (0.8%) individualsin the SSC cohort with high-functioning autism carriedde novoLGD variants in autism-associated genes, including ANK2, HIVEP3, and BAZ2B. This frequency was not significantly different from the expected frequencyof de novo variantsin thegeneral population (p=0.095, one-tailed Student’s T-test). In contrast, 20/562(3.6%) individuals with both autism andIDcarried de novo LGD variants in autism-associated genes, such as CHD8, SCN2A, and SYNGAP1, representing a 19.2-fold enrichment of de novo variants compared with theexpected rate in the general population (p=9.48×10−6).Thus, de novo LGD variants in autism genes conferred a 4.85-fold increased risk (p=0.011, 95% CI 1.43–16.42) towards comorbid ID in individuals with autism.
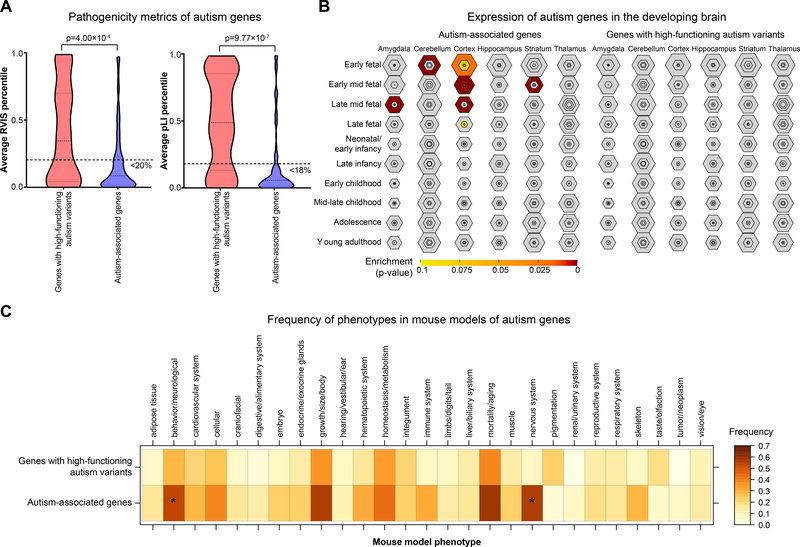
We further sought to determine the biological relevance of the 42 genes withde novoLGD variants identified in individuals with high-functioning autism, and found that these genes in aggregate had less functional relevance towards neurodevelopmentthanthe reported autism-associated genes. For example, genes with de novo LGD variants in individuals with high-functioning autism were less resistant to genetic variation than reported autism-associated genes, as measured by average Residual Variation Intolerance Score (RVIS) (average score 0.413 vs. 0.185, p=4.00×10−4, Mann-Whitney two-tailed test) and pLIpercentile (average score 0.498 vs. 0.179, p=9.77× 10−7gene metrics[23, 24] (Figure 2A). In fact, while the RVIS and pLIpercentiles of the reported autism genes were clustered below the thresholds for pathogenicity (RVIS <20th percentile and pLI<18th percentile, or raw score >0.9), genes disrupted in individuals with high-functioning autism were evenly distributed across the range of percentiles. Additionally, while autism genes wereenriched for specific expression in the cortex(p=3.13×10−4, Fisher’s Exact test with Benjamini-Hochberg correction) and cerebellum (p=0.020) during early fetal development[15], genes with de novo LGD variants in high-functioning autism individuals werenot enriched for any specific expression patterns in the developing brain(Figure 2B). Furthermore, mouse models of genes identified inindividuals with high-functioning autism were significantly less likely to manifest nervous system (12/42 genes, 28.6%, p=4.90×10−3, one-tailed Fisher’s Exact test with Benjamini-Hochberg correction) and behavioral/neurological (10/42 genes, 23.8%, p=0.037)phenotypes thanmouse models of reported autism-associated genes(behavior/neurological: 93/173 genes, 53.8%; nervous system: 96/173 genes, 55.5%) (Figure 2C). These findings suggest that genes with de novo LGD variants in individuals with high-functioning autism are less pathogenic in humans and model organisms, and therefore may not necessarily contribute towards the specific high-functioning autism phenotype.
Figure 2.
Functional analysis of genes with de novo LGD variants in individuals with high-functioning autism. (A)Genes with de novo LGD variants in individuals with high-functioning autism (SRS>75 and IQ>100) had loweraverage RVIS (left) and pLI (right) percentile scores than those for reported autism-associated genes. Thick dotted lines across the violin plots indicate thresholds for gene pathogenicity: <20th percentile for RVIS and <18th percentile for pLI(>0.9 raw score).Thin lines within the violin plots indicate the median and first and third quartiles. p-values were derived from two-tailed Mann-Whitney tests. (B) Expression of genes with de novovariants in individuals with high-functioning autism and autism-associated genes in the developing human brain. Autism-associated genes wereenriched (p<0.1, Fisher’s Exact test with Benjamini-Hochberg correction)for specific expression in the cortex and cerebellum during early development, while no enrichment was seen in the genes identifiedin individuals with high-function autism. Hexagon sizes represent the number of genes preferentially expressed in each brain tissue and timepoint, while the colors of the hexagons represent p-values for the enrichment of autism genes among each set of preferentially-expressed genes. (C) Frequency of phenotypes observed in mouse knockout models for genes with de novoLGD variants in individuals with high-functioning autism compared with reported autism-associated genes. * indicates p<0.05 (one-tailed Fisher’s Exact test with Benjamini-Hochberg correction).
DISCUSSION
Our resultssuggestthat pathogenic variants such asde novoLGD variants and CNVs contribute to autism phenotypes primarily in individuals with comorbid ID, especially if these variants disrupt a gene previously associated with autism. Several themes regarding the study of high-functioning autism have emerged from these findings. First, the consistentlyhigh degree of comorbidity between autism and ID has led to an ascertainment bias towards individuals who manifestboth disorders in large-scale sequencing cohorts, as it is difficult to exclude all individuals with comorbid disorders and still have adequate power to identify recurrent variants. Indeed, more than 80% of the SSC cohort had an IQ score less than 100, and the average IQ of the cohort (81.5) was 18.5 points below the population average.This bias has contributed towardsthe identification of genes and CNV regions related to both autism and ID, as evidenced by decreased IQ among carriers of de novo variants in these genes as well as a high incidence of comorbid phenotypes reported in published case studies. Large-scale sequencing studies still hold a high value in uncovering shared biological mechanisms that could underlie both disorders[25]. However, understanding the biological of the core autism phenotypes would require concerted efforts to recruit individuals who specifically manifest high-functioning autism without ID.
Second, individuals with high-functioning autism are less likely to carryde novo LGD variants in candidate autism genes, as each of these candidate genes were primarily associated with autism and comorbid ID. Instead,de novo variants in individuals with high-functioning autism tend to disrupt genes with less functional relevance towards neurodevelopment. These genes likely carry non-recurrent de novo LGD variants that either confer a small effect size towards autism risk on their own, or are not associated at all with neurodevelopment.We therefore propose that multiple genomic factors with varying effect sizes, such as missense variants, common variants, variants in regulatory and non-coding regions, orthe combinatorial effects of inherited variants,contribute towards autism phenotypes without ID. For example,Schaaf and colleagues performed targeted sequencing of 21 candidate autism genes in 339 individuals with high-functioning autism[26]. They found that 2% of individuals carried de novo missense variants in candidate autism genes, such as PTENand FOXP2, suggesting that allelic variants of differing severity within the same gene might contribute to distinct neurodevelopmental trajectories.Interestingly, the same study also found that 7% of individuals with high-functioning autism carried multiple inherited missense variants in candidate autism genes, potentially contributing to an oligogenic model for high-functioning autism phenotypes.Similarly, common variants have been found to contribute towards increased autism risk in individuals without ID[27, 28]. For example, Grove and colleagues recently reported that the heritability attributed to common variants, including those primarily associatedwith cognitive ability and educational attainment, was three times lower in individuals with autism and ID compared to those without ID[28]. Finally, variants that may not contribute directly towardsautism phenotypesthemselves, including de novo LGD variants observed in individuals with high-functioning autism,couldstill be responsible for subtler modification of theseverity of autism or ID phenotypes.
Overall, our results emphasize the importanceof dissecting phenotypic heterogeneity in family-based sequencing studies of complex diseases,especially those with a high frequency of comorbid disorders. While a larger cohort of individuals recruited specifically for high-functioning autism could identify associations withrecurrent genes or different types of variants, these findings should be validatedusingfunctional studies to more fully differentiate the genetic causes forhigh-functioning autism from those for autismwith comorbid ID.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank FereydounHormozdiari (UC Davis), Lucilla Pizzo (Penn State), and Vijay Kumar (Penn State) for their helpful discussions and comments on the manuscript. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet,R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind,R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C.Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K.Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J.Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). We appreciate obtaining access to phenotypic data on the Simons Foundation Autism Research Initiative (SFARI) Base. Approved researchers can obtainthe SSC data sets described in this study byapplying at https://www.base.sfari.org.
FUNDING
This work was supported by NIH R01-GM121907, SFARI Pilot Grant (#399894) and resources from the Huck Institutes of the Life Sciences to S.G., and NIH T32-GM102057 to M.J.
Footnotes
COMPETING INTERESTS
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association; 2013. [Google Scholar]
- 2.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nürnberger JI Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet 2017;18:362–76. [DOI] [PubMed] [Google Scholar]
- 3.Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, Zahorodny W, Robinson C, Rosenberg, White T, Durkin MS, Imm P, Nikolaou L, Yeargin-Allsopp M, Lee L-C, Harrington R, Lopez M, Fitzgerald RT, Hewitt A, Pettygrove S, Constantino JN, Vehorn A, Shenouda J, Hall-Lande J, Van K, Naarden Braun, Dowling NF. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ 2018;67:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak A, Kubina RM, Girirajan S. Comorbidity of intellectual disability confounds ascertainment of autism: Implications for genetic diagnosis. Am J Med Genet Part B Neuropsychiatr Genet 2015;168:600–8. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach GD, Lord C. The Simons Simplex Collection: A Resource for Identification of Autism Genetic Risk Factors. Neuron 2010;68:192–5. [DOI] [PubMed] [Google Scholar]
- 6.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE. Excess of rare, inherited truncating mutations in autism. Nat Genet 2015;47:582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM, Raskin S, Leppig K, Thiese H, Smith R, Aberg E, Niyazov DM, Escobar LF, El-Khechen D, Johnson KD, Lebel RR, Siefkas K, Ball S, Shur N, McGuire M, Brasington CK, Spence JE, Martin LS, Clericuzio C, Ballif BC, Shaffer LG, Eichler EE. Phenotypic Heterogeneity of Genomic Disorders and Rare Copy-Number Variants. N Engl J Med 2012;367:1321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW, State MW. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015;87:1215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry 2016;73:275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, Stessman HAF, Doebley AL, Bernier RA, Nickerson DA, Eichler EE. NAR Breakthrough Article denovo-db: A compendium of human de novo variants. Nucleic Acids Res 2017;45:D804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014;46:944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CL, Blake JA, Kadin JA, Richardson JE, Bult CJ. Mouse Genome Database (MGD)-2018: Knowledgebase for the laboratory mouse. Nucleic Acids Res 2018;46:D836–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, Bennet C, Bertagnolli D, Brouner K, Butler S, Caldejon S, Carey A, Cuhaciyan C, Dalley RA, Dee N, Dolbeare TA, Facer BAC, Feng D, Fliss TP, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Howard RE, Jochim JM, Kuan CL, Lau C, Lee C-K, Lee F, Lemon TA, Lesnar P, McMurray B, Mastan N, Mosqueda N, Naluai-Cecchini T, Ngo N-K, Nyhus J, Oldre A, Olson E, Parente J, Parker PD, Parry SE, Stevens A, Pletikos M, Reding M, Roll K, Sandman D, Sarreal M, Shapouri S, Shapovalova NV.,Shen EH, Sjoquist N, Slaughterbeck CR, Smith M, Sodt AJ, Williams D, Zöllei L, Fischl B, Gerstein MB, Geschwind DH, Glass IA, Hawrylycz MJ, Hevner RF, Huang H, Jones AR, Knowles JA, Levitt P, Phillips JW, Šestan N, Wohnoutka P, Dang C, Bernard A, Hohmann JG, Lein ES. Transcriptional landscape of the prenatal human brain. Nature 2014;508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 2010;38:4218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson EB, Samocha KE, Kosmicki JA, McGrath L, Neale BM, Perlis RH, Daly MJ. Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc Natl Acad Sci 2014;111:15161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuen RKC, Merico D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D’Abate L, Chan AJS, Koyanagi S, Paton T, Pereira SL, Hoang N, Engchuan W, Higginbotham EJ, Ho K, Lamoureux S, Li W, MacDonald JR, Nalpathamkalam T, Sung WWL, Tsoi FJ, Wei J, Xu L, Tasse AM, Kirby E, Van Etten W, Twigger S, Roberts W, Drmic I, Jilderda S, Modi BM, Kellam B, Szego M, Cytrynbaum C, Weksberg R, Zwaigenbaum L, Woodbury-Smith M, Brian J, Senman L, Iaboni A, Doyle-Thomas K, Thompson A, Chrysler C, Leef J, Savion-Lemieux T, Smith IM, Liu X, Nicolson R, Seifer V, Fedele A, Cook EH, Dager S, Estes A, Gallagher L, Malow BA, Parr JR, Spence SJ, Vorstman J, Frey BJ, Robinson JT, Strug LJ, Fernandez BA, Elsabbagh M, Carter MT, Hallmayer J, Knoppers BM, Anagnostou E, Szatmari P, Ring RH, Glazer D, Pletcher MT, Scherer SW. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 2017;20:602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, McCready E, Pichon O, Le Caignec C, Van Dijck A, Pope K, Voorhoeve E, Yoon J, Stankiewicz P, Cheung SW, Pazuchanics D, Huber E, Kumar V, Kember RL, Mari F, Curró A, Castiglia L, Galesi O, Avola E, Mattina T, Fichera M, Mandarà L, Vincent M, Nizon M, Mercier S, Bénéteau C, Blesson S, Martin-Coignard D, Mosca-Boidron AL, Caberg JH, Bucan M, Zeesman S, Nowaczyk MJM, Lefebvre M, Faivre L, Callier P, Skinner C, Keren B, Perrine C, Prontera P, Marle N, Renieri A, Reymond A, Kooy RF, Isidor B, Schwartz C, Romano C, Sistermans E, Amor DJ, Andrieux J, Girirajan S. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med 2019;21:816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Jia X, Wu H, Xun G, Ou J, Zhang Q, Li H, Bai T, Hu Z, Zou X, Xia K, Guo H. Genotype and phenotype correlations for SHANK3 de novo mutations in neurodevelopmental disorders. Am J Med Genet Part A 2018;176:2668–76. [DOI] [PubMed] [Google Scholar]
- 20.Al Shehhi M, Forman EB, Fitzgerald JE, McInerney V, Krawczyk J, Shen S, Betts DR, Ardle LM, Gorman KM, King MD, Green A, Gallagher L, Lynch SA. NRXN1 deletion syndrome; phenotypic and penetrance data from 34 families. Eur J Med Genet 2019;62:204–9. [DOI] [PubMed] [Google Scholar]
- 21.Torti E, Keren B, Palmer EE, Zhu Z, Afenjar A, Anderson IJ, Andrews MV.,Atkinson C, Au M, Berry SA, Bowling KM, Boyle J, Buratti J, Cathey SS, Charles P, Cogne B, Courtin T, Escobar LF, Finley SL, Graham JM, Grange DK, Heron D, Hewson S, Hiatt SM, Hibbs KA, Jayakar P, Kalsner L, Larcher L, Lesca G, Mark PR, Miller K, Nava C, Nizon M, Pai GS, Pappas J, Parsons G, Payne K, Putoux A, Rabin R, Sabatier I, Shinawi M, Shur N, Skinner SA, Valence S, Warren H, Whalen S, Crunk A, Douglas G, Monaghan KG, Person RE, Willaert R, Solomon BD, Juusola J. Variants in TCF20 in neurodevelopmental disability: description of 27 new patients and review of literature. Genet Med 2019;21:2036–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosmicki JA, Samocha KE, Howrigan DP, Sanders SJ, Slowikowski K, Lek M, Karczewski KJ, Cutler DJ, Devlin B, Roeder K, Buxbaum JD, Neale BM, MacArthur DG, Wall DP, Robinson EB, Daly MJ. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 2017;49:504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes. PLoS Genet 2013;9:e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, Karczewski KJ, Minikel EV.,Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen M, Girirajan S. Mapping a shared genetic basis for neurodevelopmental disorders. Genome Med 2017;9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A, Lewis L, Akbar H, Varghese R, Boerwinkle E, Gibbs RA, Zoghbi HY. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet 2011;20:3366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, Werge T, Hougaard DM, Taylor J, Skuse D, Devlin B, Anney R, Sanders SJ, Bishop S, Mortensen PB, Børglum AD, Smith GD, Daly MJ, Robinson EB, Bækvad-Hansen M, Dumont A, Hansen C, Hansen TF, Howrigan D, Mattheisen M, Moran J, Mors O, Nordentoft M, Nørgaard-Pedersen B, Poterba T, Poulsen J, Stevens C, Anttila V, Holmans P, Huang H, Klei L, Lee PH, Medland SE, Neale B, Weiss LA, Zwaigenbaum L, Yu TW, Wittemeyer K, Willsey AJ, Wijsman EM, Wassink TH, Waltes R, Walsh CA, Wallace S, Vorstman JAS, Vieland VJ, Vicente AM, Van Engeland H, Tsang K, Thompson AP, Szatmari P, Svantesson O, Steinberg S, Stefansson K, Stefansson H, State MW, Soorya L, Silagadze T, Scherer SW, Schellenberg GD, Sandin S, Saemundsen E, Rouleau GA, Rogé B, Roeder K, Roberts W, Reichert J, Reichenberg A, Rehnström K, Regan R, Poustka F, Poultney CS, Piven J, Pinto D, Pericak-Vance MA, Pejovic-Milovancevic M, Pedersen MG, Pedersen CB, Paterson AD, Parr JR, Pagnamenta AT, Oliveira G, Nürnberger JI, Murtha MT, Mouga S, Morrow EM, DeLuca DM, Monaco AP, Minshew N, Merikangas A, McMahon WM, McGrew SG, Martsenkovsky I, Martin DM, Mane SM, Magnusson P, Magalhaes T, Maestrini E, Lowe JK, Lord C, Levitt P, Martin CL, Ledbetter DH, Leboyer M, LeCouteur AS, Ladd-Acosta C, Kolevzon A, Klauck SM, Jacob S, Iliadou B, Hultman CM, Hertz-Picciotto I, Hendren R, Hansen CS, Haines JL, Guter SJ, Grice DE, Green JM, Green A, Goldberg AP, Gillberg C, Gilbert J, Gallagher L, Freitag CM, Fombonne E, Folstein SE, Fernandez B, Fallin MD, Ercan-Sencicek AG, Ennis S, Duque F, Duketis E, Delorme R, DeRubeis S, DeJonge M V.,Dawson G, Cuccaro ML, Correia CT, Conroy J, Conceição IC, Chiocchetti AG, Celestino-Soper PBS, Casey J, Cantor RM, Cafe C, Brennan S, Bourgeron T, Bolton PF, Bölte S, Bolshakova N, Betancur C, Bernier R, Beaudet AL, Battaglia A, Bal VH, Baird G, Bailey AJ, Bader JS, Bacchelli E, Anagnostou E, Amaral D, Almeida J, Buxbaum JD, Chakravarti A, Cook EH, Coon H, Geschwind DH, Gill M, Hakonarson H, Hallmayer J, Palotie A, Santangelo S, Sutcliffe JS, Arking DE. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 2017;49:978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, Awashti S, Belliveau R, Bettella F, Buxbaum JD, Bybjerg- Grauholm J, Bækvad-Hansen M, Cerrato F, Chambert K, Christensen JH, Churchhouse C, Dellenvall K, Demontis D, De Rubeis S, Devlin B, Djurovic S, Dumont AL, Goldstein JI, Hansen CS, Hauberg ME, Hollegaard M V., [Google Scholar]; Hope S, Howrigan DP, Huang H, Hultman CM, Klei L, Maller J, Martin J, Martin AR, Moran JL, Nyegaard M, Nærland T, Palmer DS, Palotie A, Pedersen CB, Pedersen MG, DPoterba T, Poulsen JB, Pourcain BS, Qvist P, Rehnström K, Reichenberg A, Reichert J, Robinson EB, Roeder K, Roussos P, Saemundsen E, Sandin S, Satterstrom FK, Davey Smith G, Stefansson H, Steinberg S, Stevens CR, Sullivan PF, Turley P, Walters GB, Xu X, Wray NR, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Air TM, Andlauer TFM, Bacanu SA, Beekman ATF, Bigdeli TB, Binder EB, Blackwood DHR, Bryois J, Buttenschøn HN, Cai N, Castelao E, Clarke TK, Coleman JRI, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV.,Dunn EC, Eley TC, Escott-Price V, Kiadeh FFH, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Goes FS, Gordon SD, Hall LS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga JJ, Ising M, Jansen R, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Li Y, Lind PA, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, Nivard MG, Nyholt DR, O’Reilly PF, Oskarsson H, Owen MJ, Painter JN, Peterson RE, Pettersson E, Peyrot WJ, Pistis G, Posthuma D, Quiroz JA, Rice JP, Riley BP, Rivera M, Mirza SS, Schoevers R, Schulte EC, Shen L, Shi J, Shyn SI, Sigurdsson E, Sinnamon GCB, Smit JH, Smith DJ, Streit F, Strohmaier J, Tansey KE, Teismann H, Teumer A, Thompson W, Thomson PA, Thorgeirsson TE, Traylor M, Treutlein J, Trubetskoy V, Uitterlinden AG, Umbricht D, Van der Auwera S, van Hemert AM, Viktorin A, Visscher PM, Wang Y, Webb BT, Weinsheimer SM, Wellmann J, Willemsen G, Witt SH, Wu Y, Xi HS, Yang J, Zhang F, Arolt V, Baune BT, Berger K, Boomsma DI, Cichon S, Dannlowski U, de Geus EJC, DePaulo JR, Domenici E, Domschke K, Esko T, Grabe HJ, Hamilton SP, Hayward C, Heath AC, Kendler KS, Kloiber S, Lewis G, Li QS, Lucae S, Madden PAF, Magnusson PK, Martin NG, McIntosh AM, Metspalu A, Müller-Myhsok B, Nöthen MM, O’Donovan MC, Paciga SA, Pedersen NL, Penninx BWJH, Perlis RH, Porteous DJ, Potash JB, Preisig M, Rietschel M, Schaefer C, Schulze TG, Smoller JW, Tiemeier H, Uher R, Völzke H, Weissman MM, Lewis CM, Levinson DF, Breen G, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hromatka BS, Huber KE, Kleinman A, Litterman NK, McIntyre MH, Mountain JL, Noblin ES, Northover CAM, Pitts SJ, Sathirapongsasuti JF, Sazonova OV.,Shelton JF, Shringarpure S, Tung JY, Vacic V, Wilson CH, Stefansson K, Geschwind DH, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Neale BM, Daly MJ, Børglum AD. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019;51:431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.