TO THE EDITOR
The evolutionary and functionally conserved Hippo signaling pathway is essential for the regulation of cell growth, proliferation, and organ development (Moya and Halder, 2019). Activation of the Hippo pathway leads to phosphorylation and inactivation of the transcriptional co-activator and major effector Yes-associated protein 1 (YAP1). Upon turning off the Hippo pathway, YAP is dephosphorylated which allows translocation to the nucleus to act as a coactivator with the TEAD family of transcription factors to drive the expression of genes essential for cell growth and proliferation. There are 4TEAD (TEAD1–4) genes in mammals which may have redundant or non-redundant functions depending on specific cell/tissue and developmental context (Lin et al., 2017). Double knockout of Tead1 and Tead2 in mice results in embryonic lethality by day E9.5 with growth impairments and failure to form a notochord and somites (Sawada et al., 2008). Tead1 knockout embryos survive later into development (E11–12) but result in heart defects (Chen et al., 1994). Tead4 loss in mice is also embryonic lethal due to an inability to differentiate into trophoblast cells which prevents embryo implantation (Yagi et al., 2007). Since TEADs are necessary for embryonic development, it is unclear the role of the TEAD factors in adult tissue. In skin, Yap1 overexpression in the nucleus results in epidermal hyperplasia while conditional knockout leads to hypoplasia, impaired terminal differentiation and loss of barrier function (Schlegelmilch et al., 2011, Zhang et al., 2011). These effects are mediated through Yap interactions with Tead transcription factors however it is not known which specific TEAD(s) are important for this process (Beverdam et al., 2013, Schlegelmilch et al., 2011). The YAP pathway is also necessary for basal (BCC) and squamous (SCC) cell carcinoma initiation which further illustrates the importance of understanding the specific TEADs that are necessary to mediate YAP’s impact on epidermal growth (Debaugnies et al., 2018).
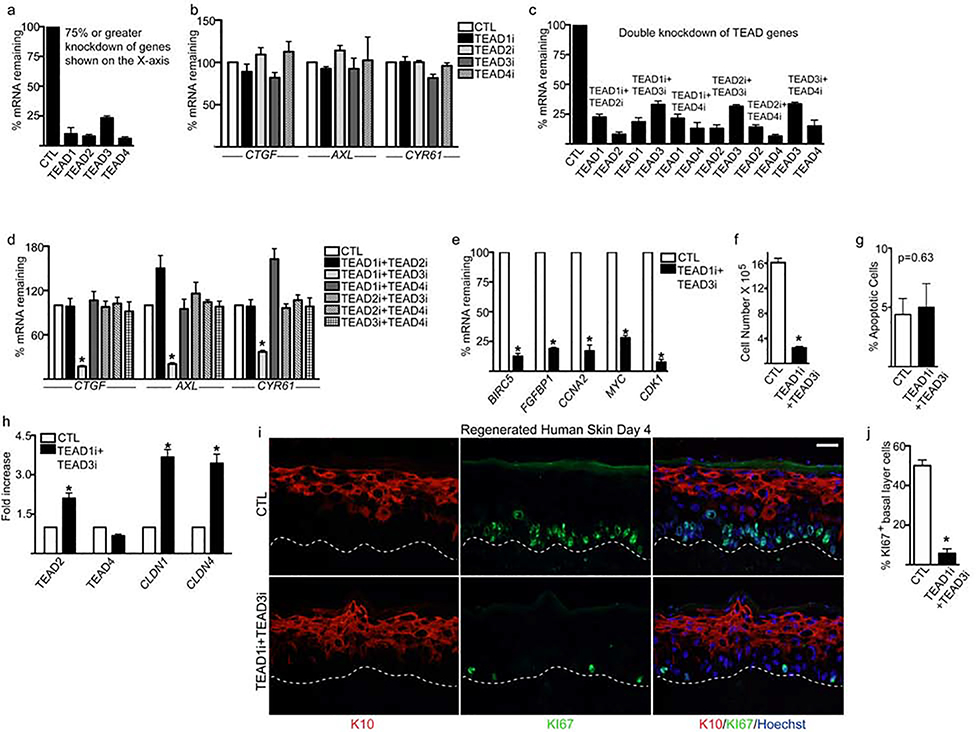
To determine the expression patterns of the TEADs in proliferating primary human keratinocytes, we used our previously generated RNA-Seq data and found that TEAD1 had the highest expression (Supplementary Figure 1a)(Li et al., 2019). RNA-Seq data from human skin (human protein atlas) showed that TEAD3 had the highest expression followed by TEAD1 (Supplementary Figure 1b). To establish the functions of TEADs in human keratinocytes, we first knocked down individual TEADs with siRNAs, and obtained ≥75% knockdown of genes as measured by RT-QPCR (Figure 1a). However, we did not observe a significant change in the mRNA levels of CTGF, AXL, or CYR61 which are the well-known YAP/TEAD downstream target genes and are factors critical for cell proliferation and survival (Figure 1b) (Zhang et al., 2011, Zhao et al., 2008). These data suggested TEADs could function redundantly and compensate for one another in keratinocytes.
Figure 1. TEAD1 and TEAD3 are necessary for epidermal growth.
(a) Primary human keratinocytes were knocked down with control (CTL) or TEAD1–4 siRNAs (TEAD1–4i) and the remaining mRNA levels were measured by RT-QPCR. (b) RT-QPCR for YAP target genes CTGF, AXL, and CYR61 in CTL and TEAD1–4i cells. QPCR results were normalized to L32 levels. (c) Combinations of TEADs were knocked down in keratinocytes and the remaining mRNA levels were measured by RT-QPCR. (d) Measurement of CTGF, AXL, and CYR61 mRNA levels in cells with different combinations of TEADs double knockdown. (e) RT-QPCR for YAP/TEAD downstream target genes as well as proliferation/growth genes in CTL and TEAD1i+TEAD3i cells. QPCR results were normalized to L32 levels. (f) CTL and TEAD1i+TEAD3i cells were seeded at 300,000 and counted 4 days later. (g) Annexin V staining was used to determine the percentage of apoptotic cells in CTL and TEAD1i+TEAD3i cells cultured for 4 days in proliferation conditions. (h) RT-QPCR for TEAD2, TEAD4 and tight junction genes in CTL and TEAD1i+TEAD3i cells. (i) Human epidermis was regenerated using three-dimensional organotypic cultures with CTLi or TEAD1i+TEAD3i cells. Expression of Keratin 10 (K10:red) and proliferation marker (Ki67:green) were characterized by immunostaining at day 4. White scale bar= 40μm. (j) Quantitation of the %KI67 positive cells in the basal layer. N=3 independent experiments performed in Figure 1. Mean values are shown with error bars=SD. *p <0.05 (t-test).
To dissect which TEADs are essential for epidermal progenitor cell function, we performed combinational TEAD knockdown and achieved efficient depletion of these genes (Figure 1c). Interestingly, we found that only double knockdown of TEAD1 and TEAD3 resulted in a significant loss of CTGF, AXL, and CYR61 mRNAs (Figure 1d). Furthermore, knockdown of TEAD1 and TEAD3 resulted in downregulation of critical self-renewal and proliferation genes such as BIRC5, FGFBP1, CCNA2, MYC, CDK1 and dramatically inhibited cell proliferation. (Figure 1e–f). There was also no significant increase in the percentage of apoptotic cells upon TEAD1/TEAD3 knockdown (Figure 1g). Depletion of TEAD1/TEAD3 did not impact TEAD4 levels but increased TEAD2 and the tight junction genes CLDN1 and CLDN4 (Figure 1h). Control and double knockdown cells were also used to regenerate human skin by seeding the cells on devitalized human dermis. This allows the cells to establish cell-cell and cell-basement membrane contact in 3 dimensions, which promotes the stratification of the tissue and the faithful representation of the gene expression program of human epidermis (Li et al., 2019). The proliferative capacity of TEAD1 and TEAD3 knockdown basal layer cells were abolished 4 days after seeding the cells onto dermis which can be seen from the dramatic decrease in KI67 staining (Figure 1i–j). Terminal differentiation proteins necessary for barrier formation such as filaggrin (FLG) and loricrin (LOR) were also lost in double knockdown tissue similar to Yap1 knockout in murine skin (Supplementary Figure 1c) (Schlegelmilch et al., 2011). 6 days after seeding the double knockdown cells onto dermis, the epidermis was extremely hypoplastic with its thickness less than half of control tissue (Supplementary Figure 1d–e). Importantly, single knockdown of TEAD1 or TEAD3 in regenerated human skin did not impact proliferation of the basal layer cells, expression of terminal differentiation genes (FLG, LOR), or affect expression of YAP target genes (CTGF, AXL, or CYR61), or (Supplementary Figure 2a–b).
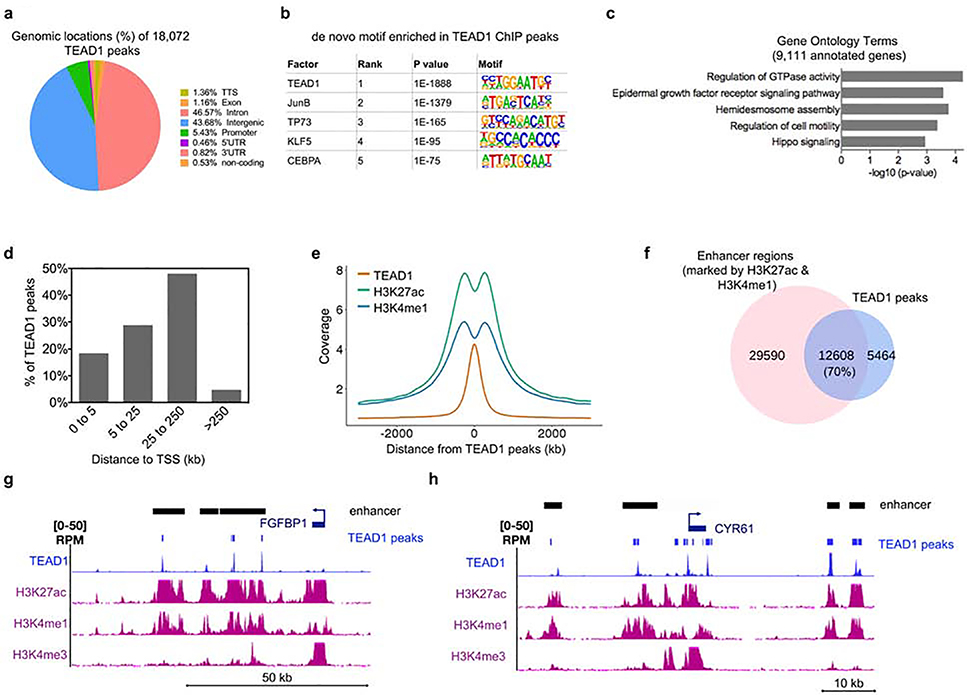
To gain insight into TEAD genomic binding profiles in primary human keratinocytes, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) with a TEAD1-specific antibody. This led to the identification of 18,072 TEAD1 binding sites (based on replicates with matching inputs with a cutoff p-value at 0.01 and IDR cutoff at 0.01) (Figure 2a and Supplementary Table 1). High reproducibility was observed between the replicate data with a Pearson correlation coefficient (PCC) of 0.97 (Supplementary Figure 2c). The vast majority of the TEAD1 peaks mapped to intergenic and intronic regions (~44% and ~47% respectively) while only ~5% were found at promoters (Figure 2a). A de novo motif search showed TEAD1 as the most significant binding motif followed by JunB, p73, KLF5, and CEBPA (Figure 2b). The 18,072 TEAD1 bound regions mapped back to 9,111 genes which were enriched for Gene Ontology terms such as epidermal growth factor receptor signaling pathway, regulation of GTPase activity, and Hippo signaling (Figure 2c and Supplementary Table 1).
Figure 2. ChIP-Seq analysis of TEAD1 genome-wide binding sites shows that TEAD1 is primarily associated with active enhancers.
(a) The 18,072 TEAD1 significantly bound peaks were mapped back to various regions of the genome. The percentage of the TEAD1 peaks found within each region of the genome is shown. (b) De novo motif search for TEAD1 bound peaks. (c) The 18,072 TEAD1 bound peaks were mapped back to 9,111 genes. Gene ontology analysis of the 9,111 annotated genes was performed using Enrichr. (d) Bar graph from GREAT analysis showing the percent distribution of TEAD1 peaks in relation to the transcription start site (TSS) of genes. Distance to TSS is shown in kilobases (kb). (e) Distribution of H3K27ac and H3K4me1 centered around TEAD1 peaks. Distance to TEAD1 peaks is shown in kb. (f) Overlap between TEAD1 peaks and active enhancer regions (marked by the presence of both H3K27ac and H3K4me1). (g-h) Gene tracks showing TEAD1 (blue), H3K27ac (purple), H3K4me1 (purple), and H3K4me3 (purple) binding. Active enhancer (shown as black bars) regions are shown as regions that have H3K27ac and H3K4me1 peaks without the presence of H3K4me3. The presence of H3K4me3 is used to rule out gene promoters and gene bodies.
Since the majority of TEAD1 peaks were found outside of promoter regions, we mapped the distance between TEAD1 bound regions to the nearest transcriptional start site (TSS) of genes. >50% of TEAD1 bound regions were ≥ 25kb away from a TSS (Figure 2d). This distal pattern of occupancy suggested that TEAD1 may exert its effects via binding enhancers. To evaluate whether these distal binding sites occur within functional regions such as enhancers, we compared TEAD1 bound sites with histone marks of active enhancers (H3K27ac and H3K4me1)(Creyghton et al., 2010, Heintzman et al., 2007). H3K27ac and H3K4me1 binding in primary human keratinoyctes derived from ENCODE data was plotted as a distance from TEAD1 peaks (Gerstein et al., 2012). We observed a bimodal distribution of H3K27ac and H3K4me1 around the TEAD1 peak summit (Figure 2e). There were 42,198 regions with both H3K27ac and H3K4me1 histone marks in keratinocytes. Intersecting these active enhancers with TEAD1 binding sites revealed that ~70% of the TEAD1 peaks overlapped with enhancer regions (Figure 2f). These results suggest that TEAD1 binds primarily at active enhancers to regulate downstream target genes. Analysis of TEAD1 binding at FGFBP1, CYR61, CTGF, and AXL showed co-localization of TEAD1 with active enhancers (H3K27ac and H3K4me1) (Figure 2g–h and Supplementary Figure 2d–e). TEAD1 and TEAD3 knockdown also resulted in the diminished expression of these direct target genes suggesting TEAD1 and TEAD3 binding are necessary for their transcription (Figure 1d–e). It is also interesting to note that for these classical YAP pathway downstream genes (CYR61, CTGF, and AXL) TEAD1 binding was also found in their promoter regions (Figure 2h and Supplementary Figure 2d–e). However, this accounts for only a small minority of cases as only ~5% of TEAD1 binding occurs at promoters.
Strong nuclear YAP expression has been observed in human BCCs and SCCs(Debaugnies et al., 2018). In human SCCs stronger nuclear YAP expression correlated with poorly differentiated SCCs(Debaugnies et al., 2018). TEAD genes have also been found to be overexpressed in a variety of cancers including gastric, prostate, and colorectal(Holden and Cunningham, 2018). However, it is currently not known whether TEAD genes are also overexpressed in skin cancer and whether specific TEADs are necessary for YAP mediated skin tumorigenesis. Our data would suggest that TEAD1 and TEAD3 may couple with YAP to promote skin cancer since these are the TEADs critical for epidermal growth and would be an exciting area of future investigation. Taken together, our findings suggest that TEAD1 and TEAD3 function redundantly to promote human epidermal growth by binding enhancers to drive the expression of proliferation related genes.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIH R01AR072590 and R01CA225463) to G.L. Sen.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
DATA AVAILABILITY
The TEAD1 ChIP-Seq data has been deposited in GEO with the following accession number: GSE138727 at the following site: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138727
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beverdam A, Claxton C, Zhang X, James G, Harvey KF, Key B. Yap controls stem/progenitor cell proliferation in the mouse postnatal epidermis. J Invest Dermatol 2013;133(6):1497–505. [DOI] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev 1994;8(19):2293–301. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010;107(50):21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaugnies M, Sanchez-Danes A, Rorive S, Raphael M, Liagre M, Parent MA, et al. YAP and TAZ are essential for basal and squamous cell carcinoma initiation. EMBO Rep 2018;19(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012;489(7414):91100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39(3):311–8. [DOI] [PubMed] [Google Scholar]
- Holden JK, Cunningham CN. Targeting the Hippo Pathway and Cancer through the TEAD Family of Transcription Factors. Cancers (Basel) 2018;10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Xu X, Jones J, Tiwari M, Ling J, et al. HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs. Nature communications 2019;10(1):4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KC, Park HW, Guan KL. Regulation of the Hippo Pathway Transcription Factor TEAD. Trends Biochem Sci 2017;42(11):862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 2019;20(4):211–26. [DOI] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol 2008;28(10):3177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 2011;144(5):782–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007;134(21):3827–36. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A 2011;108(6):2270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22(14):1962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.