Abstract
The use of immunotherapy to treat patients with myelodysplastic syndromes (MDS) shows promise but is limited by our incomplete understanding of the immunologic milieu. In solid tumors, CD141Hi conventional dendritic cells (CD141Hi cDCs) are necessary for anti-tumor immunosurveillance and the response to immunotherapy. Here, we found that CD141Hi cDCs are reduced in MDS bone marrow and based on the premise established in solid tumors, we hypothesized that reduced numbers of CD141Hi cDCs are associated with inferior overall survival in MDS patients. We found that MDS patients with reduced numbers of CD141Hi cDCs, but not other DC populations, showed reduced overall survival. To examine the basis for reduction in CD141Hi cDCs, we found fewer numbers of progenitors committed to DC differentiation in the MDS bone marrow and these progenitors expressed lower levels of interferon regulatory factor-8 (IRF8), a master regulator of CD141Hi cDC differentiation. To rescue impaired CD141Hi cDC differentiation, we used pharmacologic inhibition of lysine-specific demethylase 1A (LSD1) to promote CD141Hi cDC differentiation by MDS progenitors. These data reveal a previously unrecognized element of the MDS immunologic milieu. Epigenetic regulation of CD141Hi cDC differentiation offers an intriguing opportunity for intervention and a potential adjunct to immunotherapy for patients with MDS.
Introduction
More than 10,000 people in the US are diagnosed with myelodysplastic syndromes (MDS) each year [1]. These bone marrow failure syndromes cause defective clonal hematopoiesis resulting in anemia, thrombocytopenia, and increased infection risk. About a third of patients with MDS progress to acute myeloid leukemia (AML) [2]. For those patients with high risk disease, the hypomethylating agents (HMAs) decitabine and azacitidine are standard-of-care [3]. HMAs produce significant rates of clinical response (43–62%) but these responses do not last and failure of HMA therapy is associated with poor rates of 5-year survival (<12%) [4].
Recent studies demonstrate that HMAs have immune modulatory activities and we and others have combined HMAs with immunotherapies, such as immune checkpoint inhibitors and cellular therapies, to treat patients with MDS and other cancers [5]. In prior pre-clinical work, we found that HMAs induce expression of the tumor-associated antigen NY-ESO-1 in AML and MDS patients. This led to our recent Phase I clinical trial in which we tested the ability of a vaccine against NY-ESO-1 to induce an adaptive immune response in 9 MDS patients [6]. Our vaccine was targeted to DEC-205+ antigen-presenting cells and we found antigen-specific responses to vaccination were associated with the quantity of a specific DEC-205+ conventional dendritic cell (cDC) population marked by high-expression of CD141 (CD141Hi cDC) [6]. These data suggested a connection between CD141Hi cDCs and response to an immunotherapy in MDS.
CD141Hi cDCs are relatively a relatively rare population (between 0.05 – 0.08% of mononuclear cells in the peripheral blood, bone marrow or tonsil) but a series of studies in solid tumors have demonstrated that these cells reside at the epicenter of the immune response to cancer (reviewed in [7]). Pathological specimens from patients with different solid tumor diagnoses have shown that tumor infiltration with CD141Hi cDCs is associated with increased survival [8–10]. Moreover, the murine homolog to CD141Hi cDCs (marked by expression of CD8, CD103, or CD24) initiates robust cytotoxic T-cell responses against immunogenic tumors [8,9,11–13]. CD141Hi cDCs produce IL-12 which activates natural killer (NK) cells and suppresses tumor metastases [14]. These studies establish that CD141Hi cDCs are required for immunosurveillance. CD141Hi cDCs are also required for optimal efficacy of immune checkpoint inhibitors and adoptive T cell therapy in mouse models[15–17] and are associated with superior response to immune checkpoint inhibitor therapy in patients [10]. Importantly, increasing the number of CD141Hi cDCs in mouse models through pharmacologic approaches can improve the response to immunotherapy, suggesting that such approaches could benefit patients [16].
In MDS, the role of CD141Hi cDCs is less well defined [18]. While our initial study examined the CD141Hi cDC population in the context of response to an immunotherapy, we found an overall decrease in CD141Hi cDCs in the peripheral blood of MDS patients compared to age-matched healthy donors [6]. Based on the scientific premise established in solid tumor models that CD141Hi cDCs could impact disease progression and survival as well a response to therapy, we hypothesized that decreased quantity and/or quality of CD141Hi cDCs in MDS patients would adversely impact overall survival. We now show in a larger cohort, that MDS patients have fewer CD141Hi cDC in the bone marrow compared with age-relevant healthy donors and that this deficiency is associated with inferior overall survival. MDS patients have fewer myeloid progenitors committed to dendritic cell differentiation, specifically monocyte-DC (MDP) and common DC progenitors (CDP), which may explain reduced CD141Hi cDCs. Decreased expression of the master transcriptional regulator of CD141Hi cDCs differentiation, Interferon Regulatory Factor-8 (IRF8), in MDS MDPs is associated with decreased numbers of descendant CDPs and CD141Hi cDCs. Finally, we rescued differentiation of CD141Hi cDCs from MDS progenitors using pharmacologic inhibition of Lysine-Specific Demethylase 1A (LSD1). Together, these results suggest a paradigm in which the CD141Hi cDC population impacts survival in MDS and provide a potential therapeutic approach for restoring this population to benefit MDS patients.
Methods
Human Bone Marrow Specimens
Bone marrow (BM) cells from MDS patients (or patients with AML and MDS related changes; n = 81 individual patients) were collected prior to HMA treatment (baseline). Survival data were evaluable for 66 patients. BM cells from healthy donors (HD; n = 29 individual donors) defined as absence of hematologic malignancy) were obtained during the collection of products for use in bone marrow transplant or from patients undergoing hip-replacement surgery. For all samples, buffy coats were cryopreserved following Ficoll centrifugation (GE Healthcare, Uppsala, Sweden). Samples were collected in accordance with the Declaration of Helsinki and their use approved under Internal Review Board (IRB) approved protocols at the Roswell Park Comprehensive Cancer Center (Roswell Park) and University at Buffalo. All patients had provided written informed consent for collection of sample material and its retrospective use under IRB approved protocols at Roswell Park. Clinical characteristics are described in Supplemental Table S1. Details on drug treatments and in vitro differentiation are described in Supplemental Methods.
Murine Studies
Studies were performed under protocols approved by the Institutional Animal Care and Use Committee of Roswell Park. c-kit+ cells were isolated from bone marrow cells collected from All studies used bone marrow cells collected from female B6(Cg)-Irf8tm2.1Hm/J (Irf8-eGFP), B6(Cg)-Irf8tm1.2Hm/J (Irf8-KO) and littermate wild-type (WT) mice (range 12 – 16 weeks of age). Details on drug treatments and in vitro differentiation are described in Supplemental Methods.
Flow Cytometry
Bone marrow cells were stained as previously described with primary antibodies and secondary reagents (Supplemental Table S2) [14]. Intracellular IRF8 staining was performed as per manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). Live cells were determined using LIVE/DEAD Fixable Blue Dead Cell Stain (Thermo Fisher Scientific, Eugene, OR, USA). Cells were analyzed using an LSR II flow cytometer (BD Biosciences, San Jose, CA, USA). Gating strategies for DC and hematopoietic stem and progenitor cell populations are shown in Supplemental Figures 1A and 1B respectively. Median fluorescent intensity of the anti-IRF8 antibody signal was normalized to the average isotype signal of HD or MDS samples respectively and log2 transformed. Flow cytometry data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR, USA).
Statistical Analysis
We dichotomized CD141Hi cDC percentages at the upper tertile and estimated the survival function using the Kaplan-Meier estimator. We employed Cox’s regression model to estimate the effect across R-IPSS strata, verified the proportional hazards assumptions graphically, and tested with Grambsch-Therneau’s method [19]. Where IRF8 protein levels were measured by flow cytometry, median fluorescent intensity of the anti-IRF8 antibody signal was normalized to the average isotype signal of HD or MDS samples respectively and log2 transformed. Model-based clustering was used to identify two groups of MDS patients based on a threshold of IRF8 expression of 2.0 (calculated as described above). The choice of two groups was determined by the Bayesian Information Criterion. All other statistical analyses were performed using non-parametric Mann-Whitney tests or parametric two-sided unpaired t-tests based on the appropriate assumptions regarding distribution and variance of the data (Graph Pad Prism 7, GraphPad Software, San Diego, CA, USA).
Results
Bone marrow CD141Hi cDC numbers in patients with MDS correlate with survival
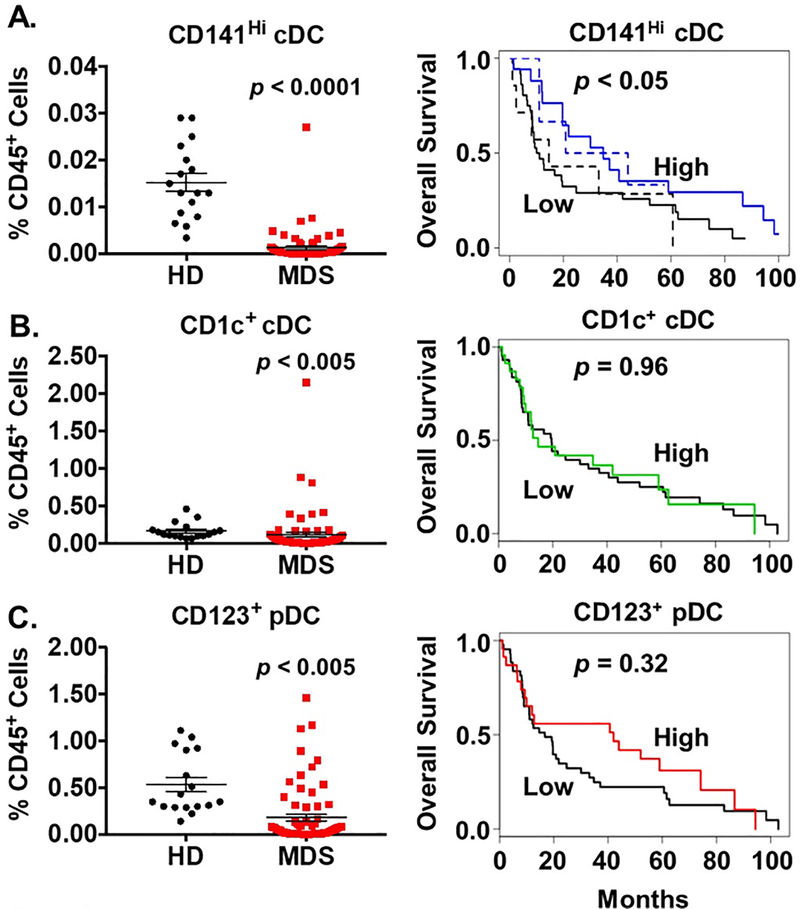
We assessed the number of different DC populations using flow cytometry (see Supplemental Figure 1A for gating strategy) in bone marrow specimens from a cohort of MDS patients across multiple risk groups as defined by the revised International Prognostic Scoring System (r-IPSS) [20]. We compared our MDS specimens to a cohort of specimens from age-relevant healthy donors (HD, median age 48 years; Supplemental Table 1). Patients with MDS were sampled prior to disease modifying therapy and had significantly fewer bone marrow CD141Hi cDCs compared to HD (Figure 1A). The number of CD141Hi cDCs was inversely correlated with total bone marrow cellularity (Supplemental Figure 2A). This suggests that the reduced quantity of CD141Hi cDCs is not solely linked to the total quantity of bone marrow cells.
Figure 1. Lower Numbers of CD141Hi Conventional Dendritic Cells are Associated with Decreased Overall Survival of MDS Patients.
(A) Left: Average frequency of CD141Hi conventional dendritic cells (cDC) in CD45+ bone marrow of healthy donors (HD, n = 17, black circles) and MDS patients (n = 71, red squares). Gating strategy is shown in Supplemental Figure 1. Right: Kaplan-Meier plot of overall survival based on numbers of CD141Hi cDCs in the entire patient cohort (solid lines) and among patients receiving an HMA (dotted lines). Patients whose follow-up data were available were stratified into upper (High, n = 23, blue lines) versus lower tertiles (Low, n = 43, black lines). (B) Left: Average frequency of CD1c+ cDCs in HD and MDS bone marrow. Right: Kaplan-Meier plot of overall survival in the entire patient cohort based on numbers of CD1c+ cDCs. (C) Left: Average frequency of plasmacytoid dendritic cells (pDC) in HD and MDS bone marrow. Right: Kaplan-Meier plot of overall survival in the entire patient cohort based on numbers of pDCs. Data are presented as values for individual patients. Horizontal bars represent the mean value and error bars represent standard error of the mean. P values were determined using the non-parametric Mann Whitney t-test or Cox’s regression model.
MDS patients with the highest number of CD141Hi cDCs (those in the upper tertile) had superior survival (controlled for risk category by r-IPSS; Figure 1A). Among MDS patients receiving an HMA, those with the highest number of CD141Hi cDCs also showed superior survival. (Figure 1A). While MDS patients also had fewer CD1c+ cDCs and CD123+ plasmacytoid DCs (pDC), no survival differences were seen in patients stratified by these dendritic cell populations (Figures 1B and 1C). Our data mirror outcome studies in patients with solid tumors and suggest that deficiency of CD141Hi cDCs may contribute to inferior outcome in MDS patients.
In melanoma, NK cells producing FLT3 ligand (FLT3L) provide support for CD141Hi cDCs in the tumor microenvironment and enhance the response to immunotherapy [10]. Since NK cells are reported to be deficient in patients with MDS, we interrogated this relationship in our population using retrospectively available data from clinical flow cytometry testing (Supplemental Figure 2B) [21]. Prior to the start of therapy, MDS patients demonstrated a positive correlation between bone marrow CD141Hi cDCs and NK cells (p < 0.05). Neither CD1c+ cDC nor CD123+ pDC numbers correlated with NK cells.
We performed RNA-sequencing on sorted MDS and HD CD141Hi and CD1c+ cDCs and analyzed gene expression signatures for two pathways fundamental to CD141Hi cDC function; the toll-like receptor 3 signaling pathway and the antigen cross-presentation pathway (Supplemental Figure 2C and Supplemental Table S3) [22,23]. CD141Hi cDCs from our tested MDS patients showed pathway gene expression signatures similar to HD CD141Hi cDCs, suggesting their competency.
Bone marrow specimens from MDS patients have fewer DC progenitors
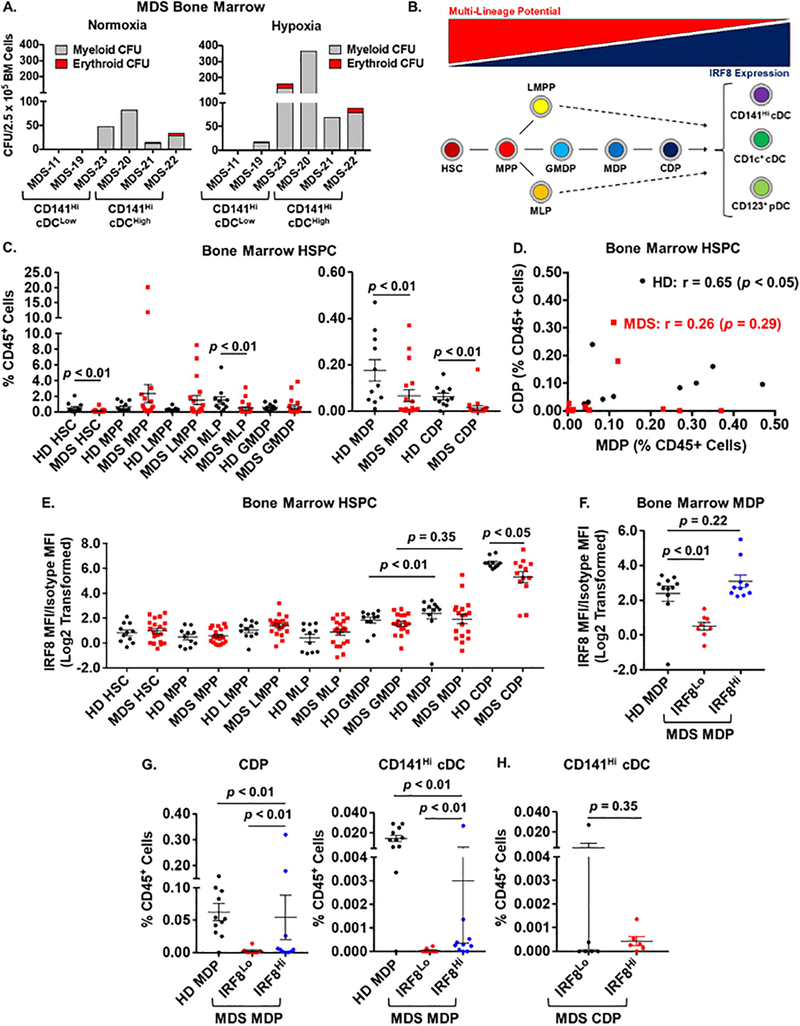
We assessed the colony-forming potential of hematopoietic stem and progenitor cells (HSPCs) in MDS specimens that had low and high numbers of CD141Hi cDC (Figure 2A). Under normoxic conditions, 4/6 samples showed growth of colonies (1/3 CD141Hi cDCLow and 3/3 CD141Hi cDCHigh). We then tested whether these specimens would respond to hypoxic conditions (1% O2) [24]. We observed that 5/6 samples showed the expected increase in colony formation compared to normoxia. These results suggest that progenitors from CD141Hi cDCLow and cDCHigh specimens retain colony-forming capacity. Using flow cytometry we then quantified specific HSPC populations, defined by immunophenotype, in bone marrow samples from our cohort of MDS patients (Figure 2B): we quantified hematopoietic stem cells (HSC), multipotent progenitors (MPP), lymphoid-myeloid primed progenitors (LMPP), multi-lymphoid progenitors (MLP), granulocyte-monocyte-DC progenitors (GMDP), monocyte-DC progenitors (MDP), and common DC progenitors (CDP) [25] (see Supplemental Figure 1B for gating strategy). We found that compared to HDs, patients with MDS have fewer HSCs, MLPs, MDPs, and CDPs in their bone marrow (Figure 2C). MLPs, MDPs, and CDPs all have the potential to differentiate into CD141Hi cDCs. We further found that there was a correlation between MDPs and CDPs in HD specimens but not in MDS specimens (Figure 2D).
Figure 2. MDS Patients Exhibit Decreased Numbers of Dendritic Cell Committed Progenitors and Decreased Expression of IRF8.
(A) Frequency of myeloid and erythroid colony-forming units (CFU) in MDS bone marrow specimens with low (CD141Hi cDCLow) or high numbers (CD141Hi cDCHigh) of CD141Hi cDCs as defined in Figure 1. For each group, n = 3 specimens from different patients. CFU assays were performed under normoxic and hypoxic (1% O2) conditions as described in Supplemental Methods. (B) Diagram depicting inverse relationship between multi-lineage potential (red triangle) and IRF8 expression (blue triangle) in specific hematopoietic stem and progenitor (HSPC) populations as defined by immunophenotype (described in the text). Intermediate stages in the differentiation of LMPPs and MLPs to DCs are shown as dotted lines. (C) Average frequency of different hematopoietic stem and progenitor (HSPC) populations in healthy donors (HD, black circles, n = 11) and MDS patients (red squares, n = 19). Monocyte-DC progenitors (MDP) and common DC progenitors (CDP) are displayed on a separate graph for clarity. Gating strategy and immunophenotypes are described in Supplemental Figure 1. p values were determined using the Mann Whitney t-test. (D) Correlations between numbers of MDPS and CDPs in HD (black circles) and MDS (red squares) bone marrow as determined using non-parametric Spearman’s rank-order correlation (r). (E) Average IRF8 expression as measured by flow cytometry in specific HD (black circles) and MDS (red squares) HSPC populations. (F) Average IRF8 expression in HD MDPs (n = 9, black circles) and in MDS MDPs separated based on IRF8 expression: IRF8Lo (< 2.0; n = 8, red circles); IRF8Hi (> 2.0; n = 10, blue circles). For (E) and (F), p values were obtained using unpaired t-test. (G) Average frequency of CDPs (Left) and CD141Hi cDCs (Right) in HD specimens and MDS specimens with IRF8Hi versus IRF8Lo MDPs. (H) Average frequency of CD141Hi cDCs in MDS specimens with IRF8Lo (n = 6) versus IRF8Hi (n = 6) CDPs. Patients were stratified based on median expression of IRF8 in MDS CDPs (IRF8Lo < 5.6, IRF8Hi > 5.6). Data are presented as values for individual patients. For (G) and (H), p values were determined using the Mann Whitney t-test. For all panels, horizontal bars represent the mean value and error bars represent standard error of the mean.
MDS DC progenitors express reduced levels of IRF8
IRF8 is a master transcription factor driving differentiation and function of DCs including CD141Hi cDCs [25–27]. We and others have shown that IRF8 expression is reduced in bulk progenitors from patients with myeloid malignancies [28–31]. We compared expression of IRF8 in HD versus MDS HSPC populations. In normal hematopoiesis, IRF8 expression is lowest in HSCs and MPPs and increases as progenitors commit to the DC lineage (Figure 2B). Compared to HD, MDS patients did not show a significant increase in IRF8 expression between the GMDP and MDP stages (Figure 2E). This result, combined with that presented in Figure 2D, indicated the need for further analysis of the MDP population. We identified two groups within the population of MDS patients based on IRF8 expression in MDPs; IRF8Hi > 2.0 and IRF8Lo < 2.0 (Figure 2F). MDS patients with IRF8Hi MDPs demonstrated IRF8 expression similar to that observed in HD MDPs, while those in the IRF8Lo group had significantly lower IRF8 expression. MDS patients with IRF8Lo MDPs produced fewer CDPs and CD141Hi cDCs compared to those with MDPs expressing higher levels of IRF8 (IRF8Hi; Figure 2G). By contrast, there was no difference in CD141Hi cDC numbers in MDS patients when stratified based on IRF8 expression in CDPs even though overall IRF8 expression was significantly lower in CDPs from patients with MDS compared to HD (Figure 2H). In addition, patients with IRF8Hi MDPs also produced fewer CD141Hi cDCs compared to WT, suggesting that additional mechanisms besides expression of IRF8 contribute to decreased production of CD141Hi cDCs in MDS patients.
Inhibition of LSD1 increases CD141Hi cDC differentiation of MDS CD34+ progenitors
These results suggest that induction of IRF8 expression might enhance DC differentiation of MDS progenitors offering a strategy to increase the number of CD141Hi cDCs in patients with MDS. LSD1 is a histone demethylase that acts primarily as a transcriptional repressor. Inhibition of LSD1 induces differentiation of myeloid leukemia cells and several clinical trials are currently underway to test the efficacy of LSD1 inhibitors in myeloid malignancy [32–35]. Pharmacologic inhibition of LSD1 induces expression of IRF8 in mouse and human leukemia cells [36–39]. We hypothesized that inhibition of LSD1 in HD and MDS CD34+ progenitors would promote CD141Hi cDC differentiation.
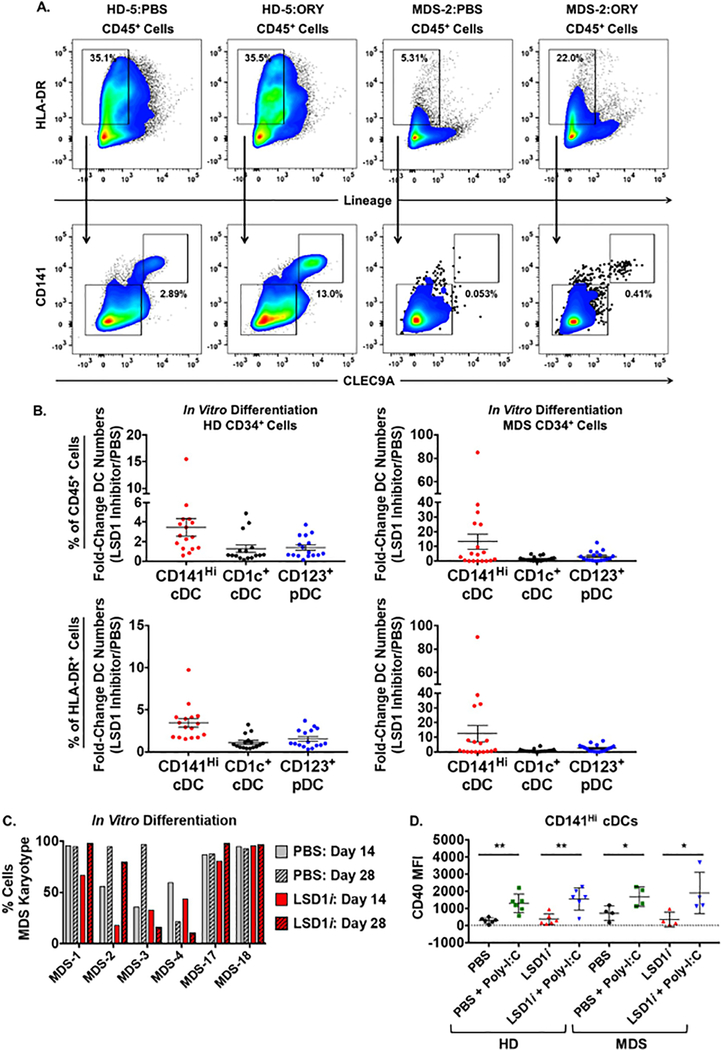
We tested this hypothesis using an in vitro model of DC differentiation and used two previously described compounds, GSK2879552 (GSK) and ORY-1001 (ORY), as pharmacologic tools to inhibit LSD1 activity [34,40–42]. Pharmacologic inhibition of LSD1 in HD CD34+ cells increased the number of terminally differentiated CD141Hi cDCs in 88% of specimens (Figures 3A and 3B, Supplemental Figure 3; n = 16 different donor specimens). This was observed using both LSD1 inhibitors (combined average 3.5-fold change compared to PBS). Similarly, LSD1 inhibition in MDS CD34+ cells increased the number of CD141Hi cDCs (n = 18 different patient specimens) in 61% of patient specimens (combined average 13.2-fold change compared to PBS). For both HD and MDS specimens, LSD1 inhibition increased the number of CD141Hi cDCs when measured as a percentage of human CD45+ or HLA-DR+ cells. Some individual specimens demonstrated expansion of multiple DC populations which may be due to LSD1 inhibition inducing a general increase in monocyte-DC lineage differentiation (Supplemental Figure 3). Since MDS is an oligo-clonal disorder and primary cultures of patient samples might contain both malignant and normal progenitor populations, we assessed the presence of MDS-defining cytogenetic abnormalities in samples before and after in vitro DC differentiation. Critically, differentiated cells exposed to LSD1 inhibitors during culture contained both normal karyotype (i.e. healthy) and cytogenetically abnormal clones (Figure 3C). We further tested whether LSD1 inhibition might affect the response of CD141Hi cDCs to stimulation. Following differentiation, we treated cultures with poly-I:C and measured expression of CD40, a molecule up-regulated during DC maturation that is critical for their activation [43]. CD141Hi cDCs differentiated in the presence of an LSD1 inhibitor responded appropriately to stimulation with poly-I:C by up-regulating CD40 suggesting that these cells can mature normally (Figure 3D). Together, these data suggest that inhibition of LSD1 can promote the differentiation of mature CD141Hi DCs from progenitors derived from both HD and patients with MDS.
Figure 3. LSD1 Inhibition Enhances CD141Hi cDC Differentiation of MDS CD34+ Progenitors.
CD34+ cells from healthy donors (HD, n = 12 different donors) and MDS patients (n = 12 different specimens) were cultured as described in the “Methods” section. Measurement of CD141Hi cDC differentiation was performed using flow cytometry at the end of the culture period (28 days). CD141Hi cDCs were defined by the immunophenotype: human CD45+, lineage-negative (lin-), HLA-DR+, CD14-, CD1c-, CD141Hi, CLEC9A+. (A) Flow cytometry analysis of Lin-, HLA-DR+ (Top) and CD141Hi, CLEC9A+ (Bottom) populations following culture of HD-5 (left) and MDS-2 (right) CD34+ cells treated with PBS or ORY. (B) Fold-change in CD141Hi cDCs (red circles), CD1c+ cDCs (black circles) and CD123+ pDCs (blue circles) numbers following in vitro differentiation of HD (left) and MDS (right) CD34+cells treated with GSK or ORY compared to PBS. Fold-change was calculated based on the percentage of CD141Hi cDCs in total human CD45+ cells (Top) or HLA-DR+ cells (Bottom). Data are presented as values for individual specimens. Specimens treated with GSK or ORY have been pooled together. Some specimens were treated with GSK and ORY (not in the same culture) and the superior effect is shown. See Supplemental Figure 3 for results of individual cultures. 3–5 technical replicates were performed for each sample and averaged together. Horizontal bars represent the mean value and error bars represent standard error of the mean. (C) Percentage of cells with an MDS-defining karyotype during in vitro differentiation of CD34+ cells (treated with PBS (gray bars) or GSK/ORY (LSD1i: red bars) (red bar). MDS karyotypes were defined as Del(−5q)/Monosomy 5 (MDS-1, MDS-2, MDS-17), Del(−7q)/Monosomy 7 (MDS-3, MDS-18) or trisomy 8 (MDS-4). Karyotypes were detected using FISH as described in “Methods” after 14 days (post-expansion) and 28 days (post-differentiation). 100 total cells were scored for all treatment and timepoints with the following exceptions: MDS-3 (LSD1i, Day 28) = 55 cells; MDS-4 (PBS, Day 14) = 48 cells; MDS-4 (LSD1i, Day 14) = 52 cells. (D) Flow cytometry analysis of CD40 expression in in vitro differentiated CD141Hi, CLEC9A+ cells following stimulation with poly-I:C. HD (n = 6, left) and MDS (n = 4, right) CD34+ cells were treated with PBS or GSK as described in the “Methods” section. Horizontal bars represent the mean value and error bars represent standard deviation. p-values were obtained using unpaired t-test: * = p < 0.05; ** = p < 0.01.
Inhibition of LSD1 promotes IRF8 expression in human progenitors
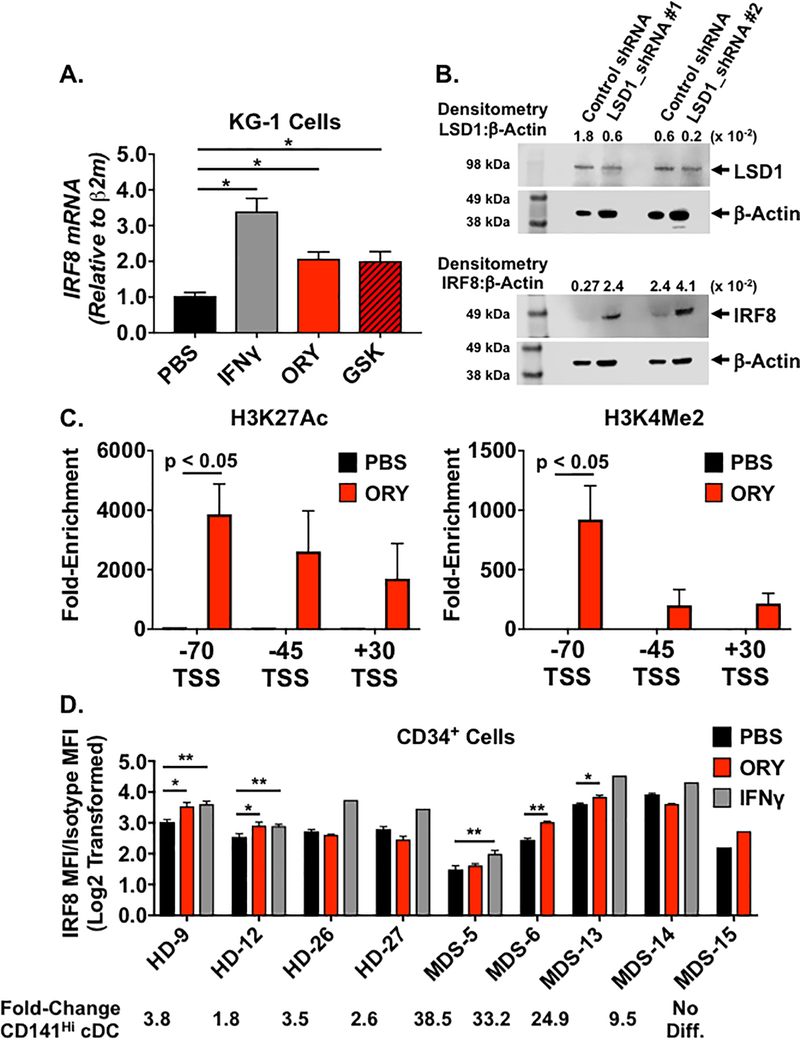
Based on previous studies demonstrating that LSD1 inhibition induced IRF8 expression, we tested whether induction of IRF8 expression in HD and MDS progenitors was associated with increased CD141Hi cDC differentiation. Treatment of KG-1 cells with either GSK or ORY increased IRF8 expression (Figure 4A and Supplemental Table S4). A similar effect was observed using shRNA to target LSD1 (Figure 4B; see Supplemental Figures 4A for uncropped images). Densitometry analysis showed a reciprocal relationship between decreased LSD1 protein and increased IRF8 protein (Figure 4B and Supplemental Figure 4B). To determine whether LSD1 inhibition directly altered epigenetic marks at the IRF8 locus, we performed chromatin immunoprecipitation studies using publicly-available data to identify regions in the IRF8 locus with high levels of LSD1 binding (Supplemental Figure 4C) [37]. This analysis revealed potential regulatory elements located −70, −45, and +30 kb relative to the transcription start site (TSS). Treatment of KG-1 cells with ORY resulted in increased H3K27 acetylation (H3K27Ac) and H3K4 dimethylation (H3K4Me2) at the −70 region, which demonstrated the highest level of LSD1 binding (Figure 4C and Supplemental Table S4).
Figure 4. LSD1 Inhibition Enhances IRF8 Expression and Promotes Histone Acetylation and Demethylation at the IRF8 Locus.
(A) Average IRF8 mRNA levels in KG-1 cells treated with the LSD1 inhibitors GSK2879552 (GSK, Left) or ORY-1001 (ORY, right). Cells were treated for 72 hours with PBS vehicle (black bar), 1 μM GSK or 0.5 μM ORY (red bar), or 50 units/ml interferon-gamma (IFNγ, gray bar), n = 3 – 5 for each condition. IRF8 mRNA was quantified using qPCR and normalized to Beta-2-microglobulin (β2M) mRNA. (B) LSD1 (Top) and IRF8 protein levels in KG-1 cells transduced with scramble control shRNA, LSD1_shRNA #1, or LSD1_shRNA #2 lentiviral vectors. Protein was measured using Western blot analysis. Separate blots for LSD1 and IRF8 were run in parallel using the same protein lysate for each condition. β-Actin protein acted as a loading control. All blot images have been cropped for clarity and the uncropped images are shown in Supplemental Figure 4. Bands corresponding to LSD1 (≈ 94 kDa), IRF8 (50 kDa), and Actin (43 kDa) are marked with black arrowheads. Densitometry values showing the ratio of LSD1:β-Actin and IRF8:β-Actin are shown. (C) Average fold-enrichment of H3K27 acetylation (H3K27Ac, Left) and H3K4 dimethylation (H3K3Me2) at regions located −70, −45, and +30 kb relative to the IRF8 transcriptional start site in KG-1 cells. H3K27Ac and H3K4Me2 levels were quantified using chromatin immunoprecipitation (ChIP) followed by quantitative PCR and were calculated by normalizing anti-H3K27Ac or antiH3K4Me2 to isotype controls. ChIP assays were performed on KG-1 cells 72 hours after treatment with PBS (black bar) or ORY (red bar). (D) Average IRF8 protein levels (calculated as described under “Methods”) in healthy donor (HD, 4 different donors) and MDS (5 different patients) CD34+ cells treated with PBS vehicle, 0.5 μM ORY, or 100 units/ml IFNγ (n = 3 replicates per individual specimen unless noted otherwise). For MDS-6 and MDS-15, cell numbers limited the treatments to PBS or ORY. Analysis of MDS-15 was limited to a single replicate. For HD-26, HD-27, MDS-13, and MDS-14, cell numbers limited the treatment with IFNγ to a single replicate. IRF8 protein was quantified using flow cytometry and normalized to isotype control. For all samples, the fold-changes in CD141Hi cDC numbers following treatment with LSD1 inhibitor compared to PBS (see Supplemental Figure 3) are depicted below the graph. Horizontal bars represent the mean value and error bars represent standard deviation. p-values were obtained using unpaired t-test: * = p < 0.05; ** = p < 0.01.
We then tested whether LSD1 inhibitors induced expression of IRF8 in primary human HD and MDS specimens. Treatment with ORY increased IRF8 expression in 50% of HD CD34+ cells (n = 4 different donors) and 60% of MDS CD34+ cells (n = 5 different patients; Figure 4D). IFNγ induced IRF8 expression in all tested samples from both HD and MDS specimens, providing a positive control for the capacity for IRF8 induction. We observed intra-specimen variation in the response to LSD1 inhibition [44]. Among both HD and MDS cohorts, there were individual specimens that did not show increased IRF8 expression following LSD1 inhibition despite the fact that these specimens exhibited a similar increase in CD141Hi cDCs (e.g. MDS-5 and MDS-6). MDS-15 showed increased IRF8 expression but did not exhibit differentiation of CD141Hi cDCs. Together, these data suggest that LSD1 inhibition can increase expression of IRF8 in primary MDS specimens, but in some cases, this may not be sufficient to induce CD141Hi cDC differentiation [45].
Enhanced CD141Hi cDC differentiation by LSD1 inhibition is dependent on IRF8
We tested whether the effect of LSD1 inhibition on cDC differentiation was dependent on IRF8. The role of IRF8 in cDC differentiation is conserved between human and mouse and IRF8 expression is necessary for differentiation of CD24+ cDCs, the mouse homolog of CD141Hi cDCs [26,27]. To test our hypothesis, we compared the effect of LSD1 inhibition on bone marrow c-kit+ cells (analogous to human CD34+ cells) from Irf8 knock-out mice (Irf8-KO) and littermate controls (WT). We established an in vitro model of mouse DC differentiation in which we used low-dose Flt3L in order to produce a ratio of conventional DC subsets similar to that observed in humans (Supplemental Figure 5).
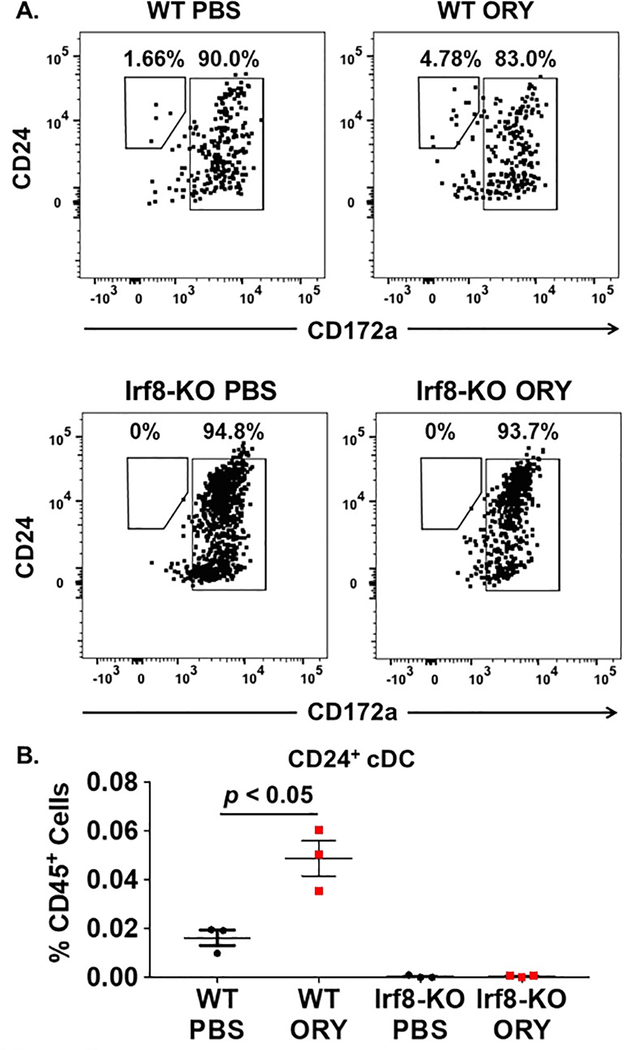
We cultured bone marrow c-kit+ cells from WT and Irf8-KO mice and treated these cells with vehicle or ORY. As expected, treatment of WT c-kit+ cells with ORY resulted in an increased number of CD24+ cDCs following our in vitro differentiation experiment (Figures 5A and 5B). By contrast, c-kit+ Irf8-KO cells treated with ORY failed to differentiate into CD24+ cDCs. These data demonstrate that LSD1 inhibition drives CD24+ cDC differentiation through IRF8 in normal c-kit+ cells.
Figure 5. LSD1 inhibition requires IRF8 for effect on CD24+ cDC differentiation.
C-kit+ cells were isolated from mouse bone marrow and cultured on MS-5 stroma cells for 14 days in media supplemented with 10 ng/ml Flt3 ligand as described in the “Methods” section. Cultures were treated with PBS vehicle or ORY (0.1 μM) for the 1st 7 days of the culture period. Measurement of CD24+ cDC differentiation was performed using flow cytometry at the end of the culture period. CD24+ cDCs were defined by the immunophenotype mouse CD45+, lineage-negative (lin-), MHC-II+, CD11c+, CD24+, CD172-. (A) Flow cytometry analysis of CD24+ cDC populations following culture of wild-type (WT; top) or Irf8−/− (Irf8-KO; bottom) c-kit+ cells treated with PBS vehicle (left panels) or ORY (right panels). (B) Numbers of CD24+ cDCs following differentiation of normal WT or Irf8-KO c-kit+ cells treated with PBS or ORY. 3–5 technical replicates were performed for each sample and averaged together. Horizontal bars represent the mean value and error bars represent standard deviation. p-values were obtained using unpaired t-test.
Discussion
Immune dysregulation in MDS is proposed to play a causal role in development and progression of disease. Excessive stimulation of innate immune signaling pathways impairs hematopoietic differentiation of MDS progenitors [46]. Our work and that of others suggests that the immune dysregulation in MDS is not limited to pro-inflammatory states. MDS patients with high risk disease have more immune suppressive regulatory T cells and myeloid-derived suppressor cells [47,48]. Decreased number and function of NK cells is also a hallmark of MDS [21]. These studies establish the scientific premise for immune dysregulation in MDS.
We know less about how defects in immune activating populations impact MDS progression and development [18]. We hypothesize a role for CD141Hi cDCs based on the established function of CD141Hi cDCs in suppressing disease progression in solid tumor models. Saft, et al., reported that patients with MDS have decreased numbers of cDCs in the bone marrow [49]. Advances in the field allowed us to build upon this work and quantify specific in situ DC populations in patients with MDS across a spectrum of conventional risk categories. We previously reported a small cohort of MDS patients with deficient peripheral blood CD141Hi cDCs [6]. We now propose the importance of the CD141Hi cDC population for patients with MDS based on our observations that decreased bone marrow CD141Hi cDCs are associated with decreased survival, even when controlled for risk category. In addition, we have found that rationally designed epigenetic therapy can improve differentiation of CD141Hi cDCs from MDS progenitors.
As reported in solid tumors, our data show that decreased numbers of CD141Hi cDCs are associated with reduced overall survival in MDS [8,10]. This survival impact is independent of r-IPSS risk group, suggesting that the immunologic status of our patients may contribute to disease progression. Preliminary data show that following an in vitro differentiation program, CD141Hi cDCs induce expression of CD40 following stimulation, suggesting, but not proving that the function of these cells may be intact. In our Phase I trial in patients with MDS, we found that patients with the highest number of CD141Hi cDCs showed the most robust humoral and adaptive immune response to vaccination against the NY-ESO-1 antigen [6]. These data, combined with the analysis of gene expression signatures associated with TLR3 signaling and antigen presentation, suggest that given the proper stimulus, CD141Hi cDCs from patients with MDS can effectively activate immune responses in MDS patients. Further studies are required to definitively demonstrate a causal role for CD141Hi cDCs in activation of immune responses to regulate disease progression and response to therapy in MDS [50].
Our data indicating decreased numbers of DC progenitors in the bone marrow from patients with MDS suggest one potential mechanisms for the decreased numbers of CD141Hi cDCs in these patients. MDS patients with higher expression of IRF8 at the MDP progenitor stage exhibited greater numbers of descendant CDPs and CD141Hi cDCs compared to patients with lower levels of IRF8, suggesting that IRF8 expression may be one factor that regulates CD141Hi numbers in MDS. Among HD and MDS specimens that exhibited similar expression of IRF8 at the MDP stage, CD141Hi cDC numbers in HD specimens were increased compared to MDS. This observation, combined with our finding that CD141Hi cDC differentiation is not necessarily linked to IRF8 expression following LSD1 inhibition in vitro, suggests that additional intrinsic and extrinsic mechanisms regulate CD141Hi cDC numbers in MDS [27,51]. Our observation of a positive correlation between NK cells and CD141Hi cDCs in MDS suggests the possibility that the decreased numbers of CD141Hi cDCs and their association with inferior survival may be due to interactions between multiple cell populations [10].
We found that pharmacologic inhibition of LSD1 enhanced CD141Hi cDC differentiation from HD and MDS progenitors. These data support prior work demonstrating that LSD1 inhibition drives differentiation programs in myeloid leukemia cells [33,34,37–39,44]. We confirm prior reports that LSD1 inhibition enhances IRF8 expression by modulating local chromatin structure but it is possible that LSD1 inhibition may also interact with IRF8 at other loci to regulate the chromatin structure during DC lineage differentiation [37]. Using a mouse genetic knockout model of normal hematopoiesis, we show that IRF8 expression is necessary for LSD1 inhibition to promote CD24+ cDC differentiation. We found that LSD1 inhibition had variable effects on IRF8 expression and CD141Hi cDC differentiation in primary MDS specimens. Recently, Duy, et al., also showed variable responses in long-term cultures of primary AML specimens exposed to an LSD1 inhibitor. In that study, 80% of AML specimens showed a response compared to 61% of MDS specimens reported here. These investigators also showed that TET2 mutations were associated with the greatest response to the combination of LSD1 inhibition and 5-azacitidine, suggesting a hypothesis that the response to LSD1 inhibition is partly regulated by the MDS mutational landscape.
Immunotherapy for cancer has shown promise, but thus far limited success in myeloid cancer [52]. Our prior experience suggests that understanding of the immunologic milieu in MDS is required to maximize such responses [6]. CD141Hi cDCs are recognized to regulate the efficacy of immunotherapies such as immune checkpoint inhibitors and adoptive T cell transfer in solid tumors [10,15–17]. Approaches to increase CD141Hi cDCs in patients with MDS, such as LSD1 inhibitors, might therefore be hypothesized to enhance response to immune therapies and even HMAs. Pre-clinically, LSD1 inhibition has already been demonstrated to improve the efficacy of anti-PD-1 therapy [53]. Further elucidation of the mechanisms governing development and function of CD141Hi cDCs in MDS patients is essential to translate these studies to the clinical benefit of our patients.
Supplementary Material
Acknowledgements
First, we thank our patients and their families. We acknowledge the contributions of the Hematologic Procurement Resource at Roswell Park: Laurie Ann Ford, Tara Cronin, Linda G. Lutgen-Dunckley, Brandon L. Martens and Joseph R. Moberg. We thank Philip L. McCarthy, George L. Chen, Maureen Ross, Barbara J. Bambach, Stephen Schinnagel, and Mary Bayers-Thering for sourcing de-identified healthy donor specimens. We thank our research coordinators Krista Belko and Justin Kocent. We thank Renae Holtz for assistance with shRNA studies and Scott Portwood and Eunice S. Wang for assistance with hypoxia studies. We thank Kelvin Lee for KG-1 cells. We acknowledge Tim Somervaille for helpful discussions. We thank David Eifrig and Charles Flippen for editorial assistance. This work was funded by the Roswell Park Alliance Foundation (EAG and MJN), the Rapaport Foundation (EAG and MJN), NIH grant 5T32 CA085183-17 (ST), and NIH grant R01 CA172105 (SIA). This work was supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Flow and Image Cytometry, Bioinformatics, Biostatistics, Laboratory Animal, and Genomics Shared Resources.
Conflict-of-Interest Statement
Elizabeth A. Griffiths: Advisory Board/Honoraria: Celgene (Relevant), Boston Scientific, Persimmune, New Link Genetics, Astex/Otsuka (Relevant), Partner Therapeutics, Inc., Alexion Pharmaceuticals, Abbvie, Novartis. Research Funding/Clinical Trials: Astex Pharmaceuticals (clinical trial PI), Celgene (clinical trial PI, research funding), Genentech (research funding), Appelis pharmaceuticals (clinical trial PI)
Michael J. Nemeth: Genentech (research funding)
References
- 1.Ma X, Does M, Raza A, Mayne ST. Myelodysplastic syndromes: incidence and survival in the United States. Cancer. 2007;109:1536–42. [DOI] [PubMed] [Google Scholar]
- 2.Nachtkamp K, Stark R, Strupp C, Kündgen A, Giagounidis A, Aul C, et al. Causes of death in 2877 patients with myelodysplastic syndromes. Ann Hematol. 2016;95:937–44. [DOI] [PubMed] [Google Scholar]
- 3.Nieto M, Demolis P, Béhanzin E, Moreau A, Hudson I, Flores B, et al. The European Medicines Agency Review of Decitabine (Dacogen) for the Treatment of Adult Patients With Acute Myeloid Leukemia: Summary of the Scientific Assessment of the Committee for Medicinal Products for Human Use. Oncologist. 2016;21:692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jabbour E, Garcia-Manero G, Batty N, Shan J, O’Brien S, Cortes J, et al. Outcome of patients with myelodysplastic syndrome after failure of decitabine therapy. Cancer. 2010;116:3830–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daver N, Garcia-Manero G, Basu S, Boddu PC, Alfayez M, Cortes JE, et al. Efficacy, Safety, and Biomarkers of Response to Azacitidine and Nivolumab in Relapsed/Refractory Acute Myeloid Leukemia: A Nonrandomized, Open-Label, Phase II Study. Cancer Discov. 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths EA, Srivastava P, Matsuzaki J, Brumberger Z, Wang ES, Kocent J, et al. NY-ESO-1 Vaccination in Combination with Decitabine Induces Antigen-Specific T-lymphocyte Responses in Patients with Myelodysplastic Syndrome. Clin Cancer Res. 2018;24:1019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2019;144:646–18. [DOI] [PubMed] [Google Scholar]
- 8.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, et al. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell. 2016;30:324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat Med. 2018;24:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CMT, Pryer N, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittal D, Vijayan D, Putz EM, Aguilera AR, Markey KA, Straube J, et al. Interleukin-12 from CD103+ Batf3-Dependent Dendritic Cells Required for NK-Cell Suppression of Metastasis. Cancer Immunol Res. 2017;5:1098–108. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez-Paulete AR, Cueto FJ, Martínez-López M, Labiano S, Morales-Kastresana A, Rodríguez-Ruiz ME, et al. Cancer Immunotherapy with Immunomodulatory Anti-CD137 and Anti-PD-1 Monoclonal Antibodies Requires BATF3-Dependent Dendritic Cells. Cancer Discov. 2016;6:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell. 2017;31:711–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerkhoff N, Bontkes HJ, Westers TM, de Gruijl TD, Kordasti S, van de Loosdrecht AA. Dendritic cells in myelodysplastic syndromes: from pathogenesis to immunotherapy. Immunotherapy. 2013;5:621–37. [DOI] [PubMed] [Google Scholar]
- 19.Grambsch PM, Therneau TM. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika. 1994;81:515–26. [Google Scholar]
- 20.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Solé F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epling-Burnette PK, Bai F, Painter JS, Rollison DE, Salih HR, Krusch M, et al. Reduced natural killer (NK) function associated with high-risk myelodysplastic syndrome (MDS) and reduced expression of activating NK receptors. Blood. 2007;109:4816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hémont C, Neel A, Heslan M, Braudeau C, Josien R. Human blood mDC subsets exhibit distinct TLR repertoire and responsiveness. J Leukoc Biol. 2013;93:599–609. [DOI] [PubMed] [Google Scholar]
- 23.Chiang M-C, Tullett KM, Lee YS, Idris A, Ding Y, McDonald KJ, et al. Differential uptake and cross-presentation of soluble and necrotic cell antigen by human DC subsets. Eur J Immunol. 2016;46:329–39. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JE, Conlon JP, Yang X, Sanchez PV, Carroll M. Enhanced growth of myelodysplastic colonies in hypoxic conditions. Exp Hematol. 2007;35:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Zhou YJ, Ma W, Zhang W, Aljoufi A, Luh T, et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol. 2017;18:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M, et al. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity. 2016;45:626–40. [DOI] [PubMed] [Google Scholar]
- 27.Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring JF, et al. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood. 1998;91:22–9. [PubMed] [Google Scholar]
- 29.Waight JD, Banik D, Griffiths EA, Nemeth MJ, Abrams SI. Regulation of the interferon regulatory factor-8 (IRF-8) tumor suppressor gene by the signal transducer and activator of transcription 5 (STAT5) transcription factor in chronic myeloid leukemia. J Biol Chem. 2014;289:15642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, da Silva Ferreira M, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21:1172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaillard C, Surianarayanan S, Bentley T, Warr MR, Fitch B, Geng H, et al. Identification of IRF8 as a potent tumor suppressor in murine acute promyelocytic leukemia. Blood Adv. 2018;2:2462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–87. [DOI] [PubMed] [Google Scholar]
- 34.Maes T, Mascaró C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, et al. ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell. 2018;33:495–511. [DOI] [PubMed] [Google Scholar]
- 35.Cusan M, Cai SF, Mohammad HP, Krivtsov A, Chramiec A, Loizou E, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPα-dependent enhancers in AML. Blood. 2018;131:1730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Reports. 2018;22:3641–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barth J, Abou-El-Ardat K, Dalic D, Kurrle N, Maier A-M, Mohr S, et al. LSD1 inhibition by tranylcypromine derivatives interferes with GFI1-mediated repression of PU.1 target genes and induces differentiation in AML. Leukemia. 2019;33:1411–26. [DOI] [PubMed] [Google Scholar]
- 39.Bell CC, Fennell KA, Chan Y-C, Rambow F, Yeung MM, Vassiliadis D, et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat Comm. 2019;10:2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Breton G, Aljoufi A, Zhou YJ, Puhr S, Nussenzweig MC, et al. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J Immunol Methods. 2015;425:21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouault-Pierre K, Mian SA, Goulard M, Abarrategi A, Di Tulio A, Smith AE, et al. Preclinical modeling of myelodysplastic syndromes. Leukemia. 2017;31:2702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell. 2015;28:57–69. [DOI] [PubMed] [Google Scholar]
- 43.Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duy C, Teater M, Garrett-Bakelman FE, Lee TC, Meydan C, Glass JL, et al. Rational Targeting of Cooperating Layers of the Epigenome Yields Enhanced Therapeutic Efficacy against AML. Cancer Discov. 2019;9:872–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurotaki D, Kawase W, Sasaki H, Nakabayashi J, Nishiyama A, Morse HC, et al. Epigenetic control of early dendritic cell lineage specification by the transcription factor IRF8 in mice. Blood. 2019;133:1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganan-Gomez I, Wei Y, Starczynowski DT, Colla S, Yang H, Cabrero-Calvo M, et al. Deregulation of innate immune and inflammatory signaling in MDS. Leukemia. 2015; 29:1458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kordasti SY, Ingram W, Hayden J, Darling D, Barber L, Afzali B, et al. CD4+CD25high Foxp3+ regulatory T cells in myelodysplastic syndrome (MDS). Blood. 2007;110:847–50. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Eksioglu EA, Zhou J, Zhang L, Djeu J, Fortenbery N, et al. Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest. 2013;123:4595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saft L, Björklund E, Berg E, Hellström-Lindberg E, Porwit A. Bone marrow dendritic cells are reduced in patients with high-risk myelodysplastic syndromes. Leuk Res. 2013;37:266–73. [DOI] [PubMed] [Google Scholar]
- 50.Kline DE, MacNabb BW, Chen X, Chan W-C, Fosco D, Kline J. CD8α+ Dendritic Cells Dictate Leukemia-Specific CD8+ T Cell Fates. J Immunol. 2018; 201:3759–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott CL, Soen B, Martens L, Skrypek N, Saelens W, Taminau J, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med. 2016;213:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Bewersdorf JP, Stahl M, Zeidan AM. Immunotherapy in acute myeloid leukemia and myelodysplastic syndromes: The dawn of a new era? Blood Rev. 2019;34:67–83. [DOI] [PubMed] [Google Scholar]
- 53.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell. 2018;174:549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.