Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoma, and front line therapies have not improved overall outcomes since the advent of immunochemotherapy. By pairing DNA and gene expression data with clinical response data, we identified a high-risk subset of non-GCB DLBCL patients characterized by genomic alterations and expression signatures capable of sustaining an inflammatory environment. These mutational alterations (PIM1, SPEN, and MYD88 [L265P]) and expression signatures (NF-κB, IRF4, and JAK-STAT engagement) were associated with proliferative signaling and were found to be enriched in patients treated with RCHOP that experienced unfavorable outcomes. However, patients with these high-risk mutations had more favorable outcomes when the immunomodulatory agent lenalidomide was added to RCHOP (R2CHOP). We are the first to report the genomic validation of a high-risk phenotype with a preferential response towards R2CHOP therapy in non-GCB DLBCL patients. These conclusions could be translated to a clinical setting to identify the approximately 38% of non-GCB patients that could be considered high-risk and would benefit from alternative therapies to standard RCHOP based on personalized genomic data.
Introduction
As the most common form of aggressive non-Hodgkin lymphoma (NHL) afflicting nearly 30,000 patients in the USA each year, diffuse large B-cell lymphoma (DLBCL) represents a significant challenge in hematology/oncology1, 2. New DLBCL treatments remain a clinical need despite the success of rituximab combined with CHOP chemotherapy (cyclophosphamide, doxorubicin, vincristine, and predisone), RCHOP, which results in durable responses in 60-70% of patients3. Those refractory to, or who relapse following, first-line therapy have a very poor outcome, with only 20% surviving beyond 5 years despite second-line therapies4-6. Rationally-targeted frontline strategies are needed, especially for those with high-risk disease or early clinical failure7. Herein, we present the case for precision targeting of a genetically-distinct population of RCHOP-insensitive DLBCL patients with targeted therapy.
Since 2002, DLBCL has traditionally been divided into two cell of origin (COO) subcategories based on tumor gene expression profiles (GEP): Activated B-cell (ABC) and Germinal Center B-cell (GCB). Patients with ABC tumors are characterized by a more aggressive profile and active NF-κB and BCR signaling pathways8, 9 while GCB cases are associated with alterations that drive aberrant chromatin-modification, PI3K signaling, and the upregulation of MYC and BCL2 through translocations or copy number gains10, 11. Recently, using tumor samples from patients treated with RCHOP, new classification models have focused on DNA alterations, including previously-identified drivers of aggressive disease such as MYD88 (L265P), CARD11, and TNFAIP312-19. MYD88 L265 mutations have specifically been shown to be enriched in aggressive ABC DLBCL cases16, 20, 21. Key downstream effects of oncogenic MYD88 include activation of the NF-κB, JAK/STAT, and upregulation of inflammatory cytokines, often augmented by the loss of the inhibitory TNFAIP3 gene19. The NOTCH signaling pathway has also been characterized in aggressive cases of DLBCL22. To date, none of these studies have impacted the design of phase III trials for untreated DLBCL patients; rather, these trials have focused on selecting and randomizing patients based on the International Prognostic Index (IPI) and tumor GEP. However, this strategy has failed to show benefit of adding novel agents to RCHOP based on these classifiers, and higher intensity therapies such as DA-EPOCH, ibrutinib, idelalisib, obinutuzumab, and bortezomib have all failed to improve outcomes in clinical trials21, 23-27. The field is now focused on designing trials based on personal tumor signatures12, 17, 18. Building on new genetic profiling studies to personalize clinical treatment could allow clinicians to add targeted therapies to the RCHOP backbone based on individual tumor signatures. Wilson et. al. showcased this methodology of precision medicine by identifying that the driver mutations of a BCR-driven, non-GCB tumor (CD79A, CD79B) can be successfully treated with ibrutinib as a single agent and RCHOP + Ibrutinib (in patients younger than 60) when they would have likely failed standard RCHOP treatment21, 27, 28.
Aiolos/Ikaros-degrading immunomodulatory drugs (IMiDs®) such as thalidomide, lenalidomide, and pomalidomide play a pivotal role in the treatment of multiple myeloma29-31. More recently, a role for lenalidomide has been reported in phase II studies for the treatment of aggressive DLBCL32-34. The results of two clinical trials comparing RCHOP combined with lenalidomide (R2CHOP) versus RCHOP alone were recently reported35, 36. The phase III ROBUST trial was restricted to ABC-type DLBCL and failed to show significant clinical benefit of R2CHOP, but the phase II ECOG-ACRIN1412 trial was open to patients of all COO subtypes and showed significantly superior event free and overall survival benefits for those treated with R2CHOP. Early research highlighted the ability of lenalidomide to exploit synthetic lethality in ABC cell lines by deregulating oncogenic programs37. Lenalidomide binds Cereblon resulting in rapid degradation in transcriptional repressors Aiolos and Ikaros which leads to upregulation of interferon-stimulated genes such as IRF7 and apoptosis in B-cells, particularly those associated with the ABC-DLBCL subtype38. In T-cells, the result is activation and IL-2 production39. These immunomodulatory effects made lenalidomide a prime candidate for treating aggressive cases of DLBCL, especially those with an inflammatory microenvironment. Herein, we report the profile of a high-risk ABC/non-GCB subset of DLBCL driven by genomic alterations in inflammatory genes that are susceptible to front-line treatment R2CHOP but continue to experience poor outcome with RCHOP alone33. These results showcase the success of personalized RCHOP + X therapy application based on the genetic signature and biological profile of patient tumors.
Materials and Methods
Study Population
A total of 196 patients with DLBCL were studied. Forty-seven patients with Ann Arbor stages II-IV were treated with R2CHOP from an investigator-initiated, open-label, single-arm phase II study ( NCT00670358) were included in the study33, 40. 149 newly diagnosed DLBCL cases treated with RCHOP, or R-immunochemotherapy (herein called RCHOP), and followed prospectively through the Molecular Epidemiology Resource (MER) of the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence (SPORE) were used as a matched contemporary cohort. Full details of this prospective cohort study of lymphoma outcomes have been previously published41. All patients provided written consent at enrollment into the clinical trial or MER for use of their clinical samples. Disease progression, relapse, unplanned re-treatment after initial immunochemotherapy, and death from any cause were verified through medical record review. Cell of origin (COO) was determined in the available R2CHOP samples by the Lymph2Cx assay (nanoString, N=45)42. For the MER RCHOP cases, COO was determined by GEP (N=36), nanoString (N=68), or Hans (N=35)43. For this study, DLBCL were categorized into GCB and non-GCB (ABC and Unclassified) groups. Baseline clinical characteristics of all patients in this study are shown in Table 1 and detailed clinical information for each cohort is provided in Supplemental Table 1.
Table 1.
DLBCL Patient Characteristics by Treatment
| RCHOP (N=149) | R2CHOP (N=47) | P-Value | |
|---|---|---|---|
| Age median (range) | 64 (26-89) | 61 (19-87) | 0.15* |
| IQR | 56-72 | 56-71 | |
| Age ≥ 60 | 96 (64.4%) | 27 (57.4%) | 0.39† |
| Male | 91 (61.1%) | 29 (61.7%) | 0.99† |
| PS 2+ | 21 (14.1%) | 5 (10.6%) | 0.63† |
| Ann arbor stage III-IV | 93 (62.4%) | 42 (89.4) | <0.0001† |
| 2+ extranodal sites | 29 (19.5%) | 14 (29.8%) | 0.16† |
| LDH > ULN | 76 (58.0%) # | 29 (61.7%) | 0.73† |
| IPI | |||
| 0 - 1 | 46 (30.9%) | 16 (34.0%) | 0.52‡ |
| 2 | 45 (30.2%) | 12 (25.5%) | |
| 3 | 43 (28.9%) | 11 (23.4%) | |
| 4 - 5 | 15 (10.1%) | 8 (17.0%) | |
| COO | |||
| ABC | 50 (33.6%) | 14 (29.8%) | 0.86‡ |
| GCB | 78 (52.3%) | 27 (57.4%) | |
| Unclassified | 11 (7.4%) | 4 (8.5%) | |
| N-miss | 10 (6.7%) | 2 (4.3%) | |
| EFS24 Achieved | 100 (67.1%) | 34 (78.7%) | 0.15† |
Abbreviations: R2CHOP, lenalidomide added to RCHOP; IQR, interquartile range; PS, performance score; IPI, international prognostic index; COO, cell of origin; EFS24, event-free survival over 24 months
Unpaired t test
Fischer’s exact test
Chi square test
18 cases without data
Whole Exome Sequencing
For the identification of DNA alterations in the RCHOP and R2CHOP cases, whole exome sequencing (WES) was performed. RCHOP-treated cases (N=149) were sequenced as previously described19. R2CHOP-treated cases (N=47) were sequenced as follows: DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) DLBCL tumors using the QIAamp DNA FFPE Tissue Kit (Qiagen GmbH, Hilden, Germany) in the Mayo Biospecimens Accessioning and Processing Core. Prior to isolation, tumor blocks were reviewed by a Mayo Clinic hematopathologist, tumor areas were circled, and four 1 mm cores were used for DNA isolation. The minimum tumor purity for study was 30%. WES was performed at the Mayo Clinic Genome Analysis Core. Sequencing was carried out on an Illumina HiSeq 2000 at a depth of ~100 million 100 bp paired-end reads per sample. Data from all cases were mapped to human genome reference build 38 using BWA-MEM. Quality control was performed by FASTQC (v0.11.3). After realignment and recalibration by GATK (v3.4-46), SNV and INDELs from individual germline and tumor samples were called by GATK haplotype caller (v3.4-46). Variants were annotated using the Mayo Clinic in-house annotation tool BioR. Sample inclusion required a minimum of 20x coverage. Mutation data was visualized as a waterfall plot using the “complex heatmaps” R program tool44. Copy number alterations were determined and annotated for available cases as performed in Wenzl et. al. but not included in the overall analysis as no significant differences were seen between the RCHOP and R2CHOP samples. All WES data is available upon request.
Gene Expression Analysis
RNA from non-GCB RCHOP (N=104) and R2CHOP (N=42) treated tumors were assayed with the nCounter® PanCancer Pathways Panel with 730 cancer pathways genes (nanoString, Seattle WA), and analyzed with GenePattern tools from the Broad institute, including GENE-E, Comparative Marker Analysis, and Gene Neighbor analysis45, 46. GEP was normalized using standardized NanoString protocols (Positive Control Normalization and CodeSet Content Normalization). RNA from non-GCB RCHOP (N=45) and R2CHOP (N=14) treated tumors was analyzed in a second analysis.
Statistical Analysis
Graphpad Prism software and GenePattern tools were used to plot and format figures, analyze data, and calculate statistical significance46. Comparison of quantitative data between groups was done by Student’s t-test or one-way ANOVA test. Gene ontology analyses and protein-protein interactomes were generated using ToppGene Suite tools47. EFS was defined as time from diagnosis to progression or relapse, unplanned re-treatment after initial immunochemotherapy, or death from any cause. The primary clinical outcome metric used was event-free survival at 24 months (EFS24). Since most relapses in DLBCL patients occur in the first 24 months, EFS24 is an early predictor of long-term outcome7. RCHOP and R2CHOP populations were divided on the basis of EFS24 status to calculate favorability enrichment percentages for DNA alterations and T statistic metrics for gene expression among populations. Specifically, favorability enrichment percentages for DNA alterations were calculated as follows: (number of EFS24 achieved patients with a mutation/total number of EFS24 achieved patients) – (number of EFS24 failed patients with a mutation/total number of EFS24 failed patients. A positive value indicates mutation is enriched in EFS24 achieved patients and a negative value indicates mutation is enriched in EFS24 failed patients. EFS between groups were compared using the Kaplan-Meier method and the log-rank test. Multiple clinical factors were analyzed by a Chi-square test in Graphpad Prism. All reported P values were two-sided. P values less than 0.05 were considered statistically significant.
Results
Specific DNA Mutations Predict EFS24 and R2CHOP Response in DLBCL
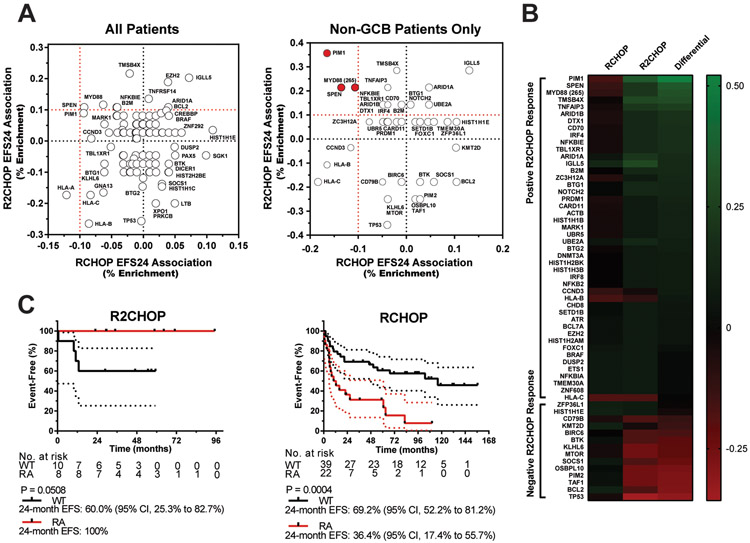
The high prevalence of MYD88 mutations combined with the promising clinical results of R2CHOP in ABC DLBCL suggest that there are underlying biologic and genomic differences that may correlate with tumor-specific responses18, 33. To identify a genomic signature for high-risk DLBCL that was lenalidomide-responsive, we first used WES data to determine the mutation profile of both R2CHOP and RCHOP cases and then looked for enrichment of variants in the R2CHOP responsive tumors. 47 patients with DLBCL that met eligibility criteria were enrolled in the R2CHOP cohort and were evaluated against 149 RCHOP comparison patients (Table 1). The outcome data are summarized for each treatment cohort in Table 2. The treatment cohorts did not significantly differ on the basis of either overall EFS or OS (overall survival) (Supplemental Figure 1). IPI and COO were significantly associated with outcome for the RCHOP population, but not for the R2CHOP population (Supplemental Figure 2). Mutations in known driver genes in both the R2CHOP and RCHOP cohorts, based on designation by one of the major DLBCL analyses12, 17, 18 are documented in Supplemental Figure 3 (N=211). We first identified genes that were mutated in both cohorts (N=186) and then calculated the presence of the alteration in populations that achieved or failed EFS247. Taking the difference between these values revealed whether R2CHOP treatment resulted in an increased association of a gene towards achieving EFS24. These results were plotted against each other in an XY scatter plot for all patients (Figure 1A; left panel). This analysis in all GCB and non-GCB patients together did not reveal any genes that met the criteria for R2CHOP outcome improvement (10% enrichment in RCHOP EFS24 failure and 10% R2CHOP achievement), although SPEN, PIM, and MYD88 trended towards association. In a secondary analysis of non-GCB cases only, mutations in genes (N=60) were explored due to the specific benefits of R2CHOP seen in these patients (Figure 1A; right panel) (Supplemental Table 2)7. Three genes were enriched in the RCHOP cases that failed EFS24 but achieved EFS24 with R2CHOP: PIM1, SPEN, and MYD88 (L265P) (Supplemental Figure 4; Table 3). PIM1 and MYD88 mutations have previously been observed to occur together, but SPEN mutations were observed to be almost entirely independent from both48. A separate analysis of GCB DLBCL was also performed and no genes were associated with and EFS24 R2CHOP response. A heatmap showing differential enrichment of genes associated with non-GCB R2CHOP EFS24 response is shown in Figure 1B. Together, PIM1, SPEN, or MYD88 (L265P) mutations were present in 38.0% of all non-GCB cases (N=30/79) and are herein referred to as R2CHOP EFS24 responder alterations (RA).
Table 2.
Treatment Summary by COO
| RCHOP GCB | RCHOP non-GCB | R2CHOP GCB | R2CHOP non-GCB | |
|---|---|---|---|---|
| N | 78 | 61 | 27 | 18 |
| EFS24 | 74.4% | 57.4% | 81.5% | 77.8% |
| OS24 | 82.1% | 72.1% | 96.3% | 88.9% |
|
EFS HR 95%CI |
Reference | 1.67* 1.03 to 2.72 |
Reference | 0.709 0.237 to 2.12 |
|
EFS OS 95%CI |
Reference | 2.18** 1.27 to 3.74 |
Reference | 0.738 0.152 to 3.58 |
Abbreviations: EFS, event-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval
Figure 1: Identification of DNA Alterations that Predict High-risk non-GCB DLBCL that Respond to R2CHOP.
(A) Association of individual genes with EFS24 response to RCHOP (x-axis) and R2CHOP (y-axis) are compared on an XY scatter plot. The data from both GCB and non-GCB patients are plotted in the left panel; data from non-GCB patients only are plotted in the right panel. (B) Heatmap showing differential enrichment of alterations in non-GCB patients who fail EFS24 with RCHOP but achieve with R2CHOP. Scale shown represents percentage enriched in EFS24 population. Genes with 10% or lesser favorability in RCHOP and 10% or greater favorability in R2CHOP were designated as EFS24 responder alterations (RA). (C) Kaplan-Meier curves for event free survival of R2CHOP (N=18) and RCHOP (N=61) treated cases. 95% CI ranges are shown as dotted lines.
Table 3.
EFS24 Responder Alteration Details
| RCHOP Achieved |
RCHOP Failed |
R2CHOP Achieved |
R2CHOP Failed |
Proportional Difference |
Pathway | |
|---|---|---|---|---|---|---|
| PIM1 | 5/35 | 8/26 | 5/14 | 0/4 | 0.522 | Cell Survival/Proliferation, Somatic Hypermutation |
| SPEN | 2/35 | 5/26 | 3/14 | 0/4 | 0.349 | NOTCH |
| MYD88 (L265P) | 3/35 | 5/26 | 3/14 | 0/4 | 0.321 | NFKB |
Results are reported in decimal porportions on the basis of EFS24
We next analyzed the overall EFS of R2CHOP and RCHOP treated non-GCB cases with or without RA. In the R2CHOP cases, patients with a RA had a better overall EFS (P = 0.051) compared to wild type patients (no RA) (Figure 1C, left panel). In contrast, RCHOP treated non-GCB patients with a RA had a significantly worse overall EFS (P = 0.0004) compared to patients without a RA (Figure 1C right panel). Patients incurring a single RA were not significantly different than those incurring multiple RA, suggesting that a mutation in any one of the RA genes may predict response to R2CHOP (Supplemental Figure 5). In a secondary analysis, R2CHOP and RCHOP samples were clustered based on criteria from Chapuy et. al (Supplemental Table 2). While the sample size was small, these data suggest that the C1 and C5 subtypes were responsive to R2CHOP (Supplemental Figure 6). This is most likely due to the fact that C1 is enriched for SPEN mutations and C5 is enriched for MYD88 and PIM1 mutations12, 18.
Gene Ontology Analysis Reveals Unique Signature of R2CHOP Response Program
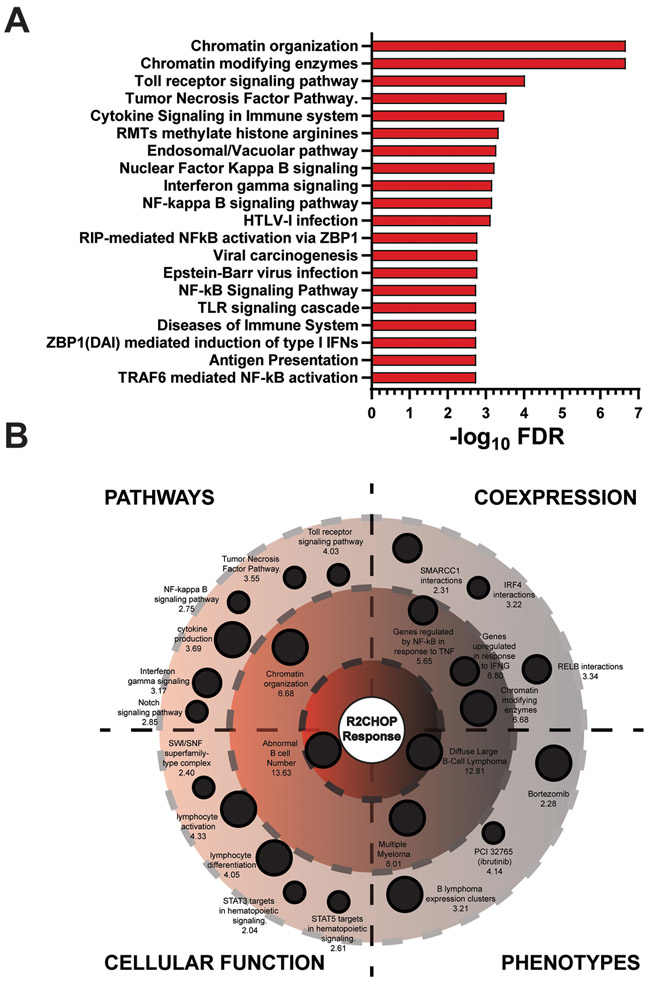
To explore the genetic programs and pathways that were susceptible to R2CHOP, gene ontology analysis was performed. Of the 60 driver genes with mutations in both RCHOP and R2CHOP non-GCB patient cohorts, 46 were more associated with achieving EFS24 in patients treated with R2CHOP. Gene ontology analysis revealed the top 20 cellular pathways associated with these genes (Figure 2A)47. Specific genetic programs associated with the highlighted pathways are shown in Figure 2B. The cases achieving EFS24 were enriched for mutations in genes associated with chromatin modification, cytokine production, IRF4, TLR signaling, IFNG signaling, and the NOTCH and NF-κB pathways (Supplemental Table 3). These results provide insights towards high-risk pathways activated in DLBCL that are vulnerable to the addition of IMiDs.
Figure 2: Pathway Enrichment of DLBCL Cases that Respond to R2CHOP.
(A) The top 20 pathway enrichments for genes associated with an improved R2CHOP EFS24 response are represented in a bar graph through −log10 false discovery rate (FDR) significance values. (B) An orbital diagram is divided into four quadrants representing ontology characteristics of R2CHOP favorable gene sets. Ontologies are labelled with their title and their −log10 FDR value. More significant FDR values approach the center of the orbital. The following thresholds designate −log10 FDR orbitals: outer orbit = 1.33 to 5, intermediate orbit = 5 to 10, and inner orbit = 10+. Size of the ontology is determined by a normalized threshold of relative genes in the hit list compared to the total number of input genes.
Genetic Expression Programs with Specific Favorable Response to R2CHOP Identified
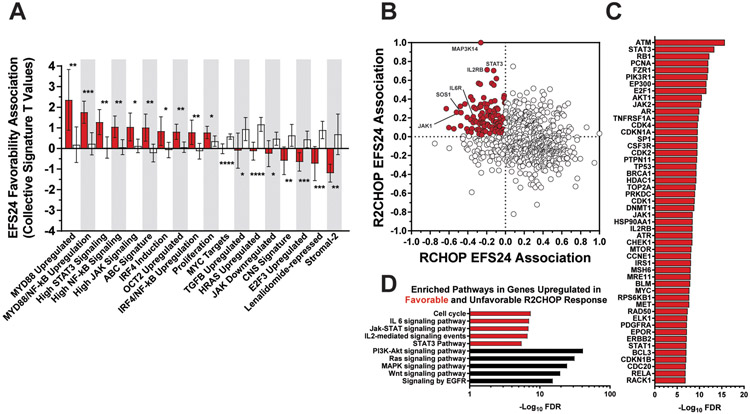
We next analyzed gene expression data from the PanCan panel of 730 B-cell-related genes to determine gene expression profiles characteristic of the high-risk/R2CHOP-profile. Gene expression data was available on 59 non-GCB DLBCL cases (45 RCHOP; 14 R2CHOP). A two-sided comparative marker analysis T statistic test was applied to assess what genes displayed differential expression based on achieving EFS24 in both populations (Supplemental Table 4)45. Each gene was assigned a T statistic value based on this analysis. Positive values indicated greater RNA expression in the patient population that achieved EFS24, and negative values indicated greater RNA expression in the patient population that failed EFS24. The collective R2CHOP and RCHOP EFS24 T values were significantly different for 18 previously-defined gene expression signatures (Figure 3A). Non-GCB patients treated with R2CHOP had superior EFS24 when expressing genes associated with high MYD88, NF-κB, STAT3, JAK, IRF4, and OCT2 induction. The ABC and proliferation signatures were also associated with achieving EFS24 when treated with R2CHOP instead of RCHOP. IRF4 dysregulation has been previously highlighted as a mechanism of lenalidomide response37. In contrast, cases that were associated with EFS24 failure when treated with R2CHOP expressed genes associated with the Stromal-2, lenalidomide-repressed, CNS, HRAS, TGFB, E2F3, JAK-downregulated, and MYC upregulation signatures.
Figure 3: RNA Expression Analysis Reveals R2CHOP Response Pathways.
(A) Mean values of signature gene EFS24 association T statistics for each treatment are plotted as bar graphs for R2CHOP (red) and RCHOP (white) favorability. Positive T values represent greater gene expression in patients that achieved EFS24. Negative T values represent greater gene expression in patients that failed EFS24. Error bars represent the 95% confidence interval. Difference in means was observed with a student’s t test with significance achieved at α = 0.05. (B) Individual genes are plotted in an XY scatter plot based on their EFS24 T value associations with RCHOP and R2CHOP. T statistics have been normalized to facilitate presentation. R2CHOP responder expressers are highlighted in red (N=113). (C) Top interacts of the 113 responder expressers are displayed by their significance. (D) Pathway enrichment analyses were performed for the top and bottom 5% of genes that displayed a shift in expression from poor EFS24 RCHOP cases to favorable EFS24 R2CHOP cases and vice versa. Ontologies composed of genes enriched for R2CHOP success (red) and failure (black) are documented on the basis of −log10 FDR value.
To visualize gene expression relationships, normalized T statistics for R2CHOP and RCHOP were plotted against one another in an XY scatter plot. The difference between the two was used to assess whether a gene was more associated with achieving EFS24 if treated with R2CHOP. In total, 113 genes were identified; the top three were MAP3K14, IL2RB, and STAT3 on the basis of T statistic differential (Figure 3B). Other notable genes include JAK1, SOS1, and IL6R. The 113 gene set was next used to assess gene-gene interaction enrichment. The top 50 partners of this network are shown in Figure 3C. The top and bottom 5% (37/730) of genes were isolated for gene ontology analysis. The R2CHOP cases that achieved EFS24 were enriched for genes involved in cell cycle, IL6, JAK-STAT, IL2, and STAT3 pathways (Figure 3D). Of note, patients who failed R2CHOP were enriched for genes involved in PI3K-AKT, RAS, MAPK, WNT, and EGFR signaling, pathways recently highlighted as mechanisms of lenalidomide resistance in multiple myeloma31. IL2RB and STAT3 transcript levels were significantly negatively correlated with the RAS signaling components RASGRP2 and HRAS (Supplemental Figure 7). Analysis of tumor microenvironment components through xCell RNA analysis revealed that a low CD8+ T-cell signature was associated with R2CHOP patients that failed EFS24 (Supplemental Figure 8). This aligns with lenalidomide’s mechanism of action involving T cell activation resulting in tumor cell cytotoxicity.
Gene Expression Neighbor Analysis of R2CHOP Responders Reveal Mechanisms of Lenalidomide
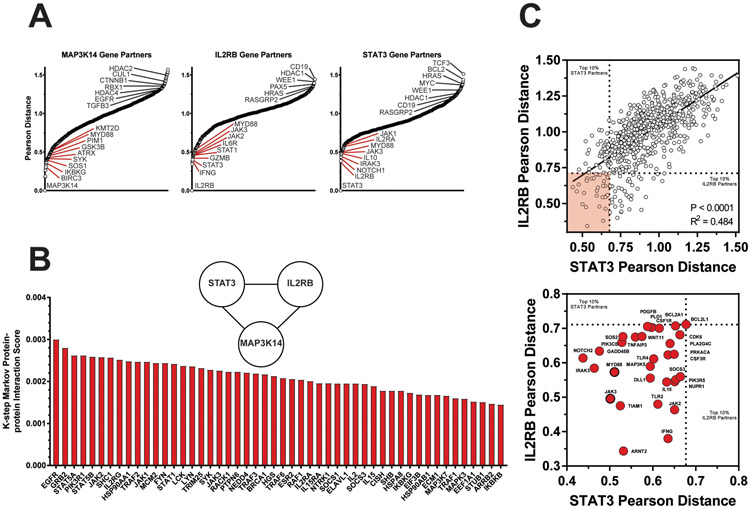
From the 113 genes that predicted differential EFS24 response between RCHOP and R2CHOP, 3 genes displayed the greatest associations with R2CHOP EFS24 achievement and RCHOP EFS24 failure: MAP3K14, IL2RB, and STAT3. To identify genes that were closely associated with these targets, Genetic Pearson Distance was calculated. Genes with a smaller (closer) distance to a target gene most closely match the expression patterns of that gene. Genetic distance was calculated and plotted against all 730 genes for MAP3K14, IL2RB, and STAT3 (Figure 4A). The MAP3K14 plot highlights its key role as a nonconical NF-κB activator, sharing matching expression profiles with genes such as SOS1, GSK3B, PIM1, and MYD88. Next, a protein-protein interaction network was generated using ToppGene suite tools47. The top 50/359 protein-protein interaction partners between MAP3K14, IL2RB, and STAT3 based on K-step Markov prioritization are displayed (Figure 4B). However, the most specific correlative relationship emerged between IL2RB and STAT3. These two genes are so closely linked that IL2RB is the top expression neighbor to STAT3 (Figure 4C, left panel). This close relationship highlights several gene expression neighbors that are shared between IL2RB and STAT3. Genes within the top 10% of genetic distance of both are highlighted (N = 30) in the zoomed figure (Figure 4C, right panel). The MYD88 and JAK3 genes were also within the top 10% of MAP3K14 genetic distance. Other notable genes that shared expression patterns with both IL2RB and STAT3 included NOTCH2, IRAK3, JAK2, and IFNG.
Figure 4: Nearest Neighbor Analysis of Top Responder Genes Reflect NF-κB, JAK/STAT, and Cytokine Signaling Programs.
(A) Plots illustrate ranked Pearson distance between responder genes and the other 729 PanCan genes. For each plot, genes to the left of the X-axis and designated in red represent genes with similar expression profiles as the target gene. Those with greater Pearson distance are plotted to the right and exhibit dissimilar expression patterns to the target gene. (B) A bar graph highlights the top 50 protein-protein interaction partners between MAP3K14, IL2RB, and STAT3 based on k-step Markov distance. (C) A scatter plot documents the correlative genetic partners of IL2RB and STAT3 for all genes. Dotted lines designate limits for top 10% closest genes to each. 23/730 genes meet these criteria and are shaded in red. A zoomed view of these gene neighbors is displayed in the bottom plot. MYD88 and JAK3 are also top 10% gene partners with MAP3K14 and highlighted with a dark border.
Responder Alterations are Associated with Specific Patterns of Gene Expression
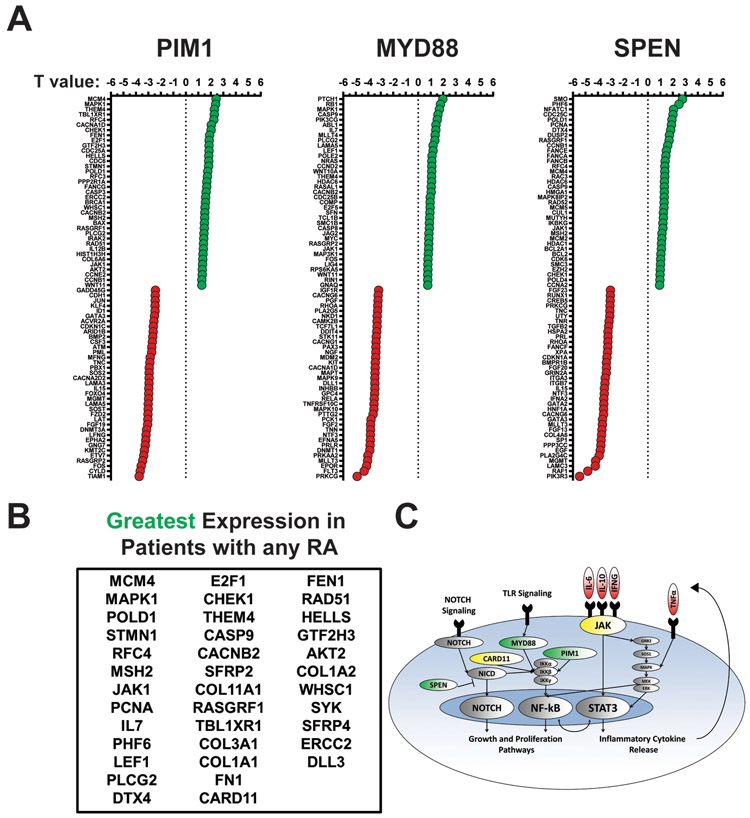
To integrate the genomic and transcriptomic data described above and identify lenalidomide sensitive mechanisms, the relationships between the RA (PIM1, SPEN, and MYD88) and the RNA pathway genes associated with R2CHOP EFS24 were explored. 15 of the 44 non-GCB RCHOP patients with paired DNA and gene expression data had a RA. T statistic differential expression analyses were performed for all 730 PanCan genes in those cases with or without RA. The top 5% genes with overall increases or decreases in patients with individual RA are noted (Figure 5A). The top 5% genes with overall increases in patients with any of the RA are listed in Figure 5B. A graphic summarizes the signaling pathways associated with the RA and highlight a potential mechanism of lenalidomide action, including inhibition of the NOTCH, NF-κB, and JAK-STAT pathways.
Figure 5: Responder Alterations Correspond to Distinct RNA Expression Profiles.
Combined data from 44 cases that had paired WES and PanCan data was used for analysis, R2CHOP (N=13) RCHOP (N=31). 15 patients had at least one RA. (A) The top 5% and bottom 5% (N=37 each) of genes associated with the presence of each RA are visualized through dot plots. Genes with greater differential expression in the presence of an RA are closer to the top (greater T value; green) and genes with lesser expression in the presence of an RA are closer to the top (lower T value; red). Each RA is individually documented. (B) The table documents genes associated with greater differential expression in the presence of any RA. (C) A graphic summarizes the DNA alterations and the hypothesized high-risk phenotype. RA are designated green, genes associated with greater expression in yellow, and hypothesized cytokines in red.
Discussion
Non-GCB DLBCL cases have been linked with inferior rates of overall survival as a result of aggressive activation of survival and proliferation pathways9, 49-51. This includes constitutive activation of the NF-κB transcription factor family via dysregulation of genes such as MYD88 or NOTCH and results in a chronic inflammatory cytokine milieu14, 16, 22. Past work has highlighted the potential benefit of the immunomodulatory drug IMiD® lenalidomide in this patient population, however much of this work has focused on either in vitro or clinical studies, with lack of insight on the impact of lenalidomide on patient derived samples. We have extended upon these observations by aligning clinical data with DNA and gene expression analysis from patients treated on trial with R2CHOP, and we are the first to identify a high-risk profile of non-GCB patients that benefit from the addition of lenalidomide to RCHOP. These results highlight the potential of precision medicine strategies in DLBCL to identify a vulnerable patient population, validate a genetic phenotype, and apply a personalized therapy that elicits a favorable response when standard therapy likely would not.
We report three gene alterations to be predictive of high-risk DLBCL that fail standard RCHOP yet are susceptible to R2CHOP: PIM1, SPEN, and MYD88 (L265P). Each have well-documented roles in non-GCB DLBCL, with MYD88 being supported by numerous studies16, 52, 53. The high incidence of MYD88 mutations combined with the clinical results of R2CHOP in ABC DLBCL suggest that there are underlying biologic and genetic differences that may account for ABC-specific responses18, 33. Mutations in MYD88 have also been specifically linked with a supportive cytokine milieu that sustains an inflammatory JAK-STAT phenotype20. Other work has highlighted the self-sustaining capabilities of active STAT3 signaling and its reliance on NF-κB-triggering mechanisms54. PIM1 alterations and those that activate NOTCH signaling (SPEN) have also been documented to induce an inflammatory cytokine environment22, 55-57. In support of our finding, MYD88 and PIM1 alterations were clustered into the unfavorable C5 and MCD genomic subtypes and are associated with aggressive NF-κB signaling12, 18. SPEN has also been identified as an unfavorable marker in the non-GCB phenotype and is present in the C1 and BN2 clusters, which resemble a novel non-GCB phenotype more reliant on NOTCH and immune escape12, 17, 18. Together these studies suggest that PIM1, SPEN, and MYD88 (L265P) are associated with high-risk non-GCB DLBCL and drive activation of inflammatory TLR signaling, NOTCH, and IRF437. While these are not known direct targets of lenalidomide, they are expressed in tumors that have vulnerabilities to lenalidomide.
Building on the genomic studies, our PanCan analysis was able to identify gene expression patterns that reflected the high-risk profile and R2CHOP susceptibility. Inflammatory pathways such as JAK-STAT, NF-κB, IRF4, MYD88, and OCT2 displayed significantly greater association with achieving EFS24 when treated with R2CHOP compared to RCHOP. High-risk patients are likely receiving benefit from lenalidomide’s interference with continuous loops of IRF4, NF-κB, and STAT transcription factor signaling. Conversely, signatures associated with R2CHOP failure included the Stromal-2 survival, lenalidomide-repressed genes, CNS lymphoma, E2F3, HRAS, TGFB, and MYC upregulation signatures. These pathways likely identify a subset of non-GCB patients that rely on a supportive tumor microenvironment (TME) and proliferative signaling.
Upon further analysis of the gene expression data we identified MAP3K14, IL2RB, and STAT3 as predictors of high-risk disease on the basis of EFS24. MAP3K14, also known as NIK, emerged as a key component of sustained noncanonical NF-κB signaling associated with MYD88, and equally interesting was the highly-correlated pair of IL2RB and STAT3 expression20, 58-60. This partnership has been previously observed, as IL2RB, IL6, and STAT3 displayed significantly responsive expression and were significantly associated with STAT3 ChIP-seq peaks54. JAK1, JAK2, JAK3, IL6R, SOS1, MYD88, NOTCH2, and IFNG were also expressed at high levels in unfavorable RCHOP cases22, 54-57, 61. Based on these results, IL2RB and STAT3 could prove to be useful predictive markers for high-risk disease and R2CHOP response. The JAK-STAT pathway and resulting cytokine signaling profile was specifically highlighted by the rrDLBCL study by Morin and colleagues, making this observation all the more fitting as a high-risk pathway20. Integrative analysis of the DNA and gene expression data revealed how each RA (PIM1, SPEN, and MYD88) uniquely influences an inflammatory phenotype. Several genes associated with NF- κB engagement displayed greater expression in patients with any of the alterations (CARD11, PLCG2, and JAK1), but the SPEN profile was uniquely associated with microenvironment and NOTCH genes, highlighting its presence in the novel C1 and BN2 genetic classifications.
In conclusion, our combined analysis of DNA and RNA across R2CHOP and RCHOP treatment cohorts identifies a high-risk non-GCB phenotype that is capable of sustaining JAK-STAT and NF-κB signaling, and is sensitive to R2CHOP. This phenotype encompasses approximately 38% of non-GCB patients, and the positive results of the ECOG-ACRIN1412 highlight the clinical success of R2CHOP. Our data supports the hypothesis that R2CHOP has activity in tumors reliant on IRF4, NF-κB, and STAT transcription factors, leading to a loss of proliferative feedback systems. Although promising and highly relevant due to the use of early phase clinical trial samples and a large comparison cohort, these conclusions require additional validation that can be done as R2CHOP trial samples with existing tissue and long term follow up become available. The RA signature generated in this work could retrospectively identify patients in larger studies (such as the ROBUST trial) that would most likely benefit from the combination. Combined with prior studies on RCHOP + ibrutinib21, 28, 62, the groundwork for a precision therapy approach in DLBCL in which DNA or RNA profiles can be used to identify patients early in treatment who may not benefit from the current standard of care, RCHOP, and who would benefit from the addition of lenalidomide or other targeted agents.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health (P30 CA015083 to T.E.W., P50 CA097274 to J.R.C. and A.J.N; R01 CA212162 to A.J.N and J.R.C.) and the Henry J. Predolin Foundation, Inc. (to T.E.W.).
Footnotes
Competing Interests
Consultant or Advisory Role: Grzegorz S. Nowakowski, Celgene (U); Thomas E. Witzig, Celgene (U) Research Funding: Thomas E. Witzig, Celgene; Anne J. Novak, Celgene
The online version of this article contains a data supplement.
References
- 1.Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. 2011. p. 5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019. [DOI] [PubMed] [Google Scholar]
- 3.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. The New England Journal of Medicine 2002; 346(4): 235–242. [DOI] [PubMed] [Google Scholar]
- 4.Crump M, Neelapu SS, Farooq U, Van Den Neste E, Kuruvilla J, Westin J, et al. Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood 2017; 130(16): 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farooq U, Maurer MJ, Thompson CA, Thanarajasingam G, Inwards DJ, Micallef I, et al. Clinical heterogeneity of diffuse large B cell lymphoma following failure of front-line immunochemotherapy. Br J Haematol 2017. October; 179(1): 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Den Neste E, Schmitz N, Mounier N, Gill D, Linch D, Trneny M, et al. Outcome of patients with relapsed diffuse large B-cell lymphoma who fail second-line salvage regimens in the International CORAL study. Bone Marrow Transplantation 2016; 51(1): 51–57. [DOI] [PubMed] [Google Scholar]
- 7.Maurer MJ, Ghesquières H, Jais JP, Witzig TE, Haioun C, Thompson CA, et al. Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy. Journal of Clinical Oncology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kB in diffuse large B-cell lymphoma. Nature 2009; 459(7247): 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis RE, Ngo VN, Lenz G, Tolar P, Young R, Romesser PB, et al. Chronic Active B Cell Receptor Signaling in Diffuse Large B Cell Lymphoma. Nature 2010; 463(7277): 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aukema SM, Siebert R, Schuuring E, Van Imhoff GW, Kluin-Nelemans HC, Boerma EJ, et al. Double-hit B-cell lymphomas. 2011. p. 2319–2331. [DOI] [PubMed] [Google Scholar]
- 11.Erdmann T, Klener P, Lynch JT, Grau M, Vočková P, Molinsky J, et al. Sensitivity to PI3K and AKT inhibitors is mediated by divergent molecular mechanisms in subtypes of DLBCL. Blood 2017; 130(3): 310–322. [DOI] [PubMed] [Google Scholar]
- 12.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:1290–1. [DOI] [PubMed] [Google Scholar]
- 13.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009; 459(7247): 712–716. [DOI] [PubMed] [Google Scholar]
- 14.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 Mutations in Human Diffuse Large B Cell Lymphoma. Science 2008; 319(5870): 1676–1679. [DOI] [PubMed] [Google Scholar]
- 15.Manso BA, Wenzl K, Asmann YW, Maurer MJ, Manske M, Yang ZZ, et al. Whole-exome analysis reveals novel somatic genomic alterations associated with cell of origin in diffuse large B-cell lymphoma. Blood Cancer Journal 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011; 470(7332): 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. New England Journal of Medicine 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenzl K, Manske MK, Sarangi V, Asmann YW, Greipp PT, Schoon HR, et al. Loss of TNFAIP3 enhances MYD88L265P-driven signaling in non-Hodgkin lymphoma. Blood Cancer Journal 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clinical Cancer Research 2016; 22(9): 2290–2300. [DOI] [PubMed] [Google Scholar]
- 21.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nature Medicine 2015; 21(8): 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karube K, Enjuanes A, Dlouhy I, Jares P, Martin-Garcia D, Nadeu F, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartlett NL, Wilson WH, Jung SH, Hsi ED, Maurer MJ, Pederson LD, et al. Dose-Adjusted EPOCH-R Compared With R-CHOP as Frontline Therapy for Diffuse Large B-Cell Lymphoma: Clinical Outcomes of the Phase III Intergroup Trial Alliance/CALGB 50303. J Clin Oncol 2019. July 20; 37(21): 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunleavy K, Pittaluga S, Czuczman MS, Dave SS, Wright G, Grant N, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 2009; 113(24): 6069–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fruman DA, Cantley LC. Idelalisib — A PI3Kδ Inhibitor for B-Cell Cancers. New England Journal of Medicine 2014; 370(11): 1061–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romejko-Jarosinska J, Rymkiewicz G, Paszkiewicz-Kozik E, Dabrowska-Iwanicka AP, Borawska A, Domanska-Czyz K, et al. R- DAEPOCH as a first line treatment for high grade B cell lymphoma and diffuse large B cell lymphoma with unfavorable features. Hematological Oncology 2017. [Google Scholar]
- 27.Younes A, Sehn LH, Johnson P, Zinzani PL, Hong X, Zhu J, et al. Randomized Phase III Trial of Ibrutinib and Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone in Non-Germinal Center B-Cell Diffuse Large B-Cell Lymphoma. J Clin Oncol 2019. May 20; 37(15): 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younes A, Thieblemont C, Morschhauser F, Flinn I, Friedberg JW, Amorim S, et al. Combination of ibrutinib with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for treatment-naive patients with CD20-positive B-cell non-Hodgkin lymphoma: a non-randomised, phase 1b study. Lancet Oncol 2014. August; 15(9): 1019–1026. [DOI] [PubMed] [Google Scholar]
- 29.Gribben JG, Fowler N, Morschhauser F. Mechanisms of Action of Lenalidomide in B-Cell Non-Hodgkin Lymphoma. Journal of Clinical Oncology 2015; 33(25): JCO.2014.2059.5363–JCO.2014.2059.5363-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson GH, Davies FE, Pawlyn C, Cairns DA, Striha A, Collett C, et al. Lenalidomide maintenance versus observation for patients with newly diagnosed multiple myeloma (Myeloma XI): a multicentre, open-label, randomised, phase 3 trial. The Lancet Oncology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu YX, Shi C-X, Bruins LA, Wang X, Riggs DL, Porter B, et al. Identification of lenalidomide resistance pathways in myeloma and targeted resensitization using cereblon replacement, inhibition of STAT3 or targeting of IRF4. Blood Cancer Journal 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowakowski GS, Chiappella A, Witzig TE, Spina M, Zhang L, Flament J, et al. Randomized, phase III trial of the efficacy and safety of lenalidomide plus R-CHOP vs R-CHOP in patients with untreated ABC-type diffuse large B-cell lymphoma. 2015; 33(15_suppl): TPS8600–TPS8600. [Google Scholar]
- 33.Nowakowski GS, LaPlant B, Macon WR, Reeder CB, Foran JM, Nelson GD, et al. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of non-germinal center B-cell phenotype in newly diagnosed diffuse large B-cell lymphoma: A phase II study. Journal of Clinical Oncology 2015. [DOI] [PubMed] [Google Scholar]
- 34.Witzig TE, Vose JM, Zinzani PL, Reeder CB, Buckstein R, Polikoff JA, et al. An international phase II trial of single-agent lenalidomide for relapsed or refractory aggressive B-cell non-Hodgkin’s lymphoma. Annals of Oncology 2011; 22(7): 1622–1627. [DOI] [PubMed] [Google Scholar]
- 35.Nowakowski GS. 15th International Conference on Malignant Lymphoma Palazzo dei Congressi, Lugano (Switzerland) 18 - 22 June, 2019 Hematological Oncology 2019. 2019/June/01; 37(S2): 5–9. [DOI] [PubMed] [Google Scholar]
- 36.Vitolo U 15th International Conference on Malignant Lymphoma Palazzo dei Congressi, Lugano (Switzerland) 18 - 22 June, 2019 Hematological Oncology 2019. 2019/June/01; 37(S2): 5–9. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Shaffer AL, Emre NCT, Ceribelli M, Zhang M, Wright G, et al. Exploiting Synthetic Lethality for the Therapy of ABC Diffuse Large B Cell Lymphoma. Cancer Cell 2012; 21(6): 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagner P, Chiu H, Ortiz-Estevez M, Biyukov T, Brachman C, Trneny M, et al. Lenalidomide exhibits activity in mantle cell lymphoma through increased NK cell mediated cytotoxicity. Blood 2015. [Google Scholar]
- 39.Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4 CRBN. British Journal of Haematology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowakowski GS, Laplant B, Habermann TM, Rivera CE, Macon WR, Inwards DJ, et al. Lenalidomide can be safely combined with R-CHOP (R2CHOP) in the initial chemotherapy for aggressive B-cell lymphomas: Phase i study. Leukemia. 2011;25:1877–81. [DOI] [PubMed] [Google Scholar]
- 41.Cerhan JR, Link BK, Habermann TM, Maurer MJ, Feldman AL, Syrbu SI, et al. Cohort Profile: The Lymphoma Specialized Program of Research Excellence (SPORE) Molecular Epidemiology Resource (MER) Cohort Study. International Journal of Epidemiology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, Jaffe ES, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004. [DOI] [PubMed] [Google Scholar]
- 44.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016. September 15; 32(18): 2847–2849. [DOI] [PubMed] [Google Scholar]
- 45.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics 2006. [DOI] [PubMed] [Google Scholar]
- 46.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0 [2]. 2006. p. 500–501. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubois S, Viailly PJ, Bohers E, Bertrand P, Ruminy P, Marchand V, et al. Biological and Clinical Relevance of Associated Genomic Alterations in MYD88 L265P and non-L265P-Mutated Diffuse Large B-Cell Lymphoma: Analysis of 361 Cases. Clin Cancer Res 2017. May 1; 23(9): 2232–2244. [DOI] [PubMed] [Google Scholar]
- 49.Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nature Reviews Immunology 2015; 15(3): 172–184. [DOI] [PubMed] [Google Scholar]
- 50.Lenz G, Wright GW, Emre NCT, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proceedings of the National Academy of Sciences of the United States of America 2008; 105(36): 13520–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staudt LM, Dave S The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005; 87:163–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin LH, Kim J, Kim-Ha J, Cho HS, Kim B, Choi JK, et al. Requirement of Split ends for Epigenetic Regulation of Notch Signal-Dependent Genes during Infection-Induced Hemocyte Differentiation. Molecular and Cellular Biology 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RSK, Küppers R, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001; 412(6844): 341–346. [DOI] [PubMed] [Google Scholar]
- 54.Ouyang Z, Hardee J, Kundaje A, Zhang Y, Snyder M, Lacroute P. STAT3 Targets Suggest Mechanisms of Aggressive Tumorigenesis in Diffuse Large B-Cell Lymphoma. G3: Genes∣Genomes∣Genetics 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colombo M, Mirandola L, Chiriva-Internati M, Basile A, Locati M, Lesma E, et al. Cancer cells exploit notch signaling to redefine a supportive cytokine milieu. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin S, Mutvei AP, Chivukula IV, Andersson ER, Ramsköld D, Sandberg R, et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene 2013. 2013/October/01; 32(41): 4892–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shirogane T, Fukada T, Muller JMM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 1999. [DOI] [PubMed] [Google Scholar]
- 58.Aaronson D, Horvath C. Roadmap for those who dont know JAK-STAT. Science 2002; 296(May): 1653–1655. [DOI] [PubMed] [Google Scholar]
- 59.Sheng L, Zhou Y, Chen Z, Ren D, Cho KW, Jiang L, et al. NF-κB-inducing kinase (NIK) promotes hyperglycemia and glucose intolerance in obesity by augmenting glucagon action. Nature Medicine 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkB. Genes and Development 2007; 21(11): 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aue G, Sun C, Liu D, Park J-H, Pittaluga S, Tian X, et al. Activation of Th1 Immunity within the Tumor Microenvironment Is Associated with Clinical Response to Lenalidomide in Chronic Lymphocytic Leukemia. J Immunol. 2018; 201(7):1967–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaidya R, Witzig TE. Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Annals of Oncology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.