Abstract
Developing prognostic biomarkers for specific cancer types that accurately predict patient survival is increasingly important in clinical research and practice. Despite the enormous potential of prognostic signatures, proposed models have found limited implementations in routine clinical practice. Herein, we propose a generic, RNA sequencing platform-independent, statistical framework named Whole Transcriptome Signature for Prognostic Prediction (WTSPP) to generate prognostic gene signatures. Using ovarian cancer and lung adenocarcinoma as examples, we provide evidence that our prognostic signatures over-perform previous reported signatures, capture prognostic features not explained by clinical variables and expose biologically relevant prognostic pathways, including those involved in the immune system and cell cycle. Our approach demonstrates a robust method for developing prognostic gene expression signatures. In conclusion, our statistical framework can be generally applied to all cancer types for prognostic prediction and might be extended to other human diseases. The proposed method is implemented as an R package (PanCancerSig) and is freely available on GitHub (https://github.com/Cheng-Lab-GitHub/PanCancer_Signature).
Keywords: Clinical variables, Survival analysis, Gene expression profile, Prognostic signature
Introduction
Cancer is a major cause of morbidity and mortality worldwide, accounting for more than 8 million deaths each year [1]. In the United States alone, an estimated 1.8 million new cases will be diagnosed in 2020, and more than one third of these cases will succumb to the disease [2]. As a multifarious disease, the prognosis of patients with a specific type of cancer varies substantially [3]. Consequently, developing models for specific cancer types that accurately predict patient survival using prognostic biomarkers is increasingly important in clinical research and practice since they optimize treatment options.
Currently, prognosis is predominantly predicted based on clinical variables. The Tumor Node Metastasis (TNM) staging system has been developed for most solid tumors types and is based on tumor size, regional lymph node status and metastasis [4]. Although TNM staging is prognostic in many cancer types, certain cancer stages have high variation in terms of clinical outcomes, which limits the application of this staging system in directing therapeutic practice. For example, overall survival in stage III melanoma ranges from 93% to 32% depending on node involvement [5]. Additionally, for certain cancer types, few additional prognostic clinical variables are available. In ovarian cancer for example, cytoreductory status is one of the few other clinical variables utilized. The main treatment for this disease is surgery followed by platinum-based chemotherapy [6]. Prognosis prediction is primarily based on surgery status (or debulking), which is classified as optimal for complete tumor resection or suboptimal for incomplete resection and predicts poor prognosis for suboptimal cytoreductory status [7]. Thus, clinical variables certainly play important roles in achieving useful prognostic prediction. However, further improvement can be anticipated by including other information, such as genomic features.
Genomic data, especially transcriptomic profiles, provide a comprehensive characterization of the intrinsic molecular features of tumor samples, making them promising tools for predicting cancer prognosis [8]. Indeed, gene expression-based signatures have been extensively explored and have in some cases led to biomarkers that significantly improve the prediction of patient outcome. For example, expression-based tests such as MammaPrint or Oncotype DX can accurately predict patient outcome in ER+ breast cancer patients and are increasingly being implemented into clinical practice [9–11]. For many other cancer types, clinically relevant prognostic signatures have been proposed as well. Two strategies are generally utilized for creating genomic prognostic signatures: some signatures are based on prior knowledge, including signatures based on hypoxia-associated genes [12,13], EMT genes [14], or frequently mutated genes such as TP53 [15,16] and BRCA1 [17], whereas other prognostic signatures are based on supervised selection of genes that are statistically associated with patient prognosis [18–21].
In spite of the enormous potential of prognostic signatures, proposed models have reached limited implementations in routine clinical practice [22]. For example, a multitude of prognostic signatures have been developed for ovarian cancer [23–25], but none have reach the clinic yet. In general, proposed signatures tend to be validated through cross-validation or in a small subset of all available independent datasets [26]. This results in low reproducibility in new datasets and impedes the prognostic robustness needed for clinical application, as noted in ovarian cancer, lung cancer, melanoma and other cancer types [26–28]. In addition, gene expression data generated by both microarrays and RNAseq platforms should be amenable to signature application. Thus, there is a need for a standardized framework to develop prognostic gene signatures with high reproducibility from gene expression data in a straightforward fashion.
In this study, we propose a generic, RNA sequencing platform-independent, statistical framework named Whole Transcriptome Signature for Prognostic Prediction (WTSPP) to generate prognostic gene signatures. Our platform relies on signature generation from a training dataset based on Cox regression and consequent application of this signature in multiple independent test datasets. To exemplify the utility of our platform, we utilized TCGA data from ovarian cancer and lung adenocarcinoma to define tumor-specific prognostic signatures and systematically tested the predictive power of these signatures in a comprehensive list of independent tumor gene expression datasets with matched patient survival information. Our ovarian cancer signature (OV signature) outperformed 14 published gene signatures and was prognostic in all collected datasets. In lung cancer, we could significantly predict survival in almost all collected datasets, even after adjustment of clinical variables. In conclusion, our statistical framework can be generally applied to all cancer types for prognostic prediction and might be extended to other human diseases. To facilitate future utility, we already defined prognostic signatures for 13 cancer types based on the TCGA and developed these signatures into the PanCancerSig R package.
Materials and methods
Datasets and pre-processing
TCGA datasets for ovarian serous cystadenocarcinoma (OV) and lung adenocarcinoma (LUAD) were obtained from TCGA on FireBrowse (gdac.broadinstitute.org/). These datasets included Level 3 RNAseq samples with matched clinical information and provided RSEM normalized gene expression for 20,502 genes. Matched one-channel (Affymetrix) and two-channel (Agilent) microarray Ovarian cancer datasets were also obtained from TCGA on FireBrowse for cross-platform validation. The one-channel dataset was log10 transformed and the two-channel microarray dataset was log2 transformed. Additional gene expression datasets were obtained from Gene Expression Omnibus (GEO) as Series Matrix files. Only datasets with more than 40 samples were considered in our analysis. This resulted in the inclusion of 21 validation datasets (see Suppl. Table 5 and 6).
Definition of signatures
Tumor-specific whole-gene signatures for prognostic prediction (WTSPPs) were generated utilizing univariate Cox regression models. In this study, we used the TCGA OV and LUAD datasets to define OV- and LUAD-signatures, respectively. For each dataset, a Cox regression model was fit for each gene to evaluate its effect on overall survival. The proportional hazards assumption was checked for each gene using the cox.zph function from the R survival package. The Benjamini-Hochberg (BH)-adjusted p-values ranged from 0.29 to 1.0 for the OV signature. Three genes LCE4A, OR8U1, and SNORA36B) did not meet the proportional hazards assumption at a BH-adjusted p-value of 0.001. Since these genes comprised less than 0.02% (3/20501) of the gene weights and had very small weights (0.0002 for each of the three genes), we assumed that their influence on the final score would be negligible. We assumed that each p-value reflected the association of this gene with overall survival and utilized log10 transformed p-values as gene weights. Weights exceeding 10 were trimmed to 10 if necessary to reduce the effect of outliers. Subsequently, all transformed p-values were scaled from 0 to 1 and this vector represented the initial weight profile (wp) in which this scaled p-value represented the weight (w) of each gene (i). The wp was separated into a protective (wpp) and hazardous (wph) wp, based on the hazard ratio of each gene-specific Cox model; genes with hazard ratios <1 were considered to be protective, whereas genes with hazard ratios >1 were classified as hazardous. Gene weights of hazardous genes in wpp were set to zero and vice versa for protective genes in wph. The resulting two weight profiles, wpp and wph, represented the WTSPP. Identical steps were followed to generate RFS-based signatures, but now using RFS as the dependent variable instead of OS.
Calculation of prognostic scores
Prognostic scores (PSs) were calculated using the BASE algorithm [29], which has been made available in prior publications [30,31]. BASE calculates the enrichment of protective and hazardous genes for each patient, based on the WTSPP and patient gene expression data. First, one channel microarray or RNAseq gene expression datasets were median normalized by BASE, whereas two channel microarray datasets were not. Second, patient gene expression profiles were ranked in a descending order. Third, the distribution of two cumulative functions, the foreground (f) and background (b) functions, were calculated separately for the protective (wpp) and hazardous (wph) genes. These functions are given by:
where the weight w and rank g of gene i are evaluated. To obtain an enrichment score for protective genes in a patient sample, weights from wpp were inputted into these functions and a cumulative foreground and background distribution was defined. In a scenario in which protective genes were relatively highly expressed in a patient sample, the high ranks gi and large contributions of gene weights (wi) to the foreground distribution f(i) but low ranks gi and small contributions (1-wi) to the background distribution b(i) would increase f(i) more than b(i). The enrichment score of all protective genes (PSp) was defined by the maximum deviation of the foreground and background distributions and PSp would thus be high if protective genes were relatively highly expressed in a patient sample. The same enrichment procedure was repeated 1000 times for each sample with randomized patient expression profiles to generate a patient-specific null distribution for PSp. For each patient, the normalized PSp was then generated by dividing PSp by the mean of the absolute value of the permuted, randomized PSp values. An identical process was repeated with the hazardous genes in wph to establish an enrichment score for hazardous genes (PSh). Lastly, the final PS was determined by subtracting PSh from PSp. This generated low PSs for patients with enriched expression of hazardous genes and high PSs for patients with predominant expression of protective genes. Concordantly, high PS predicted good prognosis and low PS predicted poor prognosis.
Survival analysis
Survival analyses were performed using the R “survival” package. Log-rank tests were performed to evaluate overall or relapse free survival probabilities of PS-high and PS-low groups, using the “survdiff” function. Additionally, Cox proportional hazards models were performed on continuous PS, using the “coxph” function to determine the association with overall or relapse free survival. Reported p-values were derived from each Cox proportional hazards model and calculated using a two-sided Wald test. Concordance indices were also determined by the “coxph”. The “survfit” function was utilized to create Kaplan-Meier plots that visualize differences in survival probabilities.
Correlation analysis
The Spearman correlation coefficient (SCC) was reported for all correlation analyses as the assumptions underlying the Pearson correlation (i.e., normal distribution, homoscedasticity or linearity) were not met. SCC was calculated using the R function “cor”. All analyses were conducted in R (version 3.4.2).
R package
An R package, “PanCancerSig”, has been made available to facilitate the application of the presented prognostic signatures. Users can input clinical gene expression datasets of interest, calculate patient-specific PS, and perform survival analyses. PanCancerSig is available on GitHub at github.com/Cheng-Lab-GitHub/PanCancer_Signature.
Results
WTSPP: a new statistical framework for predicting cancer prognosis
WTSPP provides a generic statistical framework to define prognostic signatures based on tumor gene expression data. In contrast to previous methods that focus on the selection of prognostic genes, the prognostic signatures defined by WTSPP contain all genes. A weight is assigned to each gene based on its positive or negative association with prognosis. These signatures are then used as a reference to gauge the expression profiles of a new tumor dataset by computing prognostic scores (PSs) for each patient. The PS is a statistical summary that quantifies the relative expression of genes that are associated with prognosis, i.e., highly weighted genes in the prognostic signature. In general, a sample will have a high PS when genes associated with good prognosis tend to be highly expressed and genes associated with poor prognosis are lowly expressed. Thus, although all genes are included in the prognostic signature, the PSs of tumor samples are driven by genes with the highest absolute weights.
Application of WTSPP to prognostic prediction in ovarian cancer
To demonstrate the utility and performance of WTSPP-derived signatures, we utilized ovarian cancer as an example due to the limited prognostic tools available for this disease currently. We first developed a prognostic gene signature based on the TCGA ovarian cancer RNAseq dataset (Table 1, Suppl. Table 1 and 2). Gene Ontology enrichment analysis indicated that genes with high positive weights (associated with prolonged survival) were involved in immune responses, while genes with high negative weights (associated with shorter survival) were associated with extracellular matrix deposition (Suppl. Table 3). As an initial validation of this signature, we applied it to the original TCGA ovarian cancer RNAseq data and, as expected, the resulting PSs were significantly associated with overall survival (P=1E-17, Cox regression) (Suppl. Table 4).
Table 1.
Summary of OV and LUAD TCGA datasets.
| Cancer | Samplea | Eventb | Event %b | # Samples with RNAseq data | Univariate Cox regressionc | |||
|---|---|---|---|---|---|---|---|---|
| HR<1 | HR>1 | HR<1* | HR>1* | |||||
| OV | 591 | 298 | 51.7 | 302 | 11 | 37 | 0 | 0 |
| LUAD | 522 | 123 | 24.5 | 496 | 243 | 244 | 7 | 9 |
Sample refers to number of samples with clinical information.
Event and percentage refer to the number and percent of death occurrences, respectively.
Number of genes indicated by Hazard Ratio (HR)>1 or HR<1 are significantly associated with overall survival (p<0.001).
Number of genes indicated by HR>1* or HR<1* are significantly associated with overall survival after multiple hypotheses correction using the Benjamini-Hochberg procedure (FDR<0.001).
In addition to TCGA ovarian cancer RNAseq data, we also derived OV signatures from TCGA ovarian cancer gene expression profiles measured by Affymetrix (one-channel array) and Agilent (two-channel array) to examine the applicability of an RNAseq-derived signature to microarray platforms and vice versa. The signatures from three independent platforms were similar with an average Spearman Correlation Coefficient (SCC) of 0.63 (Suppl Fig S1). Although the prognostic performance of the three OV signatures were comparable, we found that the signature derived from the RNAseq dataset achieved the best prognostic ability (Suppl. table 4), presumably due to the superior sensitivity of RNAseq platform.
We subsequently applied the RNAseq-derived TCGA OV signature to a collection of 6 OV microarray datasets from previous publications, each containing at least 100 tumor samples (Suppl. table 5). For each dataset, we calculated PSs for all patients. Cox regression analysis indicated that PSs were significantly associated with overall (Fig. 1a) and/or relapse-free survival (Fig. 1b) in all datasets (HR < 1, P < 0.05, Cox regression model), even after adjusting for clinical variables (Table 2). Furthermore, we stratified patients into PS-high and PS-low groups using median PS as threshold and compared survival times. In all 6 datasets, patients from the PS-high group exhibited significantly longer overall survival than those from the PS-low group (P < 0.05, log rank test) (Fig. 1a). For the two datasets (GSE9891 and GSE17260) with relapse-free survival information, we also observed significantly longer relapse-free survival in the PS-high patient groups (P < 0.05, log rank test) (Fig. 1b). However, a recurrence-free survival (RFS)-based signature was better able to predict RFS in two out of three datasets, suggesting that prediction of recurrence could be further improved with an RFS-based signature (Suppl. Fig 1d). Taken together, these results indicate that the OV signature defined from TCGA RNAseq data by our WTSPP framework is predictive of patient prognosis in a platform independent manner. The PSs resulting from this signature are predictive of survival in all collected ovarian cancer expression datasets.
Figure 1: The OV signature is predictive of patient survival in 6 independent ovarian cancer gene expression datasets.
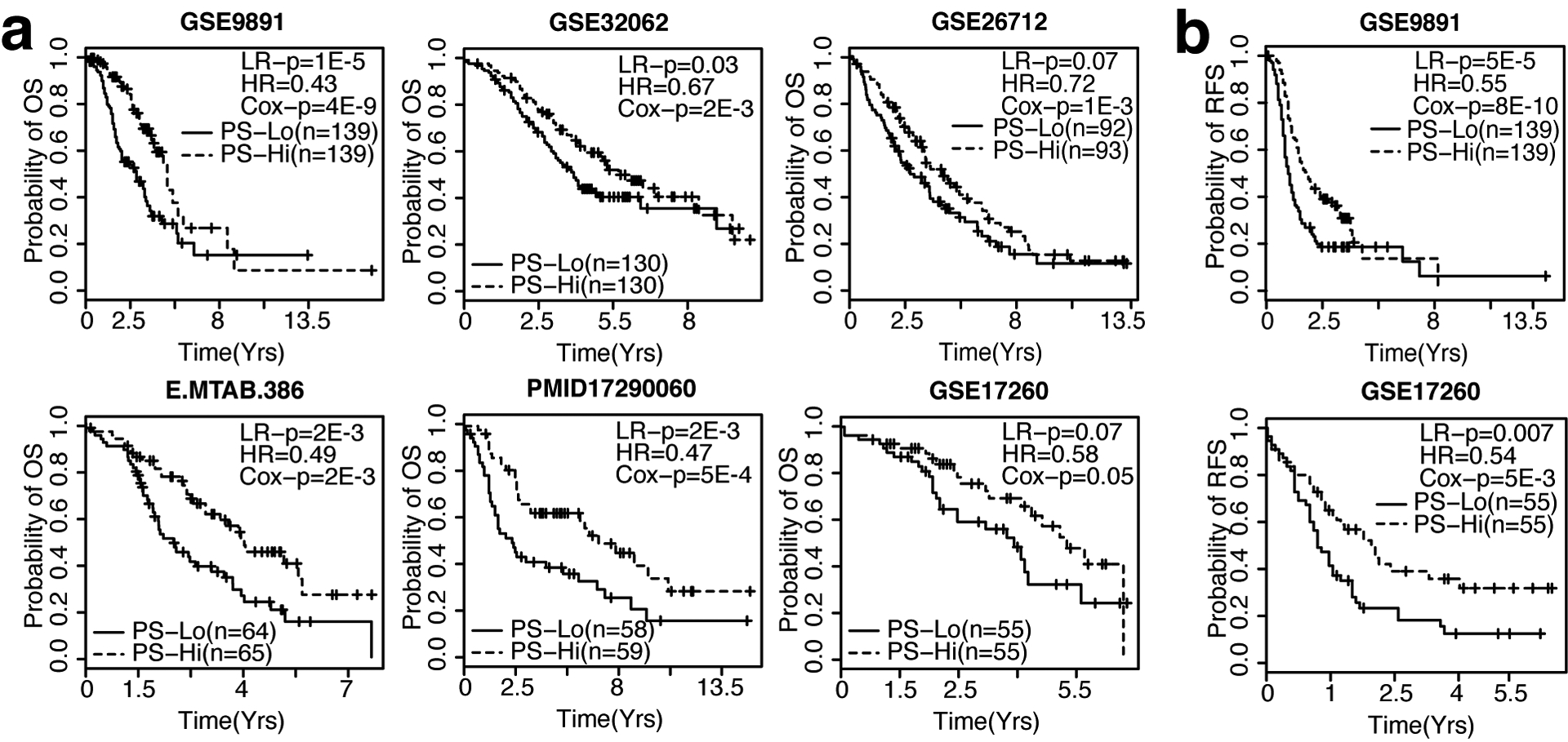
a. Kaplan-Meier curves depicting overall survival (OS) probability over time, ordered by sample size (n). b. Kaplan-Meier curves depicting recurrence-free survival (RFS) probability over time, ordered by n. For all Kaplan-Meier curves, patients were dichotomized into PS-high (red) and PS-low (blue) using median PS as threshold. LR-p = p-value of Log Rank test, HR = Hazard Ratio, Cox-p = p-value calculated from the univariate Cox regression model in which PS is used as the only variable.
Table 2.
Univariate Cox regression and C-indices using PS as a continuous variable.
| Study | Sample Sizea | PS only | PS + Clin. variables | ||||
|---|---|---|---|---|---|---|---|
| P-val | HR | C-index | P-val | HR | C-index | ||
| TCGA | 289 | 6.6E-12 | 0.92 | 0.66 | 1.5E-8 | 0.93 | 0.68 |
| GSE9891 | 278 | 4.2E-9 | 0.97 | 0.7 | 4.6E-8 | 0.97 | 0.74 |
| GSE32063 | 260 | 2.1E-3 | 0.88 | 0.6 | 0.01 | 0.9 | 0.64 |
| GSE26712 | 185 | 1.1E-3 | 0.98 | 0.61 | 0.03 | 0.99 | 0.66 |
| E.MTAB.386 | 129 | 2.3E-3 | 0.97 | 0.6 | 2.5E-3 | 0.96 | 0.63 |
| PMID17290060 | 117 | 4.7E-4 | 0.97 | 0.66 | 7.4E-4 | 0.97 | 0.67 |
| GSE17260 | 110 | 0.05 | 0.99 | 0.6 | 0.09 | 0.99 | 0.66 |
Sample size indicates number of gene expression samples used in analysis. PS = prognostic score. HR = hazard ratio. C-index = concordance index. Clin. variables include all clinical variables (stage, grade, age and/or debulking) available for each dataset (See Supp. Table 5).
The OV signature provides additional prognostic value over Clinical variables
Clinical variables have been used to predict prognosis and direct therapeutic strategies in ovarian cancer [32]. One of the most utilized prognostic clinical variables is surgery status (optimal versus suboptimal) with suboptimal cytoreductory status predicting poor prognosis [7]. To examine if our signature could provide additional prognostic information to surgery status and other clinical variables in ovarian cancer, we applied a multivariate Cox regression analysis including our OV signature and several clinical variables across 12 collected ovarian cancer datasets (Suppl. Table 5). Here, datasets GSE9891 and E-MTAB-386 were chosen as examples for illustration due to their complete clinical information. Although surgery status was not associated with survival (p>0.05, Cox regression model), high PSs were protective in both datasets and was the only consistently significant variable associated with prognosis (Fig 2a, b).
Figure 2: The OV signature provides additional prognostic value over clinical variables.
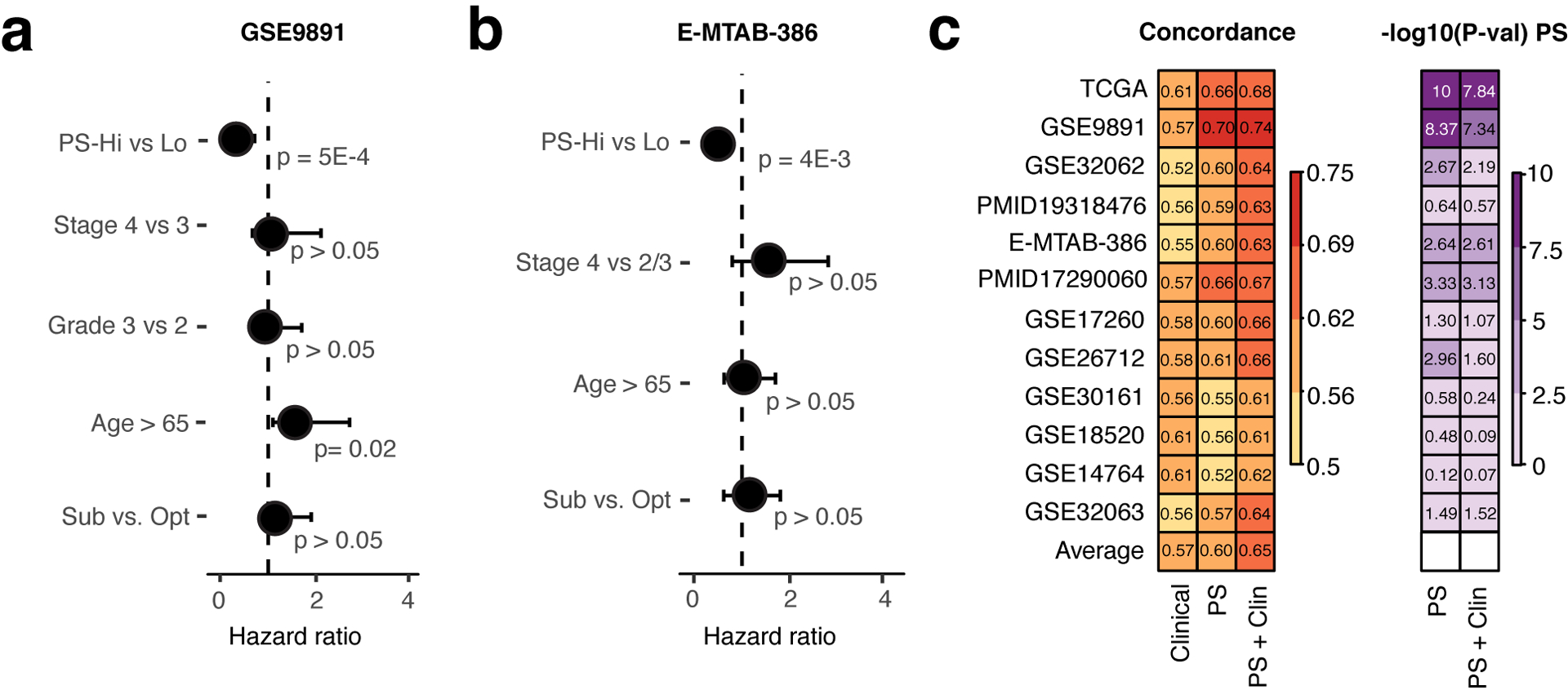
Forest plots depicting the prognostic significance of PS and clinical variables in a. the GSE9891 and b. the E-MTAB-386 dataset (multivariate Cox regression model). c. Heatmaps of concordance indices (left) and -log10(P-values) (right) of Cox regression models including clinical variables alone, PS alone, and PS adjusted by clinical variables in 12 ovarian cancer datasets.
As clinical variables have been suggested to be prognostic in ovarian cancer, we wondered if our OV signature could provide additional prognostic value. For each of the 12 ovarian cancer datasets, we calculated the concordance index of a multivariate Cox model including clinical variables only (age, stage, grade, surgery status), a univariate Cox model including PSs only, and a multivariate model including clinical variables and PSs. A comparison between concordance-indices (C-indices) showed that our signature indeed improved the prognostic ability of clinical variables in all 12 ovarian cancer datasets (Fig. 2c). Although clinical variables alone were prognostic as indicated by an average C-index of 0.57, our OV signature performed slightly better, showing an average C-index of 0.60 (p > 0.05, paired t-test). Adjusting our signature for clinical variables further increased the prognostic performance of our model (average C-index = 0.65) and showed significant improvement of predictive ability compared to clinical variables alone (p=1e-4, paired t-test). Thus, our signature captured tumor characteristics not explained by clinical variables and could significantly improve the predictive ability of clinical variables in ovarian cancer.
Since PSs could substantially improve the collective prognostic ability of clinical variables, we were curious if PSs were associated with individual clinical variables. The GSE9891 dataset was used as an example for illustration due to its large sample size and rich clinical annotations. Ovarian cancer staging is currently a major predictor of prognosis, since 80% of late-stage ovarian cancer patients treated with Platinum-based therapies experiences incurable recurrence [32]. We indeed observed that patients diagnosed at later stages (stage 3 and 4) had significantly lower PSs than patients with early stage disease (stage 1 and 2, P=0.004, t-test) (Fig 3a) and that lower PSs were associated with recurrence (p=0.01, t-test) (Fig 3b). Additionally, low PSs were more often observed in patients with suboptimal cytoreduction status compared to patients classified as having optimal surgery status (p=6e-4, t-test) (Fig 3c). We next stratified patients with optimal or suboptimal status based on PSs. In both optimal and suboptimal patient groups, patients with high PSs had significantly longer survival than patients with low PSs (P=6e-5, log-rank test, HR=0.42 for suboptimal, p=4e-6, log-rank test, HR=0.33 for optimal) (Fig 3d). Notably, patients with optimal surgery status but low PSs had identical survival probabilities as patients with suboptimal cytoreductory status and high PSs. Similar results were observed in other ovarian cancer datasets (Suppl. Fig S2a–e). These results indicate that our OV signature could improve the prognostic ability of individual clinical variables.
Figure 3: The OV signature can be used to stratify individual clinical variables.
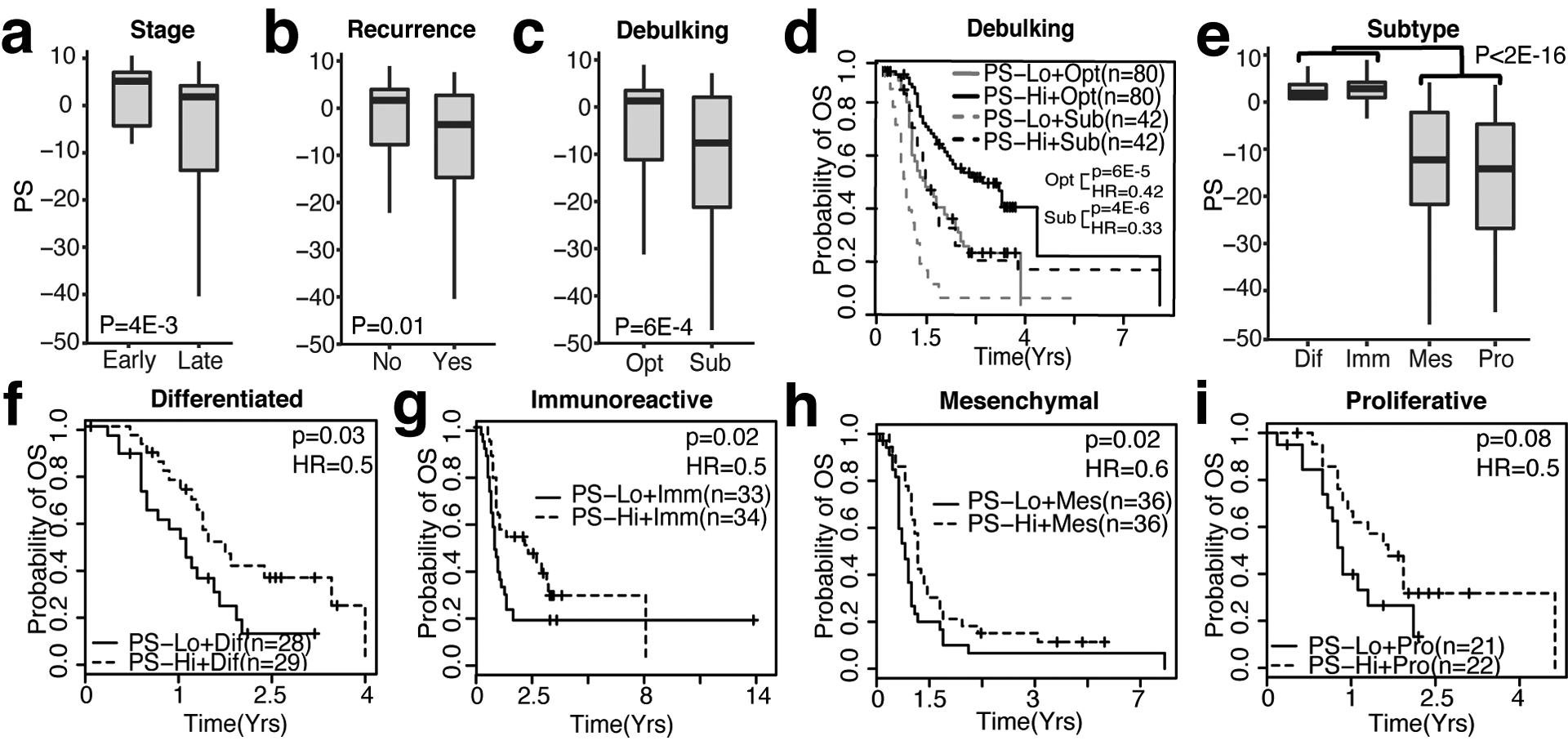
a. Box plot of PSs in samples from early (1 and 2) and late stages (3 and 4), Wilcoxon Rank-Sum Test. b. Box plot of PSs comparing samples with non-recurrent and recurrent events, Wilcoxon Rank-Sum Test. c. Box plot of PSs in samples classified as optimal (Opt) and suboptimal (Sub), Wilcoxon Rank-Sum Test. d. Kaplan-Meier plot depicting survival probability of optimal and suboptimal patients stratified by high-PS or low-PS (based on median PS), Log-rank Test. e. Box plot of PS in differentiated (Dif), Immune (Imm), Mesenchymal (Mes), and Proliferative (Pro) OV subtypes, Wilcoxon Rank-Sum Test. Kaplan-Meier plots depicting survival probability of PS-high and PS-low samples in f. Differentiated, g. Immunoreactive, h. Mesenchymal, and i. Proliferative OV subtypes, Log-rank Test.
Ovarian cancer is a heterogeneous disease and the TCGA has identified 4 distinct molecular ovarian cancer subtypes [33]. Each subtype presents itself with a different prognosis [34]. Generally, patients with Mesenchymal (Mes) and Proliferative (Pro) have poorer survival compared to patients with Differentiated (Dif) and Immune (Imm) molecular subtypes [34]. In concordance with this, patients with Mes and Pro ovarian cancer subtypes had significantly lower PS than patients with Dif and Imm subtypes in two out of three datasets (Fig 3e, Suppl. Fig. 2f, 2h). Additionally, within ovarian cancer subtypes, patients in PS-high groups had longer survival in three out of four ovarian cancer subtypes, including Dif (p=0.03, log-rank test, HR=0.5) (Fig 3f), Imm (p=0.02, log-rank test, HR=0.5) (Fig 3g), and Mes (p=0.02, log-rank test, HR=0.6) (Fig 3h, Suppl. Fig 2h–i). The same trend was also observed in the Pro subtype, but this association did not reach statistical significance (p=0.08, log-rank test, HR=0.5) (Fig 3i), which might be due to a smaller sample size, since the Pro subtype could be stratified significantly in an independent dataset with a larger sample size (Suppl. Fig 2f–g). Thus, our OV signature can stratify molecular subtypes further based on PSs.
The OV signature outperforms previously published ovarian cancer gene signatures
After showing that our OV signature has clinical relevance, we wanted to define the robustness and performance of our WTSPP framework. We assessed whether (i) TCGA is the best dataset to generate signatures from, and (ii) compared the prognostic ability of our signature to published ovarian cancer specific gene signatures.
First, we derived prognostic signatures from each of the 11 collected ovarian cancer datasets by applying our WTSPP framework to each dataset. We then evaluated the prognostic ability of these signatures using C-indices (Fig 4a). Although all datasets could be used to create prognostic signatures, variability between the prognostic abilities of signatures was observed. The TCGA-derived signature had the best, overall performance among the 12 signatures and performed well in almost all datasets (Fig. 4a, left). The TCGA signature was followed in prognostic ability by signatures derived from the GSE9891 and GSE32063 datasets, each with an average C-index of 0.59 and 0.58, respectively (Fig 4a, right). There seemed to be a trend between dataset sample size and prognostic ability, where datasets with high sample sizes (e.g. TCGA and GSE9891) tended to produce more robust prognostic signatures, compared to datasets with lower sample size (e.g. GSE14764 and GSE32063).
Figure 4: The OV signature outperforms 14 published ovarian cancer specific gene signatures.
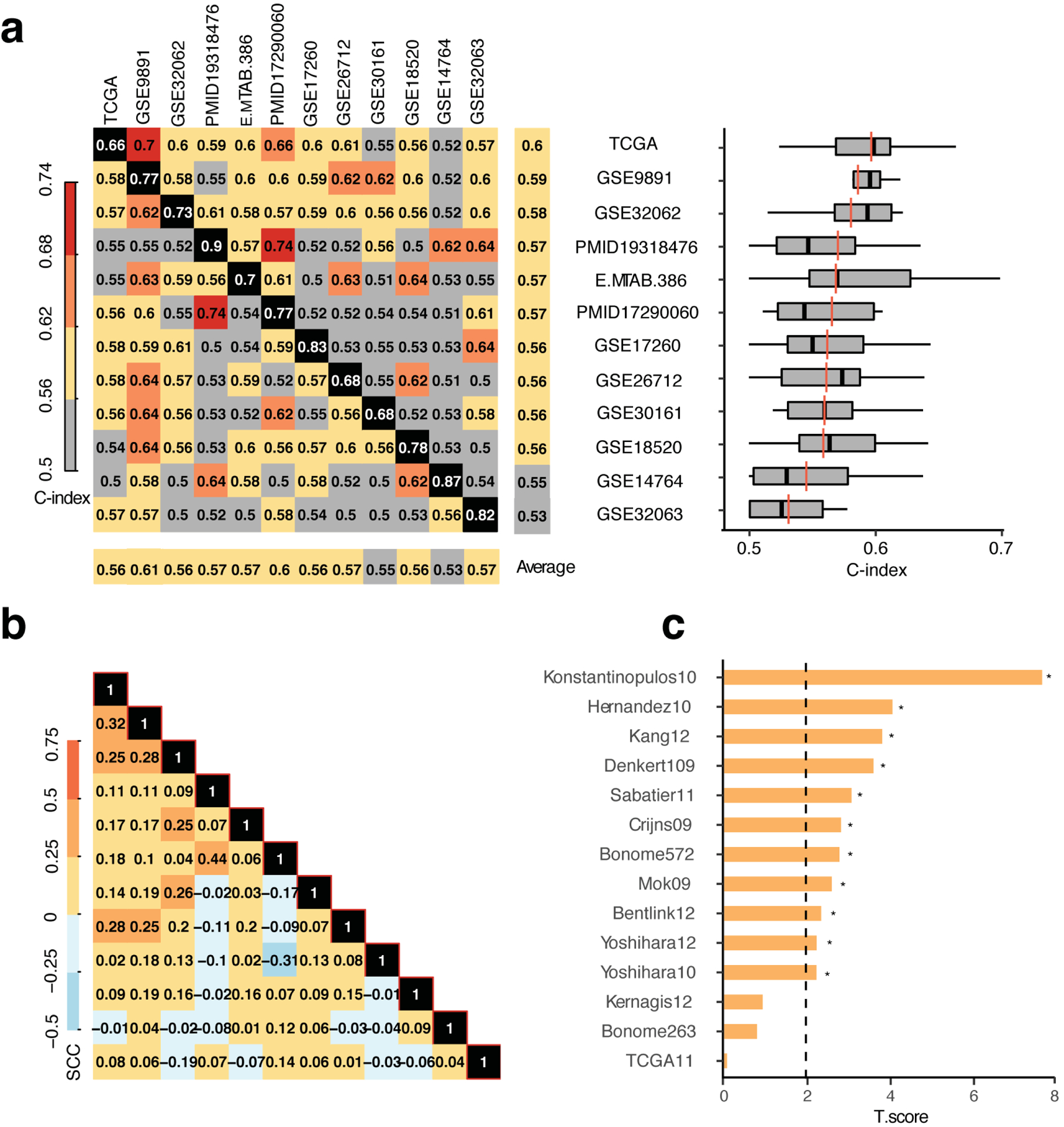
a. Left: concordance matrix with signatures derived from each dataset (horizontal labels) and applied to each other dataset (vertical labels). Numbers indicate C-indices. Black panels indicate self-concordance, which was excluded in all averages. Right: Box plot of C-indices displayed in rows of the concordance matrix. Vertical back lines indicate average including self-C-index, red lines indicate average excluding the self-C-index, outliers not indicated. b. Correlation matrix of prognostic genes across datasets based on the transformation -I(HR<1)*log10(P-value) for each gene in the signatures, where I indicates the indicator function. SCC = Spearman correlation coefficient. c. T statistics of C-indices reported in [26] compared to our TCGA-based C-indices (paired t-test). Dashed line indicates cutoff for significance of p<0.05.
To assess the similarities between the 12 derived OV signatures, we calculated the pairwise correlation between each of the signatures and observed relatively low correlations between datasets (Fig. 4b). This indicated that the prognostic genes identified by different datasets generally have low consistency. Despite this, the WTSPP framework can still achieve consistent prognostic prediction across different datasets.
Lastly, since several gene expression-based prognostic signatures have been proposed in the literature for high-grade ovarian cancer, we compared the prognostic ability of our OV signature to published gene signatures. Waldron et al. recently evaluated 14 prognostic signatures in publicly available datasets [26]. We evaluated the performance of our OV signature in the same datasets and conducted paired t-tests on the C-indices of our signature and those reported by Waldron. Our signature outperformed all 14 published OV signatures reported in the review, as judged using the t-statistic, which indicated the direction of higher concordance scores, higher t-statistic meaning higher concordance scores produced by our OV signature (Fig. 4c). The largest difference in prognostic ability was observed with the Konstantinopulous signature, which was clearly outperformed by our signature (P=1e-4, paired t-test). Interestingly, our signature performed slightly better than a published TCGA-derived signature. This TCGA signature was derived from a TCGA OV microarray dataset and adapted a similar Cox regression approach but selected the most significant genes to create a signature, rather than the entire transcriptome [33]. This finding confirmed the superiority of TCGA data in defining prognostic signatures. Thus, our TCGA-derived OV signature outperformed all evaluated published signatures for ovarian cancer.
Application of WTSPP to prognostic prediction in lung adenocarcinoma
To exemplify the potential use of the WTSPP platform in other cancer types, we extended our analysis to lung adenocarcinoma. We used the lung adenocarcinoma (LUAD) TCGA dataset (Table 1) to generate a prognostic signature and applied the signature to a collection of LUAD microarray datasets from previous publications (Supp Table 6). For each dataset with at least 40 samples, we calculated PSs for all patients. Cox regression analysis indicated that PSs were significantly associated with overall survival in 8 out of 10 independent datasets (Fig. 5a, Suppl. Fig 3a) (HR < 1, P < 0.05, Cox regression model). Furthermore, by stratifying patients into PS-high and PS-low groups using median PS as threshold, patients in the PS-high group exhibited significantly longer overall survival than those from the PS-low group (P < 0.05, log rank test) (Fig. 5a). In addition, our signature was also predictive of recurrence free survival (RFS) in 3 out of 4 datasets (Suppl. Fig 3b). Datasets c00182 and GSE31210 provided extensive clinical information, which we utilized to preform multivariate Cox regression including PSs as a predictor. We observed that PSs are still prognostic in both datasets, even after adjusting for clinical variables such as stage, age and smoking status (Fig. 5b). Taken together, these results indicate that the LUAD signature defined from TCGA RNAseq data by our WTSPP framework is predictive of patient prognosis in independent datasets.
Figure 5: The LUAD signature is predictive of patient survival in independent lung adenocarcinoma gene expression datasets.
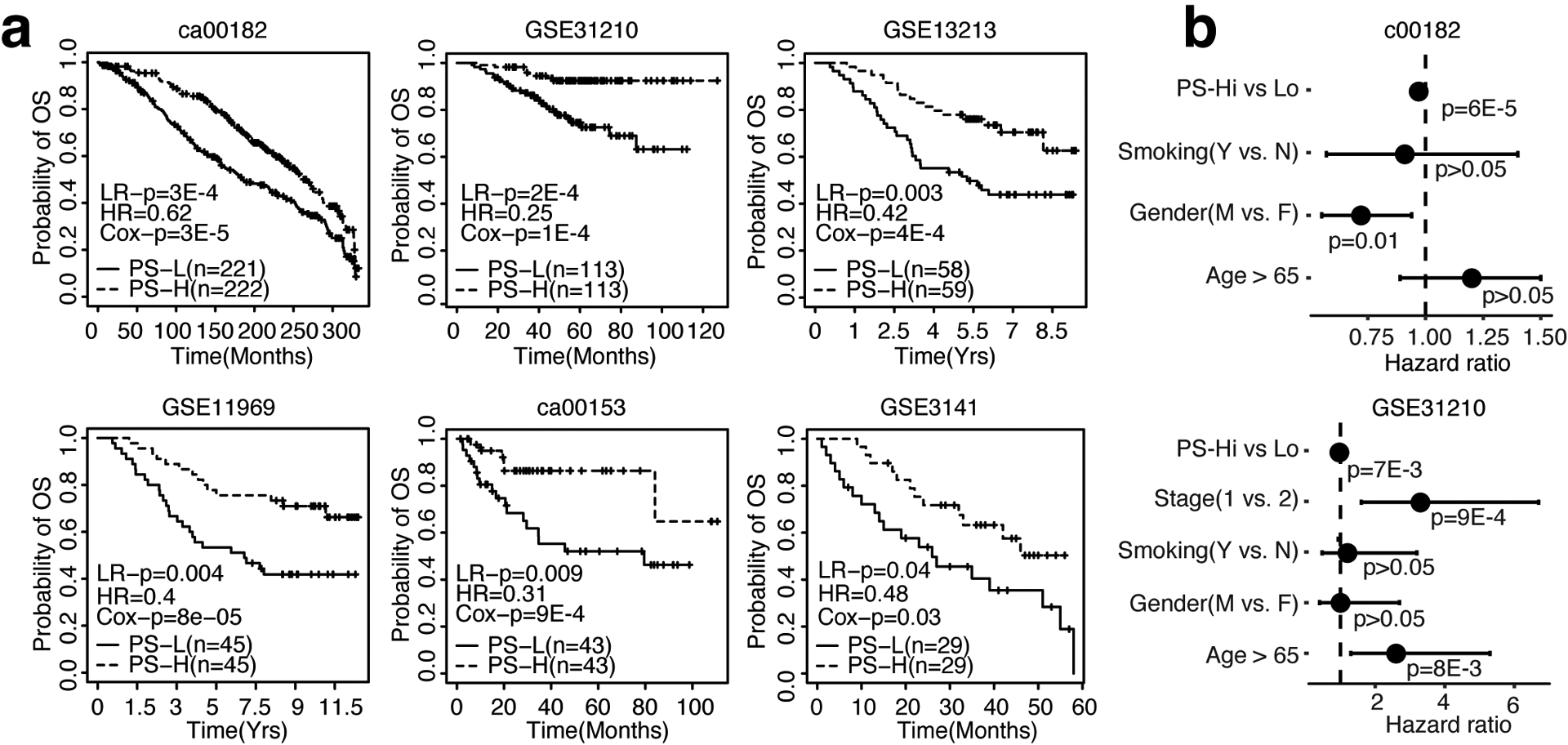
a. Kaplan-Meier curves depicting overall survival (OS) probability over time, ordered by sample size (n). b. Forest plots showing results of multivariate coxph model including all variables depicted. For all Kaplan-Meier curves, patients were dichotomized into PS-high (red) and PS-low (blue) using median PS as threshold. LR-p = p-value of Log Rank test, HR = Hazard Ratio, Cox-p = p-value calculated from the univariate Cox regression model in which PS is used as the only variable.
Discussion
Considerable effort has been devoted to understanding transcriptional deregulation in cancer. Consequently, a large number of prognostic signatures have been proposed based on gene expression levels, with varying levels of prognostic ability. However, a prognostic platform that can generate prognostic signatures on the pan-cancer level has been lacking. In this study, we presented a prognostic framework that generates cancer-specific signatures and evaluated the OV and LUAD signatures extensively.
Various proposed prognostic signatures are created by selecting a number of highly associated mRNA transcripts as determined by a method of preference, or by the inclusion of genes known to take part in a relevant biological process. The key assumption of these studies is that prognosis can be captured by a relatively small number of genes. However, as we have shown here, a much larger number of transcripts may be involved in tumor prognosis prediction, with each individual gene having a weak association with patient outcome. As an example, none of the genes in the OV signature were significantly associated with prognosis after multiple testing correction (Table 1). We did attempt to reduce the prognostic signatures to contain a smaller number of genes by only including the top x% prognostic genes, were x was ranged from 90 to 20 (data not shown). Signatures were relatively stable in predicting survival when excluding up to 50% of the genes, but the C-indices dropped when excluding more genes. In addition, the complete signature preformed best in the majority of dataset and the smallest variance in C-indices was observed with the complete signature. Additionally, when comparing the prognostic genes amongst ovarian cancer signatures (Fig 3c), we observed heterogeneity between signatures. By using a rank-based approach that includes all genes, we are still able to capture enough prognostic genes to predict prognosis. Thus, the inclusion of all genes in our prognostic signatures generates a robust framework.
Using ovarian cancer as an example, we showed that our OV signature captures tumor characteristics that cannot be accounted for by clinical variables. Importantly, clinical variables can further enhance the predictive power of our signature. Multiple, individual clinical variables were associated with PSs, where high-PS predicted lower stage, non-recurrence, optimal debulking, and less aggressive ovarian cancer subtypes. PS could also further separate optimal and suboptimal debulking groups, showing that patients with suboptimal cytoreduction status and high PS had a similar prognosis as optimal cytoreductory patients with low PS. These findings are of unique importance, as TMN staging and cytoreductory status are currently the only prognostic indications for ovarian cancer [32]. Notably, our OV-WTSPP signature outperformed 14 published prognostic OV signatures, indicating the strength of our WTSPP framework in generating prognostic signatures. In addition, application of the framework in a different cancer type, LUAD, showed a similar association with prognosis, indicating the extended application of our prognostic framework, as it can be used to generate prognostic signatures for multiple cancer types.
The WTSPP framework generates RNA sequencing platform-independent signatures, which can be applied to RNAseq, one-channel and two-channel microarray datasets without modification (Supp table 3). This is an important aspect, because the difference in sensitivity and data distribution between these two techniques results in an incompatibility of analysis methods and hinders the utility of applications designed for certain platforms [35]. We showed the superiority of using RNAseq TCGA data over other published datasets (Fig 3a). This superiority is likely due to at least two reasons, (i) RNAseq is more sensitive than microarray studies and generates more appropriate weights for the signature, (ii) the number of genes is higher in RNAseq datasets, which captures tumor characteristics better than microarray datasets that contain a more limited number of genes. Thus, we show that TCGA data has prognostic value and can be at the basis of robust signatures that are prognostic in independent datasets.
Although our study proposes a new prognostic framework, several limitations are noted here. First, our analysis was conducted retrospectively. To validate the clinical utility of our signatures, they should be tested in a prospective manner under conditions that simulate clinical application of the signature to determine whether the use of the signature results in patient benefit. Second, most subjects in TCGA received standard treatments, which might limit the use of our signature for prognostic prediction for patients treated with newly developed treatments, such as immunotherapy or targeted therapies. Third, we utilized overall survival to determine a gene’s association with prognosis in TCGA datasets, however, disease-specific survival would likely improve the prognostic accuracy of our framework. Finally, our framework only assesses mRNA expression levels, which might not capture all prognostic variables present in a tumor. Additional omics technologies could be added to our signatures to improve prediction accuracy.
In conclusion, we have established a generic framework that has been well validated and can easily be applied to other datasets. Additionally, WTSPP can be further extended to other diseases. We have made an R package available to facilitate the use of our signatures.
Supplementary Material
Acknowledgements
This work is supported by the Cancer Prevention Research Institute of Texas (CPRIT) (RR180061 to CC) and the National Cancer Institute of the National Institutes of Health (1R21CA227996 to CC). CC is a CPRIT Scholar in Cancer Research.
Footnotes
Disclosure of Potential Conflicts of Interests
The authors declare they have no competing interests.
Supplementary information
Supplementary information is available at Laboratory Investigation’s website.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–386. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller, Kimberly D, Ahmedin Siegel J. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, et al. , editors. AJCC Cancer Staging Manual. 8th ed Springer International Publishing; 2017. [Google Scholar]
- 5.Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:472–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortez AJ, Tudrej P, Kujawa KA, Lisowska KM. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol 2018;81:17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang S-J, Hodeib M, Chang J, Bristow RE. Survival impact of complete cytoreduction to no gross residual disease for advanced-stage ovarian cancer: a meta-analysis. Gynecol Oncol 2013;130:493–8. [DOI] [PubMed] [Google Scholar]
- 8.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev 2011;25:534–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beumer IJ, Persoon M, Witteveen A, Dreezen C, Chin S-F, Sammut S-J, et al. Prognostic Value of MammaPrint® in Invasive Lobular Breast Cancer. Biomark Insights 2016;11:139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. New England Journal of Medicine 2004;351:2817–26. [DOI] [PubMed] [Google Scholar]
- 11.Tsai M, Lo S, Audeh W, Qamar R, Budway R, Levine E, et al. Association of 70-Gene Signature Assay Findings With Physicians’ Treatment Guidance for Patients With Early Breast Cancer Classified as Intermediate Risk by the 21-Gene Assay. JAMA Oncol 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toustrup K, Sørensen BS, Metwally MAH, Tramm T, Mortensen LS, Overgaard J, et al. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. Acta Oncol 2016;55:1091–8. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Forker L, Irlam JJ, Pillay N, Choudhury A, West CML. Validation of a hypoxia related gene signature in multiple soft tissue sarcoma cohorts. Oncotarget 2018;9:3946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial to mesenchymal transition. Clin Cancer Res 2016;22:609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Varn FS, Cai G, Xiao F, Amos CI, Cheng C. A P53-Deficiency Gene Signature Predicts Recurrence Risk of Patients with Early-Stage Lung Adenocarcinoma. Cancer Epidemiol Biomarkers Prev 2018;27:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S, Moriya T, Ishida T, Shibata H, Sasano H, Ohuchi N, et al. Prediction of breast cancer prognosis by gene expression profile of TP53 status. Cancer Sci 2008;99:324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Ung MH, Cantor S, Cheng C. Computational Investigation of Homologous Recombination DNA Repair Deficiency in Sporadic Breast Cancer. Scientific Reports 2017;7:15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh SC, Sohn BH, Cheong J-H, Kim S-B, Lee JE, Park KC, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nature Communications 2018;9:1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia 2012;26:2406–13. [DOI] [PubMed] [Google Scholar]
- 20.O’Mara TA, Zhao M, Spurdle AB. Meta-analysis of gene expression studies in endometrial cancer identifies gene expression profiles associated with aggressive disease and patient outcome. Sci Rep 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian S Identification of Subtype-Specific Prognostic Genes for Early-Stage Lung Adenocarcinoma and Squamous Cell Carcinoma Patients Using an Embedded Feature Selection Algorithm. PLOS ONE 2015;10:e0134630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res 2012;72:6097–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentink S, Haibe-Kains B, Risch T, Fan J-B, Hirsch MS, Holton K, et al. Angiogenic mRNA and microRNA Gene Expression Signature Predicts a Novel Subtype of Serous Ovarian Cancer. PLOS ONE 2012;7:e30269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabatier R, Finetti P, Bonensea J, Jacquemier J, Adelaide J, Lambaudie E, et al. A seven-gene prognostic model for platinum-treated ovarian carcinomas. Br J Cancer 2011;105:304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kernagis DN, Hall AHS, Datto MB. Genes with bimodal expression are robust diagnostic targets that define distinct subtypes of epithelial ovarian cancer with different overall survival. J Mol Diagn 2012;14:214–22. [DOI] [PubMed] [Google Scholar]
- 26.Waldron L, Haibe-Kains B, Culhane AC, Riester M, Ding J, Wang XV, et al. Comparative Meta-analysis of Prognostic Gene Signatures for Late-Stage Ovarian Cancer. J Natl Cancer Inst 2014;106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst 2010;102:464–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tímár J, Gyorffy B, Rásó E. Gene signature of the metastatic potential of cutaneous melanoma: too much for too little? Clin Exp Metastasis 2010;27:371–87. [DOI] [PubMed] [Google Scholar]
- 29.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics 2007;8:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varn FS, Andrews EH, Mullins DW, Cheng C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat Commun 2016;7:10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varn FS, Wang Y, Mullins DW, Fiering S, Cheng C. Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment. Cancer Res 2017;77:1271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers 2016;2:16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhaak RGW, Tamayo P, Yang J-Y, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 2013;123:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel-MacDonnell JR, Usset J, Goode EL, Fridley BL. Assessment of data transformations for model-based clustering of RNA-Seq data. PLOS ONE 2018;13:e0191758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


