Abstract
The N-Methyl-D-aspartate (NMDA) receptor is a crucial mediator of pathological glutamate-driven excitotoxity and subsequent neuronal death in acute ischemic stroke. Although the roles of the NMDAR’s composite GluN2A-C subunits have been investigated in this phenomenon, the relative importance of the GluN2D subunit has yet to be evaluated. Herein, GluN2D−/− mice were studied in a model of ischemic stroke using MALDI FT-ICR mass spectrometry imaging to investigate the role of the GluN2D subunit of the NMDA receptor in brain ischemia. GluN2D−/− mice underwent middle cerebral artery occlusion (MCAO) and brain tissue was subsequently harvested, frozen, and cryosectioned. Tissue sections were analyzed via MALDI FT-ICR mass spectrometry imaging. MALDI analyses revealed increases in several calcium related species, namely vitamin D metabolites, LysoPC, and several PS species, in wild type mouse brain tissue when compared to wild type. In addition, GluN2D−/− mice also displayed an increase in PC, as well as a decrease in DG, suggesting reduced free fatty acid release from brain ischemia. These trends indicate that GluN2D−/− mice show enhanced rates of neurorecovery and neuroprotection from ischemic strokes compared to wild type mice. The cause of neuroprotection may be the result of an increase in PGP in knockout mice, contributing to greater cardiolipin synthesis and decreased sensitivity to apoptotic signals.
Keywords: Imaging, Bioanalytical methods, Mass Spectrometry
Graphical Abstract
Introduction
N-methyl-D-aspartate receptors (NMDAR) are glutamate-gated ion channels, which play a critical role in cellular signaling in the mammalian nervous system [1], contributing to synaptic plasticity, memory, and learning functions [2]. However, these receptors are also involved in processes related to neurodegenerative diseases and brain trauma [1, 3]. Composed of two GluN1 subunits and two GluN2 subunits, most NMDA receptors are heterotetramers, with both GluN1 and GluN2 subunits consisting of different splice variants. GluN1 subunits may be expressed by eight different splice variants (a-h), while GluN2 subunits are expressed by four different splice variants (A-D) [1, 4]. These variations contribute to the great diversity of NMDA receptors present in mammalian neurons, each with distinct biochemical properties [5, 6]. Because each subunit variant exhibits their own unique chemical properties, the type of subunit present in the NMDA receptor determines the receptors’ functional properties and its involvement in biological processes, including its role in neurological diseases [7, 8].
Previous studies have shown that following an induced ischemic stroke, ischemia is attenuated in GluN2A−/− and GluN2A−/−/GluN2B−/− mice when compared to wild-type mice [9]. In addition, previous studies in the Castellino laboratory have found that mice that do not express the GluN2C subunit displayed decreased cerebral edema and an enhanced rate of neurological recovery following ischemic stroke when compared to WT mice [10]. These neuroprotective changes corresponded to decreased expression of Fyn kinase and thus decreased phosphorylation of the GluN2B subunit. Although many of the ischemic roles of the GluN2A, GluN2B, and GluN2C subunits have been heavily studied, the role of the GluN2D subunit, with respect to ischemic strokes, has not been thoroughly investigated. Here, we investigate the effects of ischemic strokes on GluN2D/- mice using MALDI FT-ICR mass spectrometry imaging (MSI).
MALDI MSI is an effective tool for mapping proteins [11, 12, 13], peptides [13, 14], lipids [15, 16], and metabolites [17, 18, 19] in biological samples, allowing detection of these species while gaining valuable information about their spatial localization within a tissue sample. In addition, MALDI MSI enables determination of molecular and spatial information for a variety of species without prior knowledge of their presence, effectively allowing one to screen hundreds of species simultaneously during a single acquisition [20, 21]. The objective of this study is to investigate metabolomic and lipidomic changes in GluN2D−/− mice after an ischemic stroke using MALDI MSI. By doing so, we gain valuable information about the GluN2D subunit of the NMDA receptor and its effect on brain ischemia.
To accomplish this goal, we utilized the annotation platform, Metaspace. Metaspace is a platform which hosts a search engine for metabolite annotation of mass spectrometry imaging data, allowing users to search their data and obtain peak annotations from several public databases simultaneously. By doing these searches, Metaspace provides the user with several false discover rate (FDR)-controlled annotations for their imaging data within a matter of minutes [22, 23]. In addition, the Metaspace workflow can be utilized to annotate metabolites and lipids from a number of different samples and from many different organisms [24, 25, 26]. Imaging data may be from both 2D and 3D datasets acquired in both negative and positive ion modes [27]. By utilizing MALDI MSI coupled with a Metaspace workflow, we detected and identified metabolomic and lipidomic changes in ischemic mice and organized them by location within the healthy or damaged brain tissue in order to understand the off target effects of the GluN2D subunit during brain ischemia.
Materials and Methods
Chemicals and Materials
Alpha-cyano-4-hydroxycinnamic acid (CHCA) matrix was purchased from Sigma Aldrich (St. Louis, MO). ITO-coated glass slides were purchased from Delta Technologies (Loveland, CO). All unspecified reagents were purchased from Sigma Aldrich.
Mice
Male postnatal (day 21–33) wild type mice (C57B1/6J; Jackson Laboratories, Bar Harbor, ME) and GluN2D knockout/nβ-galactosidase knock in mice (GluN2D−/−) (NIH). Mice were housed on a 12 hour light-dark cycle with ad libitum access to food and water. All animal procedures were to accordance with the University of Notre Dame animal care committee’s regulations.
Stroke Induction
Mice were anesthetized with isofurane before the main carotid and internal carotid arteries were ligated with a 6–0 suture to prevent blood flow. Coated-tip sutures (#602334, Doccol Corporation, Sharon, MA) were then inserted to occlude the middle carotid artery via the external carotid artery. After 90 minutes, mice were anesthetized again to remove the occluder and to allow reperfusion. Mice were then sacrificed 24 hours after stroke induction.
Tissue Preparation
Brain samples from GluN2D−/− and wildtype mice that had and had not undergone ischemic strokes were collected and flash frozen using liquid N2 immediately following sacrifice. Tissue from B (frontal lobe and temporal lobe) and C (parietal lobe and temporal lobe) regions of the brain were then sectioned. All mouse brain tissues were sectioned into consecutive 15 μm slices using a cryostat sectioning device at −20°C. Mouse brain tissue sections were thaw mounted onto ITO-coated glass slides and coated with 10 mg/mL CHCA matrix using an HTX Imaging M5 TM-Sprayer (Chapel Hill, NC). Matrix was applied at 80°C for 4 passes over the sample. The flow rate of matrix was 0.1 mL/min at a velocity of 1000 mm/min, track spacing of 2 mm, pressure 10 psi, gas flow rate of 3 L/min, nozzle height 40 mm, and a drying time of 20 seconds between each pass. Tissue slides were stored in a vacuum desiccator until MALDI FT-ICR mass spectrometry imaging analysis.
MALDI MSI
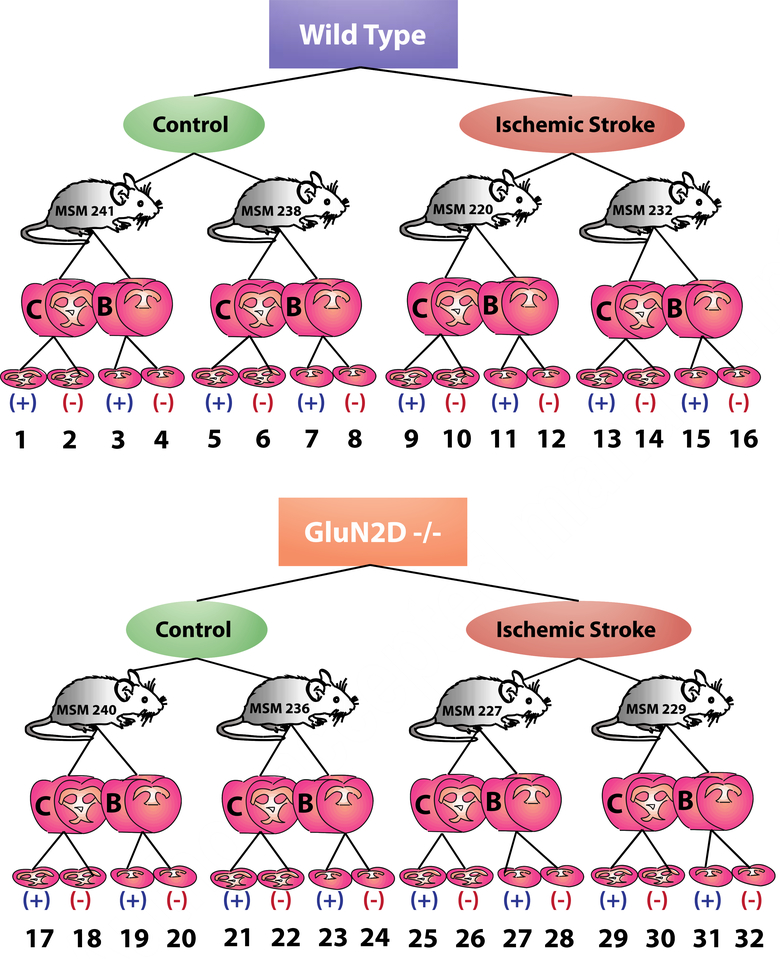
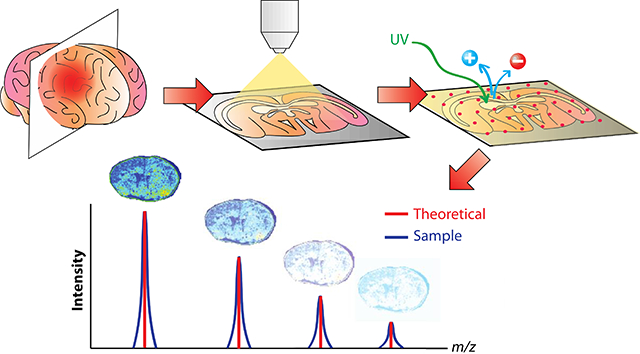
A 15T Solarix FT-ICR (Bruker Daltonics, Billerica, MA) was operated in positive and negative ion modes with a mass range set to acquire at m/z 100–1000. The laser spot size was set to medium with the raster distance set to 100 μm along both the x and y axes. Laser power was adjusted to just above the MALDI threshold to avoid fragmentation. To obtain optimal signal intensity, each pixel is the result of 500 consecutive laser shots with external calibration performed using a red phosphorous mixture on a CHCA-coated slide. The images were processed using ftmsControl 2.1, flexImaging 4.1, and SCiLS Lab 2016b (Bruker Daltonics, Billerica, MA). All spectra were normalized against total ion count, defined as the sum of all intensities in the mass range analyzed to reduce influences by matrix hot spots. To generate reliable comparisons, one tissue section from each condition (knockout stroke, knockout control, wild-type stroke, wild-type control) in the same relative regions of the brain (B region) was analyzed at the same time to minimize day-to-day variability. To generate intensity plots and ion maps, SCiLS Lab 2016b was used to compare four tissue sections that were run on the same day, each from a different condition, as representative sections to compare intensities. Consecutive tissue sections from all four conditions were analyzed in both positive and negative ion modes for both B and C regions of the brain, resulting in 32 total tissue sections analyzed by MALDI-FT-ICR MSI (Figure 1).
Metaspace
All 32 mouse brain tissue sections were processed in SCiLS Lab and exported to Metaspace for lipid and metabolite annotation. Detection parameters for Metaspace annotation used a resolving power of 200,000 at m/z 500. Annotations were collected by searching the mass spectrometry imaging data against the Human Metabolome Database (HMDB) v4, BraChem Database 2018, and the Pseudomonas aeruginosa Database (PAMDB) v1.0. All annotations were acquired in MS1 and thus correspond to Level 2 of the Metabolomics Standards Initiative Guidelines [28].
Results
Comprehensive Analysis of Metaspace Results
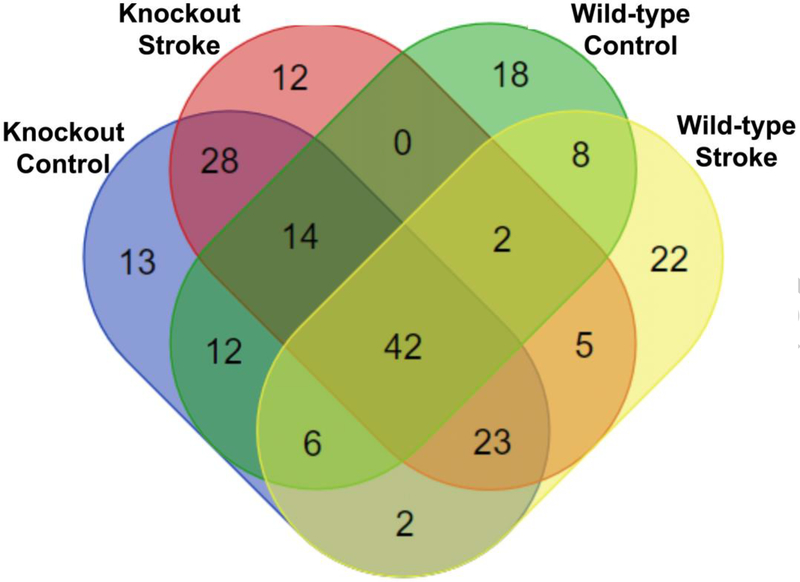
Resulting annotations from Metaspace were compared and the results are provided in Figure 2. Metabolite annotations from all conditions show correlation, as 42 of the 207 total annotations are found in mice from all four conditions. These species mostly consist of PE, PC, and other lipids (see Electronic Supplementary Material (ESM) Table S1). The high number of species that were detected in all four conditions indicates the reproducibility of the Metaspace annotation workflow, even in samples from different conditions.
Several species were detected in only one of the four conditions. The knockout control mice had 13 unique annotations (ESM Table S2), knockout stroke mice had 12 unique annotations (ESM Table S3), wild-type control mice 18 unique annotations (ESM Table S4), and wild-type stroke mice had 22 unique annotations (ESM Table S5). When comparing the lists of uniquely annotated species in the four different conditions, knockout control mice have a greater number of annotated phosphoinositide (PI) lipids, while knockout stroke mice had a greater number of unique phosphatidylglycerol (PG) annotations than the other three conditions.
Discussion
It is important to note that although a species may not have been annotated in a given sample, this absence does not necessarily indicate that it is not present in the sample. In order to annotate a given analyte, Metaspace assigns an MSM score to a set of peaks detected in the sample. MSM scores are assigned based on spatial, spectral, and chaos parameters [22]. Spatial scores are assigned based on whether or not a set of peaks match up spatially to form a substructure, while spectral scores are assigned by relative isotope intensities. It is possible for an analyte to be present, but not necessarily be annotated due to limited isotope intensity or structure within the tissue sample. Thus, the presence or absence of annotated analyte was determined by comparing intensity plots for an analyte’s respective parent ion.
Comparing annotations from B and C sections of the same mouse showed very little overlap (ESM Fig. S1), suggesting both structural and molecular differences between the two regions of the brain. However, comparing annotations from all mouse sections from the same cohort showed improved agreement, as seen in ESM Fig. S2, with control mice having significantly more overlap than mice that had undergone a stroke. This difference in overlap was attributed to stroke mice having varying degrees of neurological deficit. Comparing annotations even further, knockout mice, as a whole, displayed significantly greater overlap than wildtype mice as a whole (ESM Fig. S3). Knockout mouse sections had nearly twice as many annotations in common as wild type mouse sections when comparing stroke versus control. This suggests that knockout stroke mice are more similar to knockout control mice and that wild type control mice and wild type stroke mice are quite different.
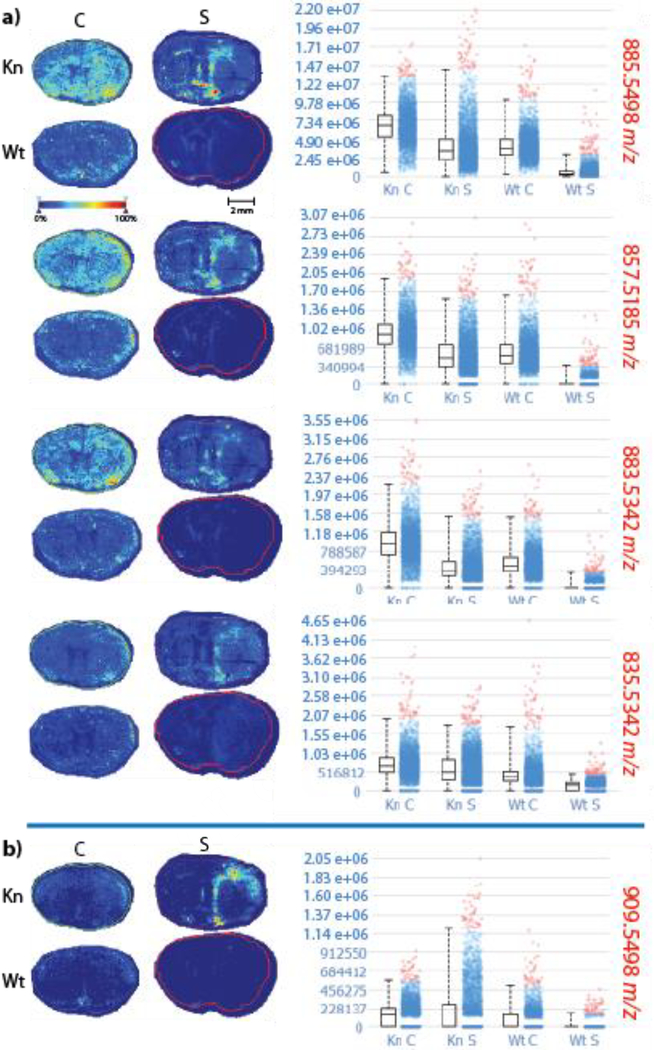
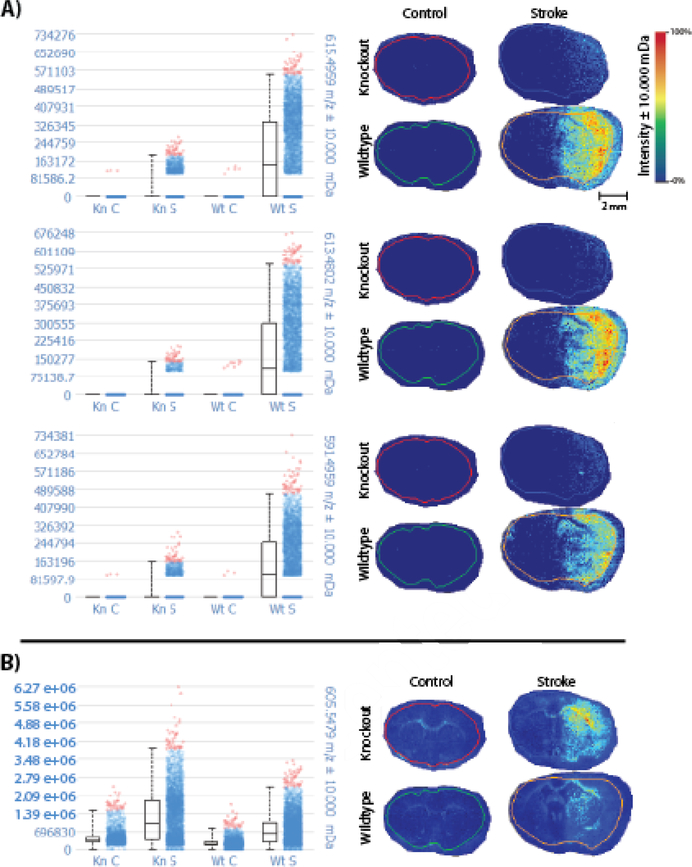
Although knockout control mice appeared to have unique PI annotations with parent ions at m/z 909.5498 and 835.5342, comparing intensity plots for these parent ions show that m/z 909.5498 is actually much more intense in the knockout stroke model than in the other three conditions, while m/z 835.5342 exhibits relatively equal intensities in all four conditions (Figure 3). In addition, when comparing intensity plots for all five PI annotations obtained in Metaspace, we see that PI species in general appear to be relatively equal in abundance in all four conditions, with the exception of m/z 909.5498. The annotations for all five PI species are displayed in ESM Table S6. When comparing the intensity plots for all PI species, m/z 909.5498 appears to be much more intense in knockout stroke mouse brains when compared to the other three conditions. PI plays an important role in membrane function and trafficking by regulating the activity of many integral membrane proteins and ion channels [29]. PI variation between the knockout and wild type mice is logical, as the knockout mouse model did not express the GluN2D subunit of the NMDA receptor, which is a calcium ion channel.
Metaspace Identification of Calcium-Related Species
During ischemic strokes, high blood flow rates cause sudden neuron depolarization [30, 31]. This depolarization results in a sudden influx of calcium to enter the cells through ion channels such as the NMDA receptor. Elevated intracellular calcium then begins to overload the mitochondria, reducing its capacity for oxidative phosphorylation [32]. Because of these calcium-related effects that occur during ischemia, several calcium-related species identified using Metaspace were specifically examined in this study.
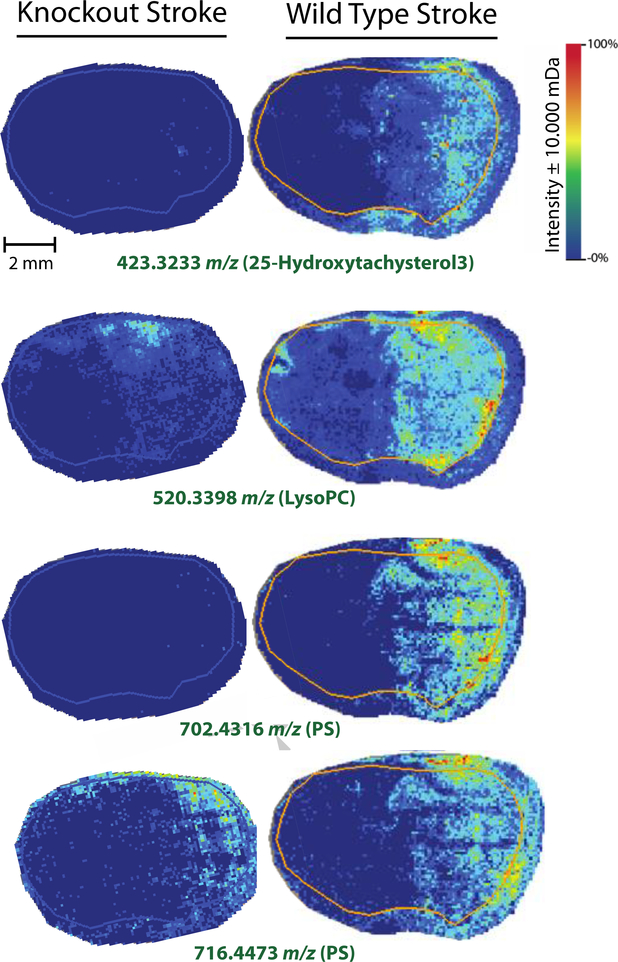
Of the 207 unique annotations retrieved via Metaspace, several calcium-related species were identified. The presence and absence of these species were expected to vary in wild type and knockout mouse models, as the knockout model is missing a subunit of a calcium ion channel. The collected data supported this hypothesis. Annotations for 25-hydroxytachysterol3, lysophosphatidylcholine (LysoPC), and several phosphatidylserine (PS) species were obtained using Metaspace. Heat maps for these species in knockout and wild type stroke mouse brains are displayed in Figure 4. The first species, 25-hydroxytachysterol3, was only detected in the damaged half of wild type stroke brain tissue. In addition, this species was completely absent in knockout stroke mouse brain tissue. Hydroxytachysterol3 is a biologically active metabolite of vitamin D3. When biologically active, hydroxytachysterol3 is characterized by its ability to elevate serum calcium concentrations [33, 34, 35]. In addition, previous studies have shown that serum calcium levels can be used as a prognostic indicator of ischemic strokes, as its presence directly correlates with ischemia in patients [36]. It is worth noting that accurate mass isotope matching gives several other possible annotations for this species, many of which, are also vitamin D metabolites such as alfacalcidol and calcidiol. The presence of these vitamin D metabolites in wild type mouse brains after ischemic stroke agree with the results of previous studies. In addition, the absence of these metabolites in ischemic knockout mouse brains suggest an enhanced rate of neurological recovery following ischemic stroke, as previously observed in GluN2C−/− mice [10].
A similar trend was also observed when analyzing lysoPC levels in knockout and wild type mouse brains after ischemic stroke. Although lysoPC is produced by a number of different pathways, one of the main biological methods of lysoPC synthesis is via hydrolysis of an oxidized phosphocholine (PC) fatty acid by lipoprotein-associated phospholipase A2 (Lp-PLA2) [37, 38] Lp-PLA2 has been shown to be a predictor for ischemic stroke severity, as well as early neurological deterioration in patients with acute ischemic strokes [39, 40, 41]. Because Lp-PLA2 is the main producer lysoPC in most biological systems, lysoPC could reasonably be an acceptable indicator of Lp-PLA2 activity. The presence of lysoPC in wild type ischemic stroke mouse brains, as well as the absence of lysoPC in knockout ischemic stroke mouse brains, agree with the previous findings that suggest that GluN2D−/− mice show similar neuroprotection from ischemia as GluN2C−/− mice.
In addition, the same trend that was observed for the vitamin D metabolites and lysoPC was also observed for several PS species, as seen in Figure 4. Three of the eight PS species annotated using Metaspace were more abundant in wildtype ischemic stroke mouse brains when compared to knockout stroke mouse brains. Two of these PS species (m/z 830.5308 and 702.4316) were detected at very low levels in the knockout stroke mouse brains. These data agree nicely with our previous findings, as PS lipid species are widely known to have the ability to chelate calcium [42, 43, 44, 45]. Interestingly enough, intracellular calcium concentration has been shown to regulate PS synthesis as well [46]. Because ischemic strokes result in an increase in intracellular calcium, eventually resulting in cell death, these results again agree that GluN2D−/− mice show some degree of neuroprotection from ischemic strokes, as PS was far lower in the knockout mouse brains when compared to the wildtype mouse brains.
Lipid Pathway Analysis
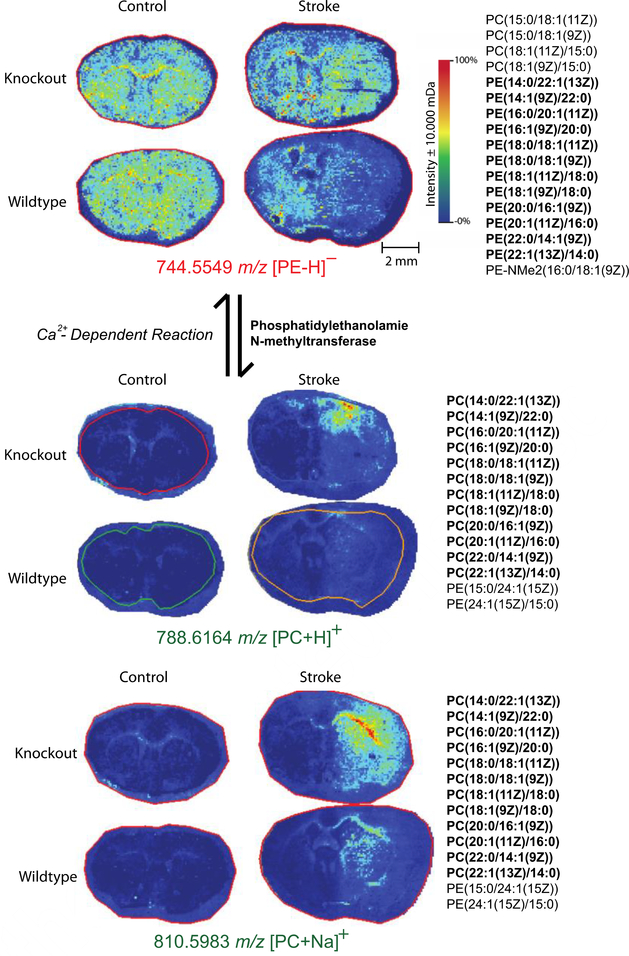
Using accurate mass annotations for the 207 unique species annotated by Metaspace, we analyzed lipid pathways by tracking specific sets of lipid side chains to gain for information about other possible effects of the GluN2D−/− knockout mutation during ischemia. One such pathway is displayed in Figure 5, where phosphatidylethanolamine N-methyltransferase catalyzes the production of phosphatidylcholine (PC) from phosphatidylethanolamine (PE) [47]. Using accurate mass isotope matching, Metaspace generated several possible side chains for both of these PE and PC species. By matching the side chains, we determined that this particular PE is converted to PC via this pathway. PE was detected to be relatively abundant in both knockout and wild type mouse brains; however, its PC counterpart is much more abundant in knockout stroke samples when compared to the other three conditions. Previous studies have shown that during ischemia, PC is broken down into free fatty acids [48, 49, 50]. These free fatty acids generate radicals that potentiate ischemic injury [51]. The absence of these PC species in wild type mice suggests greater ischemic injury in these mouse brains when compared to knockout mice. In addition, when comparing intensity plots for all 19 annotated PC species, 9 of the annotated PC species were detected with higher abundance in the knockout mice when compared to wildtype mice (ESM Fig. S4). In addition, 5 of the 19 annotated species were far less abundant in the wild type stroke mice than the other three conditions (ESM Fig. S5); the remaining four PC species were relatively equal in all four conditions. The abundance of PC in knockout mice and relative absence of PC in ischemic wild type mice further supports the hypothesis that GluN2D−/− mice show increased neuroprotetion and recovery from ischemic strokes when compared to wild type mice.
A similar trend was also observed when comparing annotated diacylglycerol (DG) species in wildtype and GluN2D−/− mouse brains. Of the four unique DG species annotated in Metaspace, three were far more abundant in wildtype stroke mouse brains when compared to the other three conditions (Figure 6). Several studies have reported an increase in DG species accompanying the increase in free fatty acids that result from PC degradation during ischemia [52, 53, 54, 55]. This observed increase in DG species in only the wildtype stroke mice supports the hypothesis of increased neuroprotection in GluN2D−/− mice.
Mitochondrial Stability
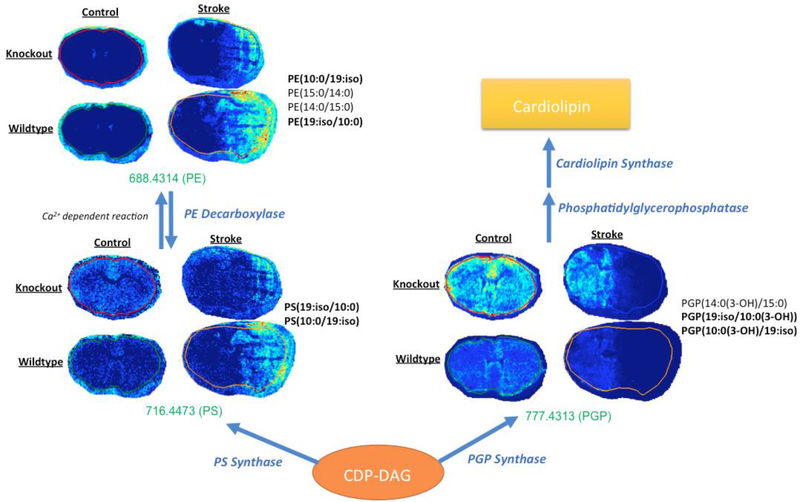
When conducting lipid pathway analysis, several trends relating to mitochondrial stability were observed as well. Of the three PS lipid species that are more abundant in wildtype stroke mouse brains, one species (m/z 716.4473) can be converted into PE (m/z 688.4314) via a calcium-dependent reaction in the inner mitochondrial membrane, catalyzed by phosphatidylserine decarboxylase [56]. When comparing both of these species, both PE and PS appear to be more abundant in the wild type stroke mouse brains (Figure 7). Previous studies report that PE and PS species translocate from the inner leaflet to the outer leaflet after ischemia [57, 58, 59] and have also found a direct correlation between non-bilayer preferring PE abundance and the degree of cell damage observed during ischemia [60]. Decreased detection of PE and PS species in the stroked regions of knockout mouse brains further support increased neuroprotection as a result of the GluN2D−/− mutation. In addition, these same lipid chains were also identified in a phosphatidyl glycerophosphate (PGP) lipid species. This particular lipid species was far more abundant in knockout mouse brains when compared to wild type.
In mammalian cells, PGP acts as one of the main precursors for cardiolipin synthesis. Phosphatidylglycerophosphatase catalyzes the conversion of PGP to phosphaidylglycerol, which is the immediate precursor of cardiolipin in a reaction catalyzed by cardiolipin synthase [61, 62]. Cardiolipin is a dimeric phosphoglycerolipid predominantly present in the inner mitochondrial membrane, but has also been reported to be present in the outer mitochondrial membrane as well [63]. In the mitochondria, cardiolipin has pivotal functions in cellular energy metabolism and the initiation of apoptotic pathways [64]. During mitochondrial dysfunction, superoxide production causes cardiolipin to become peroxidized, resulting in the release of cytochrome c [65, 66]. Previous studies have reported that cardiolipin deficient cells display increased sensitivity to apoptotic signals and accelerated rates of apoptosis [67]. This further supports the hypothesis that GluN2D−/− mice display increased neuroprotection from ischemic strokes. Increased abundance of PGP in knockout mice may contribute to increased cardiolipin synthesis, and thus, decreased sensitivity to apoptotic factors.
Conclusion
After MALDI FT-ICR MSI analysis of 32 mouse brain tissue sections, several trends relating to neuroprotection in GluN2D−/− mice were observed. When compared to wild type mice, GluN2D−/− mice displayed decreases in several calcium-related species including several possible vitamin D metabolites, LysoPC, and several PS species. GluN2D−/− mice also displayed an increase in PC abundance and a decrease of DG, indicating reduced ischemic free fatty acid and DG release. In addition, an increase in PGP abundance was observed in all knockout mice, suggesting decreased sensitivity to apoptotic factors. All of these observations suggest an enhanced rate of neurological recovery and neuroprotection from ischemic strokes when compared to wild type mice, just as previously observed in GluN2C−/− mice. Furthermore, these observations in MALDI MSI data that contribute to neuroprotection could not be detected through microscopy alone. Future directions for this project will be to directly analyze cardiolipin content in GluN2D−/− and wildtype mice to further investigate the causes of neuroprotection in the GluN2D−/− mouse model.
Supplementary Material
Acknowledgements
ABH was supported by NIGMS award R01-GM110406 and WTA was supported by NIA R21-AG062144. The authors would like to acknowledge the Campus Chemical Instrument Center at the Ohio State University for providing instrument usage, as well as Arpad Somogyi for helpful discussions and support. The 15 T Bruker SolariX FT-ICR instrument was supported by NIH Award Number Grant S10 OD018507.
Appendix
Figure 1.
Schematic of experimental setup to investigate ischemic effects of the GluN2D subunit of the NMDA receptor using MALDI-FT-ICR MSI. GluN2D−/− and wild type mice either did or did not experience ischemic strokes. Both B and C regions of the brain were isolated from each mouse and sectioned using a cryostat. Resulting consecutive tissue sections were then analyzed using MALDI-FT-ICR MSI in both positive (+) and negative (−) ion modes, resulting in 32 tissue sections for analysis. These 32 different sets of imaging data were then annotated using Metaspace.
Figure 2.
Comprehensive comparison of all annotations from 32 mouse brain tissue sections annotated using Metaspace. Annotations from all tissue sections collected from all mice of the same condition were pooled together to compare annotations from knockout control, knockout stroke, wild type control, and wild type stroke mice.
Figure 3.
MALDI-MSI heat maps for phosphatidylinositol (PI) species in knockout (Kn) control (C), knockout stroke (S), wild type (Wt) control, and wild type stroke mouse brains, annotated using Metaspace. All heat maps were acquired using a 15T FT-ICR mass spectrometer with a mass window of ±10 mDa. Mass-to-charge ratios in red indicate that samples were acquired in negative ion mode. PI species appeared to be in a) relatively equal abundance in all four conditions b) with the exception of m/z 909.5498.
Figure 4.
MALDI MSI heat maps for calcium related species: 25-hydroxytachysterol3 (m/z 423.3233), lysoPC (m/z 520.3398), and two PS species (m/z 702.4316 and 716.4473). Ion intensity bar and scale bar are displayed in the top panel. All four calcium-related species appeared to be more abundant in the wild type stroke mouse brains when compared to the knockout stroke mouse brains. An example of PS species detected exclusively in wild type stroke tissue (m/z 702.4316) and detected in both mouse stroke models (m/z 716.4473) are displayed. MALDI MSI analysis was conducted using a 15T FT-ICR mass spectrometer with a mass window of ±10 mDa. Mass-to-charge ratios in green indicate that samples were acquired in positive mode.
Figure 5.
MALDI MSI heat maps for phosphatidylethanolamine and phosphatidylcholine for lipids with identical side chains. Both PE and PC species appear to be more abundant in knockout mouse models than wild type mouse models. Possible identifications for each m/z are listed to the right of the ion heat maps. MALDI MSI analysis was conducted using a 15T FT-ICR mass spectrometer with a mass window of ±10 mDa. Mass-to-charge ratios in green indicate that samples were acquired in positive mode, while red indicates negative mode. Ion intensity bar and scale bar are displayed in the top panel
Figure 6.
MALDI MSI ion heat maps and intensity plots for DG species in knockout and control mouse brain tissue that have and have not undergone ischemic stroke. A) Three of the DG species annotated in Metaspace are far more intense in wildtype stroke mouse brains when compared to the other three conditions. B) One of the four DG species annotated in Metaspace appears to be relatively equal in abundance in both wild type and knockout mice after ischemia. Experiments were conducted on a15T FT-ICR mass spectrometer with a mass window of ±10 mDa Ion intensity bar and scale bar are displayed in the top panel
Figure 7.
Lipid pathway analysis and MALDI MSI heat maps for particular PE, PS, and PGP species annotated using Metaspace. PE and PS species appeared to be more intense in wild type stroke mouse brains, while PGP appeared to be more intense in knockout mouse brains. Possible side chain identifications are listed to the right of each respective heat map. Identifications that match other lipid species are bolded. Experiments were conducted on a15T FT-ICR mass spectrometer with a mass window of ±10 mDa. Mass-to-charge ratios in green indicate that samples were acquired in positive mode.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci 2013. 14, 383–400. [DOI] [PubMed] [Google Scholar]
- 2.Nakazawa K, McHugh TJ, Wilson MA, Tonegawa S. NMDA receptors, place cells and hippocampal spatial memory. Nat. Rev. Neurosci 2004, 5:361–372. [DOI] [PubMed] [Google Scholar]
- 3.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev 1999, 51:7–61. [PubMed] [Google Scholar]
- 4.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R, Glutamate receptor ion channels: structure, regulation, and function, Pharmacol. Rev 2010, 62, 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheriyan J, Balsara RD, Hansen KB, Casetellino FJ. Pharmacology of triheteromeric N-methyl-d-aspartate receptors. Neurosci Lett. 2016, 617, 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994, 12, 529–540. [DOI] [PubMed] [Google Scholar]
- 7.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Science STKE. 2004, re16. [DOI] [PubMed] [Google Scholar]
- 8.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994, 368, 144–147. [DOI] [PubMed] [Google Scholar]
- 9.Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011, 71, 1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes A, Zhou N, Donahue DL, Balsara R, Castellino FJ. A deficiency of the GluN2C subunit of the N-methyl-D-aspartate receptor is neuroprotective in a mouse model of ischemic stroke. Biochem Biophys Res Commun. 2018, 495, 136–144. [DOI] [PubMed] [Google Scholar]
- 11.Dudley E MALDI Profiling and applications in medicine. Adv Exp Med Biol. 2019, 1140, 27–43. [DOI] [PubMed] [Google Scholar]
- 12.Ahmen M, Broeck G, Baggerman G, Schildermans K, Pauwels P, Van Craenenbroeck AH, Dendooven A. Next-generation protein analysis in the pathology department. J Clin Pathol. 2019, doi: 10.1136/jclinpath-2019-205864. [DOI] [PubMed] [Google Scholar]
- 13.Berghmans E, Van Raemdonck G, Schildermans K, Willems H, Boonen K, Maes E, Mertens I, Pauwels P, Baggerman G. MALDI mass spectrometry imaging linked with top-down proteomics as a tool to study the non-small-cell lung cancer tumor microenvironment. Methods and Protocols. 2019, 2, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews WT, Skube SB, Hummon AB. Magnetic bead-based peptide extraction methodology for tissue imaging. Analyst. 2017, 143, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallah K, Quanico J, Raffo-Romero A, Cardon T, Aboulouard S, Devos D, Kobeissy F, Ziabara K, Salzet M, Fournier I. Matrix-assisted laser desorption/ionization-mass spectrometry imaging of lipids in experimental model of traumatic brain injury detecting acylcaritines as injury related markers. Anal Chem. 2019, doi: 10.1021/acs.analchem.9b02633. [DOI] [PubMed] [Google Scholar]
- 16.Tobias F, Olson MT, Cologna SM. Mass spectrometry imaging of lipids: untargeted consensus spectra reveal spatial distributions in Niemann-Pick disease type C1. J Lipid Res. 2018. 12, 2446–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Flinders C, Mumenthaler SM, Hummon AB. MALDI mass spectrometry imaging for evaluation of therapeutics in colorectal tumor organoids. J Am Soc Mass Spectrom. 2018. 29, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Hummon AB. Chemical imaging of platinum-based drugs and their metabolites. Sci Rep. 2016, doi: 10.1038/srep38507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eveque-Mourroux MR, Emans PJ, Zautsen RRM, Boonen A, Heeren RMA, Cilero-Pastor B. Spatially resolved endogenous improved metabolite detection in human osteoarthritis cartilage by matrix assisted laser desorption ionization mass spectrometry imaging. Analyst. 2019, doi: 10.1039/c9an00944b. [DOI] [PubMed] [Google Scholar]
- 20.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev 2011, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med 2001, 7, 493–496. [DOI] [PubMed] [Google Scholar]
- 22.Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, Kovalev V, Fuchser J, Nikolenko S, Pineau C, Becker M, Alexandrov T. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nature Methods. 2016, 14, 57–60. [DOI] [PubMed] [Google Scholar]
- 23.Alexandrov T, Ovchnnikova K, Palmer AJ, Kovalev V, Tarasov A, Stuart L, Nigmetzianov R, Fay D. METASPACE: A community-populated knowledge base of spatial metabolomes in health and disease. BioRxiv. 2019, doi: 10.1101/539478. [DOI] [Google Scholar]
- 24.Ellis SR, Paine MRL, Eijkel GB, Pauling JK, Husen P, Jervelund MW, Hermansson M, Ejsing CS, Heeren RMA. Automated, parallel mass spectrometry imaging and structural identification of lipids. Nature Methods. 2018, 5, 515–518. [DOI] [PubMed] [Google Scholar]
- 25.Vaysse PM, Heeren RMA, Porta T, Balluff B. Mass spectrometry imaging for clinical research-latest developments, applications, and current limitations. Analyst. 2017, 142, 2690–2712. [DOI] [PubMed] [Google Scholar]
- 26.Gemperline E, Horn HA, DeLaney K, Currie CR, Li L. Imaging with mass spectrometry of bacteria on the exoskeleton of fungus-growing ants. ACS Chem. Biol 2017, 12, 8, 1980–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duenas ME, Essner JJ, Lee YJ. 3D MALDI mass spectrometry imaging of a single cell: spatial mapping of lipids in the embryonic development of zebrafish. Sci Rep. 2017, 7, 14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summer LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007, 3(3), 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkenburger BH, Jensen JB, Dickson EJ, Suh BC, Hille B. Phosphoinositides: lipid regulators of membrane proteins. J Physiol. 588 (2010). 3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heuser D, Guggenberger H. Ionic changes in brain ischemia and alterations produced by drugs. Brit J Anesthesia. 57(1). (1985). 22–33. [DOI] [PubMed] [Google Scholar]
- 31.White BC, Wiegenstein JG, Winegar CD. Brain ischemia and anoxia: Mechanisms of injury. JAMA 251 (1984). 158–690. [PubMed] [Google Scholar]
- 32.Farber JL, Chien KR, Mittnacht S Jr. Myocardial ischemia: the pathogenesis of irreversible cell injury in ischemia. Am J Pathol. 102(2). (1981). 271–81. [PMC free article] [PubMed] [Google Scholar]
- 33.Blunt JW, DeLuca HF. The synthesis of 25-hydroxycholecalciferol. A biologically active metabolite of vitamin D3. Biochemistry. 8(2). (1969). 671–675. [DOI] [PubMed] [Google Scholar]
- 34.Kopic S, Geibel JP. Gastric acid, calcium absorption, and their impact of bone health. Physiol Rev. 93(1). (2013). 189–268. [DOI] [PubMed] [Google Scholar]
- 35.Sturkie PD. Hormonal Regulation of Calcium Metabolism. Basic Physiology. (1981). Springer-Verlag New York Incorporated; New York, NY: 414–418. [Google Scholar]
- 36.Borah M, Dhar S, Gogoi DM, Ruram AA. Association of serum calcium levels with infarct size in acute ischemic stroke: observations from northeast India. J Neurosci Rural Pract. 7(S1). (2016). S41–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canning P, Kenny BA, Prise V, Glenn J, Sarker MH. Hudson N, Brandt M, Lopez FJ, Gale D, Luthert PJ, Adamson P, Turowski P, Stitt AW. Lipoprotein-associated phospholipase A2 (Lp-PLA2) as a therapeutic target to prevent retinal vasopermeability during diabetes. Proc Natl Acad Sci U. S. A 113(26). (2016). 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, Ke LY. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol. 20(5). (2019). 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou F, Liu Y, Huang Q, Zhou J. Relation between lipoprotein-associated phospholipase A2 mass and incident ischemic stroke severity. Neurol Sci. 39(9). (2018). 1591–1596. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Hu S, Ren L, Lan T, Cai J, Li C. Lp-PLA2 as a risk factor of early neurological deterioration in acute ischemic stroke with TOAST type of large arterial atherosclerosis. Neurol Res. 41(1). (2019). 1–8. [DOI] [PubMed] [Google Scholar]
- 41.Ding CY, Cai HP, L Ge H, Yu LH, Lin YX, Kang DZ. Assessment of lipoprotein-associated phospholipase A2 level and its changes in the early stages as predictors of delayed cerebral ischemia in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. (2019). 1–7. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, Nagata S. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 288(19). (2013). 13305–13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bull RK, Jevons S, Barton PG. Complexes of prothrombin with calcium ions and phospholipids. J Biol Chem. 247(9). (1972). 2747–2754. [PubMed] [Google Scholar]
- 44.Vallabhapurapu SD, Blanco VM, Sulaiman MK, Vallabhapurapu SL. Chu Z, Franco RS, Qi X. Variation in human cancer cell external phosphatidylserine is regulated by flippase activity and intracellular calcium. [DOI] [PMC free article] [PubMed]
- 45.Martin-Molina A, Rodriguez-Beas C, Faraudo J. Effect of calcium and magnesium on phosphatidylserine membranes: experiments and all-atomic simulations. Biophys J. 102(9). (2012). 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aussel C, Pelassy C, Mary D, Breittmayer JP, Cousin JL, Rossi B. Calcium-dependent regulation of phosphatidylserine synthesis in control and activated Jurkat T cells. J Lipid Mediators Cell Signaling. (1991). 3(3). 267–281. [PubMed] [Google Scholar]
- 47.Zinrajh D, Horl G, Jurgens G, Marc J, M Sok C Cerne. Increased phosphatidylethanolamine N-methyltransferase gene expression in non-small-cell lung cancer tissue predicts shorter patient survival. Oncol Lett. 7(6). (2014). 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muralikrishna Adibhatla R, Hatcher JF, Larsen EC, Chen X, Tsao FHC. CDP-choline significantly restores phosphatidylcholine levels by differentially affecting phospholipase A2 and CTP: phosphocholine cytidylyltransferase after stroke. J Biol Chem. 281 (2005). 6718–6725. [DOI] [PubMed] [Google Scholar]
- 49.Nishida A, Emoto K, Shimizu M, Uozumi T, Yamawaki S. Brain ischemia decreases phosphatidylcholine-phospholipase D but not phosphatidylinositol-phospholipase C in rats. Stroke. 25(6). (1994). 1247–1251. [DOI] [PubMed] [Google Scholar]
- 50.Sabogal-Guaqueta AM, Villamil-Ortiz JG, Arias-Londono JD, Cardona-Gomez GP. Inverse phosphatidylcholine/phosphatidylinositol levels as peripheral biomarkers and phosphatidylcholine/lysophosphoethanolamine-phosphatidylserine as hippocampal indicator of postischemic cognitive impairment in rats. Frontiers in Neuroscience. 21 (2018). 10.3389/fnins.2018.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali Mousavi F S. Khorvash, T. Hoseini. The efficacy of citroline in the treatment of ischemic stroke and primary hypertensive intracereal hemorrhage; a review article. ARYA Atherosclerosis. 6(3). (2010). 122–125. [PMC free article] [PubMed] [Google Scholar]
- 52.Moto A, Hirashima Y, Endo S, Takaku A. Changes in lipid metabolites and enzymes in rat brain due to ischemia and recirculation. Mol Chem Neuropathol. 14(1). (1991). 35–51. [DOI] [PubMed] [Google Scholar]
- 53.Hattori T, Nishimura Y, Sakai N, Yamada H, Kameyama Y, Nozawa Y. Effects of pentobarbital on brain lipid metabolism during global ischemia. Neurol Surg. 38(6). (1986). 585–591. [PubMed] [Google Scholar]
- 54.Matthys E. Patel Y, Kreisberg J, Stewart JH. Venkatachalam M. Lipid alterations induced by renal ischemia: pathogenic factor in membrane damage. Kidney International. 26(2). (1984). 153–161. [DOI] [PubMed] [Google Scholar]
- 55.Sun GY, Lu FL, Lin SE, Ko MR. Decapitation ischemia-induced release of free fatty acids in mouse brain. Relationship with diacylglycerols and lysophospholipids. Mol Chem Neuropathol. 17(1). (1992). 39–50. [DOI] [PubMed] [Google Scholar]
- 56.van der Veen JN, Kennelly JP, Wan S, Vance JE, Vance DE, Jacobs RL. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochimica et Biophysica Acta (BBA)- Biomembranes. Volume 1859 Issue 9 Part B. (2017). 1558–1572. [DOI] [PubMed] [Google Scholar]
- 57.Nilanjana M, Kagan VE, Tyurin VA, Das DK. Redistribution of phosphatidylethanolamine and phosphatidylserine precedes reperfusion-induced apoptosis. American Journal of Physiology: Heart and Circulatory Physiology. (1998). H242–H248. [DOI] [PubMed] [Google Scholar]
- 58.Kawai H, Chaudhry F, Shekhar A, Petrov A, Nakahara T, Tanimoto T, Kim D, Chen J, Lebeche D, Blackenberg FG, Pak KT, Kolodgie FD, Virmani R, Sengupta P, Narula N, Hajjar RJ, Strauss HW, Narula J. Molecular imaging of apoptosis in ischemia reperfusion injury with radiolabeled duramycin targeting phosphatidylethanolamine. Effective target uptake and reduced nontarget organ radioation burden. Journal of the American College of Cardiology: Cardiovascular Imaging. 11(12). (2018). DOI: 10.1016/j.jcmg.2017.11.037 [DOI] [PubMed] [Google Scholar]
- 59.Schabitz WR, Giuffrida A, Berger C, Aschoff A, Schwaninger M, Schwab S, Piomelli D. Release of fatty acid amides in a patient with hemispheric stroke, a microdialysis study. Stroke. 33 (2002). 2112–2114. [DOI] [PubMed] [Google Scholar]
- 60.Post JA, Bivelt JJ, Verkleij AJ. Phosphatidylethanolamine and sarcolemmal damage during ischemia or metabolic inhibition of heart myocytes. American Journal of Physiology: Heart and Circulatory Physiology. 268(2). (1995). H773–H780. [DOI] [PubMed] [Google Scholar]
- 61.Hirabayashi T, Larson TJ, Dowan W. Membrane-associated phosphatidylglycerophosphate synthetase from Escherichia coli: purification by substrate affinity chromatography on cytidine 5’-diphospho-1,2-diacyl-sn-glycerol sepharose. Biochemistry. 15(24). (1976). 5205–5211. [DOI] [PubMed] [Google Scholar]
- 62.Morita SY, Terada T. Enzymatic measurement of phosphadiylglycerol and cardiolipin in cultured cells and mitochondria. Scientific Reports. 5 (2015). doi: 10.1038/srep11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, Rehling P, Meisinger C, Ryan MT, Wiedemann N, Greenberg ML, Pfanner N, Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Current Biology. 19(24). (2009). 2133–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. The EMBO Journal. 29(12). (2010). 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C. Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem. 274(2). (1999). 1156–1163. [DOI] [PubMed] [Google Scholar]
- 66.Orrenius S, Zhivotovsky B. Cardiolipin oxidation sets cytochrome c free. Nat Chem Biol. 1(4). (2005). 188–189. [DOI] [PubMed] [Google Scholar]
- 67.Choi SY, Gonzalvez F, Jenkins GM, Slominanny C, Chretien D, Arnoult D, Petit PX, Frohman MA. Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell Death and Differentiation. 14 (2007). 597–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.