Abstract
Background:
Autism Spectrum Disorder (ASD) is highly familial, with a positively skewed male to female ratio that is purported to arise from the so-called “female protective effect” (FPE). A serious implication of FPE is that familial ASD liability would be expected to aggregate asymptomatically in sisters of affected probands and would incur elevated rates of ASD among their offspring. Currently, there exists no data on second-generation recurrence rates among families affected by ASD.
Methods:
We analyzed data from the Swedish National Patient Register and the Multi-Generation Register for a cohort of children born between 2003 and 2012. ASD was ascertained in both the child and parental generations.
Results:
Among 847,732 children, 13,103 children in the cohort (1.55%) were diagnosed with ASD. Among their maternal and paternal aunts/uncles, 1,744 (0.24%) and 1,374 (0.18%) were diagnosed with ASD. Offspring of mothers with a sibling(s) diagnosed with ASD had higher rates of ASD than the general population RR=3.05 (95% CI, 2.52-3.64), but not more than would be predicted for second degree relatives within a generation, and only slightly more than was observed for fathers with siblings with ASD RR=2.08 (95% CI, 1.53-2.67). Models adjusting for temporal trends and for psychiatric history in the parental generation did not alter the results.
Conclusions:
These findings establish a robust general estimate of ASD transmission risk for siblings of individuals affected by ASD, the first ever reported. Our findings do not suggest female protective factors as the principal mechanism underlying the male sex bias in ASD.
Keywords: Autism, Female Protective Effect, Sex Bias, Epidemiology, Population- based, Psychiatry
INTRODUCTION
Autism Spectrum Disorder (ASD) is a heritable, genetically heterogeneous neuro- developmental disorder (1, 2). A vast share of the population-attributable risk for this condition is accounted for by polygenic inheritance, as evidenced by (1) accumulated genetic epidemiological research demonstrating heritability of 0.80 or greater (1, 3, 4); and (2) several recent molecular genetic analyses indicating that the majority of causal genetic variation for ASD is additive (5, 6).
A well-established epidemiological feature of ASD is the elevated prevalence in males, with a usual ratio of approximately triple the number of males versus females, depending on the method of ascertainment (7). This phenomenon is not strictly accounted for by classic sex-chromosome-linked disease genes, because evidence from human genetic studies suggests that a relatively minor proportion of genetic risk for ASD is resolvable to genes on sex chromosomes (8). Although community-based ASD diagnoses are somewhat more likely for males than females at a given level of symptoms (8), the magnitude of the described diagnostic bias likely accounts for a minority of the observed male predominance of ASD. Several studies of toddlers, including prospective infant sibling studies (9) and general population screening studies (10, 11) have confirmed a true sex disparity manifest long before puberty (12).
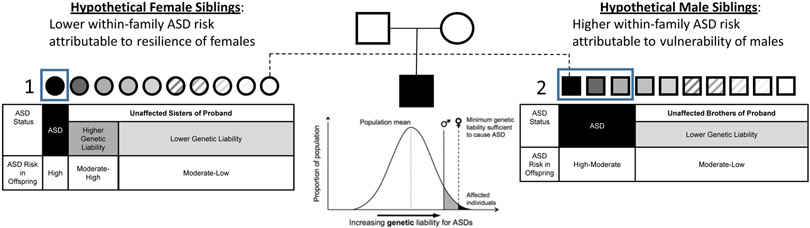
Sex-specific modulation of the expression of heritable ASD liability could involve either or both of two general mechanisms: a) protective factors conferring a higher liability threshold in females or b) susceptibility factors conferring a lower susceptibility threshold in males, as shown as difference in liability thresholds (Figure 1). Several recent findings have suggested a potential role for female protective factors. For example, highly penetrant autosomal de-novo CNVs causing ASD tend to be larger and to contain more genes in affected females versus males (13, 14). Similarly, a recent analysis of exome sequence data from over 27,000 trios affected by neurodevelopmental disorders revealed that 6.5% of affected females harbored a de novo mutation in a gene more commonly disrupted in affected females than in affected males, whereas 2.5% of males harbored a de novo mutation in “male enriched” genes (15). Some family studies have suggested elevated genetic burden in cohorts enriched for ASD females, for example, through greater ASD recurrence rates (16), and higher autistic trait scores in co-twins of affected females than in the co-twins of affected males (17). While these findings do not directly compare male versus female liability thresholds for ASD, in aggregate, they are consistent with females requiring greater genetic ASD risk to manifest a categorical diagnosis. Should such a “female protective effect” account for the ASD sex ratio, it would imply that unaffected females with a family history of ASD may carry and silently transmit proportionally greater genetic liability than unaffected males, amplifying recurrence rates in their male offspring, in particular. This possibility poses a significant public health issue, particularly in light of the international increase in ASD prevalence within the past two decades (18, 19), specifically rendering sisters of individuals with ASD particularly concerned about risk of transmitting autism to their own future offspring. The prevalence shift has complicated reliable estimation of epidemiological risk to children in the second generation of ASD-affected families, and there exists no pre-conceptional guidance for the unaffected siblings of individuals with autism who have reached childbearing age.
Figure 1. Sex-specific liability thresholds and the expected ASD liability under the "female protective effect" (FPE).
The expected ASD liability under the "female protective effect" (FPE). Example: An individual has ASD and, hypothetically, he has 10 sisters (left, circles) and 10 brothers (right, squares). Under FPE, only the sisters with very high genetic liability will be diagnosed with ASD (1). For the brothers, there will be more ASD diagnosed cases (2), in this example a 3:1 male-to-female-ratio. In the next generation (not shown in the pedigree), since it is more likely that un-diagnosed sisters carry a moderate-to-high genetic liability, the relative risk of ASD among offspring to the un-diagnosed siblings is expected to be higher among those to the sisters than those to the brothers.
Given evidence for the sex-specific modulation of ASD and widespread familial liability in the general population, quantifying silent maternal transmission of ASD represents a key step towards specification of second-generation recurrence risk in families, as well as risk stratification and identification of children most likely to benefit from early intervention. The complex polygenic nature of most cases of ASD constrains the capacity of molecular genetic methods to specify individual risk, particularly because the most robust associations of single variants with autism has involved highly deleterious mutations that arise de novo in the germline. By definition, de novo variation makes no contribution whatsoever to inherited risk in families, and can confound studies of familial risk by virtue of the fact that different de novo variants may amplify ASD liability in different ASD-affected member of the same multiplex family (20). Thus, population- level quantification of transmitted ASD liability provides a more relevant and actionable appraisal of average inherited risk to offspring of siblings, when clinical genetic information is not available or has not identified one or more variants contributing to autism in an affected family member.
The aim was therefore to examine the maternal transmission of ASD liability by testing if ASD risk differs between maternal and paternal lineage, where a higher risk from the maternal lineage is hypothesized under a female protective effect. Specifically, we compared whether ASD is more prevalent among second-generation offspring with an aunt diagnosed with ASD to those with an uncle diagnosed with ASD, noting that among such families that offspring affectation represents an incident offamilial recurrence, and such families thereby comparable to prior research involving multiplex families. The study effectively implements a within-family design within an epidemiologic two- generation sampling frame, which confers an advantage analogous to the transmission disequilibrium test in molecular genetic research; both are conservative methods for accommodating heterogeneity in genetic causation across families in the estimation of main effects for the population.
METHODS
Study population
The study population includes all children live born in Sweden between 1st January 2003 and 31st December 2012 identified from the Swedish Medical Birth Register (MBR). The register links the children with their mother and covers 99% of all births nation-wide since 1973 (21, 22). Paternity is assumed to be the husband or the male acknowledged by the mother and adoption or other non-biological relations are flagged. Fathers, siblings, cousins, uncles and aunts were identified through linkage with the unique Swedish Multi-Generation Register (23), an example family pedigree is given in Figure S1 to illustrate how offspring and uncle/aunt pairs were identified on maternal and paternal lineage. The Multi-Generation Register includes identifiers for all Swedish citizens 15 years old or younger and their parents from 1947 and onwards. To be included in the register index persons had to be alive in 1961 (when the register was computerized) or thereafter. We only included children of parents with a full sibling.
ASD and Psychiatric Diagnoses
In Sweden, all infants and preschool children regularly undergo routine medical and developmental examinations. At age 4 years, a mandatory developmental assessment (motor, language, cognitive, and social development) is conducted. Children with suspected developmental disorders are referred for further assessment by a specialized team in a child psychiatry unit or habilitation service. Diagnostic information is reported to the Swedish National Patient Register (NPR) (24). The NPR includes all inpatient psychiatric diagnoses in Sweden since 1973 and outpatient visits from 2001 with almost complete national coverage from 2005. The diagnoses in NPR are coded using the International Coding of Diseases (ICD) versions 8, 9 and 10 and are assigned by clinical specialists. NPR has been subject to extensive validation efforts (24) and frequently used in high impact journals (25). ASD was defined by a clinical diagnosis of Autistic Disorder, Asperger syndrome or PDD-NOS (Table S1). Our database includes diagnoses in NPR until 31 December 2017.
Covariates
We considered several factors which potentially could bias the results. The prevalence of childhood psychiatric disorders has increased over time. Therefore, to adjust for potential temporal trends, we included birth year and sex from the MBR and calculated age at first diagnosis of ASD and using the NPR. We defined psychiatric history as presence of a maternal or paternal psychiatric diagnosis of at least one diagnosis among 10 ICD disorder categories (Table S1) at the time of offspring birth. Similarly, we defined ASD-exposed individual as presence of an ASD diagnosis (Table S1) in aunt/uncle at the time of offspring birth. Besides an assessment of 'any psychiatric diagnosis' we also defined covariates for parental psychiatric history at birth for 10 different psychiatric diagnostic groups and disorders (Table S1) (25). Using the same procedure as for the parents we also defined aunt and uncle psychiatric history. To adjust for differences in length of follow-up we used date of death and date of emigration from Sweden using data from Statistics Sweden, the Swedish government bureau for official statistics.
Statistical analysis
A key biological hypothesis tested is whether ASD transmission from one generation to the next occurs disproportionately through females, among whom inherited liability is less expressed, or phenotypically 'silenced', in comparison to males. We tested this hypothesis by using the presence or absence of ASD in the parental generation as an indirect measure of parental exposure to genes conferring increased ASD risk (by virtue of relation to probands in Figure 1). The actual degree of transmitted risk was measured by the rate of ASD in the offspring of the parental generation (subjects C1, C2 in Figure S1).
To directly address the underlying biological hypothesis, we estimated the relative risk (RR) of ASD in individuals with an ASD-diagnosed aunt or uncle compared to offspring for whom an aunt or uncle were not diagnosed with ASD and then separately estimated RR for offspring when (a) an aunt or (b) an uncle had ASD, in comparison to offspring for whom aunts or uncles were not diagnosed with ASD. We estimated the RR for maternal-lineage aunt/uncles and for paternal-lineage aunt/uncles separately. We fitted Cox proportional hazards regression models and calculated hazard ratios as measures of RR together with two-sided 95% confidence intervals (CIs) corresponding to tests of statistical hypotheses on the two-sided 5% significance level. Each child was followed for an outcome of ASD from age 2 until death, emigration from Sweden or end of follow-up 31 December 2017, whichever came first.
We fitted a sequence of Cox regression models with increasing degree of adjustment for potentially confounding factors. First, we fitted 'crude models', only including a covariate for the exposure group (maternal ASD-diagnosed aunt or uncle). Thereafter, we adjusted for temporal trends by adding parameters for maternal birth year, birth year of the maternal aunt or uncle and birth year in the offspring generation using natural cubic splines (26); Finally, we included indicators of any mental illness (yes/no) of the mother and the uncle and aunt (27). We repeated the models above separately for paternal offspring. Lastly, we repeated the approach above by first refitting the models only considering uncles as exposures and then only considering aunts as exposures.
Since each child can occur several times in the calculations and since siblings and cousins can be assumed to be correlated, we estimated the CIs using bootstrap techniques (28, 29). In the bootstrap we resampled each child with replacement, so each child has the probability of being selected more than once as a representative of the overall study population. We examined the assumption of proportional hazards by visua inspection of weighted Schoenfelt residuals (30). Our database, using data from the national registers, was essentially free from any missing values. We used SAS version 14.2 on a Linux RedHat 7.2 server for all calculations including PROC PHREG for Cox regression.
We also performed several sensitivity analyses. (1) To test the specificity of RR associations we repeated analyses with Autistic Disorder (AD) in the offspring instead of ASD and (2) with male and female offspring separately. (3) We extended the diagnoses corresponding to familial liability for ASD/AD in the parental uncle/aunt to include schizophrenia, intellectual disability or schizoid personality disorder. In the parental generation, when ASD was less well-recognized, these outcomes may have lower specificity but increased sensitivity for inherited risk of ASD. (4) In our analyses, data were at offspring-aunt/uncle pair level (see Online Material-Model assumption for example), where some individuals were included in more than one cousin-uncle/aunt comparison, allowing individuals from larger families to contribute more to the sample size. Therefore, as a test of robustness, we fitted additional models where the oldest cousin pairs were drawn within 'families'. (5) To test the specificity of the findings relative to overall familial liability to psychiatric illness (parental generation), we repeated analyses on the subgroup of families with no psychiatric history other than ASD. (6) While adjusting for covariates, instead of parameters for 'any mental health', we included indicators for specific psychiatric disorders (intellectual disability, depression, anxiety disorders, substance use disorders, Bipolar disorder, compulsive disorder, Attention-Deficit Hyperactive Disorder (ADHD), affective disorders, schizophrenia, schizoid personality disorder) of the mother and the uncle/aunt (27). (7) To verify that our results were not biased due to sparse data which potentially can result in biased estimates, we performed a supplementary analysis using the Firth correction for monotone likelihood to adjust for the case-control imbalance (31, 32).
RESULTS
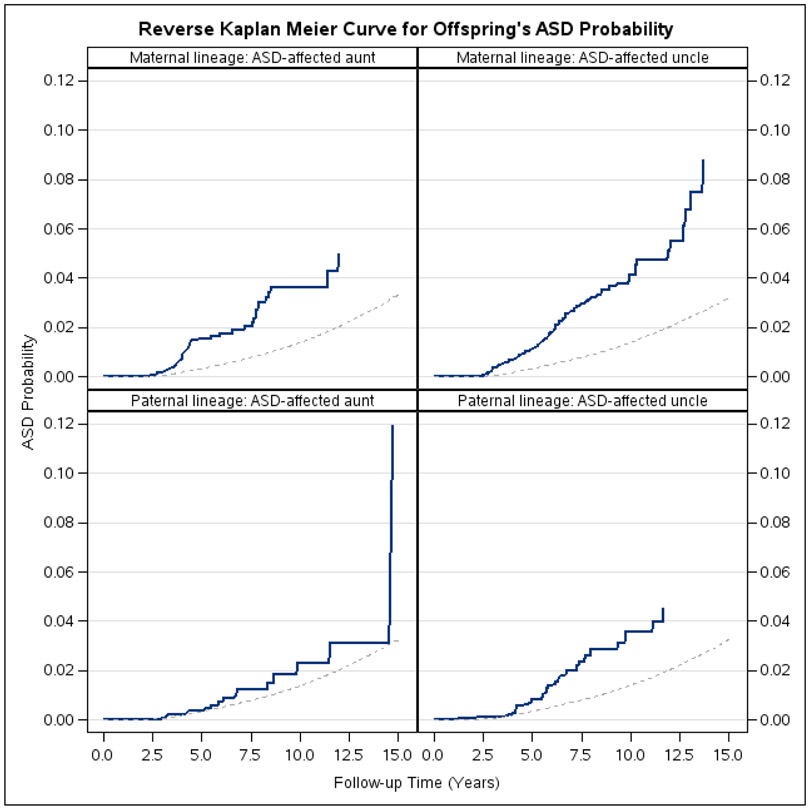
The study cohort included a total of 847,732 children, 51.43% male, followed for ASD from 5 to 15 years of age. There were 13,103 (1.55%) with ASD diagnosis, of which 8,216 (0.97%) were diagnosed with AD. The median age on onset was estimated at 7.72 years for ASD. Among the cohort children, 742,125 (87.5%) had a maternal aunt or uncle, 742,813 (87.6%) had a paternal aunt or uncle (Table 1); of which 29,646 and 20,616 person-years of follow-up were obtained for children with ASD-affected uncle(s)/aunt(s) from maternal and paternal lineage (Table 2). Age-specific ASD prevalence for offspring with uncles and aunts diagnosed with ASD from maternal lineage and paternal lineage are presented in Figure 2.
Table 1.
Study Characteristics.
| Variable | Participants (offspring) |
Maternal lineage | Paternal lineage | |||||
|---|---|---|---|---|---|---|---|---|
| Total | Male to Female Ratio |
Mothers | Uncles | Aunts | Fathers | Uncles | Aunts | |
| Number of individuals |
847,732# | 1.06 | 475,669 | 379,365 | 356,244 | 479,373 | 386,103 | 364,467 |
| ASD, n (%) | 13,103 (1.55) | 2.89 | 344 (0.07) | 1,158 (0.31) | 586 (0.16) | 227 (0.05) | 898 (0.23) | 479 (0.13) |
| AD, n (%) | 8,216 (0.97) | 3.07 | 41 (0.01) | 387 (0.10) | 208 (0.06) | 37 (0.01) | 308 (0.08) | 145 (0.04) |
| Asperger’s, n (%) |
2,506 (0.30) | 3.07 | 219 (0.05) | 604 (0.16) | 251 (0.07) | 156 (0.03) | 451 (0.12) | 251 (0.07) |
| PDD-NOS, (%) |
3,511 (0.41) | 2.39 | 121 (0.03) | 319 (0.08) | 194 (0.05) | 58 (0.01) | 258 (0.07) | 153 (0.04) |
| Age of 1st ASD diagnosis, median (Q1-Q3) |
7.72 (5.32-10.32) | 0.93 | 31.79 (25.44-38.23) | 26.75 (19.41-34.35) | 28.33 (20.41-35.92) | 34.41 (28.09-41.54) | 29.17 (20.57-37.68) | 30.55 (23.33-38.32) |
| Birth year, median (Q1-Q3) |
2008 (2005-2010) |
1.00 | 1976 (1972- 1981) |
1977 (1971-1983) |
1977 (1971-1983) |
1974 (1970- 1979) |
1974 (1967- 1981) |
1974 (1967- 1981) |
| Psychiatric history, n (%) | 38,553 (8.11) | 26,145 (6.89) | 29,620 (8.31) | 24,979 (5.21) | 26,301 (6.81) | 30,186 (8.28) | ||
Notes: n: number of individuals; Qi: The first quartile (25% of the data distribution is found below this value); Q3: The third quartile (25% of the data distribution is found above this value); ASD: Autism Spectrum Disorder; AD: autistic disorder; PDD-NOS: pervasive developmental disorder-not otherwise specified. ASD, AD, Asperger’s, and PDD-NOS were taken from the ICD-10 diagnosis.
Conditions of parental generation were measured at the birth date of the youngest offspring.
There were 435,982 (51.43%) males among the 847,732 participants.
Table 2.
Relative risks for ASD among participants with ASD-affected uncle(s)/aunt(s) compared to participants with uncle(s)/aunt(s) free from ASD diagnosis.
| Exposure | Person-years of follow-up |
Rates of ASD per 100,000 person- years |
Relative risk (95% Confidence interval)# | ||||
|---|---|---|---|---|---|---|---|
| Exposed | Unexposed | Exposed | Un- exposed |
Crude | Adjusted 1 | Adjusted 2 | |
| Maternal lineage | |||||||
| ASD-affected uncle(s) | 19,662 | 6,675,534 | 401.79 | 151.07 | 3.22 (2.54-3.91) | 2.75 (2.15-3.36) | 1.92 (1.49-2.37) |
| ASD-affected aunt(s) | 9,984 | 6,337,615 | 340.54 | 152.71 | 2.73 (1.88-3.73) | 2.36 (1.62-3.21) | 1.73 (1.19-2.35) |
| ASD-affected uncle(s)/aunt(s) |
29,646 | 13,013,149 | 381.16 | 151.87 | 3.05 (2.52-3.64) | 2.62 (2.17-3.14) | 1.88 (1.54-2.26) |
| Paternal lineage | |||||||
| ASD-affected uncle(s) | 13,274 | 6,723,265 | 301.35 | 153.23 | 2.39 (1.72-3.13) | 2.09 (1.50-2.74) | 1.63 (1.16-2.16) |
| ASD-affected aunt(s) | 7,342 | 6,292,408 | 190.68 | 151.99 | 1.52 (0.79-2.30) | 1.33 (0.69-2.01) | 1.07 (0.56-1.63) |
| ASD-affected uncle(s)/aunt(s) |
20,616 | 13,015,674 | 261.94 | 152.63 | 2.08 (1.53-2.67) | 1.82 (1.32-2.34) | 1.44 (1.05-1.86) |
Notes: ASD: autism spectrum disorder; Adjusted 1: Adjusted for birth year of the participant, the mother, and the uncle/aunt; Adjusted 2:
Adjusted for covariates in Adjusted 1 and any mental illness (yes/no) of the mother and the uncle/aunt;
Bootstrapped 95% confidence interval: 2.5%-97.5% percentiles of estimates from 1000 bootstrapped samples. ASD-affected: Individual with a clinical diagnosis of ASD.
Figure 2.
Inverse Kaplan-Meier curves for ASD probability among participants with ASD-affected uncle(s)/aunt(s), compared to participants with uncle(s)/aunt(s) free from ASD diagnosis, by maternal and paternal lineage
Note: Exposed groups (participants with ASD-affected aunt/uncle) were plotted in solid lines; Unexposed groups (participants with aunt/uncle free from ASD diagnosis) were plotted in dash lines.
In analyses without covariate adjustment, presence of ASD diagnosis in a maternal uncle or aunt was associated with an increased risk of ASD compared with maternal uncle or aunt without ASD diagnosis; RR=3.05 (95% CI: 2.52-3.64) while the RRs for the paternal side was estimated at RR=2.08 (95% CI: 1.53-2.67). After adjustment for confounding, including adjustment for temporal trends and family psychiatric history, the RRs were slightly diluted: RR=1.88 (95% CI: 1.54-2.26) for maternal lineage and RR=1.44 (95% CI: 1.05-1.86) for paternal lineage respectively (Table 2).
The RRs were similar for ASD in aunts and in uncles, for both the maternal lineage and the paternal lineage (Table 2): for adjusted models, maternal uncles RR=1.92 (1.49-2.37), maternal aunts RR=1.73 (1.19-2.35); paternal uncles RR=1.63 (1.16-2.16) and paternal aunts RR=1.07 (0.56-1.63).
Complementary analyses
The results remained robust across a set of sensitivity analyses (online appendix 1): For maternal and paternal uncles and aunts with AD (Table S2a, S2b); for maternal and paternal uncles and aunts diagnosed with ASD/Schizophrenia/Intellectual disability or Schizoid personality disorder (Table S2a, S2b); for offspring AD (Table S3a, S3b); for male and female offspring evaluated separately (Table S4). The results were also robust when adjusting for family size, and for potential correlations within families (Table S5, Table S6). The results also remained in the subgroup of families with aunts and uncles free from psychiatric history, other than ASD (Table S7); as well as when adjusting for psychiatric history in eight different psychiatric disorders instead of 'any' psychiatric history (Table S8); and when excluding offspring to mothers with ASD from the analysis (TableS 9). Our large sample size allowed us to consider the subgroup of offspring to the 344 mothers with an ASD diagnosis at time of delivery. For this subgroup the RR of ASD in offspring with an uncle or aunt compared to offspring with uncle or aunt without an ASD diagnosis was estimated at RR=5.23 (1.94, 14.11) (Table S10). The results in Table 2 remained unchanged when applying Firth's adjustment for monotone likelihood (Table S11).
DISCUSSION
To our knowledge, this work represents the first epidemiological study comparing transmission of maternally- versus paternally mediated ASD risk in second-generation offspring of parents having siblings with ASD.
When estimating second-generation ASD risk, we observed that offspring of females with a sibling diagnosed with ASD did not exceed what would be expected on the basis of autism recurrence rates in large-scale population-based twin and family studies: 0.8 for identical twins, 0.2 for non-identical siblings, and 0.04 for second degree relatives (33). Additionally, while point estimates were numerically higher in models examining maternal lineages, overlapping confidence intervals indicated no difference in recurrence risk between offspring of females versus males with a sibling diagnosed with ASD. Within the second generation, ASD risk estimates did not differ between male and female offspring, contrary to the expected elevation for males under a female protective effect. Given these observations, an overarching inference is that the sex disparity in ASD may not be primarily derived from a female protective effect.
There are other potential models to explain the male-predominance in ASD, first the corollary that male sex confers heightened sensitivity and therefore greater phenotypic expression of a given inherited liability. This would result in unaffected brothers and sisters of ASD probands carrying comparable levels of sub threshold liability, with males at the upper extreme being affected and not contributing to risk among second generation offspring. A second possibility relates to a recently-published observation among ASD-affected monozygotic twins (predominantly male-male pairs) that there can exist pronounced variability in severity between identical co-twins (34). If such variability is more pronounced in male than in female carriers of ASD liability, this too could contribute to the observed sex ratio. Greater variability among males (GVM) would predict that more males than females would manifest higher severity of autistic features arising from the same level of inherited liability. From a statistical genetic standpoint, FPE and male sensitivity hypotheses refer to sex-specific genetic thresholds of expression of liability, while GVM relates to sex-specific variance in phenotypic expression. To directly test the GVM hypothesis—which is as yet unproven—a population-level liability modeling and quantification of either genetic risk or phenotypic expression would be required—both are beyond the scope of the data available to this study. It is also important to note that FPE, male sensitivity, and GVM are not mutually exclusive events and could hypothetically co-exist; the key conclusion from this analysis is that if FPE exists, it does not do so to an extent that substantially raises risk to second- generation offspring of sisters of ASD probands over what would be expected for second degree relatives of affected individuals.
We also examined whether these estimates differed based on the time period of ASD diagnosis and family history of psychiatric diagnoses in the parental generation. These adjusted models, which accounted for potential bias due to temporal trends in ASD diagnosis and confounding due to environmental factors correlated with psychiatric illness, exhibited only in a slight attenuation of estimated effects. A sensitivity analysis of individuals without a family history of psychiatric diagnosis confirmed elevated recurrence risk independent of overlapping familial liability for ASD and other psychiatric conditions. Similar recurrence risk estimates were also observed both when restricting to the oldest cousin pair within families with ASD diagnosis and when directly comparing families with and without maternal siblings with ASD, demonstrating the robustness of the findings. In addition, the similar magnitude of risk estimates, whether ascertaining ASD or AD in the parental generation, suggests this pattern of sex modulation applies across the continuum of ASD severity, consistent with a polygenic, additive genetic model of ASD.
Among neurodevelopmental disorders, ASD is distinguished by a well-documented increase in prevalence over the past two decades (18) . In spite of advances in early screening, the median diagnostic age remains well after two years (35), an initial age exhibiting diagnostic stability (36) as illustrated by this sample’s median diagnostic age of 7 years. For pediatricians representing the first-line resource for ASD screening, current guidelines do not articulate standards for evaluating family history of ASD in the parental generation. These findings suggest that, irrespective of sex, offspring of parents with a sibling diagnosed with ASD warrant especially diligent screening, including family history, to inform risk stratification and planning. As knowledge of cross-generational ASD risk factors advances, family history in the parental generation may be integrated with genotyping of affected family members, quantitative autistic trait measures, and review of psychosocial factors associated with increased offspring risk, (e.g., advanced parental age), leading to more comprehensive risk assessment and improved clinical guidance.
In population-based studies, social and potentially gender-related effects on the assessment of psychiatric diagnoses cannot be ruled out. Especially, there may be a risk for reversed causation if individuals in the parental generation seek psychiatric care after an ASD diagnosis in the offspring generations. To address this issue, we chose to consider ASD and other psychiatric diagnoses in the parental generation, including aunt and uncle, only at the birth of the offspring generation participants. Therefore, aunt/uncle’s bias in ASD assessment after an ASD diagnosis of the offspring will not affect our results.
Study strengths include a large, epidemiologic sample in a health system with equal access and near-complete follow-up of clinically ascertained diagnoses, which allowed ASD diagnoses in the offspring generation based on up-to-date ICD-10 criteria. Regarding limitations, our multi-generational cohort necessitated a parental generation which matured prior to widespread awareness of ASD and was thus prone to under-ascertainment, as suggested by that generation’s low ASD prevalence in our sample. Thus, these results likely underestimate risk for silent parental transmission: future studies that are fully representative of contemporary ASD criteria would be warranted to confirm findings in the context of milder forms of ASD in the parental generation. This concern is mitigated, however, by the lack of differences observed in transmission rates among families of probands diagnosed with ASD versus Autistic Disorder in the parental generation. Furthermore, even though we present one of the largest and most detailed epidemiological examinations of maternal transmission a study such as ours cannot detect the true underlying causes and mechanisms of transmission (or non-transmission) of autism. For this purpose, other types of studies are required, e.g. including genomic analysis with SNPs data or genomic sequencing to obtain both copy number and point mutation data (37). We note, however, that at this stage of science established molecular genetic correlates of ASD account for only a fraction of known inherited risk, and our study was focused on within-family transmission.
Although we performed a very detailed adjustment for temporal trends and familial psychiatric conditions, we acknowledge residual confounding may remain. The determination of ASD diagnoses would also likely have been susceptible to sex biases inherent in ASD diagnostic criteria (8). In future studies, quantitative autistic trait measures, which were not widely available early in the parental generation, could be used to identify undiagnosed females with clinically elevated autistic traits.
CONCLUSION
We found similar ASD risk in offspring from maternal and paternal lineages. The risk of ASD in offspring to siblings of ASD probands, equals, but does not exceed what has been observed for second-degree relatives within a single generation. These findings do not suggest female protective factors as the principal mechanism underlying the male sex bias in ASD.
While these results mitigate concern for amplification of maternally transmitted ASD risk, they affirm the importance of heightened surveillance for ASD in second-generation offspring. Given the benefits of early intervention, these results support incorporating second-degree family history of ASD in pediatric practice, as well as future studies involving behavioral phenotyping and genotyping to advance individualized estimates of ASD recurrence risk.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type |
Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. |
Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. |
Include any additional information or notes if necessary. |
| Antibody | Not applicable | |||
| Bacterial or Viral Strain | Not applicable | |||
| Biological Sample | Not applicable | |||
| Cell Line | Not applicable | |||
| Chemical Compound or Drug | Not applicable | |||
| Commercial Assay Or Kit | Not applicable | |||
| Deposited Data; Public Database | Not applicable | |||
| Genetic Reagent | Not applicable | |||
| Organism/Strain | Not applicable | |||
| Peptide, Recombinant Protein | Not applicable | |||
| Recombinant DNA | Not applicable | |||
| Sequence-Based Reagent | Not applicable | |||
| Software; Algorithm | SAS version 9.4.6 on Linux 64 bit server | See supplementary online appendix for additional information and computer code | ||
| Transfected Construct | Not applicable | |||
| Other |
ACKNOWLEDGMENTS
This work was supported by grant HD087011 (the Intellectual and Developmental Disabilities Research Center at Washington University in St. Louis) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) to Dr. Constantino. We would also like to acknowledge the following funding sources: K08MH112891 from National Institute of Mental Health and Autism Science Foundation Post-Doctoral Fellowship for Dr. Marrus and EU grant H2020-SC1: PM04-2016 for Dr. Sandin.
Footnotes
FINANCIAL DISCLOSURES
Dr. Bai reported no biomedical financial interests or potential conflicts of interest. Dr. Marrus receives funding from K08 MH112891 and has been the recipient of an Autism Science Post-Doctoral Fellowship. She is employed at Washington University School of Medicine. Dr. Yip reported no biomedical financial interests or potential conflicts of interest. Dr. Reichenberg reported no biomedical financial interests or potential conflicts of interest. Dr. Constantino reported no biomedical financial interests or potential conflicts of interest. Dr. Sandin reported no biomedical financial interests or potential conflicts of interest.
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report, or in the decision to submit the paper for publication. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication. The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A (2017): The heritability of autism spectrum disorder. Jama. 318:1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yip BHK, Bai D, Mahjani B, Klei L, Pawitan Y, Hultman CM, et al. (2017): Heritable Variation, With Little or No Maternal Effect, Accounts for Recurrence Risk to Autism Spectrum Disorder in Sweden. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tick B, Bolton P, Happe F, Rutter M, Rijsdijk F (2016): Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry. 57:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai D, Yip BHK, Windham GC, Sourander A, Francis R, Yoffe R, et al. (2019): Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. (2014): Most genetic risk for autism resides with common variation. Nature Genet. 46:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, et al. (2012): Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomes R, Hull L, Mandy WPL (2017): What Is the Male-to-Female Ratio in Autism Spectrum Disorder? A Systematic Review and Meta-Analysis. J Am Acad Child Adolesc Psychiatry. 56:466–474. [DOI] [PubMed] [Google Scholar]
- 8.Dworzynski K, Ronald A, Bolton P, Happe F (2012): How Different Are Girls and Boys Above and Below the Diagnostic Threshold for Autism Spectrum Disorders? Journal of the American Academy of Child & Adolescent Psychiatry. 51:788–797. [DOI] [PubMed] [Google Scholar]
- 9.Estes A, Zwaigenbaum L, Gu H, St John T, Paterson S, Elison JT, et al. (2015): Behavioral, cognitive, and adaptive development in infants with autism spectrum disorder in the first 2 years of life. Journal of neurodevelopmental disorders. 7:24–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nygren G, Sandberg E, Gillstedt F, Ekeroth G, Arvidsson T, Gillberg C (2012): A new screening programme for autism in a general population of Swedish toddlers. Res Dev Disabil. 33:1200–1210. [DOI] [PubMed] [Google Scholar]
- 11.Pierce K, Gazestani VH, Bacon E, Barnes CC, Cha D, Nalabolu S, et al. (2019): Evaluation of the Diagnostic Stability of the Early Autism Spectrum Disorder Phenotype in the General Population Starting at 12 Months. JAMA pediatrics. 173:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messinger DS, Young GS, Webb SJ, Ozonoff S, Bryson SE, Carter A, et al. (2015): Early sex differences are not autism-specific: A Baby Siblings Research Consortium (BSRC) study. Mol Autism. 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, et al. (2011): Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 70:886–897. [DOI] [PubMed] [Google Scholar]
- 14.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. (2011): Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 70:863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner TN, Wilfert AB, Bakken TE, Bernier RA, Pepper MR, Zhang Z, et al. (2019): Sex-Based Analysis of De Novo Variants in Neurodevelopmental Disorders. The American Journal of Human Genetics. 105:1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werling DM, Geschwind DH (2015): Recurrence rates provide evidence for sex-differential, familial genetic liability for autism spectrum disorders in multiplex families and twins. Molecular Autism. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson EB, Lichtenstein P, Anckarsater H, Happe F, Ronald A (2013): Examining and interpreting the female protective effect against autistic behavior. Proc Natl Acad Sci U S A. 110:5258–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atladottir HO, Gyllenberg D, Langridge A, Sandin S, Hansen SN, Leonard H, et al. (2015): The increasing prevalence of reported diagnoses of childhood psychiatric disorders: a descriptive multinational comparison. Eur Child Adolesc Psychiatry. 24:173–183. [DOI] [PubMed] [Google Scholar]
- 19.Christensen DL, Braun KVN, Baio J, Bilder D, Charles J, Constantino JN, et al. (2018): Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. Morbidity and mortality weekly report Surveillance summaries (Washington, DC: 2002). 65:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, et al. (2015): Whole-genome sequencing of quartet families with autism spectrum disorder. Nature medicine. 21:185–191. [DOI] [PubMed] [Google Scholar]
- 21.Axelsson O (2003): The Swedish Medical Birth Register. Acta Obstet Gynecol Scand. 82:491–492. [DOI] [PubMed] [Google Scholar]
- 22.Cnattingius S, Ericson A, Gunnarskog J, Kallen B (1990): A QUALITY STUDY OF A MEDICAL BIRTH REGISTRY. Scand J Soc Med. 18:143–148. [DOI] [PubMed] [Google Scholar]
- 23.Ekbom A (2011): The Swedish Multi-generation Register In: Dillner J, editor. Methods in Biobanking. Totowa: Humana Press Inc, pp 215–220. [DOI] [PubMed] [Google Scholar]
- 24.Ludvigsson JF, Andersson E, Ekbom A, Feychting M, Kim J-L, Reuterwall C, et al. (2011): External review and validation of the Swedish national inpatient register. BMC Public Health. 11:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandin S, Schendel D, Magnusson P, Hultman C, Suren P, Susser E, et al. (2016): Autism risk associated with parental age and with increasing difference in age between the parents. Mol Psychiatr. 21:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Friedman J (2013): The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; New York. [Google Scholar]
- 27.Viktorin A, Levine SZ, Altemus M, Reichenberg A, Sandin S (2018): Paternal use of antidepressants and offspring outcomes in Sweden: nationwide prospective cohort study. BMJ. 361:k2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burr D (1994): A Comparison of Certain Bootstrap Confidence Intervals in the Cox Model. J Am Stat Assoc. 89:1290–1302. [Google Scholar]
- 29.Zelterman D, Le CT, Louis TA (1996): Bootstrap techniques for proportional hazards models with censored observations. Statistics and Computing. 6:191–199. [Google Scholar]
- 30.Grambsch PM, Therneau TM (1994): Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 81:515–526. [Google Scholar]
- 31.Firth D (1993): Bias Reduction of Maximum Likelihood Estimates. Biometrika. 80:27–38. [Google Scholar]
- 32.Heinze G, Schemper M (2001): A solution to the problem of monotone likelihood in Cox regression. Biometrics. 57:114–119. [DOI] [PubMed] [Google Scholar]
- 33.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A (2014): The Familial Risk of Autism. JAMA-J Am Med Assoc. 311:1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castelbaum L, Sylvester CM, Zhang Y, Yu Q, Constantino JN (2019): On the Nature of Monozygotic Twin Concordance and Discordance for Autistic Trait Severity: A Quantitative Analysis. Behav Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lord C, Risi S, DiLavore PS, Shulman C, Thurm A, Pickles A (2006): Autism from 2 to 9 years of age. Arch Gen Psychiatry. 63:694–701. [DOI] [PubMed] [Google Scholar]
- 36.Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, et al. (2018): Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. Morbidity and mortality weekly report Surveillance summaries (Washington, DC: 2002). 67:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. (2020): Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 180:568–584×523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.