Abstract
The secreted protein Noggin1 was the first discovered natural embryonic inducer produced by cells of the Spemann organizer. Thereafter, it was shown that vertebrates have a whole family of Noggin genes with different expression patterns and functional properties. For example, Noggin1 and Noggin2 inhibit the activity of BMP, Nodal/Activin and Wnt-beta-catenin signalling, while Noggin4 cannot suppress BMP but specifically modulates Wnt signalling. In this work, we described and investigated phylogeny and expression patterns of four Noggin genes in lampreys, which represent the most basally divergent group of extant vertebrates, the cyclostomes, belonging to the superclass Agnatha. Assuming that lampreys have Noggin homologues in all representatives of another superclass of vertebrates, the Gnathostomata, we propose a model for Noggin family evolution in vertebrates. This model is in agreement with the hypotheses suggesting two rounds of genome duplication in the ancestor of vertebrates before the divergence of Agnatha and Gnathostomata.
Subject terms: Evolutionary developmental biology, Pattern formation
Ermakova et al. report four Noggin genes in lampreys and using phylogenetics, gene synteny analysis, and in situ hybridization, suggest that the Noggin gene underwent two rounds of duplication and evolved specific functions before the divergence of vertebrate and lamprey lineages. These findings offer insight into early vertebrate genome and developmental evolution.
Introduction
The divergence of Agnatha (jawless fishes) and Gnathostomata (gnathostomes) occurred ~535–462 MYA, i.e., at the earliest stages of the evolution of vertebrates in the Palaeozoic Era1–3. These data allow one to consider jawless fishes, including extant lampreys and hagfishes, as the most basally divergent group of vertebrates. In turn, this early separation of cyclostomes from other vertebrates makes them a recognized model for studying the genetic innovations that led to the evolutionary emergence and development of unique traits of the vertebrates, such as the telencephalon, neural crest cells, the epimorphic regeneration ability of the body appendages, and adaptive immunity4–6. It is generally accepted that the genetic basis of such innovations is genomic duplications; however, the number and timing of these duplications still need to be clarified7,8. One possible approach to resolve this issue involves comparing the evolution, expression and functions of genes that regulate basic processes of vertebrate development in representatives of cyclostomes and other vertebrates. In the present work, we performed such analysis for the genes of the Noggin family, which encode secreted factors participating in the regulation of the development of many organs during vertebrate embryogenesis9–16.
For a long time, it was believed that vertebrates have only one “classical” Noggin gene, which functions in vertebrate development by suppressing the BMP (bone morphogenetic protein) signalling pathway by sequestering BMP ligands9,10. However, first in amphibians and then in other gnathostomes, homologues of the “classical” Noggin, namely, Noggin2 and Noggin4 were described17,18. Moreover, it was shown that the role of these Noggin proteins in the development of vertebrates was not limited to the previously described inhibition of the BMP signalling pathway but also includes suppression of Nodal/Activin and Wnt signalling15,16,19,20. Thus, as modulators of these three basic intracellular signalling pathways, Noggin proteins can play important roles in many processes of early tissue differentiation.
The presence of several Noggin genes also in cyclostomes was noticed for the first time for the sea lamprey Petromyzon marinus21. However, since this work was focused on the genome-wide analysis of genes lost in the early mammalian evolution, these lamprey Noggins, while included in common phylogenetic analysis, were not described and investigated in detail. In the present work, we revealed that lampreys have four Noggin genes and then studied their phylogeny, local genomic synteny, expression patterns and the ability to induce secondary body axes, including the head. Establishing that lampreys have orthologues of Noggin1, Noggin2 and Noggin4 in gnathostomes, we consider their possible evolution in the context of the existing models of genome duplications in the early vertebrate history. The obtained results incline us to the hypothesis suggesting two rounds of genome duplication in the common ancestor of vertebrates before divergence of jawless and gnathostomes.
Results
Phylogenetic analysis of lamprey Noggin genes
The available lamprey genome databases (whole-genome shotgun contigs of the Arctic lamprey, Lethenteron camtschaticum, and sea lamprey, P. marinus) were used to search for putative lamprey homologues of known Noggin family genes. We were able to simplify the screening because the lamprey Noggin coding sequences presumably should not contain introns, since the absence of introns was previously shown for all known Noggin genes in both invertebrates and vertebrates. As a result, we revealed in the L. camtschaticum and the P. marinus genomes the following four genes, which indeed had no introns, with protein products that demonstrated homology with the Noggin1, Noggin2 and Noggin4 described in gnathostomes9,17,18.
NogginA (L. camtschaticum contig 015835, sequence ID: APJL01043027.1; P. marinus isolate animal number 11 contig 46719, Sequence ID: AEFG01046720.1);
NogginB (L. camtschaticum contig 045160, sequence ID: APJL01075429.1; P. marinus isolate animal number 11 contig 47771, sequence ID: AEFG01047772.1);
NogginC (L. camtschaticum unplaced genomic scaffold scaffold00115, sequence ID: KE993786.1; P. marinus isolate animal number 11 contig 36440, sequence ID: AEFG01036441.1); and
NogginD (L. camtschaticum contig 002783, sequence ID: APJL01035689.1; P. marinus isolate animal number 11 contig 9676, sequence ID: AEFG01009677.1).
Based on these sequences, we designed primers and cloned the full-length cDNAs of four Noggin genes of European river lamprey (Lampetra fluviatilis) Noggin genes. According to the available literature and our own data, this species is extremely close to L. camtschaticum in terms of genomic sequences and developmental traits, while its embryos are much more accessible, at least in our circumstances than those of the latter.
The alignment of the protein sequences encoded by these genes showed that all of them have conserved cysteine residues known to be important for the formation of functional Noggin dimers16,22 (Supplementary Fig. 1).
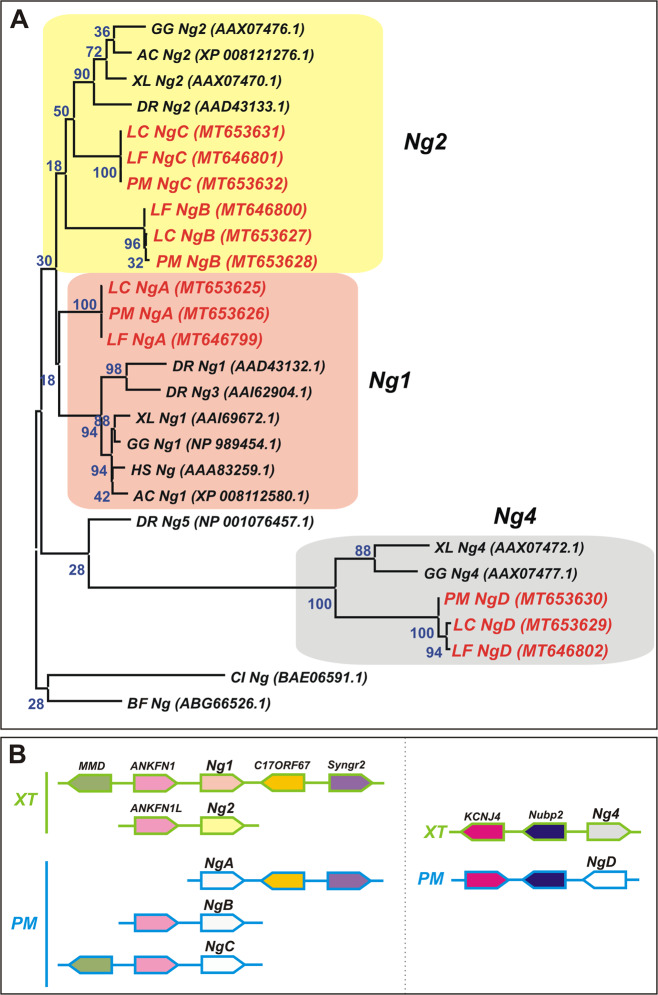
As we also revealed, by maximum likelihood (ML) protein analysis, NogginA appeared to be closer to gnathostome Noggin1, while NogginB and NogginC appeared to be closer to Noggin2. However, as the bootstrap value justifying such clustering is quite low (<50%) in our opinion, it would be more correct to speak of a cluster or cloud homology of the lamprey NogginA/B/C proteins on the one hand and jawed vertebrates’ Noggin1/2 on the other. (Fig. 1a). Adding of Echinodermata Noggin genes to the tree or using the Neighbor-joining (NJ) algorithm doesn’t clear up the picture (Supplementary Fig. 2). At the same time, NogginD was confidently grouped with Noggin4. Moreover, NogginD, the lamprey orthologue of Noggin4, had amino acid substitutions in positions critical for the binding of BMP, which presumably would prevent its binding with these ligands (Supplementary Fig. 1)16,22.
Fig. 1. Phylogenetic analysis (a) and synteny (b) of lamprey Noggins.
A - Phylogenetic tree of Noggins is constructed using the Maximum Likehood algorithm. AC Anolis carolinensis, BF Branchiostoma floridae, CI Ciona intestinalis, DR Danio rerio, GG Gallus gallus, HS Homo sapiens, LF Lampetra fluviatilis, LC Lethenteron camtschaticum, PM Petromyzon marinus XL Xenopus laevis, XT Xenopus tropicalis. B - synteny of Noggin genes of sea lamprey P. marinus (PM) and western clawed frog X. tropicalis (XT).
Hoping to clarify the phylogenetic relationship of lamprey and jawed vertebrates’ Noggin genes, we analysed their local genomic synteny in the genomes of the lamprey P. marinus and the frog Xenopus tropicalis. As a result, we found that NogginA of P. marinus and Noggin1 of X. tropicalis both have at least two common genes flanking their 3′ end, and in the X. tropicalis genome, they are designated C17ORF67 (ENSXETG00000007759) and synaptogyrin2 (Syngr2, ENSXETG00000007946). This finding indicates that both of these Noggin genes could derive from a common ancestor, as was predicted by the clustering analysis.
At the same time, NogginB and NogginC also have neighbouring genes in common with X. tropicalis Noggin1, but flanking the 5′ end. Interestingly, although NogginC has two gene homologues of X. tropicalis ANKFN (XM_018097804) and MMD (ENSXETG00000007598), NogginB adjoins with only ANKFN. Notably, the paralogue of the latter, ANKFN1L (XM_002932447.4), was found near the 5' end of X. tropicalis Noggin2. Taken together, these results suggest that NogginB and NogginC of P. marinus, as well as Noggin2 of X. tropicalis, likely have a common ancestor with Noggin1 and NogginA but they probably form another branch of genes derived from this ancestor. This conclusion is consistent with the results of a clustering analysis that slightly gravitates NogginA to Noggin1 in one cluster and NogginB, NogginC to Noggin2 in another cluster.
In contrast to NogginA, B and C, lamprey NogginD has no proximal genes homologous to the genes adjacent to Noggin1 in X. tropicalis. However, at least two genes, homologous to nubp2 (ENSXETG00000010354) and KCNJ4 (ENSXETG00000010341) that flank X. tropicalis Noggin4, also flank lamprey NogginD. On the basis of the data of the ML protein analysis and the fact that, similar to Noggin4, NogginD has characteristic amino acid substitutions putatively preventing its binding to BMP, one may confidently conclude that the lamprey NogginD is indeed the orthologue of Noggin4 in gnathostomes.
The expression of lamprey Noggins in early development
To compare the lamprey Noggins with their homologs of gnathostomes and to predict their physiological roles in early development, we investigated their temporal and spatial expression patterns.
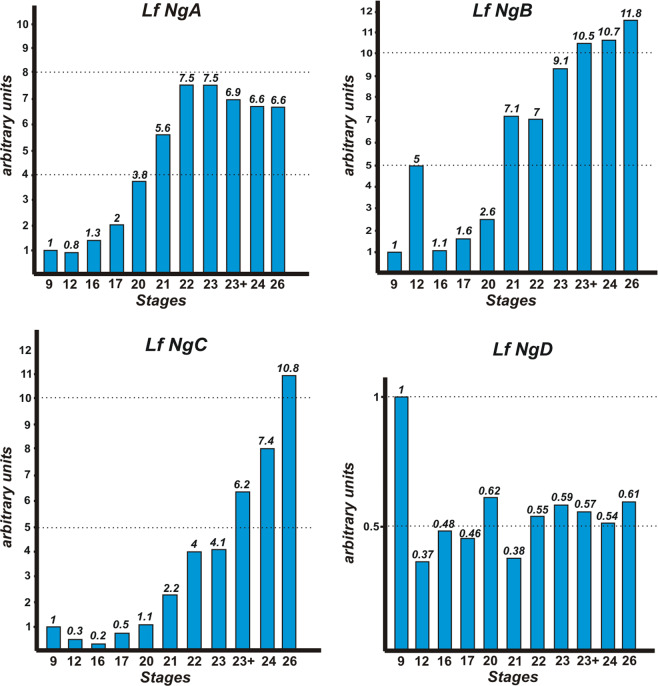
The analysis of the temporal patterns of Noggins’ expression was performed by real-time qPCR at the early stages of European river lamprey (L. fluviatilis) development, starting from stage 9 and continuing to stage 26, according to ref. 23.
As a result, we established that NogginA, NogginB and NogginC are expressed after fertilization at very low levels; however, their expression begins to increase starting from the late neurula stage - stage 20 (Fig. 2). In contrast, NogginD is expressed at approximately equal levels at all tested stages (Fig. 2).
Fig. 2. The expression dynamics of NogginA, NogginB, NogginC and NogginD in the early stages of development of L. fluviatilis.
Stage numbers are indicated according to ref. 24.
The spatial expression patterns of Noggins were analysed by whole-mount in situ hybridization. In these experiments, we analysed Noggin expression beginning from stage 11 (late blastula stage according to ref. 23) and to the stage of pre-ammocoete larvae (stage 29), using anti-sense DIG-labelled probes of mRNA for each of Noggin genes of L. fluviatilis. Sense DIG-labelled Noggin mRNAs were used as the controls.
Notably, none of the lamprey Noggin genes were expressed at the blastula or gastrula stages. In later stages, the analysis yielded the following results.
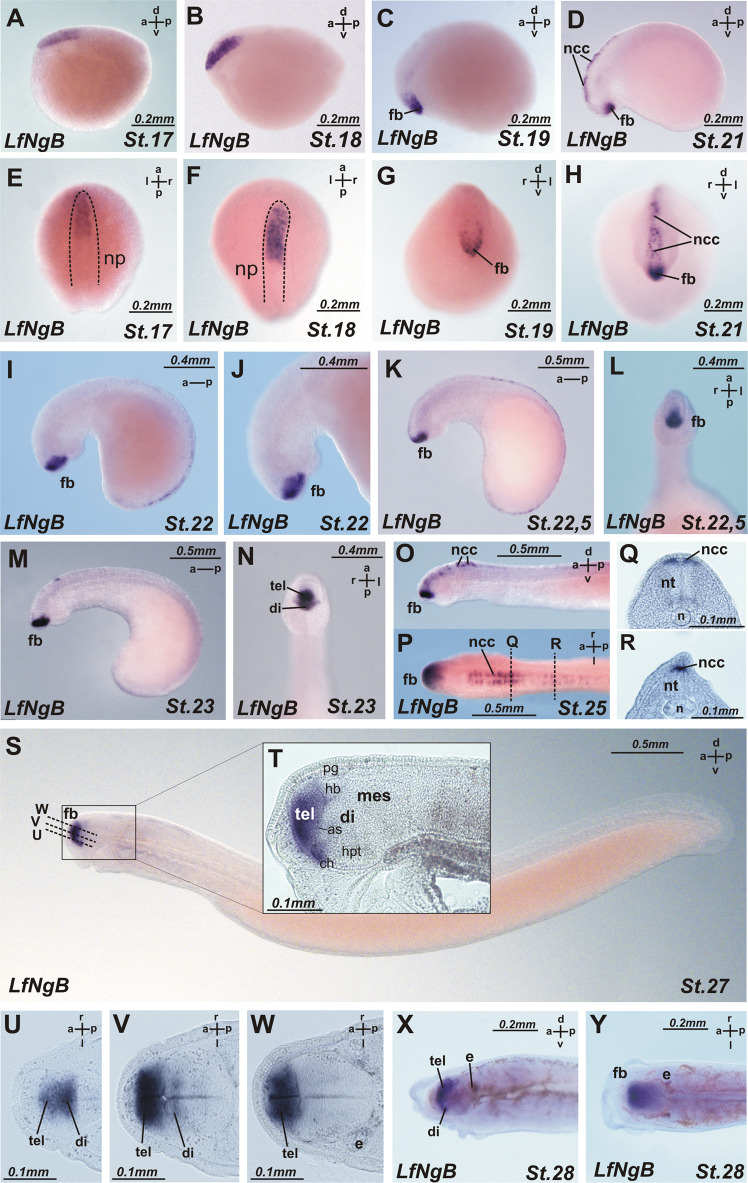
NogginA (Fig. 3a–j, Fig. 4a–j).
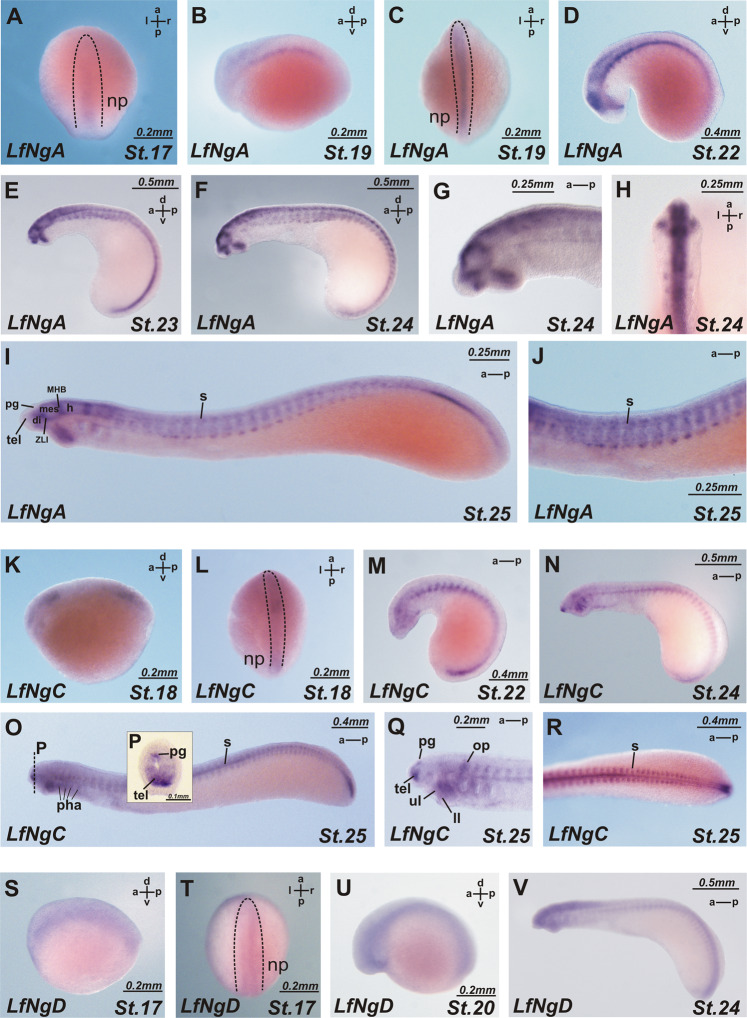
Fig. 3. Expression patterns of L. fluviatilis NogginA (a–j), NogginC (k–r) and NogginD (s–v).
a At stage 17 NogginA is expressed in the caudal chordamesoderm. b, c At stage 19 expression is in the entire chordamesoderm except for its most anterior part and in the dorsal region of the presumptive brain, excluding the most anterior part. d At stage 22 expression is in the floor and roof plate of the neural tube, in the notochord, in the somites, in the cheek process and in the diencephalon, midbrain and hindbrain. e–j At stages 23–25 expression pattern includes somites (i, j), upper and lower lips (g), notochord, in several areas of the brain: at the border of telencephalon and diencephalon, in zona limitans intrathalamica, at mid-hind brain boundary and in the hindbrain (g, i). k, l At stage 18 NogginC is expressed in the 2 chordamesoderm regions: anterior part and caudal part. m At stage 22 NogginC is found in the notochord, neural tube, pharyngeal arches, otic pits, cheek process and somites. n At stages 24 the expression of NogginC appears in the forebrain and otic visicles, upper and lower lips. o–q At stages 25 the cells of the pineal gland (transverse section on P) also express NogginC. r NogginC expression in somites at stage 25. s, t At stage 17 NogginD is expressed in the neural tube. u, v At stage 17 the expression of NogginD develops throughout the neural system in a diffuse manner and weakly in somites. as anterior intraencephalic sulcus, ch optic chiasma; di diencephalon, e eye, fb forebrain, h hindbrain, hb habenula, hpt hypothalamus, ll lower lip, ul upper lip, mes mesencephalon, MHB mid-hindbrain boundary, n notochord, ncc neural crest cells, np neural plate, nt neural tube, oc otic cup, op olfactory placode, pg pineal gland, pha pharyngeal arche, tel telencephalon, ZLI zona limitans intrathalamica.
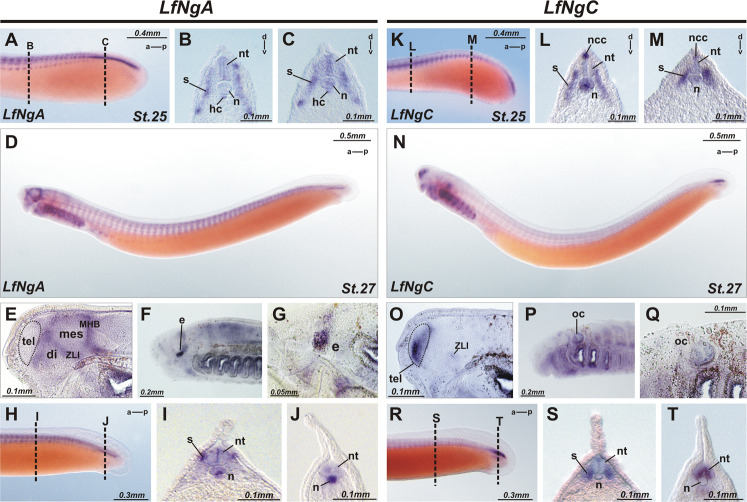
Fig. 4. Matching of expression patterns of NogginA (a–j) and NogginC (k–t).
a At stage 25 NogginA is expressed in somites, especially in dorsal and ventral angles, notochord, hypochord, neural tube. b, c Transverse sections, the section levels are shown in (a). d Whole mount expression of NogginA at stage 27. e–g Saggital section of embryo of stage 27 reveals the expression of NogginA at the border of telencephalon and diencephalon along anterior intraencephalic sulcus, in zona limitans intrathalamica and at mid-hind brain boundary and in the eyes. h–j In the tailbud the expression of NogginA increases in the notochord of the growing tail. The levels of transverse sections are shown in (h). k–m At stage 25 NogginC is expressed in somites, notochord, neural tube and premigratory neural crest cells. l, m Transverse sections, the section levels are shown in (k). n Whole mount expression of NogginC at stage27. o–q Saggital sections reveal the stronger expression in the ventral part of the telencephalon and weak expression in ZLI and in otic cups. r–t at stage 27 NogginC is expressed in somites, notochord, neural tube at the trunk level (s), but in the growing tail (t) it reveals at a high level in the central (ventricular) zone of the neural tube. The levels of transverse sections are shown in (r). For abbreviations see Fig. 3.
NogginA expression was detected for the first time at the early neurula stage (st. 17) in the caudal chordamesoderm (Fig. 3a). At the late neurula stage (st. 19–20), the expression of this gene was found in the entire chordamesoderm, except for its most anterior region (Fig. 3b, c). In addition, NogginA expression was observed in the dorsal region of the presumptive brain, excluding the most anterior part (Fig. 3b).
At the stage of head outgrowth (st. 22), the expression is clearly visible in the floor plate of the neural tube, in the notochord, in the somites, in the cheek process and in the diencephalon, midbrain and hindbrain (Fig. 3d).
The expression pattern stabilizes at stages 23–25 and includes somites, upper and lower lips, notochord (except for the anterior part) and hypochord (Figs. 3e–h, 4b, c). As observed with transverse sections, at these stages, the expression is localized at the dorsal and ventral edges of the somites. In the spinal cord, the cells of the ventricular zone are stained (Fig. 4a–c). In the brain, the expression of NogginA is found in several areas: at the border of telencephalon and diencephalon, in the region of zona limitans intrathalamica (ZLI) that separates the ventral and dorsal parts of the diencephalon and at the border of the midbrain and hindbrain (Figs. 3i, 4d, e). Importantly, since ZLI and the mid-hind brain boundary are known to be secondary organizers of the developing brain, one may suppose that NogginA plays a role as one of the factors secreted by these organizers. In addition, the expression of NogginA is seen in the borders of some rhombomeres in the hindbrain. In all these brain areas, expression is found in cells of the roof and the floor plates of the neural tube. At stage 27, the expression appears in the developing eyes and in the pharyngeal arcs (Fig. 4f, g).
Interestingly, in the course of development, the pattern of NogginA expression in the notochord changes in the following way. At the early stages (st. 17–21), this gene is expressed throughout the notochord. Then, from stage 22, the expression disappears in the anterior part of the notochord while remaining strong in the region of the hindbrain and gradually weakening towards the caudal part of the embryo. Finally, starting at the tail bud stage (st. 26–27), an increase of the expression in the notochord of the growing tail is observed (Fig. 4d, h–j). The latter expression pattern is maintained until stage 29.
NogginB (Fig. 5).
Fig. 5. Expression pattern of L. fluviatilis NogginB.
a, e At stage 17 NogginB is expressed in the anterior end of the neural tube. b, f At stage 18 its expression is enhanced in the presumptive forebrain and hindbrain and weakens in the dorsal part of the presumptive midbrain. c, d, g, h At stage 19–21 the NogginB expression focuses on the presumptive forebrain (c, g) and in the most dorsal cells of neural tube (D, H) presumably - premigratory neural crest cells. i–r At stage 22–25 NogginB expression persists in the telencephalon and in ventral diencephalon and as is shown on transverse sections (q, r) in the premigratory neural crest cells. s Whole mount expression of NogginB at stage 27. t Saggital section of embyo of stage 27 reveal NogginB expression in the dorsal and ventral parts of the telencephalon and in the preoptic region of the hypothalamus. u–w horizontal section of embyo of stage 27. The section levels are shown in S. x, y NogginB expression at stage 28. For abbreviations see Fig. 3.
The initial expression of NogginB was revealed by in situ hybridization at the early neurula stage (st. 17) in the anterior end of the neural tube (Fig. 5a, e). Presumably, these cells will develop into the neural crest or telencephalon. At the mid neurula stage (st. 18), the expression is enhanced in the entire presumptive forebrain and hindbrain and remains in the ventral part of the presumptive midbrain (Fig. 5b, f). By the late neurula stage (st. 19–20), the expression of NogginB increases in the forebrain, while its expression in the dorsal part of the hindbrain spreads to the posterior and weakens (Fig. 5c, g).
At the head outgrowth stage (st. 21–23), NogginB is expressed in the region of the presumptive forebrain and in individual cells located on the dorsal side of the neural tube from the head to the tail, presumably - premigratory neural crest cells (Fig. 5d, h, n). At stages 24–29, strong expression persists in the dorsal and ventral parts of the telencephalon and in the preoptic region of the hypothalamus (Fig. 5s–w).
In the neural crest cells on the dorsal side of the neural tube, NogginB is expressed to stage 27 symmetrically in individual cells in the mid-hindbrain and in the spinal cord (Fig. 5o–y)
In summary, one may say that NogginB is predominantly expressed in the evolutionarily younger structures, namely, in cells of the neural crest and the telencephalon, which during evolution, appeared for the first time in vertebrates.
At the neurula stage (st. 18–19), the expression of NogginC appears in two chordamesoderm domains: one in the future head and one in the caudal region (Fig. 3k, l). Subsequently, in the late neurula stage (st. 20), the expression is detected in the chordomesoderm of the trunk, in the hindbrain region and in the cells of the neural crest.
At the head outgrowth stage (st. 22), NogginC is found in the trunk part of the notochord, especially in the cells of its tail region, in the somites, pharyngeal arches, cheek process, otic pits, and cells of the neural crest (Figs. 3m, 4l, m). Later, at stages 24–25, the expression of NogginC appears in the forebrain and in the pineal gland (Fig. 3n–r). Weak staining was also observed in the ZLI region of the diencephalon.
At the tailbud stage (st. 27), the expression of NogginC in the telencephalon becomes more pronounced in the ventral part and gradually decreases towards the dorsal part (Fig. 4o). Expression in the ears was detected, but the endolymphatic duct was not stained (Fig. 4p, q). In the growing tail, a high level of expression was observed in the central (ventricular) zone of the neural tube, where neural cells are known to actively proliferate (Fig. 4r–t). This pattern of expression is maintained until stage 29.
Thus, it is notable that the expression pattern of NogginC partially overlaps the expression patterns of NogginA and NogginB. The expression in unique vertebrate structures—telencephalon and neural crest cells—was similar for NogginC and NogginB.
NogginD (Fig. 3s–v).
The expression of NogginD, similar to the expression of other lamprey Noggin genes, is first detected starting from the early neurula stage (st. 17), but in contrast to the latter, it has a diffuse character, making it uniformly distributed throughout the neural plate (Fig. 3s, t). At the next stages of development, up to the pre-ammocoete stage (st. 29), we also observed weak diffuse staining throughout the neural system and somites (Fig. 3u, v). No expression was found in the notochord.
In general, it can be noted that the expression patterns of the lamprey Noggin genes have many similarities with the expression patterns of their homologues in other vertebrates, particularly those of X. laevis17. Thus, the expression of NogginA in the diencephalon, midbrain and hindbrain, as well as in the mesoderm derivatives, notochord and somites, appears to be similar to the expression of Xenopus Noggin1. The expression of NogginB in the forebrain is similar to that of Noggin2. At the same time, the expression pattern of NogginC has many overlapping features with the expression patterns of NogginA and NogginB. Finally, the expression of NogginD, with its characteristic diffuse pattern, greatly resembles that of Noggin4.
Lamprey Noggins induce secondary body axes in X. laevis embryos
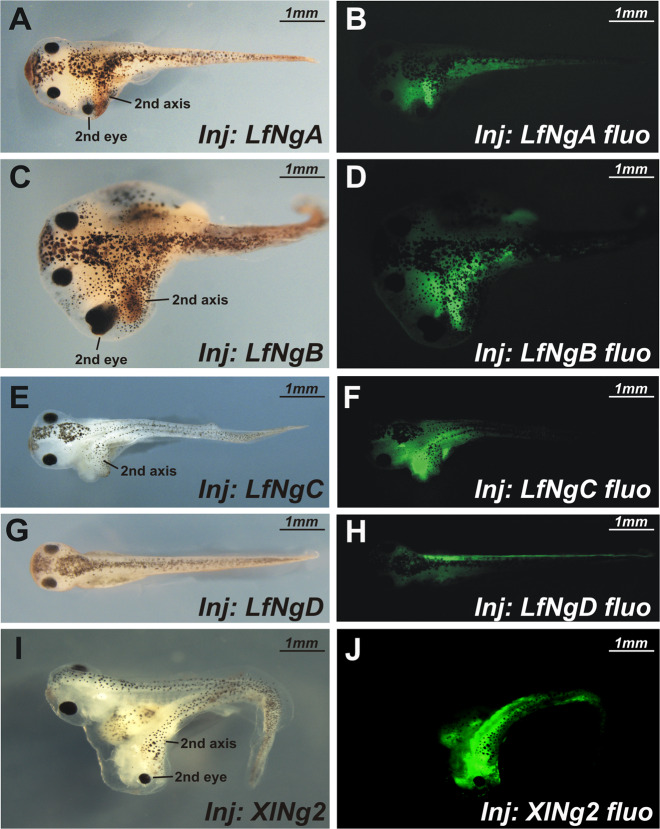
To understand the extent to which the physiological functions of the lamprey Noggins are similar to those of previously described Noggins in other vertebrates, we examined their ability to induce the formation of an additional body axis in X. laevis embryos injected ventrally at the 8-cell blastomere stage with mRNAs encoding for these four lamprey’s Noggins. The ability to form additional axial structures in the case of ventral expression in the X. laevis embryos was shown previously for Xenopus Noggin1 and Noggin215,24,25. In contrast, Noggin4 does not have this ability due to its inability to bind BMP16.
As a result, we found that NogginA, NogginB, and NogginC caused the formation of additional body axes, which in many cases, led to a forebrain with eyes (Fig. 6, Supplementary Fig. 2). Interestingly, while NogginB induced the development of a forebrain with eyes in the highest percentage of cases (13%, n = 92), NogginC generated secondary axes with a forebrain and eyes in the lowest percentage of cases (2%, n = 98), while NogginA occupied the middle position according to this criterion (5%, n = 87). These findings confirm the ability of the lamprey Noggins to induce secondary axes and are consistent with the observed homology of these proteins with the vertebrate Noggin1 and Noggin2, which have similar properties, as described previously15.
Fig. 6. Functional conservatism of Noggins in vertebrates.
Microinjections of mRNAs of lamprey NogginA (a, b), NogginB (c, d) and NogginC (e, f) in embryos of X. laevis causes the formation of secondary axes including anterior-headed structures like mRNA of X. laevis Noggin2 (i, j). For percentage of secondary axis and secondary head structures see Supplementary Figure 3. NogginD (g, h) does not induce secondary axes.
At the same time, NogginD, which is the presumptive orthologue of Noggin4, was unable to induce secondary axes, similar to the latter16 (Fig. 6g, h).
Discussion
Orthology of lamprey and gnathostomes Noggins
In this work, we described four Noggin family genes in lampreys, named NogginA, NogginB, NogginC and NogginD. As was revealed by phylogenetic analysis, NogginA gravitates to gnathostome Noggin1, NogginB and NogginC to Noggin2, and NogginD to Noggin4 (Fig. 1a). Despite the fact that the bootstrap value is rather low (50% or even less), such phylogenetic relationships of these three proteins look quite consistent with the local genomic synteny of their genes. Namely, NogginB, NogginC and Noggin2 have ANKFN1 adjoined at the 5’ end, while neither NogginA nor NogginD have this gene anywhere in their respective vicinity.
At the same time, the analysis of genomic synteny revealed a relationship of lamprey NogginA, NogginB NogginC, and gnathostome Noggin2, with gnathostome Noggin1, which is flanked by the homologue of the 5’-end neighbour of NogginB, NogginC and Noggin2 (ANKFN1) and the homologue of the 3’-end neighbour of NogginA (C17ORF67 and Syngr2) (Fig. 1b). Obviously, this finding indicates that all five genes could have been derived from a common ancestral Noggin1-like gene of vertebrates, which existed before the jawless and gnathostomes separated. In addition, the origin of NogginA, NogginB, NogginC, Noggin1 and Noggin2 from a common ancestor is confirmed by the following independent data. According to genome sequencing, P. marinus NogginA, NogginB, and NogginC are located in superscaffold 00003, 00037 and 00011, respectively. On the other hand, an analysis of the distribution of conserved syntenies in lamprey and chicken genomes, conducted by Smith et al., indicated that these superscaffolds correspond to chicken chromosome 14 and 18, which the authors believe likely emerged due to the duplication of one ancestral chromosome5. Meanwhile, chicken Noggin1 and Noggin2 genes are precisely located on chromosome 18 and 14, respectively. Thus, in good agreement with our results, these data suggest that at least three of four lamprey Noggin genes (i.e., NogginA, NogginB, and NogginC), along with gnathostome Noggin1 and Noggin2, may indeed have been derived from a single ancestral gene.
At the same time, the synteny of NogginD with Noggin4 of gnathostomes, together with the lack of their synteny with any other vertebrate Noggin gene, suggests that either these genes were derived from some other Noggin gene, which existed before the emergence of the vertebrate branch, or that the ancestor of NogginD and Noggin4 appeared as a result of a local translocation of a copy of a Noggin1- or Noggin2-type gene into different genomic locations. However, since all deuterostome invertebrates with sequenced genomes have only one Noggin, most resembling Noggin1 and Noggin2, but lack Noggin4, the second scenario, which suggests the appearance of a NogginD/Noggin4 ancestor during the initial period of vertebrate evolution, before the splitting of the jawless fish and gnathostomes, seems to us more realistic (see below).
Noggins have conserved expression patterns in vertebrates
Lamprey embryos are very attractive models for evo-devo studies because, among that of all extant vertebrates, the lamprey evolutionary branch was separated from the common trunk at the earliest stages of vertebrate evolution. Hence, it is presumed that the lamprey genes could have preserved the types of expression patterns characteristic of ancestral vertebrates.
Previous studies of the Noggin family genes in vertebrates have shown that all three Noggins identified in this group (Noggin1, Noggin2 and Noggin4) differ in their expression patterns and functional properties.
The expression of the “classic” Noggin1, as well as of that of Noggin2 and Noggin4, was studied in detail in Xenopus9,17,18. In this species, the expression of Noggin1 is detected for the first time at the beginning of gastrulation in the area of the dorsal blastopore lip, i.e., in the Spemann organizer, and then continues in the notochord, the midline of the neural plate and in the stripe of cells surrounding the anterior margin of the neural plate9. At later stages, this gene is expressed in the notochord, spinal cord, optic vesicle, branchial, pharyngeal and mandibular arches. Weaker expression is detected in the forebrain and in the head mesenchyme9.
In contrast to that of Noggin1, the expression of Noggin2 starts in X. laevis only from the early neurula stage, in the stripe of cells at the anterior margin of the neural plate; thus, it is partially superimposed with the expression of Noggin1. In contrast to Noggin1, however, no expression of Noggin2 was detected in the notochord17,18. At later stages, the expression at the highest level was observed in the dorsal region of the developing forebrain and at a lower level in the dorsal parts of the hindbrain, dorsal parts of the somites and in the forming heart. In addition, the expression of Noggin2 was detected in the derivatives of the neural crest, the pharyngeal arches17,18.
A specific feature of the expression of Noggin4 is its very diffuse distribution in embryonic tissues. Namely, in X. laevis, it is detected for the first time at the gastrula stage in the epidermal layer of the entire animal hemisphere18. At the neurula stage, Noggin4 expression is observed throughout the neural plate, with the maximum level in its anterior part, in the presumptive area of the head placodes and in the region of the future neural crest. In later stages, Noggin4 is expressed in the epidermis covering the neural tube, including the forebrain and spinal cord, in the auditory vesicles, cement gland and neural crest derivatives, including gill arches. Thin stripes of expression were also observed along the borders of the somites18.
In general, such expression patterns of Xenopus Noggin genes demonstrate many common features with the expression patterns of their lamprey orthologues.
Thus, lamprey NogginA, which begins to be expressed in lamprey somewhat later than Noggin1 in Xenopus, is also expressed in the chordamesoderm and later in the somites and head structures, including the neural crest derivatives.
NogginB, similar to its Xenopus orthologue, Noggin2, is also first detected at the early neurula stage in the anterior part of the neural tube and is subsequently expressed in the telencephalon and neural crest cells but not in the notochord.
NogginC has expression features in common both with those of both NogginA (expression in mesodermal derivatives) and NogginB (expression in telencephalon and neural crest cells).
NogginD, similar to Noggin4, has a diffuse expression pattern observed from the beginning of the neurula stage in the neural plate and later throughout the neural system.
The similarities are also revealed at the functional level. The ectopic expression of NogginA, NogginB and NogginC in X. laevis embryos induces the development of additional axes containing the forebrain with eyes, similar to those induced by ectopic expression of their Xenopus counterparts. In contrast, NogginD, like its Xenopus orthologue Noggin4, does not demonstrate this ability. Such conservatism indicates the involvement of Noggin genes in both jawless fish and gnathostomes in mechanisms regulating the early development of mainly the same anatomical structures. In turn, this observation also suggests that these mechanisms may have appeared before the divergence of the jawless and gnathostomes; thus, they are likely part of the basic regulatory network that was formed in the vertebrate ancestors and became critical for the formation of the body plan traits specific for all vertebrates. Notably, the same conclusions were previously made for some other regulatory genes after a comparative analysis of their expression patterns in lampreys and gnathostomes5,26–28.
In this respect, the expression of lamprey NogginB and Xenopus Noggin2 in such a unique vertebrate structure as the rudimentary telencephalon looks extremely exciting. Indeed, given the results of our phylogenetic analysis that indicate the origin of these genes for the first time only in vertebrates to our knowledge, one may suppose that their emergence at the beginning of vertebrate evolution could be one of the necessary prerequisites for the appearance of the telencephalon in vertebrates. Importantly, a similar role was proposed earlier for the homeobox gene Anf/Hesx1, which also emerged for the first time only in vertebrates and the expression of which is indicated during early development of all vertebrates, including lampreys, in the rudimentary telencephalon29–32. Our present data showing that the expression of another telencephalon-specific gene, NogginB/Noggin2, which also appeared for the first time in the ancestor of vertebrates, confirms that the emergence of novel genes could have indeed played an important role in acquiring unique anatomical innovations in this phylum of animals.
Hypothesis of Noggins evolution in vertebrates
As is currently discussed in the scientific community, this ancient period of vertebrate evolution included either one or two rounds of ancestral whole-genome duplication8,33,34. Initially, this supposition was inspired by the finding of four and six complexes of HOX genes in gnathostomes and lampreys, respectively35–37. At the same time, the nearest relatives of vertebrates, tunicates and cephalochordates, have only one HOX complex. Thereafter, similar results were obtained for the Pax6 gene38. These findings indicated that either two rounds of genome duplication occurred before the divergence of the jawless fish and gnathostomes, followed by an additional round of the entire or partial genomic duplications in the jawless fish, or that only one round of the duplication preceded the divergence of the jawless fish and gnathostomes, with additional rounds occurring independently in these two branches of vertebrates7,8,39.
The revealed phylogeny, synteny, expression patterns and functional analyses show that vertebrates Noggins can be confidently divided into two clusters: NogginA/B/C/1/2 and NogginD/4. At the same time, the expression pattern of lamprey NogginA has many similarities with jawed Noggin1, while NogginB has the expression pattern very similar to Noggin2. As the independent origin of so similar patterns in different lineages looks in our opinion very unlikely, one may hypothesize that the common vertebrate ancestor should have had at least three Noggin genes before the split of cyclostomes and gnathostomes. Based on these considerations, our data are not consistent with the recently proposed model suggesting only one common duplication before the split of Cyclostomes and Gnathostomes followed by additional independent duplications after separation of these lineages40. It seems more logical to suppose that the appearance of at least three common functionally diverged Noggins in the cyclostome and gnathostome common ancestor is consistent with two general scenarios: (a) one “basal” Noggin gene in vertebrate ancestor passed through two rounds of duplication before the divergence of the jawless fishes and gnathostomes; (b) two “basal” genes (obviously NogginA/B/C/1/2 and NogginD/4) passed through one round of duplication in the common ancestor. Let us consider both these models.
If two rounds of whole-genome duplications preceded the divergence of cyclostomes and gnathostomes, one may hypothesize that, during the first round, two copies of ancestral Noggin arose, followed by the evolution of one of these copies into ancestral NogginB/C/2 (Fig. 7a). The latter gene might have inherited at least two 5′ genomic neighbours of the ancestral Noggin1 (MMD and ANKFN1), which are still present in the vicinity of NogginC. Then, two copies of ancestral NogginA/1 and NogginB/C/2 could have appeared as a result of the second round of whole-genome duplication. One copy of ancestral NogginB/C/2 could have lost the homologue of MMD, thus acquiring the specific 3′ region surrounding the evolved NogginB and Noggin2 genes (the ancestor of NogginB/2). At the same time, the second copy of ancestral NogginB/C/2, which maintained both MMD and ANKFN1 near the 5′ end, became the ancestor of NogginC. Thereafter, one of two copies of ancestral NogginA/1 might have been translocated to a different genomic location, followed by specific point mutations in this translocated copy, which impaired the ability of its protein product to bind BMP. Finally, further mutagenesis could have been generated the ancestor of NogginD/4 from this translocated copy. After the subsequent separation of the jawless and gnathostomes, extant lampreys inherited all four Noggin genes: A, B, C and D. At the same time, in the vertebrate branch, the ancestor of NogginC seemingly was lost, resulting in only three Noggin genes now present in most of the extant vertebrates: Noggin1, Noggin2 and Noggin4.
Fig. 7. Three possible scenarios of the evolution of Noggin genes in vertebrates.
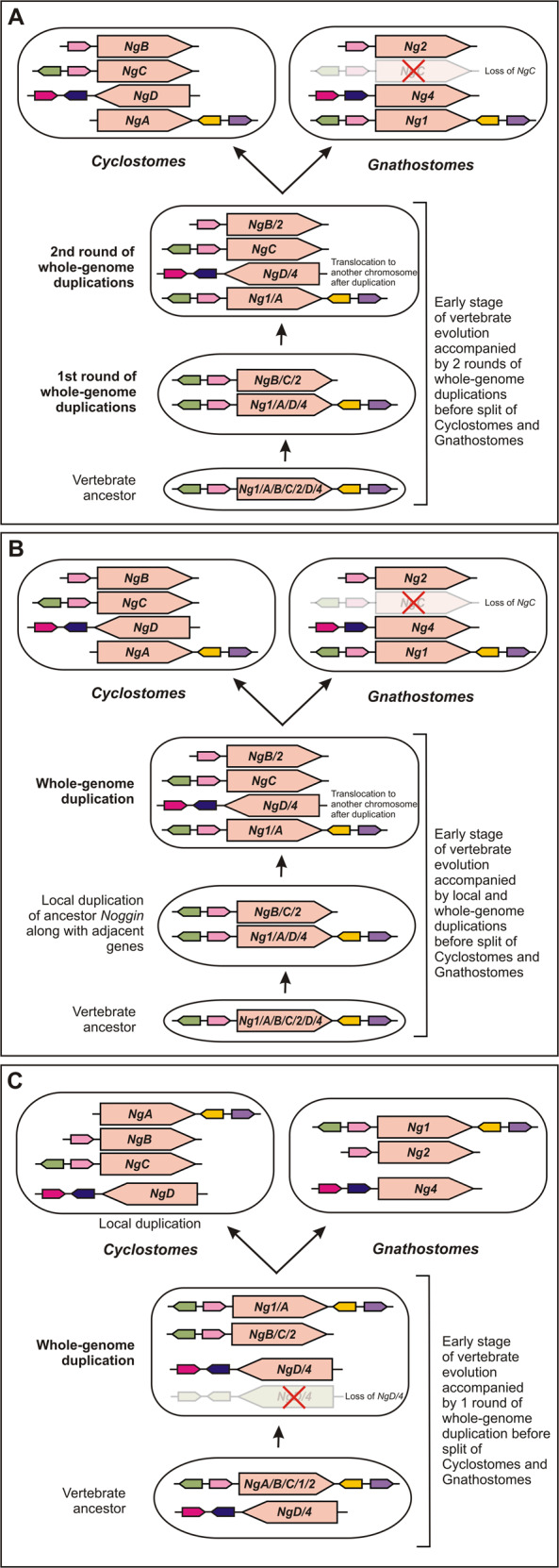
a and b scenarios suggest one ancestral Noggin gene passed through two rounds of duplication (2 rounds of whole-genome duplication in a vs local+whole-genome duplications in b). c scenario suggests two ancestral Noggins passed through one round of whole-genome duplication.
A possible variant of this scenario could include local duplication of the ancestor Noggin gene along with its adjacent genes instead of a whole-genome duplication at the first step (Fig. 7b). Based on the data of the present work, it is not possible to judge whether the first round of duplication was whole-genome or local.
In case of the one-round scenario of whole-genome duplication, one has to allow the presence of two Noggin genes in vertebrate ancestor (Fig. 7c). After the single round of genomic duplication, vertebrates could obtain NogginA/1, NogginB/C/2 and two copies of Noggin4/D genes. As all extant vertebrates have only one copy of NogginD or Noggin4 gene, another copy of this ancestor Noggin4/D had to disappear somewhere in between the moment of the whole-genome duplication and the moment of the divergence of jawless and gnathostomes. The appearance of NogginB and NogginC in this model could be the result of a local duplication and further divergence of the descendants of NogginB/C/2 in lampreys.
In support of the hypothesis of the existence of two “basic” Noggins in vertebrate ancestor one may point to the presence of multiple noggin-like genes (nlg) in the flat worm Schmidtea miditerrania41,42, which could be considered as possible predecessors of the hypothetical NogginA/B/C/1/2 and NogginD/4 in the vertebrate ancestor. As was shown by the authors using X. laevis embryos as models, at least some of the nlgs were indeed able to bind BMP, like vertebrate Noggin1 and 2, while other not, like Noggin4. However, on our opinion, it is very difficult to imagine direct phylogenetic relationship of the flat worm nlgs with vertebrate Noggin4 because of the aforementioned lack of Noggin4 homologs genes in all extant deuterostome predecessors of vertebrates that separate them from flat worms. The independent appearance of Noggin4 and noggin-like genes whose protein products lack the ability to bind BMP could be based on the emergence of multiple copies of Noggin genes in any animal lineage. In this case, one copy of Noggin may continue to retain the ability to bind BMP, while another copy(s) could escape from the pressure of stabilizing natural selection and quickly acquire point mutations in any one of the four amino acid positions, which have been shown to be critical for BMP binding14. Such mutant copy(s) of Noggin may not have been eliminated during evolution because, as we previously showed, Noggin proteins, in addition to BMP binding, can bind ligands of other signalling pathways, such as those of the Wnt and Activin/Nodal pathways, thereby participating in the regulation of other important developmental processes15,16.
Given all these considerations and being guided by Occam’s razor principle, we believe that at the present time the hypothesis of two rounds of genome duplications before the divergence of the jawless fish and gnathostomes may explain less controversially the presence of those Noggins that were revealed in lampreys and gnathostomes.
Methods
Animals
All animal experiments were performed in accordance with guidelines approved by the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry (Moscow, Russia) Animal Committee and handled in accordance with the 1986 Animals (Scientific Procedures) Act and Helsinki Declaration.
L. fluviatilis adult lampreys were collected in the Saint Petersburg district. Embryos were obtained via artificial fertilization of eggs squeezed from pregnant females. The embryos were staged as described in Tahara, 1988. For in situ hybridization, embryos were fixed in MEMFA (3,7% formaldehyde, 100 mM MOPS, 2 mM EGTA, 1 mM MgSO4), dehydrated in methanol and kept at −20C.
Cloning of lamprey Noggins cDNAs
For RT PCR we used samples of total RNA from L. fluviatilis embryos at early stages of development (st. 12–26). Total RNA from full embryos was extracted by NucleoSpin RNA XS Kit by Macherey-Nagel.
Full-length cDNAs of L. fluviatilis Noggin genes were obtained by nested PCR. For primers see Supplementary Information.
The obtained cDNA was cloned in pAL2-T vector, provided by Evrogen (www.evrogen.ru) and cDNA inserts of 3 clones were sequenced. The obtained full-length cDNAs were used for in situ hybridization and for X. laevis embryos injections.
Bioinformatics and synteny analysis
Phylogenetic analysis of protein sequences were performed via the maximum likehood43 methods using the MEGA644 program. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches45. Evolutionary distances were computed using the JTT matrix-based method46.
For synteny analysis we compared genomic scaffolds containing lamprey and amphibian Noggins by using Petromyzon marinus genome browser (https://simrbase.stowers.org/organism/Petromyzon/marinus), and Xenopus tropicals genome browser (http://www.xenbase.org/common/displayJBrowse.do?data=data/xt9_1).
RT-PCR
For qRT-PCR, three groups of the L. fluviatilis embryos were collected obtaining 50 embryos, respectively, from each of the desired stages. Total RNA was extracted using an RNA isolation kit (MASHEREY-NAGEL) according to the manufacturer’s protocol. The concentration of the extracted RNA was measured with a Qubit® fluorometer (Invitrogen), while RNA integrity was checked visually via gel electrophoresis.
First-strand preparation, qPCR parameters and primers used are described in Supplementary Information.
Two independent pairs of primers were used for each of Noggin genes to exclude unspecific signals. For the arbitrary unit in Fig. 2 we take the expression level at the earliest investigated stage - the early blastula.
Synthetic mRNA and in situ hybridization
Synthetic mRNAs of the lamprey Noggins were prepared with the mMessage Machine SP6 Kit (Ambion).
L. fluviatilis whole-mount in situ hybridization was performed as described in ref. 32.
The lamprey specimens were embedded in 20% gelatin and 35 μm sections were prepared as described in ref. 47 and observed with light microscope Rihert-Jung.
The embryos were cleared with a graded series of glycerol 25, 50, 75% in PBS and 99% at RT and observed with a Leica M205 stereomicroscope.
Statistics and reproducibility
All Noggin plasmids insert sequences were verified by Sanger sequencing of three replicates.
Phylogenetic analyses were performed in the MEGA644 program. Multiple alignment was performed via ClustalW algorithm with the default parameters.
Each qPCR sample contained total mRNA from 50 L. fluviatilis embryos of the desired stages. Three replicates of sample series were obtained from different animals. The qPCR data were analyzed using the ΔΔCt method. The geometric mean of expression of two reference housekeeping genes (ODC and EF1alpha) was used for normalization of the target gene expression levels.
The numbers of Xenopus embryos in the functional test is shown in Supplementary Fig. 3.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by RFBR grant 18-04-00015 (A.V.B.). Experiments with qRT-PCR were supported by Russian Scientific Foundation (19-14-00098). Phylogenetic analysis was supported by RFBR grant 18-29-07014 MK (A.G.Z.). X. laevis experiments were supported by RFBR grant 20-04-00675A (G.V.E.).
Author contributions
G.V.E. - embryos collection, in situ hybridization, histology, photography, figures ideas and preparation, writing of the paper; A.V.K. – L. fluviatilis animals, embryos; A.G.Z. – Xenopus embryos malformations analysis, figures ideas and preparation, writing of the paper, A.V.B. – databases analysis, cDNA cloning, qPCR, phylogenetic analysis, figures ideas and preparation, writing of the paper.
Data availability
Genome browsers used for synteny analysis: sea lamprey P. marinus genome browser (https://simrbase.stowers.org/organism/Petromyzon/marinus); western clawed frog X. tropicals genome browser (http://www.xenbase.org/common/displayJBrowse.do?data=data/xt9_1)
Lamprey Noggins cDNAs has been deposited in the GenBank database under the following accession numbers:
P. marinus NogginA MT653626, NogginB MT653628, NogginC MT653632, NogginD MT653630;
L. camtchaticum NogginA MT653625, NogginB MT653627, NogginC MT653631, NogginD MT653629;
L. fluviatilis NogginA MT646799, NogginB MT646800, NogginC MT646801, NogginD MT646802.
All the experimental data generated or analyzed during this study are present in the paper. Additional data and research materials related to this paper are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrey G. Zaraisky, Email: azaraisky@yahoo.com
Andrey V. Bayramov, Email: andrbayr@gmail.com
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-020-01234-3.
References
- 1.Janvier P. Modern look for ancient lamprey. Nature. 2006;433:921–924. doi: 10.1038/443921a. [DOI] [PubMed] [Google Scholar]
- 2.Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zool. Sci. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg TE, Mallatt J. The evolutionary and genetic origins of consciousness in the Cambrian Period over 500 million years ago. Front Psychol. 2013;4:667. doi: 10.3389/fpsyg.2013.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green SA, Bronner ME. The lamprey: a jawless vertebrate model system for examining origin of the neural crest and other vertebrate traits. Differentiation. 2014;87:44–51. doi: 10.1016/j.diff.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osório J, Rétaux S. The lamprey in evolutionary studies. Dev. Genes Evol. 2008;218:221–235. doi: 10.1007/s00427-008-0208-1. [DOI] [PubMed] [Google Scholar]
- 6.York, J. R., Lee, E. M. J. & McCauley, D. W. in Lampreys: Biology, Conservation and Control. Fish & Fisheries Series, vol 38 (ed Docker, M.) (Springer, Dordrecht, 2019).
- 7.Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015;25:1081–1090. doi: 10.1101/gr.184135.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JJ, et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018;50:270–277. doi: 10.1038/s41588-017-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 10.Knecht AK, Harland RM. Mechanisms of dorsal-ventral patterning in noggin-induced neural tissue. Development. 1997;124:2477–2488. doi: 10.1242/dev.124.12.2477. [DOI] [PubMed] [Google Scholar]
- 11.McMahon JA, et al. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes. Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunet LJ, et al. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 13.Warren SM, et al. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003;422:625–629. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 14.Botchkarev VA, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat. Cell. Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 15.Bayramov AV, et al. Novel functions of Noggin proteins: inhibition of Activin/Nodal and Wnt signaling. Development. 2011;138:5345–5356. doi: 10.1242/dev.068908. [DOI] [PubMed] [Google Scholar]
- 16.Eroshkin FM, et al. Noggin4 is a long-range inhibitor of Wnt8 signalling that regulates head development in Xenopus laevis. Sci. Rep. 2016;6:23049. doi: 10.1038/srep23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher RB, Watson AL, Harland RM. Expression of Xenopus tropicalis noggin1 and noggin2 in early development: two noggin genes in a tetrapod. Gene Expr. Patterns. 2004;5:225–230. doi: 10.1016/j.modgep.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Eroshkin FM, Ermakova GV, Bayramov AV, Zaraisky AG. Multiple noggins in vertebrate genome: cloning and expression of noggin2 and noggin4 in Xenopus laevis. Gene Expr. Patterns. 2006;6:180–186. doi: 10.1016/j.modgep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Bayramov AV, et al. Secreted protein Noggin4 participates in the formation of forebrain structures in Xenopus laevis by inhibiting the Wnt/beta-catenin signaling pathway. Russian J. Dev. Biol. 2016;47:202–206. [PubMed] [Google Scholar]
- 20.Bayramov AV, et al. Secreted protein Noggin4 - activator of Wnt/PCP signaling pathway. Russian J. Bioorg. Chem. 2017;43:216–219. [Google Scholar]
- 21.Kuraku S, Kuratani S. Genome-wide detection of gene extinction in early mammalian evolution. Genome Biol. Evol. 2011;3:1449–1462. doi: 10.1093/gbe/evr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groppe J, et al. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- 23.Tahara Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski) Zool. Sci. 1988;5:109–118. [Google Scholar]
- 24.Glinka A, Wu W, Onichtchouk D, Blumenstock C, Niehrs C. Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus. Nature. 1997;389:517–519. doi: 10.1038/39092. [DOI] [PubMed] [Google Scholar]
- 25.Glinka A, et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 26.Sugahara F, et al. Evidence from cyclostomes for complex regionalization of the ancestral vertebrate brain. Nature. 2016;531:97–100. doi: 10.1038/nature16518. [DOI] [PubMed] [Google Scholar]
- 27.Sugahara F, et al. Reconstructing the ancestral vertebrate brain. Dev. Growth Differ. 2017;59:163–174. doi: 10.1111/dgd.12347. [DOI] [PubMed] [Google Scholar]
- 28.Osorio J, Mazan S, Rétaux S. Organisation of the lamprey (Lampetra fluviatilis) embryonic brain: insights from LIM-homeodomain, Pax and hedgehog genes. Dev. Biol. 2005;288:100–112. doi: 10.1016/j.ydbio.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 29.Zaraisky AG, et al. A novel homeobox gene expressed in the anterior neural plate of the Xenopus embryo. Dev. Biol. 1992;152:373–382. doi: 10.1016/0012-1606(92)90144-6. [DOI] [PubMed] [Google Scholar]
- 30.Kazanskaya OV, et al. Anf: a novel class of vertebrate homeobox genes expressed at the anterior end of the main embryonic axis. Gene. 1997;200:25–34. doi: 10.1016/s0378-1119(97)00326-0. [DOI] [PubMed] [Google Scholar]
- 31.Ermakova GV, et al. The homeodomain factor Xanf represses expression of genes in the presumptive rostral forebrain that specify more caudal brain regions. Dev. Biol. 2007;307:483–497. doi: 10.1016/j.ydbio.2007.03.524. [DOI] [PubMed] [Google Scholar]
- 32.Bayramov AV, et al. The presence of the Anf/Hesx1 homeobox in lampreys indicates that it may play important role in telencephalon emergence. Sci. Rep. 2016;23:39849. doi: 10.1038/srep39849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacerdot C, Louis A, Bon C, Berthelot C, Roest Crollius H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 2018;19:166. doi: 10.1186/s13059-018-1559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland LZ, Ocampo Daza D. A new look at an old question: when did the second whole genome duplication occur in vertebrate evolution? Genome Biol. 2018;19:209. doi: 10.1186/s13059-018-1592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 36.Mehta TK, et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum) Proc. Natl Acad. Sci. USA. 2013;110:16044–16049. doi: 10.1073/pnas.1315760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker HJ, Bronner ME, Krumlauf R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev. Biol. 2019;453:19–33. doi: 10.1016/j.ydbio.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravi V, et al. Lampreys, the jawless vertebrates, contain three Pax6 genes with distinct expression in eye, brain and pancreas. Sci. Rep. 2019;9:19559. doi: 10.1038/s41598-019-56085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, et al. Lampreys, the jawless vertebrates, contain only two ParaHox gene clusters. Proc. Natl Acad. Sci. USA. 2017;114:9146–9151. doi: 10.1073/pnas.1704457114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simakov O, et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020;4:820–830. doi: 10.1038/s41559-020-1156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molina MD, Saló E, Cebrià F. Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr. Patterns. 2009;9:246–253. doi: 10.1016/j.gep.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Molina MD, et al. Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 2011;21:300–305. doi: 10.1016/j.cub.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 46.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 47.Oisi Y, Kakitani O, Kuratani S, Ota K. Analysis of embryonic gene expression patterns in the Hagfish. Neuromethods. 2015;99:249–262. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome browsers used for synteny analysis: sea lamprey P. marinus genome browser (https://simrbase.stowers.org/organism/Petromyzon/marinus); western clawed frog X. tropicals genome browser (http://www.xenbase.org/common/displayJBrowse.do?data=data/xt9_1)
Lamprey Noggins cDNAs has been deposited in the GenBank database under the following accession numbers:
P. marinus NogginA MT653626, NogginB MT653628, NogginC MT653632, NogginD MT653630;
L. camtchaticum NogginA MT653625, NogginB MT653627, NogginC MT653631, NogginD MT653629;
L. fluviatilis NogginA MT646799, NogginB MT646800, NogginC MT646801, NogginD MT646802.
All the experimental data generated or analyzed during this study are present in the paper. Additional data and research materials related to this paper are available from the corresponding author on reasonable request.