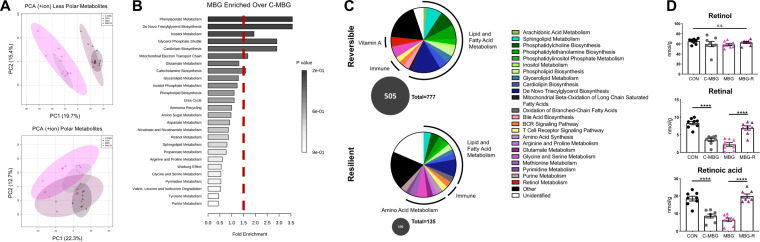
FIG 7.
Dietary reversal significantly shapes liver metabolome. (A) PCA plots of untargeted liver metabolomics via RP-UPLC–FTMS (top) and HILIC-FTMS (bottom). Data from the positive ion channel are shown. (B) Metabolite set enrichment analyses conducted with Metaboanalyst v 4.0 (SMPDB database) reported enriched metabolomic pathways in the MBG versus C-MBG liver metabolome. Metabolomic pathways to the right of the dashed red bar exhibit >1.5-fold enrichment. Fold enrichment was determined as number of observed pathway hits divided by the number of expected hits. (C) Metabolomic pathway profiles of reversible (top) and resilient (bottom) metabolites. One-way ANOVA of metabolites with post hoc Fisher’s LSD revealed metabolites significantly altered between MBG-R and MBG but not MBG-R and CON (reversible) as well as metabolites altered between MBG-R and CON but not MBG-R and MBG (resilient). Metabolites were searched in SMPDB to identify biopathway(s). The numbers of metabolites belonging to reversible or resilient pathway is shown in a gray circle; the adjacent numeric value refers to the total number of pathways within each graph. Legend lists are from SMPDB pathways. (D) Retinol, retinal, and retinoic acid levels within hepatic livers conducted by targeted LC-MS and normalized to tissue weights. Analyses were conducted from mice of the same reversal study. Bars indicate means ± SEM. Statistical significance was determined by one-way ANOVA with post hoc Tukey’s test and indicated as in the legend to Fig. 4.